Abstract
Memory CD8+ T cell development is defined by the expression of a specific set of memory signature genes (MSGs). Despite recent progress, many components of the transcriptional control of memory CD8+ T cell development are still unknown. To identify transcription factors (TFs) and their interactions in memory CD8+ T cell development, we construct a genome-wide regulatory network and apply it to identify key TFs that regulate MSGs. Most of the known TFs in memory CD8+ T cell development are rediscovered and about a dozen new TFs are also identified. Sox4, Bhlhe40, Bach2 and Runx2 are experimentally verified and Bach2 is further shown to promote both development and recall proliferation of memory CD8+ T cells through Prdm1 and Id3. Gene perturbation study identifies the mode of interactions among the TFs with Sox4 as a hub. The identified TFs and insights into their interactions should facilitate further dissection of molecular mechanisms underlying memory CD8+ T cell development.
Immunological memory refers to faster and stronger responses to re-encountering of the same antigen. The basis for this enhanced response is the persistence of more abundant and intrinsically more reactive antigen-specific memory T and B lymphocytes that are generated following the initial antigen stimulation. Memory CD8+ T cells are usually generated following antigen-stimulated T cell activation and expansion. In a typical CD8+ T cell response, naïve CD8+ T cells are activated to undergo clonal expansion when stimulated by appropriate antigen 1. The resulting T cells acquire effector functions and migratory properties that allow them to clear antigens in both lymphoid and non-lymphoid organs. As antigen is cleared, most of the effector T cells die by apoptosis and only a small fraction survive and differentiate into memory CD8+ T cells. Memory CD8+ T cells are often divided into two subsets. Effector memory T cells (TEM) are CD62LloCCR7lo and capable of rapid expression of effector functions following antigen stimulation to confer faster memory response. Central memory T cells (TCM) are CD62LhiCCR7hi and proliferate extensively upon antigen restimulation to confer stronger memory response.
Memory CD8+ T cells are developmentally programmed as they express a specific set of memory signature genes (MSGs) 2, 3, which confer them with characteristic memory phenotype and function. Like many developmental processes, memory CD8+ T cell development is ultimately controlled by transcription factors (TFs) that integrate external and internal signals to regulate the expression of the MSGs. In recent years, several studies have shed light on TFs that regulate the development of memory CD8+ T cells. T-bet (encoded by Tbx21) and Eomesodermin (encoded by Eomes), both member of the T-box family, are essential for the differentiation of effector and memory CD8+ T cells 4, 5, 6. Tcf7 is a TF downstream of the Wnt signaling. Consistent with the observation that activation of Wnt/β-catenin signaling promotes memory CD8+ T cell development by suppressing terminal differentiation of effector T cells 7, 8, Tcf7-deficiency in CD8+ T cells impairs TCM differentiation 9. Klf2 has been shown to be associated with memory CD8+ T cell development 10 probably by directly controlling the expression of cell surface receptors S1P1 and CD62L 11, 12. Id2 and Id3, the E-box-containing transcription suppressors, appear to regulate the development of memory CD8+ T cell subsets 13. Id2-knockout mice are deficient in memory CD8+ T cells, but effector T cells generated in these mice are CD127hiCD62Lhi, a phenotype similar to TCM14. Deficiency in Id3 inhibits, whereas overexpression of Id3 promotes memory CD8+ T cell development 15. The B-cell transcriptional repressor Blimp-1 (encoded by Prdm1) promotes the terminal differentiation of effector CD8+ T cells and is required for recall response of memory T cells 16, 17. Despite these progresses, the current understanding of transcriptional regulation of memory CD8+ T cell development is still limited, as additional TFs as well as their coordination are likely required to respond to external and internal signals in order to establish the MSG program for memory CD8+ T cell development.
In this study, we assemble a genome-wide regulatory network associated with the development of CD8+ T cells using publicly available gene expression data and a reverse-engineering algorithm. This regulatory network is applied to identify key TFs that regulate memory CD8+ T cell development using the master regulator analysis (MRA) of the MSGs of CD8+ T cells. The inferred TFs include most of the known TFs as well as a dozen new TFs with limited functional information in CD8+ T cell differentiation. A regulatory module controlling the MSGs is constructed and the high accuracy of the regulations in the module is verified using ChIP-PCR. Gene perturbations identify multiple regulatory motifs among the key TFs, suggesting their complex regulations during the memory CD8+ T cell development. Four of the newly identified key TFs (Sox4, Bhlhe40, Bach2, and Runx2) are experimentally validated to regulate memory CD8+ T cell development and function. Bach2 is shown to promote memory CD8+ T cell development and recall proliferation through Id3 and Prdm1. Our study represents the most comprehensive analysis of TFs and their interactions in memory CD8+ T cell development to date. The identified TFs and the insights into their mode of interactions provide a foundation for further dissecting the molecular mechanisms underlying memory CD8+ T cell development.
Results
Identification of TFs associated with memory CD8+ T cells
We collected 386 gene expression profiles of naïve, effector and memory CD8+ T cells of the mouse from 35 independent GEO datasets (Supplementary Table S1). 1,445 genes coding putative TFs 18 were manually mapped to the latest mouse genome to eliminate redundant and erroneous annotations, resulting in a total of 1,038 putative TFs. Among these putative TFs, 464 were expressed during the naïve to effector to memory CD8+ T cell development (see Methods for detail). Using a reverse-engineering algorithm CLR (context likelihood of relatedness)19, the 386 gene expression profiles and the 1,038 putative TFs, a genome-wide regulatory network was assembled. The network consisted of 107,157 interactions among 11,032 genes. 62,272 interactions (58%) were between the 276 of the 464 expressed putative TFs and 8,572 target genes, suggesting that interactions are enriched among the expressed genes (P<0.001, binomial test). Furthermore, 3,219 of these interactions involve 154 out of 196 (79%) identified MSGs 3 (Supplementary Fig. S1).
To identify key TFs that regulate MSGs, we applied the MRA to the CLR-inferred interactions (Fig. 1a, see Methods). The MRA algorithm computes the statistical significance of overlaps of all interactions of each TF (inferred by CLR) with MSGs or a control gene set by a binomial test. From the 1,038 putative TFs, MRA identified 60 MSG-specific TFs at P<0.05 (binomial test), all of which are expressed in CD8+ T cells (Supplementary Table S2). These 60 candidates were filtered by removing those whose knockout do not have any immune system phenotype as defined in MGI (Mouse Genome Informatics) 20. The positive candidates were then analyzed for enrichment of DNA-binding motifs among MSGs or differential expression among naïve, effector and memory CD8+ T cells (see Methods). This led to 21 key TFs that were ranked according to the numbers of MSG they regulate (Table 1). Text-mining of public references on these 21 TFs revealed that 8 of 12 known TFs, which have been reported to be involved in memory CD8+ T cell development and function 21, 22, 23, 24, were identified by our analysis. These results show that our systematic approach is valid for identifying TFs that regulate memory CD8+ T cell development.
Figure 1. Construction of regulatory network of memory CD8+ T cells.
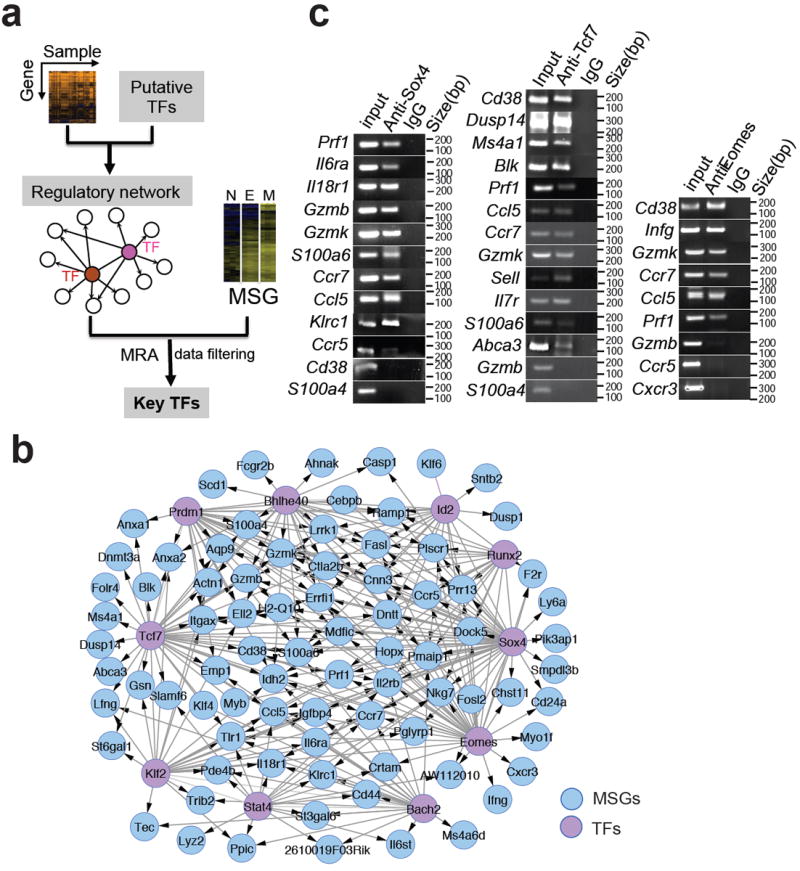
(a) Schematic diagram of regulatory network analysis for identifying key TFs. N, E, and M, naïve, effector and memory CD8+ T cells, respectively. (b) The regulatory module of the top 10 TFs (orchid circles) and their MSGs (blue). c, ChIP-PCR analysis of Sox4, Tcf7 and Emoes-regulated MSGs. ChIP was carried out with CD8+ T cells expressing the 2C TCR using antibodies specific for Sox4, Tcf7 or Eomes or control IgG antibodies. Promoter regions of the indicted genes were amplified using the precipitated DNA. Shown are PCR products after electrophoresis.
Table 1.
Ranking of the 21 key TFs identified by master regulator analysis.
| TF | #MSG | P-value | ISP* | DBM | FDR | ESG | MSG | qPCR E/N | qPCR M/N | Reference gene functions# | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sox4 | 50 | 9.55E-10 | YES | SORY | 0.0001 | YES | YES | -4.305 | -5.845 | Th2 differentiation | 50 |
|
| |||||||||||
| Tcf7 | 40 | 1.10E-06 | YES | LEFF | 0.0010 | YES | NO | -3.71 | -0.37 | Promote memory | 7, 9 |
| Eomes | 34 | 1.03E-06 | YES | BRAC | 0.0021 | YES | YES | 1.99 | 3.48 | Effector and memory | 4, 5 |
|
| |||||||||||
| Bhlhe40 | 27 | 2.50E-05 | YES | HESF | 0.3353 | YES | YES | 2.72 | 4.215 | CD8+ T cell activation | 51 |
|
| |||||||||||
| Prdm1 | 26 | 2.90E-05 | YES | PRDF | - | YES | NO | 4.185 | 4.5 | Effector/inhibit memory | 16, 17 |
| Klf2 | 24 | 0.000587 | YES | KLFS | 0.1135 | NO | NO | -2.01 | -0.63 | Homeostasis/memory | 10 |
|
| |||||||||||
| Bach2 | 22 | 0.000229 | YES | AP1R | 0.0151 | YES | NO | -3.77 | -0.935 | - | |
| Runx2 | 21 | 0.000365 | YES | HAML | 0.0031 | YES | YES | 0.19 | 2.775 | Early T cell development | 52 |
|
| |||||||||||
| Id2 | 20 | 0.000319 | YES | - | - | YES | YES | 0.265 | 2.645 | KO promotes memory | 13, 14 |
| Stat4 | 17 | 0.002650 | YES | STAT | 0.2503 | NO | NO | -2.33 | 0.935 | Effector and memory | 53, 54 |
|
| |||||||||||
| Runx3 | 15 | 0.002869 | YES | HAML | 0.0031 | NO | NO | -2.37 | -1.925 | Cooperation with Tbx21 | 55 |
|
| |||||||||||
| Id3 | 14 | 0.005424 | YES | - | - | NO | NO | -3.695 | -1.995 | Promote memory | 15 |
|
| |||||||||||
| Nfatc2 | 12 | 0.007564 | YES | NFAT | 0.0127 | NO | NO | - | - | TCR signaling | 56 |
| Gata3 | 11 | 0.012973 | YES | GATA | 0.0425 | NO | NO | - | - | Th2 differentiation | 57 |
| Ikzf1 | 11 | 0.038617 | YES | IKRS | 0.0208 | NO | NO | - | - | IL2/TCR signaling | 58 |
| Nfatc1 | 11 | 0.038617 | YES | NFAT | 0.0127 | NO | NO | - | - | PD-1/TCR signaling | 59 |
| Jun | 10 | 0.019089 | YES | AP1F | 0.0046 | NO | NO | - | - | AP-1 complex | 60 |
| Nfe2l2 | 10 | 0.019089 | YES | AP1R | 0.0151 | NO | NO | - | - | Protect memory | 61 |
|
| |||||||||||
| Tbx21 | 9 | 0.036556 | YES | BRAC | 0.0021 | NO | NO | 6.215 | 3.165 | Effector and memory | 4, 5 |
|
| |||||||||||
| Jund | 9 | 0.028085 | YES | AP1F | 0.0046 | NO | NO | - | - | AP-1 complex | 60 |
| Maf | 9 | 0.025971 | YES | AP1R | 0.0151 | YES | NO | - | - | - | |
#MSG: the total number of MSG regulated directly by the TF; * ISP: immune system phenotype; DBM, DNA binding motif; FDR, false discovery rate; ESG, effector signature genes; MSG, memory signature genes; qPCR E/N, log2 gene expression fold-changes between effector and naïve CD8+ T cells; qPCR M/N, log2 gene expression fold-changes between memory and naïve CD8+ T cells;
gene function reported in T cells. Grey marked TFs that have been reported as playing important roles in memory CD8+ T cell development or function.
Validation of a regulatory module for memory signature genes
To further explore the relative importance of the 21 identified TFs in regulating MSGs, we constructed a regulatory module using the top 10-ranked TFs (Fig. 1b). The resulting module contained 56% (86 out of 154) of MSGs that were present in the entire network (Supplementary Fig. S1). To verify this regulatory module, chromatin immunoprecipitation (ChIP) was performed for the top 3 TFs, Sox4, Tcf7 and Eomes, in CD8+ T cells followed by PCR amplification of promoter regions (within 1 kb upstream of the transcription-starting site) of randomly selected MSGs that were predicted to be regulated by Sox4 or Tcf7 or Eomes. As shown in Fig. 1c and 1d, promoter regions of 10 out of the 12 randomly selected Sox4-regulated MSGs were amplified. Similarly, 12 out of 14 randomly selected Tcf7-regulated MSGs and 6 out of 9 randomly selected Eomes-regulated MSGs were amplified. On average, 80% of the tested promoter regions were immunoprecipitated with antibodies specific for each of the three TFs (Table 2), confirming the high accuracy of the constructed regulatory module. Furthermore, when the cumulative coverage of MSGs was plotted as a function of each of the 21 TFs, the top 2 TFs, Sox4 and Tcf7, were shown to regulate 42% of MSGs (Supplementary Fig. S2).
Table 2.
Summary of the ChIP-PCR results.
| TF | Sox4 | Tcf7 | Eomes | Total |
|---|---|---|---|---|
| No. MSG target | 41 | 36 | 26 | 103 |
| No. tested target | 12 | 14 | 9 | 35 |
| No. positive | 10 (83.3%) | 12 (85.7%) | 6 (66.7%) | 28 (80%) |
Perturbation network of key TFs
Although the constructed regulatory module predicts interactions between TFs (Fig. 1b), the directions of regulation are not known. To find out these, the top 10 TFs and another two known memory-regulating TFs (Id3 and Tbx21, #12 and #19 in the list, Table 1) 6, 15, 25 were perturbed in CD8+ T cells in vitro by overexpression through retroviral transduction. The transcript level of each of the 12-selected TFs was measured by quantitative real-time PCR (Table 3). If changes in transcript level of ≥2 fold were taken as directional regulations, the perturbation results identified 41 regulations among the 12×12 matrix (31%). Notably, the top 3 TFs (Sox4, Tcf7 and Eomes) directed 19 of the 41 regulations. To verify these regulations, ChIP-PCR was performed using antibodies specific for Sox4, Tcf7 and Eomes. As shown in Fig. 2a, 18 of the 19 regulations were confirmed. ChIP-PCR also identified 4 more regulations that were not observed in the perturbation study. Thus, compared to ChIP-PCR, perturbation studies is able to identify the directional regulations with 82% sensitivity and 91% specificity.
Table 3.
Perturbation analysis.
| Gene | Sox4 | Tcf7 | Eomes | Bhlhe40 | Prdm1 | Klf2 | Bach2 | Id2 | Runx2 | Stat4 | Id3 | Tbx21* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sox4 | 7.94 | -1.38 | 1.77 | -0.62 | -3.17 | -0.28 | -0.93 | -0.48 | -0.05 | -0.70 | -0.83 | 1.21 |
| Tcf7 | -1.79 | 2.44 | -0.03 | -0.21 | -2.01 | -0.50 | 0.88 | -0.39 | 0.04 | -2.16 | 0.31 | -0.64 |
| Eomes | -2.28 | -1.11 | 5.29 | 0.38 | -0.08 | -0.26 | -0.64 | -0.10 | 0.85 | -0.55 | 0.54 | -2.31 |
| Bhlhe40 | -2.03 | 0.41 | 1.09 | 3.88 | 0.02 | 1.99 | 0.32 | 0.31 | 0.14 | -0.41 | 0.68 | 0.62 |
| Prdm1 | -0.14 | -1.48 | 2.04 | 1.13 | 5.19 | 1.34 | -1.62 | -0.17 | 0.20 | 0.08 | -0.39 | 1.30 |
| Klf2 | 0.85 | -0.78 | -1.10 | -0.55 | -1.49 | 4.06 | -1.52 | -0.73 | -0.07 | -1.33 | -1.51 | -0.57 |
| Bach2 | 1.37 | 0.75 | 0.35 | 0.00 | 0.22 | 1.40 | 4.41 | -0.18 | 0.78 | 0.01 | 0.07 | 0.15 |
| Id2 | -2.24 | 1.10 | 0.46 | 0.38 | 0.24 | 0.66 | 0.09 | 4.98 | 0.93 | -0.06 | 0.18 | 0.54 |
| Runx2 | -1.64 | 0.41 | -0.51 | -0.22 | -0.02 | 0.91 | -0.50 | -0.82 | 3.71 | -0.54 | -0.03 | -0.77 |
| Stat4 | -1.45 | -0.26 | -1.78 | 0.14 | 0.80 | 0.24 | 0.22 | -0.01 | 0.85 | 2.32 | 0.41 | -0.81 |
| Id3 | 2.61 | 0.23 | -1.32 | 0.47 | -0.23 | 0.72 | 2.02 | -1.03 | 0.25 | 0.73 | 7.44 | -1.09 |
| Tbx21 | -1.68 | -0.26 | -0.22 | 0.12 | -0.34 | 1.42 | 0.50 | 0.93 | 1.29 | -2.29 | 1.17 | 6.54 |
The highlighted 12 TFs (first row) were overexpressed individually in 2C T cells and the level of their transcripts of each TFs was quantified by real-time PCR. Expression data was normalized to the empty vector control and then log2 transformed. Changes in transcript levels for ≥2 fold are marked orange (up-regulated) or green (down-regulated). The overexpressed TFs are marked red.
Figure 2. Construction of perturbation network of TFs in CD8+ T cells.
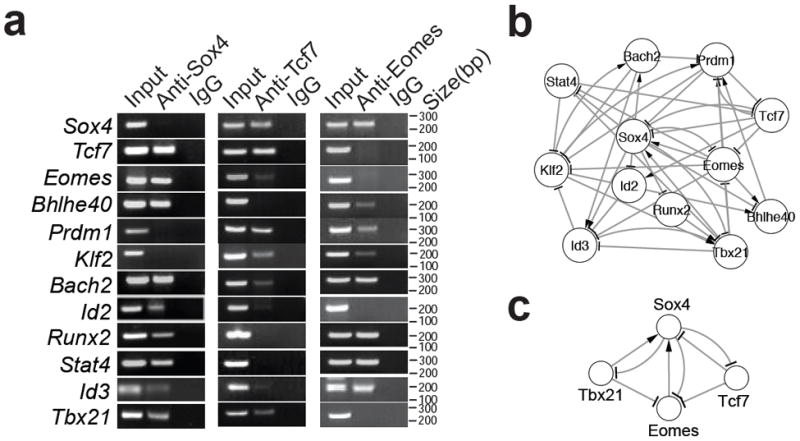
(a) ChIP-PCR analysis. ChIP was carried out with 2C T cells using antibodies specific for Sox4, Tcf7 or Eomes or control IgG antibodies. Promoter regions of the indicted genes were then amplified using the precipitated DNA. Shown are PCR products after electrophoresis. (b) Perturbation network based on Table 3. (c) An example of network motifs from the perturbation network.
We then constructed a perturbation network of the 12 TFs with directional regulations (Fig. 2b). The top 3 TFs (Sox4, Tcf7 and Eomes) and Bach2 had more downstream targets than the number of TFs that regulate them (Supplementary Fig. S3), suggesting that they are at the upstream of a regulatory structure. TFs in the perturbation network formed multiple motifs, such as feedback and feed-forward loops (Supplementary Fig. S4). For example, in a feedback motif of Sox4-Tcf7-Eomes-Tbx21 (Fig. 2c), Sox4 and Tcf7 regulate each other and they also regulate expression of Eomes and/or Tbx21. The latter regulations were further confirmed at the protein level as indicated by suppression of Eomes and Tbx21 by overexpression of Sox4 or Tcf7 (Supplementary Fig. S5). These results suggest that complex regulations involving multiple regulatory motifs among these TFs are involved in memory CD8+ T cell development.
Validation of Sox4 and Bach2 in memory CD8+ T cells
Among the top 10 TFs (Table 1), 6 are known to play important roles in memory CD8+ T cell development and/or function. We then investigated whether the other 4 TFs (Sox4, Bhlhe40, Bach2 and Runx2) are also involved in memory CD8+ T cell development/function by examining the effect of overexpression and knockdown of these TFs on the recall proliferation of memory CD8+ T cells in vitro and in vivo. CD8+ T cells expressing the 2C TCR were activated with cognate peptide SIYRYYGL (SIY) and then transduced with retroviruses expressing GFP plus Sox4, Bhlhe40, Bach2 or Runx2 or expressing GFP plus shRNA specific for one of the four TFs (Supplementary Table S3 and S4). The 2C T cells were then cultured in the presence of cytokine IL-7 to induce the development of memory CD8+ T cells (Supplementary Fig. S6). To assay recall proliferation, the in vitro memory 2C T cells were restimulated with SIY and the number of transduced (GFP+) and non-transduced (GFP-) 2C T cells were quantified on day 4 and 6. Compared to the vector control, overexpression of Sox4 or Bach2 led to a significant increase in the proportions of GFP+ cells (Fig. 3a), suggesting a higher recall proliferation. When the in vitro generated memory 2C T cells were adoptively transferred into C57BL/6 (B6) mice followed by activation through infection with influenza virus that express SIY (WSN-SIY virus) 26, a significant increase in the proportion of GFP+ 2C T cells was also observed in the draining lymph nodes (DLN) (Fig. 3b), the blood, lung and spleen (Supplementary Fig. S7) 5 days post infection (dpi) if the transduced memory T cells expressed Sox4 or Bach2. Conversely, knockdown of Sox4 or Bach2 (Supplementary Fig. S8) resulted in a significant inhibition of the recall proliferation of memory 2C T cells both in vitro and in vivo (Fig. 3c,d). Although overexpression of Bhlhe40 and Runx2 inhibited the in vivo recall proliferation of the transduced memory 2C T cells (Fig. 3b), no significant change was observed in in vitro recall response (Fig. 3a) and in knockdown assay (Fig. 3c,d). As positive controls, we tested in parallel known TFs: overexpression of Eomes promoted the recall proliferation whereas overexpression of Klf2 inhibited the recall proliferation (Supplementary Fig. S9), consistent with previous reports 9, 11. These results show that Sox4 and Bach2 likely promote the recall proliferation of memory CD8+ T cells.
Figure 3. Effect of overexpression and knockdown of TFs on memory CD8+ T-cell recall proliferation.
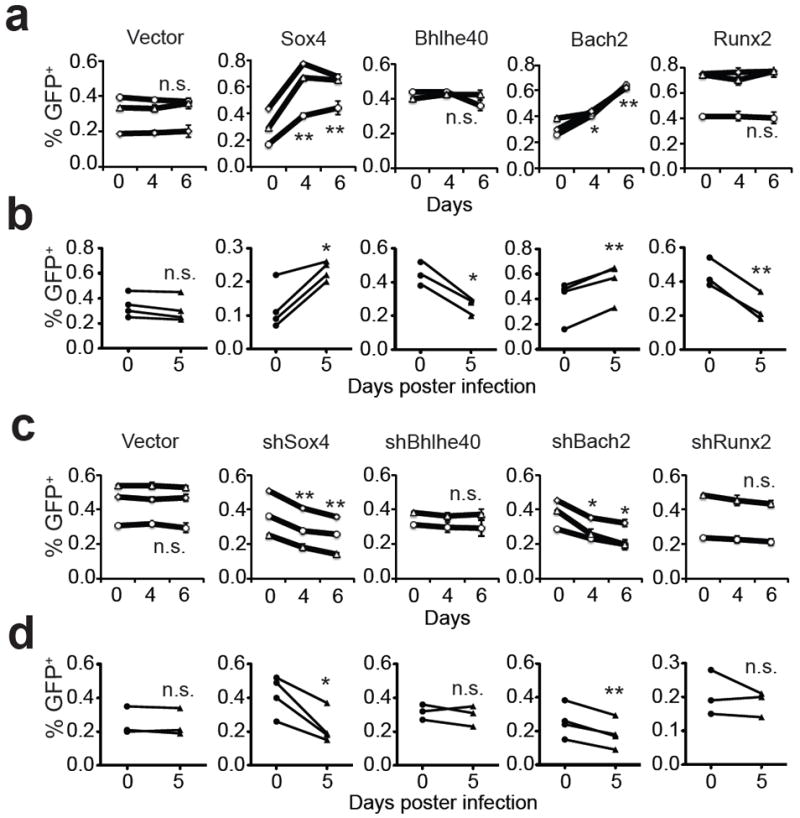
Naïve 2C T cells were activated in vitro with SIY peptide and then transduced with retroviruses expressing Sox4, Bhlhe40, Bach2 or Runx2 or expressing shRNA specific for one of the TFs. The cells were cultured in the presence of IL-7 to induce memory T cell development. The resulting memory 2C T cells were either activated in vitro with SIY peptide or transferred into mice and activated by WSN-SIY virus infection. The proportion of GFP+ (transduced) versus GFP- (non-transduced) 2C T cells was quantified 4 and 6 days post stimulation in vitro and in draining lymph node (DLN) 5 days post infection (dpi). Shown are proportion of GFP+ 2C T cells that overexpressed Sox4, Bhlhe40, Bach2 or Runx2 among total 2C T cells in vitro (a) and in vivo (b) or that expressed shRNA specific for Sox4, Bhlhe40, Bach2 or Runx2 among total 2C T cells is in vitro (c) and in vivo (d). Each line was one independent experiment with one sample per time point for the in vitro experiments and one or two mice per in vivo experiment. Data shown are mean ± s.e.m. Pairwise two-tailed t-tests were used for statistical analyses. * P<0.05; ** P<0.01.
Enhanced memory T cell development by Bach2 overexpression
To confirm the effect of overexpression of Bach2 on recall proliferation of memory CD8+ T cells, we activated 2C T cells in vitro for two days, transduced the activated T cells with retroviruses expressing GFP alone or GFP plus Bach2. The cells were cultured in the presence of IL-2 for two more days and then adoptively transferred into antigen-free B6 mice to induce in vivo memory 2C T cells (Fig. 4a). Twenty-three days after transfer, the frequency, phenotype and function of persisting 2C T cells were analyzed. Both transduced (GFP+) and non-transduced (GFP-) 2C cells persisted in the recipient mice (Fig. 4b), GFP+ 2C cells exhibited a typical memory phenotype as indicated by expression of CD62L and IL-7 receptor (IL-7R), similar to the GFP- 2C cells in the same recipient (Fig. 4c). The persisting memory 2C cells, both transduced and non-transduced, were rapidly induced to express IFNγ and TNFα following antigen stimulation (Fig. 4d). Furthermore, some recipient mice were infected with WSN-SIY virus and the recall proliferation of persisting 2C cells in the spleen and DLN were analyzed 7 days later. As shown in Fig. 4e, if the 2C cells were originally transduced with GFP-expressing retrovirus, the proportion of GFP+ versus GFP- 2C cells did not changed following WSN-SIY challenge. However, if the 2C cells were originally transduced with GFP and Bach2-expressing retrovirus, the proportion of GFP+ cells was significantly higher in both DLN and spleen, suggesting a stronger recall proliferation by Bach2-expressing memory 2C cells.
Figure 4. Bach2 promotes recall proliferation of memory CD8+ T cells.
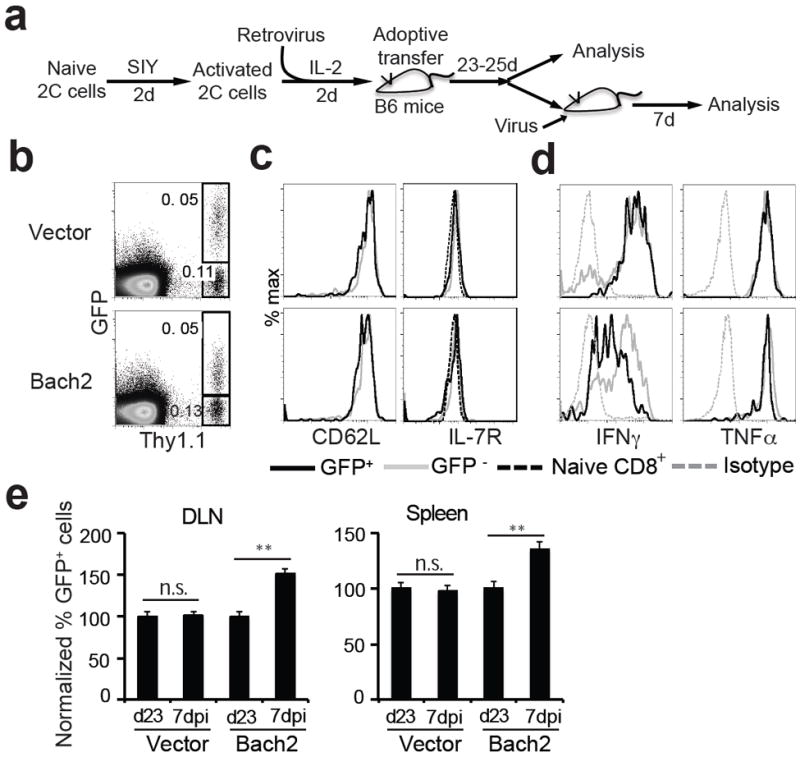
(a) Scheme of experimental protocol. (b-d) Phenotype and function of persisting memory 2C cells. Twenty-two days post transfer, single cell suspension was prepared from spleen and analyzed for CD8, Thy1.1, GFP plus CD62L or IL-7R directly or stimulated in vitro with SIY peptide for 5 hours before staining for CD8, Thy1.1, GFP plus intracellular IFNγ or TNFα. Comparison of GFP versus Thy1.1 (b) staining profiles of live cells between vector control and Bach2 overexpression group. Comparison of CD62L and IL-7R (c) or IFNγ and TNFα expression (d) between GFP+ and GFP- 2C T cells. Gray trace, nontransduced (GFP-) 2C T cells; black trace, transduced (GFP+) 2C T cells; dash gray trace, isotype control for intracellular staining; dash black trace, IL-7R staining of naïve 2C T cells. (e) Recall responses of persisting memory 2C T cells in vivo. Some recipient mice were infected with WSN-SIY 23-25 days post transfer and the proportions of GFP+ and GFP- 2C T cells in the DLN and spleen was quantified by flow cytometry 7 dpi. Comparison of proportions of GFP+ 2C T cells in the DLN and spleen before (d23) and after antigen restimulation (7dpi). Representative data from three independent experiments with 2-3 mice per group per experiment are shown as mean ± s.e.m. Two-tailed student’s t-tests were used for statistical analyses. ** P<0.01; n.s., not significant.
The observed stronger recall response by Bach2-expressing memory 2C cells could be due to the generation of more memory T cells and/or that the Bach2-expressing memory T cells are more responsive to restimulation. To investigate these possibilities, we activated 2C T cells in vitro for two days and transduced them with retroviruses expressing GFP alone (vector) or GFP plus Bach2 (Fig. 5a). Twenty-four hours later, the T cells were adoptively transferred into B6 mice followed by WSN-SIY virus infection. 2C T cell responses were analyzed by flow cytometry 7 dpi. Compared to the non-transduced 2C T cells, vector-transduced 2C T cells had the same expression profiles for CD62L, IL-7R, Klrg1 and CD27 in the same organs of the same mice (Fig. 5b, upper panel). In contrast, a significantly higher proportion of Bach2-transduced 2C T cells expressed CD62L and CD27, but a significantly lower fraction expressed Klrg1, in the DLN and spleen compared to non-transduced 2C cells in the same organs of the same recipients. When the persisting 2C T cells were analyzed 30 dpi, no significant differences in IFNγ and IL-2 expression were observed among non-transduced, vector-transduced and Bach2-transduced 2C T cells in response to restimulation in vitro (Fig. 5c and Supplementary Fig. S10). Although no difference in CD62L and IL-7R expression was detected, CD27 was higher in Bach2-transduced than vector-transduced 2C T cells in the lung and DLN. (Fig. 5c and Supplementary Fig. S10). Importantly, significantly more Bach2-transduced 2C T cells persisted in the DLN, spleen and bone marrow (Fig. 5d,e). These result suggest that overexpression of Bach2 likely promotes the generation of memory CD8+ T cells in vivo.
Figure 5. Bach2 promotes memory CD8+ T cell development.
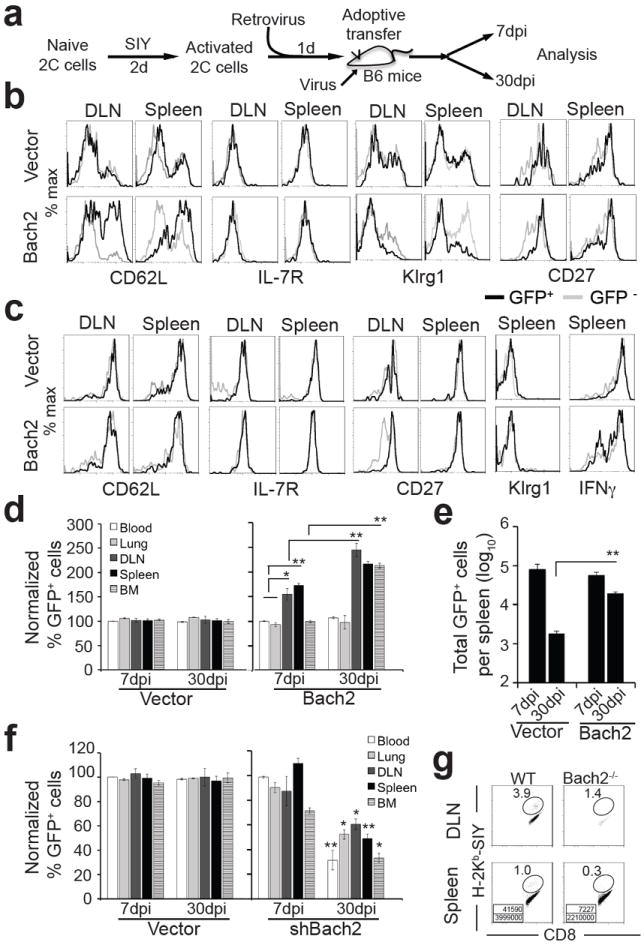
(a) Scheme of experimental protocol for b-f. (b-e) Effect of Bach2 overexpression on memory T cell development. b,c, Persistence and phenotype of transferred 2C T cells over time. Seven and 30 dpi, 2C T cells in various organs were analyzed for CD62L, IL-7R, Klrg1, CD27 and IFNγ as in Fig. 4. Shown are histograms of CD62L, IL-7R, Klrg1 and CD27 expression of Thy1.1+ CD8+ 2C T cells 7 (b) and 30 dpi (c). Gray trace, nontransduced (GFP-) 2C T cells; black trace, transduced (GFP+) 2C T cells. Representative data from 6 mice in 3 independent experiments are shown. (d) Proportion of GFP+ transduced 2C cells in different organs normalized to the average of the blood at 7 dpi. (e) Total Bach2-transduced 2C T cells (GFP+) in the spleen 7 and 30 dpi. (f) Effect of Bach2 knockdown on memory T cell development. Proportion of Bach2-knockdown 2C T cells (GFP+) in different organs normalized to the average of the blood at 7 dpi. (g) Effect of Bach2 knockout on memory T cell development. Chimera mice were constructed by injecting Bach2-/- and Bach2+/+ bone marrow cells into sublethally irradieated Rag2-/- recipient mice. Following reconstitution (3 months later), mice were infected with WSN-SIY virus and 30 dpi cells from DLN and spleen were stained for H-2Kb-SIY and anti-CD8. Shown are staining profiles of H-2Kb-SIY versus CD8. The numbers in the plots indicate percentage of SIY-specific memory CD8+ T cells. The numbers in the boxes indicate the number of SIY-specific memory CD8+ T cells (top) and total CD8+ T cells (bottom). Representative data from three independent experiments with 2-3 mice per group per experiment (d, e) and from two independent experiments with 3-4 mice per group per experiment (f) are shown as mean ± s.e.m. Two-tailed student’s t-tests were used for statistical analyses. * P<0.05; ** P<0.01.
We further investigated the effect of Bach2 overexpression in naïve 2C T cells on memory CD8+ T cell development in vivo. Bone marrow progenitor cells isolated from 2C TCR transgenic mice were transduced with retroviruses expressing GFP alone or GFP plus Bach2 and adoptively transferred into sublethally irradiated Rag2-/- mice to generate naïve 2C T cells that express GFP or GFP plus Bach2 (Fig. 6a). The resulting transduced and non-transduced naïve 2C cells were then adoptively transferred into B6 mice followed by infection with WSN-SIY virus. The frequency, phenotype and function of 2C T cells were analyzed 7 dpi. Vector-transduced and non-transduced 2C T cells from the same recipient mice had the same expression profile of CD62L, IL-7R, Klrg1, IFNγ and TNFα (Fig. 6b). While Bach2-transduced 2C T cells expressed similar levels of IFNγ and TNFα as non-transduced and vector-transduced 2C T cells, more cells expressed CD62L but fewer cells expressed Klrg1, resembling to CD62LhiKlrg1low memory precursors (Fig. 6b and Supplementary Fig. S11). By 30 dpi, no significant differences were observed among non-transduced, vector-transduced and Bach2-transduced 2C T cells in expression of CD62L, IL-7R, Klrg1, IFNγ and TNFα (Fig. 6c). However, more Bach2-transduced 2C T cells were found in the blood, lung, DLN and spleen 30 dpi (Fig. 6d,e). Together, these data suggest that overexpression of Bach2 promotes the development of memory CD8+ T cells.
Figure 6. Bach2 promotes memory CD8+ T cell development.
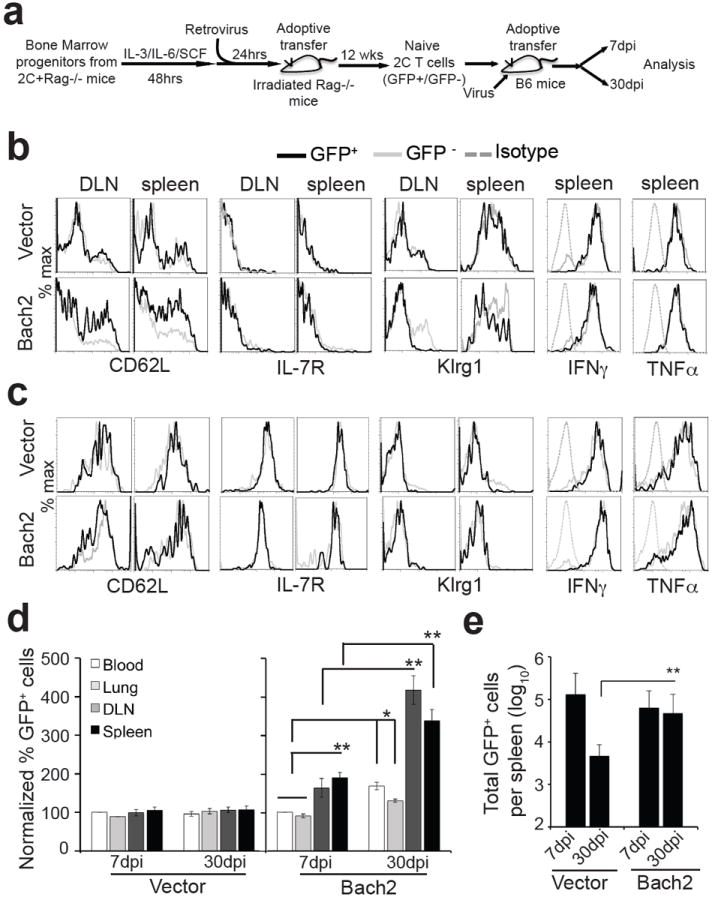
(a) Scheme of experimental protocol. (b-e) Persistence and phenotype of transferred CD8+ T cells over time. Seven and 30 dpi, 2C T cells in various organs were analyzed as in Fig. 4. Shown are histograms of CD62L, IL-7R, Klrg1, IFNγ and TNFα expression of Thy1.1+ CD8+ 2C T cells 7 (b) and 30 dpi (c). Gray trace, nontransduced (GFP-) 2C T cells; black trace, transduced (GFP+) 2C T cells; dash gray trace, isotype control for intracellular staining. Representative data from 9-12 mice in 3 independent experiments are shown. (d) Proportion of GFP+ transduced 2C cells in different organs normalized to the average of the blood at 7 dpi. (e) Total Bach2-transduced 2C T cells (GFP+) in the spleen 7 and 30 dpi. Data from three independent experiments with 3-4 mice per group per experiment are shown as mean ± s.e.m. Two-tailed student’s t-tests were used for statistical analyses. * P<0.05; ** P<0.01.
Diminished memory T cell development due to Bach2 deficiency
We also examined the effect of Bach2 knockdown on memory T cell development. The approach was the same as outlined as in Figure 5a, except the retrovirus expressed shRNA specific for Bach2. Briefly, activated 2C T cells were transduced with shRNA-expressing retrovirus and adoptively transferred into B6 mice followed by infection with WSN-SIY influenza virus. The number and frequency of GFP+ and GFP- 2C T cells in various organs were analyzed 7 and 30 dpi. No significant difference was observed among non-transduced, vector-transduced and shRNA-transduced 2C T cells in terms of CD62L, IL-7R, Klrg1, CD27 and IFNγ expression (Supplemental Fig. S12). However, the proportion of shRNA-transduced 2C T cells was reduced significantly in the blood, lung, DLN, spleen and bone marrow 30 but not 7 dpi (Fig. 5f). Consistently, the number of shRNA-transduced 2C T cells in the spleen was lower as compared to the numbers of non-transduced and vector-transduced 2C T cells 30 dpi (Supplemental Fig. S12c).
To investigate the effect of Bach2 knockout on memory T cell development, we constructed chimeric mice where T and B cells were deficient in Bach2 by adoptively transferring bone marrow cells from Bach2 knockout mice into sublethally irradiated Rag2-/- mice. Three months after reconstitution, mice were infected with WSN-SIY influenza virus and analyzed for the presence of SIY-specific memory CD8+ T cells 30 days later. The percentage of SIY-specific CD8+ T cells was lower in the DLN and spleen of chimeric mice that were reconstituted with Bach2-/- than Bach2+/+ bone marrow cells (Fig. 5g). Consistently, the number of SIY-specific CD8+ T cells in the spleen was lower in mice reconstituted with Bach2-/- than Bach2+/+ bone marrow cells. Considering that Bach2 expression was down-regulated in effector and then up-regulated in memory T cells during naïve to effector to memory cell transition (Table 1), together these results show that Bach2 promotes memory CD8+ T cell development.
Enhanced proliferation of T cells by Bach2 overexpression
To further explore the mechanism underlying the observed effect of Bach2 on memory T cell development and response, we examined whether Bach2 affects T cell proliferation. 2C T cells were activated in vitro and transduced with either vector or Bach2-expressing retroviruses. The cells were cultured in the presence of either IL-2 or IL-7 and the proportion of transduced (GFP+) versus non-transduced (GFP-) cells in the same cultures was quantified over time. In the IL-7 culture, the proportion of transduced versus non-transduced 2C cells remained stable regardless whether the 2C T cells were transduced with vector or Bach2 (Fig. 7a). Similarly, the proportion of vector-transduced versus non-transduced 2C cells remained stable in the IL-2 cultures. However, the proportion of Bach2-transduced 2C T cells increased significantly over time in the IL-2 cultures (Fig. 7a). When the cells were lableled with eFluor and followed over time, Bach2-transduced 2C T cells diluted the flourescent dye more extensively than non-transduced 2C T cells (Fig. 7b). These data suggest that Bach2 promotes proliferation of activated CD8+ T cells.
Figure 7. Bach2 promotes proliferation of CD8+ T cells.
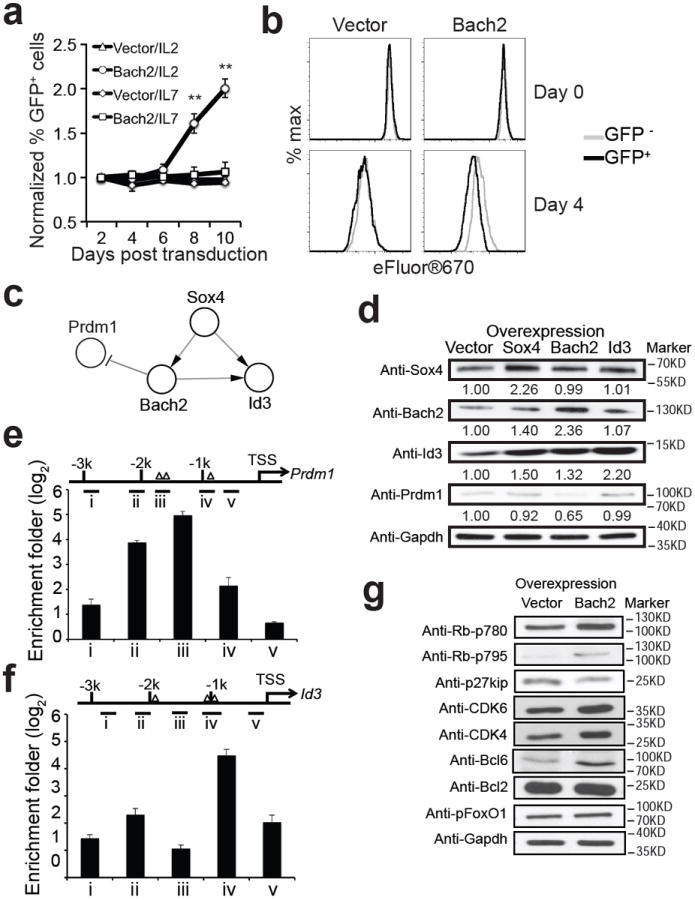
(a) 2C cells were activated in vitro and transduced with retroviruses expressing GFP alone (vector) or GFP plus Bach2 and cultured in the presence of either IL-2 or IL-7. The proportion of GFP+ 2C T cells was quantified. Shown (mean ± s.e.m.) are changes in proportion of GFP+ cells over time from four independent experiments. Two-tailed student’s t-tests were used for statistical analyses. ** P<0.01. (b) Bach2 transduced 2C T cells (day 4 in Fig. 7a) were labeled with eFluor®670 and cultured in the presence of IL-2 for four days. Shown are representative histograms of eFluor®670 from one of the two experiments. Gray trace, nontransduced (GFP-) 2C T cells; black trace, transduced (GFP+) 2C T cells. (c) Sox4-Bach2-Prdm1-Id3 regulatory motif identified in perturbation network. (d) Bach2 inhibits Prdm1 expression but promotes Id3 expression. Activated 2C T cells were transduced with retroviruses expressing GFP alone (vector) or GFP plus Bach2, Sox4 or Id3. The levels of the indicated TFs were assayed by Western blotting. Shown are representative Western blotting and the average expression level quantified from three independent experiments. (e-f) Bach2 binds to Prdm1 and Id3 promoter. Activated 2C T cells were cultured with IL-7 for 24 hours and harvested for ChIP using anti-Bach2. DNA was used to amplify different parts of the Prdm1 (e) and Id3 (f) promoter region indicated by i-v. The location of the predicted Bach2 binding motif was indicted as triangle. Data are from two independent experiments, error bar: SEM. (g) Bach2 affects Rb phosphorylation and p27kip expression. Activated 2C T cells were transduced with retroviruses expressing GFP alone (vector) or GFP plus Bach2 and cultured for 8 days (Fig. 7a). The levels of phosphorylated Rb and FoxO1, CDK4, CDK6, Bcl6, Bcl2 and p27kip in the transduced 2C T cells were assayed by Western blotting.
We noticed a feed-forward regulatory motif of Sox4-Bach2-Prdm1-Id3 in the perturbation network (Fig. 7c and Supplementary Fig. S4). We verified this regulatory motif by showing that overexpression of Bach2 suppressed expression of Prdm1 but stimulated expression of Id3 at transcriptional (Table 3) and translational levels (Fig. 7d). ChIP-PCR analysis with anti-Bach2 antibody also confirmed that Bach2 binds directly to the promoter regions of Prdm1 and Id3 (Fig. 7e, f). Further supporting the regulatory motif, overexpression of Prdm1 significantly reduced the Bach2-mediated proliferation of activated T cells (Supplementary Fig. S13). As Prdm1 is known to inhibit T cell proliferation whereas Id3 stimulates survival of effector T cells 15, 16, we determined the effect of Bach2 overexpression on molecules that regulate cell cycle and survival. Overexpression of Bach2 stimulated CDK4, CDK6 and Bcl6 expression and Rb phosphorylation at amino acid residues 780 and 795 but inhibited expression of p27kip (Fig. 7g). These results suggest that Bach2 regulates memory T cell development and recall proliferation by regulating cell cycle control possibly through Prdm1 and Id3.
Discussion
At the molecular level, development of memory CD8+ T cells is the establishment of MSG expression program, which ultimately is controlled by TFs. Although several TFs have been described to regulate memory CD8+ T cell development, for a comprehensive understanding of transcriptional regulation of memory CD8+ T cell development, it is necessary to identify most, if not all, key TFs that regulate MSGs and construct a genome-wide transcriptional network that supports memory CD8+ T cell development. Using systems biology approaches and publically available gene expression data, here we have assembled a genome-wide regulatory network associated with CD8+ T cells of the mouse. Applying MRA to this network, we have identified twenty-one key TFs, down-narrowed from 1038 putative TFs, which regulate the expression of 70% of MSGs. Our approach is valid based on the following considerations. First, our method identified eight of the twelve TFs known to be involved in memory CD8+ T cell development, including Tcf7, Eomes, Prdm1, Klf2, Id2, Stat4, Id3 and Tbx21 (Table 1). Although Bcl6, Stat3 and Myc are known to regulate memory CD8+ T cell development, they were not within the top 21 TFs identified using our methodology because CLR-inferred targets do not overlap with MSGs possibly due to limited data on MSGs or the network. NF-κb plays important roles in both effector and memory T cell development 27, 28. We found that NF-κb was a key TF that regulates effector signature genes (ESG) when MRA was applied to the network and ESGs. Second, our method identified several TFs that are not known to function in the memory CD8+ T cell development, including Sox4, Bhlhe40, Bach2 and Runx2 in the top 10 TFs (Table 1). Follow-up experimentation showed that these newly identified TFs indeed play important roles in memory CD8+ T cell development and function. Overexpression of Sox4 and Bach2 promoted a significantly higher recall proliferation of memory CD8+ T cells both in vitro and in vivo. Conversely knockdown of Sox4 and Bach2 inhibited the recall proliferation of the transduced memory T cells (Fig. 3). Overexpression of Bhlhe40 and Runx2 inhibited the in vivo recall proliferation, although no significant change was observed in in vitro recall response and in knockdown assay (Fig. 3). Further analysis showed that overexpression of Bach2 also promotes memory CD8+ T cell development (Fig. 4-6). Third, compared to the traditional method of differential gene expression analysis 2, 3, which generates a long list of candidates using fold-change-based approaches, our network methods identify and rank order TFs according to their statistical importance. Reduction of hundreds of TFs to two-dozen key TFs makes direct experimental validation more manageable. The network approach and methodologies developed here can be applied to any phenotypic transition, such as effector T cells and exhausted T cells, to identify novel transcriptional modules and TFs.
Studies have suggested that memory CD8+ T cell development is coordinately regulated by several TFs (reviewed in Refs. 22, 24), including Eomes and Tbx21 4. Our network and perturbation studies have now greatly expanded the understanding of the mode of interactions to the top 21 TFs. Our analysis reveals a dense overlapping regulation (DOR) among the key TFs (Fig. 2b). This mode of regulation is essential for sensing multiple external signals and integrate them into distinct cell fate outcomes 29. As two classes of TFs have been proposed to control the developmental potentials of effector and memory fates in a quantitative manner 22, 24, 30, the DOR might contribute to the quantitative regulations during effector to memory CD8+ T cell development. Our analysis also shows complex regulations with both feedback and feed-forward motifs among the key TFs (Table 3 and Fig. 2b). In the regulatory motif of Sox4-Tcf7-Eomes-Tbx21, Tcf7, Eomes and Tbx21 are known to be critical for memory CD8+ T cell development. The association of Sox4 with these three TFs and especially its “hub” position in this motif are intriguing. Sox4 is not known to regulate memory CD8+ T cell development. However, it stabilizes β-catenin to modulate Wnt-Tcf7 signaling 31, 32, which promotes memory CD8+ T cell development. Sox4 also regulates ‘stemness’ of cancer cells 33, a property shared by memory T cells. Furthermore, evidence suggests that Sox4 might be a direct regulatory target of TGFβ signaling 34, which is essential for the differentiation of CD8+ T cells 35. These previous observations, together with our finding of the “hub” position of Sox4 in the regulatory motif of Sox4-Tcf7-Eomes-Tbx21, suggest that Sox4 is a critical TFs regulating memory CD8+ T cell development. While this hypothesis has yet to be validated, our finding that overexpression of Sox4 promotes recall proliferation of memory T cells suggests that Sox4 is involved in memory CD8+ T cell development.
In the regulatory motif of Sox4-Tcf7-Eomes-Tbx21, we found that Tcf7 binds to Eomes promoter (Fig. 2a) and retroviral expression of Tcf7 leads to a downregulation of Eomes transcript (Table 3) and protein (Supplementary Fig. S5). The latter results contradict with the previous report showing that the level of Eomes transcript and protein are decreased in memory CD8+ T cells from Tcf7-deficient mice 9. Although we do not know the precise causes underlying the observed opposite effects, the following differences between the two studies may provide part of the explanation. First, our study was carried out in in vitro, using activated CD8+ T cells that are in transition to memory T cells, whereas the previous study used memory CD8+ T cells directly from mice. Second, in our study, we overexpressed Tcf7 for a short period (72 hrs) before assaying the effect on Eomes expression, whereas the previous study examined the accumulated effect of germline Tcf7 knockout on Eomes expression. The differences in the stage of T cells, in vitro vs. in vivo, overexpression vs. deficiency, and the length of Tcf7 overexpression or deficiency could all contribute to the observed differences in the two studies. Although the discrepancy raises concern of our approach, results from our perturbation study on the effect of Bach2 on Prdm1, Prdm1 on Tcf7, and Id2 on Id3 are all consistent with previous reports 17, 36, 37, suggesting the validity of our in vitro assay in most cases.
Our detailed analysis of the feed-forward motif of Sox4-Bach2-Prdm1-Id3 (Fig. 7c) reveals new insight into memory CD8+ T cell development. This regulatory motif includes two known TFs (Id3 and Prdm1) and two unknown TFs (Sox4 and Bach2) in memory CD8+ T cell development. Through both overexpression and knockdown/knockout in CD8+ T cells both in vitro and in mice, we provide extensive evidence showing that Bach2 promotes memory CD8+ T cell development (Fig. 4-6). One mechanism appears to be by stimulating the induction of memory T cell precursors as Bach2-expression in effector T cells leads to a phenotype of CD62LhiKlrgloCD27+ (Fig. 5b-c and Supplementary Fig. S10), which is considered as central memory T cell precursors with high proliferative potential 38, 39. Recently, two studies report that Bach2 regulates CD4+ T cell development and function by suppressing effector gene expression 40, 41. Our observation that Klrg1 is suppressed by Bach2 suggests that suppression of effector function may also be important for the development of memory CD8+ T cells. Another mechanism is by stimulating T cell proliferation. We showed that overexpression of Bach2 enhances IL-2 driven T cell proliferation in vitro and recall proliferation in vitro and in vivo. In addition, when in vitro memory 2C T cells were labeled with eFluro dye and adoptively transferred into Rag-/- mice, Bach2-transduced T cells diluted the flourescent dye more extensively than the non-transduced and vector-transduced T cells (Supplemental Fig. S14). The enhanced proliferation could lead to development and/or survival of memory T cells.
Our study further sheds light on the mechanisms by which Bach2 promotes T cell proliferation. In the regulatory motif, Bach2 promotes Id3 expression but suppresses Prdm1 expression through direct binding to their promoter regions (Fig. 7c-f), the latter is consistent with Bach2 suppression of Prdm1 expression in B cells 37. Id3 is known to promote cell cycle and recall proliferation of memory CD8+ T cells by binding to and inhibiting E proteins 15, 42. Thus, by promoting Id3 expression, Bach2 stimulates T cell proliferation. Prdm1 is known to antagonize Bcl6, which promotes cell cycle by suppressing the expression of cell cycle inhibitor p27kip 43. Bcl6-/- mice exhibit a profound deficiency of memory T cells 44, 45, whereas in the absence of p27kip, memory CD8+ T cells exhibit enhanced homeostatic and recall proliferation 46. Consistently, we show that overexpression of Bach2 promotes Bcl6 expression but inhibits p27kip expression (Fig. 7g). Thus, Bach2 also stimulates T cell proliferation by suppressing Prdm1 expression. Together, these findings suggest that Bach2 promotes memory CD8+ T cell development and recall proliferation through Id3- and Prdm1-mediated cell cycle control.
Development of memory CD8+ T cells requires integration of multiple external and internal signals to establish a new transcriptional program of MSGs that endows memory CD8+ T cells with characteristic features in phenotype, tissue distribution, homeostasis and recall potentials. In this study, we have shown that integrated systems biology approaches can be effectively used to identify key TFs and their mode of interactions that underlies memory CD8+ T cell differentiation and function. Further analysis of motifs in the regulatory network should help to elucidate in detail the molecular mechanisms underlying memory CD8+ T cell development and function.
Methods
Regulatory network and master regulator analysis
386 public microarrays related to CD8+ T cells from 35 independent GEO datasets (till September 2009) were downloaded from the NCBI database of Gene Expression Omnibus (GEO) (Supplementary Table S1). All raw image files were reprocessed to normalize the data using R program with a gcRMA method. Gene expression data was used to construct the regulatory network with the putative TFs using a reverse engineering algorithm CLR 19. Among the 1,445 putative TFs identified according to the domain predictions of protein sequences 18, 1,038 were manually mapped to the latest mouse genome. To compare gene expression in different CD8+ T cells, samples were grouped into naïve, effector and memory based the cell types from which the microarray analysis were done (Supplementary Table S1). Gene was considered as expressed in CD8+ T cells if the average gene expression level in one of the three groups was more than 8 (gcRMA values).
To identify TFs associated with memory CD8+ T cell development, we used Master Regulator Analysis (MRA) to compute the statistical significance of overlaps of all interactions of each TF (inferred by CLR) with MSGs or a control gene set by a binomial test. The MSGs were differentially expressed genes between memory CD8+ T cells and naïve/effector CD8+ T cells identified previously 3. 332 background genes were identified from the 386 gene expression profiles based on high levels of gene expression (gcRMA value >10) but minimal variation among 386 samples (variation from mean <0.5). This criterion minimizes the potential of the selected genes not being regulated by TFs in CD8+ T cells. From the 1,038 putative TFs, MRA identified 60 MSG-specific TFs at P<0.05 (binomial test), all of which are expressed in CD8+ T cells (Supplementary Table S2). These 60 candidates were filtered by removing those whose knockout does not have any immune system phenotype as defined in MGI 20. The positive candidates were analyzed for enrichment of DNA-binding motifs in the promoter regions (-2000 to -1) of the MSGs using the program MatInspector 47 or differential expression among naïve, effector and memory CD8+ T cells. This led to 21 key TFs that exhibit immune system phenotype with either an enrichment of DNA-binding motifs among MSGs or differential expression.
Mice and virus
The 2C TCR transgenic mice on Rag2-/- and C57BL/6 (B6 Thy1.1+) background (2C+Rag-/-) were maintained in the animal facility at the Massachusetts Institute of Technology (MIT). These mice express the 2C TCR on CD8+ T cells specific for SIYRYYGL peptide (SIY) in association with MHC class I Kb molecule 48. B6 and Rag2-/- mice were from the Jackson Laboratory. Mice were used at 8-16 weeks of age. All animal studies and procedures were approved by the Massachusetts Institute of Technology’s Committee for Animal Care. Recombinant WSN-SIY virus encoding the SIY epitope in the neuroaminidase stalk was constructed by plasmid-based reverse genetics and grown in Madin-Darby canine kidney cells 26. For infection, mice were anesthetized and given 100 pfu (sublethal dose) intranasally.
Flow cytometry and cell sorting
Antibodies specific for CD8α, Thy1.1, Klrg1, CD62L, CD127 (IL-7R), CD27, IFNγ, TNFα, IL-2, Eomes and T-bet (Tbx21) were purchased from BioLegend or eBiosciences and used at the recommended concentration. Single cell suspensions were prepared from spleens and mediastinal (draining) lymph nodes (DLN), peripheral blood, and lung. Splenocytes and lymphocytes were collected in 8 ml HBSS by crushing the spleen and lymph node with frosted glass slides and filtering the cell suspension through 80 μm nylon filters, respectively. Lungs were harvested and ground through a cell strainer, followed by incubation with 2 ml of digestion buffer (RPMI 1640 medium containing 3mg/ml of collagenase A (Roche), 5% FBS and 10mM HEPES) at 37°C for 1 hour. Red blood cells (RBCs) in the spleen, blood and lung were lysed with RBC lysis buffer (Gibco) and the cells were washed with complete RPMI. The cells were counted and 1-3 ×106 cells were used for surface staining. Cells were washed twice with PBS plus 2% FBS before cytometry analsysis. For intracellular staining, splenocytes were stimulated with SIY peptide for 5 hours in the presence of GolgiPlug (BD Biosciences). Cells were washed twice with PBS with 2% FBS and stained with indicated antibodies. The cells were then fixed and stained with labeled antibodies using an intracellular staining kit (Cytofix/Cytoperm kit; BD Biosciences) according to the manufacture’s instructions. Stained cells were analyzed on either a FACSCalibur or Accuri™ C6 flow cytometer (BD Biosciences). 0.5-2 × 106 events were collected and analyzed with FlowJo software. Cell sorting was carried out with a MoFlo cell sorter or FACSAria (BD Biosciences).
Retrovirus production and infection
Retroviral pMIGw-GW gateway vector was constructed by inserting a gateway cassette at EcoRI site of the pMIGw vector (Addgene #9044) using a gateway construction kit (Invitrogen). All ORFs encoding 12 TFs were amplified with primers (Supplementary Table S3) and cDNA from mouse splenocytes and cloned into pMIGw-GW using the gateway cloning technology. shRNAs for specific TFs (Supplementary Table S4) were chosen from the predicted TRC library and cloned into pMKO.1 GFP retroviral vector (Addgene #10676). Briefly, synthesized single-strand sense and antisense oligonucleotides were annealed into double-strand oligonucleotides for short hairpin RNA in the annealing buffer (10mM TrisCl (pH7.5), 50mM NaCl and 1mM EDTA). The double-strand oligonucleotides were directly treated with T4 polynucleotide kinase (NEB) and ligated into pMKO.1 GFP vector) between AgeI and EcoRI sites.
293FT cells were cultured to 60% confluency in 6-well plates. Cells were co-transfected with retroviral vector plasmid (4μg) and packing plasmids pCL-Eco (1μg) with 150μl DMEM and 15μl TransIT®-LT1 (Mirus) according to the manufacture’s instructions. On the second day, the culture was replaced with fresh medium. On the third day, supernatant was collected and filtered through a 0.45 μm low-protein binding membrane (Pall Life Science). Fresh viral supernatants were used for spin infection of CD8+ T cells in all experiments.
For infection, cells from spleens and lymph nodes were harvested from 2C+RAG-/- mice, pooled and cultured in 6-well plates in the presence of SIY peptide (1μg/ml) in the complete RPMI medium (RPMI 1640 supplemented with 10% FBS, 5mM HEPES, 2 mM glutamine, 100U/ml penicillin, 100μg/ml streptomycin and 50μM β-mercaptoethanol (Invitrogen)). Two days later, activated 2C T cells were collected, washed and resuspended at 2 × 106 cells per ml in the complete RPMI medium. 1ml fresh retrovirus supernatants and 0.25ml 2C cells with a final concentration of 5μg/ml polybrene (American Bioanalytical) were added to one well of a 24-well plate and spun for 90min at 2500rpm at 32°C to infect T cells. 24 hours later, cells were collected for direct adoptive transfer, or resuspended and cultured in 3ml fresh RPMI medium with 100U/ml IL-2 (eBioscience). After culture for 24 hours, 2C T cells were analyzed for GFP expression by flow cytometry and prepared for injection into mice to generate in vivo memory T cells or further culture to generate in vitro memory T cells.
Generation and recall proliferation of memory T cells
To generate in vitro memory T cells, activated (and transduced) 2C T cells were cultured in complete RPMI medium supplemented with 5ng/ml IL-7 (Peprotech) for 7 days with change of fresh IL-7-supplemented medium every two days. Cells were analyzed for memory phenotype by flow cytometry on day 7. To test the recall proliferation, in vitro memory 2C T cells (1×105) were cultured with B6 splenocytes (5×105) in a 12 well plate in complete RPMI medium supplemented with 1 μg/ml SIY peptide and 100 units/ml IL-2. The numbers and phenotype of 2C T cells were analyzed by flow cytometry 4 and 6 days later. Alternatively, in vitro memory 2C T cells (2×105) were transferred to B6 recipients and challenged with WSN-SIY virus. 2C cells were analyzed by flow cytometry 5 dpi.
To generate in vivo memory T cells, activated and transduced 2C T cells were adoptively transferred into B6 recipients. Twenty-third days later, the frequency, phenotype and function of persisting 2C T cells were analyzed by flow cytometry. To assay for recall response, mice were infected with 100pfu WSN-SIY virus and the number and phenotype of 2C T cells in different organs were analyzed by flow cytometry 7 dpi.
Bone marrow chimera mice
Bone marrow cells were collected from the tiba and femur of 2C+Rag-/- mice. Stem and progenitor cells were enriched using a progenitor enrichment kit (Stemcell Technologies) according to the manufacture instructions. The enriched cells were cultured for 48 hours in complete RPMI medium supplemented with IL-3 (30ng/ml), IL-6 (10ng/ml) and SCF (15ng/ml). The cells were resuspended at 2 × 106 cells per ml in complete RPMI. 600μl fresh retrovirus supernatants and 400μl cells plus a final concentration of 6 μg/ml polybrene were added to one well of a 24-well plate and spun for 90min at 2500rpm at 32°C. One the second day, cells were collected and washed and injected into Rag-/- mice that had been irradiated for 500rads 4 hours earlier. Eight weeks later, mice were bleeded to determine the reconstitution of CD8+ T cells and GFP proportion by flow cytometry. Twelve weeks later, cells were collected from spleen and analyzed for 2C T cell percentage and phenotype. Splenocytes containing 5×104 2C T cells were adoptively transferred into B6 mice followed by WSN-SIY infection. The number, phenotype and function of 2C T cells in the recipient mice were analyzed 7 and 30 dpi. To generate Bach2-/- chimeric mice, bone marrow cells from Bach2-/- mice 49 (kindly gift of Dr. Kazuhiko Igarash of Tohoko University, Japan) were directly injected into sublethally irradiated Rag2-/- mice. Eight weeks later, reconstitution of CD8+ T cells were verified by flow cytometry of peripheral blood mononuclear cells. Twelve weeks later, mice were infected with 50 pfu WSN-SIY virus and 30 dpi SIY-specific CD8+ T cells in various tissues were identified by H-2Kb DimerX (BD Biosciences) loaded with SIY peptide plus anti-CD8 by flow cytometry.
Gene perturbations and quantitative PCR
To perturb the network, selected TFs were overexpressed in CD8+ 2C T cells by retrovirus transduction as described above. Transduced 2C T cells were cultured in the presence of IL-7 for 24 hours and GFP+ 2C T cells were purified by sorting (>95% viable by PI staining). Total RNA was extracted from the purified 2C T cells using RNeasy micro kit (Qiagen) according to the manufacture’s instructions. First strand cDNA was synthesized from 1 μg total RNA using the TaqMan® Reverse Transcription Reagents (ABI). 2 μl of diluted cDNA (total 200 μl) were used as template for the quantitative PCR with LightCycler®480 SYBR Green and LightCycler®480 machine (Roche). For each TF transduced CD8+ T cells, the transcript levels of 12 TFs were measured by qPCR using gene specific primers (Supplementary Table S5). To measure the transcript level of TFs in naïve, effector and memory CD8+ T cells, naïve 2C T cells were adoptively transferred into B6 mice followed with WSN-SIY virus infection, effector and memory 2C T cells were sorted from spleen 7 dpi and 30 dpi, respectively. Total RNA was isolated from naïve, effector and memory 2C T cells and used for quantification of the transcript level of each TF by PCR.
ChIP and ChIP-PCR
A Millipore ChIP kit was used for chromatin immunoprecipitation. DNA-protein complexes were cross-linked with formaldehyde at a final concentration of 1%, sheared by sonication to 800~1000bp, followed by precipitation with nonspecific goat anti-IgG (Sigma) or rabbit anti-IgG (Cell Signaling Technology) or chromatin ChIP-grade anti-Sox4 (C-20, Santa Cruz Biotechnology), anti-Tcf7 (H-118, Santa Cruz Biotechnology), and anti-Eomes (ab23345, Abcam). DNA-protein complex was eluted, and ChIP DNA was purified by PCR purification kit (Qiagen). The promoter regions of the indicated TFs or MSGs were amplified using specific primers (Supplementary Table S6). Primers used to amplify the promoter regions were all within this 1 kb upstream of the transcription-starting site. For ChIP of Bach2 with anti-Bach2 (E-16, Santa Cruz Biotechnology), cross-linked DNA-protein complexes were digested to 400~600bp by micrococcal nuclease (Cell Signaling Technology).
Protein extraction and western blotting
Proteins were extracted from transduced 2C T cells with the CelLytic™ Lysis Reagent (Sigma). Samples containing 20μg total protein (BCA™ Protein Assay Kit, Pierce Biotechnology) were resolved on a 10% SDS-PAGE gel and electro-transferred onto a PVDF membrane (Millipore Corporation). The membrane was blocked in 5% (w/v) fat-free milk in PBST (PBS containing 0.1% Tween-20). The blot was hybridized overnight with primary antibodies: anti-GAPDH (HRP-conjugated, Cell Signaling Technology, 1:2000), anti-Sox4 (C-20, Santa Cruz Biotechnology, 1:500), anti-Bach2 (AP10133b, Abgent, 1:500), anti-Id3 (6-1, CalBioreagents, 1:2500), anti-Blimp-1 (6D3, eBioscience, 1:1000), anti-Bcl2 (BioLegend, 1:500), anti-Bcl6 (BioLegend, 1:2000), anti-pFoxO1 (Cell Signaling Technology, 1:1000), anti-CDK4 (Cell Signaling Technology, 1:1000), anti-CDK6 (Cell Signaling Technology, 1:1000), anti-Rb-p780 (Cell Signaling Technology, 1:1000), anti-Rb-p795 (Cell Signaling Technology, 1:1000) and anti-p27kip (Cell Signaling Technology, 1:1000) according to the recommended dilution in 5% fat free milk. The blot was washed twice in PBST and then incubated with HRP-conjugated secondary antibody (Cell Signaling Technology: anti-Rabbit, 1:2000; anti-mouse,1:3000. Santa Cruz Biotechnology: anti-Rat, 1:2000; anti-Goat 1:3000) in 5% fat-free milk. The membrane was washed twice in PBST and subjected to protein detection by ECL Plus Western Blotting Detection System (GE Healthcare) before being exposed to a Koda BioMax XAR film. The membrane was stripped and re-blotted with the rabbit anti-mouse HRP-conjugated anti-Gapdh antibody (Cell Signaling Technology) for protein loading control.
Statistical analysis
Statistical significance was determined with the two-tailed unpaired or paired Student’s t-test. P-values for MRA and promoter enrichment results were calculated with a binomial test. The FDRs were computed with q = p*n/i, (p = P value, n = total number of tests, i = sorted rank of P value).
Supplementary Material
Acknowledgments
The authors thank Ching-Hung Shen, Zhuyan Guo, and Camille M. Justino for technique assistance, Pete S. Bak and Herman Eisen for discussion, and Professor Kazuhiko Igarash for providing Bach-/- bone marrow cells. This work was supported in part by National Institutes of Health Grant AI69208, funds from the Singapore-MIT Alliance, and Ivan R. Cottrell Professorship and Research Fund (to J.C.), and the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute.
Footnotes
Author contributions: G.H. and C.J. conceived and designed the study. G.H. carried out all computations and experimentations. G.H. and C.J. performed data analysis and drafted the paper. All authors read and approved the final manuscript.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–261. doi: 10.1038/46218. [DOI] [PubMed] [Google Scholar]
- 2.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 3.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 5.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci U S A. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gattinoni L, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao DM, et al. Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J Immunol. 2010;184:1191–1199. doi: 10.4049/jimmunol.0901199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schober SL, Kuo CT, Schluns KS, Lefrancois L, Leiden JM, Jameson SC. Expression of the transcription factor lung Kruppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. J Immunol. 1999;163:3662–3667. [PubMed] [Google Scholar]
- 11.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 12.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 13.Yang CY, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannarile MA, et al. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 15.Ji Y, et al. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol. 2011;12:1230–1237. doi: 10.1038/ni.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 17.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 19.Faith JJ, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE Group MGD. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic acids research. 2012;40:D881–886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angelosanto JM, Wherry EJ. Transcription factor regulation of CD8+ T-cell memory and exhaustion. Immunol Rev. 2010;236:167–175. doi: 10.1111/j.1600-065X.2010.00927.x. [DOI] [PubMed] [Google Scholar]
- 22.Rutishauser RL, Kaech SM. Generating diversity: transcriptional regulation of effector and memory CD8 T-cell differentiation. Immunol Rev. 2010;235:219–233. doi: 10.1111/j.0105-2896.2010.00901.x. [DOI] [PubMed] [Google Scholar]
- 23.D’Cruz LM, Rubinstein MP, Goldrath AW. Surviving the crash: transitioning from effector to memory CD8+ T cell. Semin Immunol. 2009;21:92–98. doi: 10.1016/j.smim.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaech SM, Cui W. Transcriptional control of effector and memory CD8(+) T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen CH, Ge Q, Talay O, Eisen HN, GarcÌa-Sastre A, Chen J. Loss of IL-7R and IL-15R expression is associated with disappearance of memory T cells in respiratory tract following influenza infection. J Immunol. 2008;180:171–178. doi: 10.4049/jimmunol.180.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hettmann T, Opferman JT, Leiden JM, Ashton-Rickardt PG. A critical role for NF-kappaB transcription factors in the development of CD8+ memory-phenotype T cells. Immunol Lett. 2003;85:297–300. doi: 10.1016/s0165-2478(02)00260-2. [DOI] [PubMed] [Google Scholar]
- 28.Teixeiro E, et al. Different T cell receptor signals determine CD8+ memory versus effector development. Science. 2009;323:502–505. doi: 10.1126/science.1163612. [DOI] [PubMed] [Google Scholar]
- 29.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 30.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernard P, Harley VR. Acquisition of SOX transcription factor specificity through protein-protein interaction, modulation of Wnt signalling and post-translational modification. Int J Biochem Cell Biol. 2010;42:400–410. doi: 10.1016/j.biocel.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 32.Sinner D, et al. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Kuwahara M, et al. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat Immunol. 2012;13:778–786. doi: 10.1038/ni.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjabi S, Mosaheb MM, Flavell RA. Opposing effects of TGF-beta and IL-15 cytokines control the number of short-lived effector CD8+ T cells. Immunity. 2009;31:131–144. doi: 10.1016/j.immuni.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masson F, et al. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J Immunol. 2013;190:4585–4594. doi: 10.4049/jimmunol.1300099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochiai K, et al. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem. 2006;281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- 38.Buchholz VR, et al. Disparate individual fates compose robust CD8+ T cell immunity. Science. 2013;340:630–635. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 39.Gerlach C, et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 2013;340:635–639. doi: 10.1126/science.1235487. [DOI] [PubMed] [Google Scholar]
- 40.Roychoudhuri R, et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature. 2013;498:506–510. doi: 10.1038/nature12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsukumo S, et al. Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proc Natl Acad Sci U S A. 2013;110:10735–10740. doi: 10.1073/pnas.1306691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivera R, Murre C. The regulation and function of the Id proteins in lymphocyte development. Oncogene. 2001;20:8308–8316. doi: 10.1038/sj.onc.1205091. [DOI] [PubMed] [Google Scholar]
- 43.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 44.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ichii H, Sakamoto A, Kuroda Y, Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J Immunol. 2004;173:883–891. doi: 10.4049/jimmunol.173.2.883. [DOI] [PubMed] [Google Scholar]
- 46.Singh A, Jatzek A, Plisch EH, Srinivasan R, Svaren J, Suresh M. Regulation of memory CD8 T-cell differentiation by cyclin-dependent kinase inhibitor p27Kip1. Mol Cell Biol. 2010;30:5145–5159. doi: 10.1128/MCB.01045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cartharius K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Eisen HN, Kranz DM. A model T-cell receptor system for studying memory T-cell development. Microbes Infect. 2003;5:233–240. doi: 10.1016/s1286-4579(03)00016-9. [DOI] [PubMed] [Google Scholar]
- 49.Muto A, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 50.Kuwahara M, et al. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-beta and suppresses T(H)2 differentiation. Nat Immunol. 2012;13:778–786. doi: 10.1038/ni.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun H, Lu B, Li RQ, Flavell RA, Taneja R. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat Immunol. 2001;2:1040–1047. doi: 10.1038/ni721. [DOI] [PubMed] [Google Scholar]
- 52.Vaillant F, Blyth K, Andrew L, Neil JC, Cameron ER. Enforced expression of Runx2 perturbs T cell development at a stage coincident with beta-selection. J Immunol. 2002;169:2866–2874. doi: 10.4049/jimmunol.169.6.2866. [DOI] [PubMed] [Google Scholar]
- 53.Li Q, Eppolito C, Odunsi K, Shrikant PA. IL-12-programmed long-term CD8+ T cell responses require STAT4. J Immunol. 2006;177:7618–7625. doi: 10.4049/jimmunol.177.11.7618. [DOI] [PubMed] [Google Scholar]
- 54.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cruz-Guilloty F, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karwot R, et al. Protective role of nuclear factor of activated T cells 2 in CD8+ long-lived memory T cells in an allergy model. J Allergy Clin Immunol. 2008;121:992–999.e996. doi: 10.1016/j.jaci.2007.12.1172. [DOI] [PubMed] [Google Scholar]
- 57.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 58.Cortes M, Wong E, Koipally J, Georgopoulos K. Control of lymphocyte development by the Ikaros gene family. Curr Opin Immunol. 1999;11:167–171. doi: 10.1016/s0952-7915(99)80028-4. [DOI] [PubMed] [Google Scholar]
- 59.Oestreich KJ, Yoon H, Ahmed R, Boss JM. NFATc1 regulates PD-1 expression upon T cell activation. J Immunol. 2008;181:4832–4839. doi: 10.4049/jimmunol.181.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isakov N, Altman A. Protein kinase C(theta) in T cell activation. Annu Rev Immunol. 2002;20:761–794. doi: 10.1146/annurev.immunol.20.100301.064807. [DOI] [PubMed] [Google Scholar]
- 61.Kim HJ, Nel AE. The role of phase II antioxidant enzymes in protecting memory T cells from spontaneous apoptosis in young and old mice. J Immunol. 2005;175:2948–2959. doi: 10.4049/jimmunol.175.5.2948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


