Abstract
Alterations in xenobiotic metabolizing enzyme (XME) expression across the life course, along with genetic, nutritional, and environmental regulation, can influence how organisms respond to toxic insults. In this study, we investigated the hypothesis that in utero exposure to the endocrine active compound, bisphenol A (BPA), influences expression and epigenetic regulation of phase I and II XME genes during development. Using healthy 1st to 2nd trimester human fetal liver specimens quantified for internal BPA levels, we examined XME gene expression using PCR Array (n =8) and RNA-sequencing (n =12) platforms. Of the greater than 160 XME genes assayed, 2 phase I and 12 phase II genes exhibited significantly reduced expression with higher BPA levels, including isoforms from the carboxylesterase, catechol O-methyltransferase, glutathione S-transferase, sulfotransferase, and UDP-glucuronosyltransferase families. When the promoters of these candidate genes were evaluated in silico, putative binding sites for the E-twenty-six (ETS) and activator protein1 (AP1) related transcription factor families were identified and unique to 97% of all candidate transcripts. Interestingly, many ETS binding sites contain cytosine-guanine dinucleotides (CpGs) within their consensus sequences. Thus, quantitative analysis of CpG methylation of three candidate genes was conducted across n =50 samples. Higher BPA levels were associated with increased site-specific methylation at COMT (P <0.005) and increased average methylation at SULT2A1 (P <0.020) promoters. While toxicological studies have traditionally focused on high-dose effects and hormonal receptor mediated regulation, our findings suggest the importance of low-dose effects and nonclassical mechanisms of endocrine disruption during development.
Keywords: xenobiotic metabolism, bisphenol A, liver, DNA methylation, transcription factor
INTRODUCTION
A compound’s absorption, distribution, metabolism, and excretion via biotransformation pathways dictate its therapeutic or toxic potential. Specifically, changes in xenobiotic metabolizing enzyme (XME) gene expression and activity can alter drug efficacy and toxicity, with the greatest discrepancies observed in children compared with adults [Alcorn and McNamara, 2003]. In general, the oxidizing phase I XMEs functionalize compounds into active metabolites while the conjugating phase II XMEs increase the molecular weight of compounds for rapid excretion of toxic metabolites [Caldwell et al., 1995]. Preliminary studies indicate that relative XME expression and activity among different classes and isoforms change drastically throughout the life course [Hines, 2008]. For example, the phase II UDP-glucuronosyltransferase isoforms, UGT1A1 and UGT1A6, are both expressed at low levels in the human fetus; while UGT1A1 attains adult levels within months after birth, the UGT1A6 isoform reaches adult levels at 10 years of age [McCarver and Hines, 2002]. Therefore, the extent of metabolism of a xenobiotic chemical is dependent on age and ontogeny, or the maturation of specific XMEs [Allegaert et al., 2007]. Much of our understanding of human XME ontogeny arises from adverse pharmaceutical exposures or diseases throughout birth and infancy, or extrapolations from rodent models [Saghir et al., 2012]. Comprehensive studies examining metabolism in early human fetal development are limited; for example, recent studies characterize only a handful of XMEs, such as steroidogenic enzymes in 2nd trimester fetal liver [O’Shaughnessy et al., 2013] or cytochrome p450s (CYP) and glutathione s-transferases in fetal liver and adrenals [Wang et al., 2008]. Evaluating ontogeny, especially throughout human gestation, is thus of great importance in assessing toxicity; however, obtaining suitable human specimens may pose ethical and technical challenges.
In addition to drug metabolism, XMEs play an important role in steroid homeostasis, neuroendocrine function, and growth. Both endogenous and exogenous compounds help regulate XME ontogeny and subsequently, metabolic function. More recently, studies demonstrating the importance of hormonal regulation on the establishment of hepatic metabolism throughout pregnancy and development have emerged [Kennedy, 2008; Jeong, 2010]. Xenobiotics that can mimic endogenous hormones, such as endocrine active compounds (EAC), can potentially modify baseline hormonal regulation of xenobiotic metabolism and response to environmental stressors during critical windows of development.
Bisphenol A (BPA), a synthetic estrogen used in the production of polycarbonate plastics and epoxy resin, is a controversial EAC that is of concern primarily in the developing organism [Rubin, 2011]. Presence of BPA in fetal tissue [Cao et al., 2012; Nahar et al., 2012], reduced capacity for BPA metabolism in the fetus [Nahar et al., 2012], and the ability for BPA transfer across the placenta [Schonfelder et al., 2002; Balakrishnan et al., 2010; Jiménez-Díaz et al., 2010], place the developing human fetus at a higher risk for BPA toxicity. Although the human health consequences of BPA exposures are disputed, several rodent studies suggest that developmental exposure to BPA can alter susceptibility to disease later in life by modifying the epigenome [Ho et al., 2006; Bromer et al., 2010; Kundakovic and Champagne, 2011; Anderson et al., 2012]. Thus, we hypothesize that BPA dependent changes to epigenetic regulation of XME during development may also result in latent effects on disease susceptibility. To date, research addressing BPAs influence on XME expression and activity has been limited to animal and in vitro models. For example, rat hepatic microsome studies show that BPA inhibits CYP enzymes, including activity for CYP1A2, CYP2C11, and CYP2E1 [Hanioka et al., 2000; Pfeiffer and Metzler 2004]; yeast and human liver microsome studies associate BPA with both competitive and noncompetitive inhibition of UGT1A6 [Hanioka et al., 2008]; and human endometrial Ishikawa cell line studies show increased ALDH3A1 expression following BPA exposure [Naciff et al., 2010]. Interspecies differences in metabolism and/or relatively high BPA exposure doses, however, limit the translation of these findings for human risk assessment.
The influence of xenoestrogens on the maturation and regulation of phase I and II xenobiotic metabolizing enzymes has yet to be studied in humans. Here, utilizing a comprehensive approach evaluating XME expression, we identify multiple XME genes that are associated with physiologically relevant concentrations of total BPA, assess relative abundance of these XME mRNAs in the developing human fetal liver, and investigate DNA methylation as a potential mechanism influencing biotransformation response to BPA exposure.
METHODS AND MATERIALS
Tissue Samples
Human fetal liver samples were procured from the NIH-funded University of Washington Birth Defects Research Laboratory fetal biobank (2R24HD000836-47), and characterized for BPA concentrations by the Kannan Laboratory at the Wadsworth Center (New York State Department of Health). As previously described [Nahar et al., 2012], following surgery and consent from volunteers undergoing elective abortions during 1st and 2nd trimester of pregnancy (gestational day 74–120), healthy tissue specimens were flash frozen and immediately stored in polycarbonate-free tubing at −80°C until processed for BPA analysis and RNA/DNA extraction. No identifying clinical data were available on samples except for sex and gestational age. Total BPA concentrations measured in liver tissue ranged from below the limit of quantification at 0.071 ng/g (LOQ/√2, where LOQ =0.1 ng/g) up to 96.8 ng/g [Nahar et al., 2012].
RNA Extraction and cDNA Synthesis
Total RNA was isolated from frozen liver tissue using the AllPrep DNA/RNA/Protein kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Approximately 10 to 20 mg of homogenized tissue was added to 600 μL of Buffer RLT (containing 1% β-mercaptoethanol) in a 2 mL round bottom polypropylene eppendorf tube with a 5 mm stainless steel bead. Samples were further homogenized in solution for 2 min at 20 Hz (2×) in the TissueLyser II (Qiagen). The purity and quantity of RNA was assessed using the Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Only high quality samples with RNA integrity number (RIN) >7 were used for RT2 Profiler PCR Arrays (n =8) and high-throughput RNA-sequencing (n =12). Three samples were run on both expression platforms for a total sample size of n =17.
To produce complementary DNA for each sample, 1 μg of total RNA template was used with the iScript cDNA synthesis Kit (Bio-Rad, Hercules, CA) following the manufacturer’s protocol. The thermocycler settings for cDNA synthesis required incubation at 25°C for 5 min, 42°C for 60 min, and 90°C for 5 min.
Human Drug Metabolism RT2 ProfilerTM PCR Array
Differential mRNA expression of various xenobiotic metabolism genes were assessed by using two different cataloged human arrays from SuperArray Biosciences (Frederick, MD): the Phase I Enzyme PCR Array (PAH-068E4) and the Phase II Enzyme PCR Array (PAH-069E4). Each SYBR Green-based 96-well PCR Array platform interrogates multiple genes and isoforms relevant to a specific biological pathway along with five reference genes and several controls per sample. Based on the distribution of total BPA levels measured in 50 fetal liver samples [Nahar et al., 2012], the median BPA concentration (3.44 ng/g) was used as a cutoff for group categorization. The final samples were chosen based on the highest RIN scores and a >10-fold difference in average total BPA between low and high exposure groups. Four liver RNA samples exhibiting low total BPA concentrations (1.2–2.9 ng/g) and four liver RNA samples exhibiting high total BPA concentrations (35.4–56.1 ng/g) were submitted to the University of Michigan Sequencing Core facility and run on both PCR Array platforms using the ABI Prism 7900 HT Sequence Detection System (Applied Biosystems).
PCR Array Gene Expression Analysis
The threshold cycle (CT) was obtained for each target gene, and an average reference CT was calculated for the five endogenous reference genes: beta-2-microglobulin (B2M), hypoxanthine phosphoribosyltransferase I (HPRT1), ribosomal protein L13a (RPL13A), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and beta actin (ACTβ). Positive PCR control and reverse transcription controls were either undetected or had an extremely high CT value. Results are reported as ΔCT, which represents the difference between the CT of the target gene versus the average CT of the reference genes. The average ΔCT of the low exposure samples were subtracted from the average ΔCT of the high exposure samples to obtain the ΔΔCT value, and fold change was calculated as 2−ΔΔCT. A linear model designed for microarray analysis was utilized to increase the power to detect fold change differences in target arrays by pooling information using the limma package of Bio-conductor in R statistical software version 2.13.2 [Smyth, 2004]. Furthermore, P values were adjusted for multiple comparisons using false discovery rate [Benjamini and Hochberg, 1995]. Candidate genes were chosen based on greater than 2-fold or less than 0.5-fold change and P <0.05.
Next Generation RNA Sequencing
To characterize the relative mRNA abundance of XMEs in human fetal liver, regardless of sex or exposure, we utilized information from a second set of samples (n =12, including three samples also used in the PCR Arrays) submitted for high throughput next generation mRNA sequencing (RNA-seq) using the Illumina HiSeq 2000 platform (Illumina, San Diego, CA). General work-flow for next generation sequencing library construction consisted of enrichment of poly-A RNA, fragmentation, RNA clean up, cDNA synthesis using hexamer primers, end repair, and adaptor ligation, as previously described [Sengupta et al., 2011]. The raw sequencing image data was analyzed with the Illumina analysis pipeline, using single reads up to 150 base pairs (bp) in length. Over 438 million reads across 12 samples were obtained, with an average read number of 36.5 million per sample. The FastQC tool was used to perform quality control checks on raw data. In order to utilize reads with the best quality calls (FastQC per base sequence quality score above 28), sequences were trimmed to 70 bases in length, and reads were aligned to the hg19 human reference genome using TopHat. Parameters with the best alignment and increased search time were used in order to improve sensitivity. After removing duplicate reads resulting from PCR duplicates with SamTools, transcript abundance was measured by CuffDiff v2.0.1 software as fragments per kilobase of exon per million fragments mapped (FPKM). Total fragment reads were tabulated and normalized across 12 samples. Approximately 152 genes of the phase I and phase II genes on the PCR Arrays were identified in the RNA-seq dataset along with their normalized FPKM reads. The bottom and top quartile reads in FPKM are reported, indicative of low and high relative expression of XME genes within phase I and phase II pathways during development.
Transcription Factor Binding Site Identification
An in silico bioinformatics approach was used to identify common promoter regulatory elements within PCR Array BPA-associated candidate genes. Gene2Promoter (Genomatix, Munich, Germany) software utilizes position weight matrices to predict transcription factor binding sites (TFBS) 500 bp upstream and 100 bp downstream of transcription start sites (TSS) of candidate genes [Cartharius et al., 2005]. Using MatInspector, fasta-formatted input sequences of the PCR Array candidate gene promoters were compared, and common transcription factors were clustered together into TFBS families or “matrix families” based on similarities in binding domain or function. Regulatory families were reported when TFBS were identified in >97% of all relevant transcripts of the candidate XME genes.
Bisulfite Conversion and Epityper Methylation Analysis
To quantitatively assess methylation, high quality DNA using the Allprep kit was obtained from a larger clinical cohort (total n =50) including specimens tested for expression on the PCR Array and RNA-seq platforms. Approximately 1 μg of genomic DNA was subject to sodium bisulfite treatment using the Qiagen Epitect kit automated on the Qiagen Qiacube (Qiagen Inc., Valencia, CA) in order to produce bisulfite converted DNA for methylation analysis. Epitect methylated and unmethylated human bisulfite converted samples (Qiagen) were used as positive controls. T7 tagged primers for the moderately expressed CES2, COMT, and SULT2A1 candidate genes were designed for promoter methylation analysis using the EpiDesigner Web tool (http://www.epidesigner.-com/, Table IV). Candidate gene promoter regions containing consensus sequences for ETS related transcription factors were amplified using HotStarTaq master mix in a 30 μL PCR reaction. A standard HotStarTaq thermal cycling setting was used for the three assays with the following changes: CES2 required annealing at 56°C for 30 sec at 35 cycles, COMT required annealing at 60°C for 30 sec at 35 cycles, and SULT2A1 required annealing at 56°C for 30 sec at 40 cycles.
TABLE IV.
Sequenom Epityper Primer Assays for DNA Methylation
| Gene name | GenBank number | Strand | Primers (5′–3″) | Tm (°C) | Amplicon size | Amplicon locationa |
|---|---|---|---|---|---|---|
| Carboxylesterase 2 (CES2) | NM_003869 | Forwardb | aggaagagagTTTTGGTTTTGTATATTTGGTGAGA | 58.93 | 310 | −1,900 bp to −1,590 bp |
| Reversec | cagtaatacgactcactatagggagaaggctATTCAAACCCTAATTATCTCCCTCC | 61.01 | ||||
| Catechol-O-methyltransferase (COMT)d | NM_000754 | Forwardb | aggaagagagTTTTAGTTTTTTTATTTGGGAAGGG | 60.00 | 343 | −245 bp to +98 bp |
| Reversec | cagtaatacgactcactatagggagaaggctAACAACCCTAACTACCCCAAAAAC | 60.70 | ||||
| Sulfotransferase 2A1 (SULT2A1) | NM_003167 | Forwardb | aggaagagagAGGTTGTTTTGTATAGATGTGGGTTAT | 59.48 | 294 | −1,953bp to −1,659bp |
| Reversec | cagtaatacgactcactatagggagaaggctTCCCAACTACTCAAAAAACTAAAACA | 59.56 |
Surrounding transcription start site (TSS) where (−) represents upstream and (+) represents downstream of TSS.
All forward methylation primers contain a 10mer tag on the 5′ end, represented in lower case letters.
All reverse methylation primers include a T7 promoter tag on the 5′ end, represented in lower case letters.
Methylation designed for the MB-COMT (membrane bound) promoter.
After in vitro transcription and uracil-cleavage of amplicons as previously described [Coolen et al., 2007], promoter methylation was quantified using the mass-spectrometry based Sequenom MassArray EpiTYPER platform (Sequenom, San Diego, CA) in the University of Michigan DNA Sequencing Core. Associations between amplicon average DNA methylation and total BPA exposure were assessed using a linear mixed effects model. For this model, percent methylation was the dependent variable and CpG unit number was the random effect, while total BPA exposure, sex, and gestational age were set as fixed effects. The analysis incorporated multiple site-specific CpG units (4 for CES2 and SULT2A1; 17 for COMT) and accounted for correlations between adjacent dinucleotides for each assay. The association between CpG site-specific methylation of ETS consensus sequences and total BPA was assessed using multiple linear regression after adjusting for sex and gestational age. For each statistical analysis, regression diagnostics were used to identify samples that deviate from standard linear regression assumptions (e.g. constant variance and normal distribution of residuals). Residuals were determined to be within cook’s distance cutoff (<1); thus no data points were excluded from the final analysis. Complete methylation data was analyzed for the following number of samples: n =50 for CES2 and COMT, and n =40 for SULT2A1. Due to technical limitations, 10 samples failed the SULT2A1 Sequenom assay, and SULT2A1 CpG site #1, which falls within the ETS consensus sequence, was not resolvable on any sample via the Sequenom platform.
RESULTS
BPA Exposure Dependent XME Gene Expression
Using the RT2 ProfilerTM PCR Array, we analyzed 167 genes involved in xenobiotic metabolism (93 phase I and 74 phase II enzyme isoforms) in human fetal liver specimens. Using gene selection criteria (P <0.05 and fold change >2 or <0.5), 2 phase I genes and 12 phase II genes exhibited altered expression based on low (1.2–2.9 ng/g total; n =4) versus high (35.4–56.1 ng/g total; n =4) tissue BPA concentrations (Table I). All differentially expressed candidate genes exhibited significantly decreased expression in liver samples containing high compared with low tissue BPA concentrations. The phase I genes include the CES2 and CES5A carboxylesterase isoforms, while the phase II genes include catechol-O-methyltransferase (COMT), glutathione S-transferase (GSTA5), sulfotransferase (SULT2A1, 1B1, 6B1, 1C3), and UDP-glucuronosyltransferase (UGT1A3, 1A4, 1A6, 1A8, 1A9, 1A10).
TABLE I.
Differentially Expressed Phase I and II XME Genes by High BPA Exposure
| Xenobiotic metabolism enzyme | Symbol | Description | Fold changea | Adjusted P |
|---|---|---|---|---|
| Phase I | CES2 | Carboxylesterase | 0.186 | 0.049 |
| Phase I | CES5A | 0.124 | 0.049 | |
| Phase II | COMT | Catechol-O-methyltransferase | 0.245 | 0.025 |
| Phase II | GSTA5 | Glutathione S-transferase | 0.140 | 0.049 |
| Phase II | SULT2A1 | Sulfotransferase | 0.248 | 0.025 |
| Phase II | SULT1B1 | 0.159 | 0.026 | |
| Phase II | SULT6B1 | 0.054 | 0.026 | |
| Phase II | SULT1C3 | 0.185 | 0.026 | |
| Phase II | UGT1A3 | UDP-glucuronosyltransferase | 0.122 | 0.034 |
| Phase II | UGT1A4 | 0.121 | 0.028 | |
| Phase II | UGT1A6 | 0.249 | 0.037 | |
| Phase II | UGT1A8 | 0.048 | 0.025 | |
| Phase II | UGT1A9 | 0.094 | 0.045 | |
| Phase II | UGT1A10 | 0.187 | 0.026 |
Summary of XME genes identified as significantly altered in fetal liver with high BPA exposure compared with low BPA exposure using the human xenobiotic metabolism Phase I and Phase II PCR Array platforms.
Fold change <0.5 is downregulation; fold change >2 is upregulation.
Relative Expression Levels of XME Genes
RNA deep sequencing was employed to qualitatively characterize the relative expression of 152 of the 167 phase I and phase II XME genes contained on the PCR Arrays. While real-time polymerase chain reaction (RT-qPCR) based techniques are the gold standard for quantitative expression, RNA-seq platforms are important for simultaneous characterization of hundreds or thousands of genes; thus, this platform was utilized for exposure independent comparative expression rather than identification of differential expression. RNA-seq reads were reported as fragments per kilobase of exon per million fragments mapped (FPKM) using CuffDiff v2.0.1. Table II depicts the bottom and top quartile FPKM reads separately for phase I and phase II XME gene isoforms, with PCR Array candidate XME genes depicted in red.
TABLE II.
Bottom and Top Quartile Expression (FPKM Reads from RNA-seq) of Phase I and Phase II XME Genes in Human Fetal Liver Specimens (N =12)
| Biotransformation pathway | Gene family names | Low expression (bottom 25%) | High expression (top 25%) |
|---|---|---|---|
| Phase I | Alcohol dehydrogenase | ADH7 | ADH1A, ADH6, ADH5 |
| Aldehyde dehydrogenase | ALDH3B2, ALD1A3 | ALDH1A1, ALDH2, ALDH4A1 | |
| Carboxylesterase | CES5A, CES3 | ||
| Cytochrome P450 | CYP4F8, CYP26C1, CYP11B2 | CYP3A7, CYP27A1, CYP19A1 | |
| Epoxide hydrolase | EPHX1, EPHX2 | ||
| Flavin dependent monooxygenase | FMO2 | FMO5 | |
| Hydroxysteroid (17β) dehydrogenase | HSD17B10 | ||
| Monoamine oxygenase | MAOB | ||
| Prostaglandin synthase/cyclooxygenase | PTGS2, PTGES | ||
| Phase II | Arylalkalamine N-acetyltransferase | AANAT | |
| Acyl-CoA synthetase | ACSM1 | ACSL4, ACSL1 | |
| Guanidinoacetate N-methyltransferase | GAMT | ||
| Glutathione S-transferase | GSTA5 | GSTA1, GSTP1, GSTO1, MGST3, MGST1, MGST2 | |
| Histamine N-methyltransferase | HNMT | ||
| Arylamine N-acetyltransferase | NAT2 | ||
| Nicotinamide N-methyltransferase | NNMT | ||
| Sulfotransferase | SULT6B1, SULT1B1, SULT1C3 | SULT1A1 | |
| Thiosulfate sulfurtransferase | TST | ||
| UDP-glucuronosyltransferase | UGT2A1, UGT2B17, UGT1A1–10 | UGT2B10, UGT2B4 |
Phase I XME genes with FPKM reads <0.1 and FPKM reads >9 represent the bottom and top quartile of XME expression. For phase II XME, the genes at the bottom and top quartile of XME expression exhibit FPKM reads <0.2 and FPKM reads >12, respectively. Genes in bold are BPA-associated candidate XMEs identified from PCR Arrays. While these genes show low expression, the remaining candidate genes (CES2, COMT, and SULT2A1) show relatively moderate expression or FPKM reads.
For phase I XME genes, the epoxide hydrolase, hydroxysteroid dehydrogenase, and monoamine oxygenase gene families were expressed at high levels while carboxylesterase and cyclooxygenase gene families were primarily expressed at low levels. In several instances, an isoform of a gene family exhibited low expression while a second isoform of the same family exhibited high expression. For example, while the alcohol dehydrogenase and aldehyde dehydrogenase isoforms, ADH1A and ALDH1A1, were both expressed at high levels, the ADH7 and ALDH3B2 isoforms were both expressed at low levels. Similarly, the predominantly expressed cytochrome p450 and flavin dependent monoxygenase isoforms were CYP3A7 and FMO5, while the CYP11B2 and FMO2 isoforms were expressed at low levels.
Among the phase II genes, guandinoacetate N-methyltransferase, histamine N-methyltransferase, and thiosulfate sulfotransferase gene families were all expressed at high levels, while arylalkalamine N-acetyltransferase, arylamine N-acetyltransferase, and nicotinamide N-methyltransferase gene families were expressed at low levels. The more common conjugating families such as glutathione S-transferase, sulfotransferase, and UDP-glucuronosyltransferase displayed isoforms that were variably expressed.
Interestingly, several BPA-associated candidate XME genes identified through the PCR Arrays exhibited relatively low expression in the 1st to 2nd trimester fetal liver. They include CES5A, GSTA5, SULT6B1, SULT1B1, SULT1C3, and the UGT1A related isoforms. While none of the differentially expressed genes were expressed at high levels in early development, CES2, COMT, and SULT2A1 showed moderate expression and were subsequently assayed for epigenetic analysis.
Identification of Common Transcription Factors in Candidate Genes
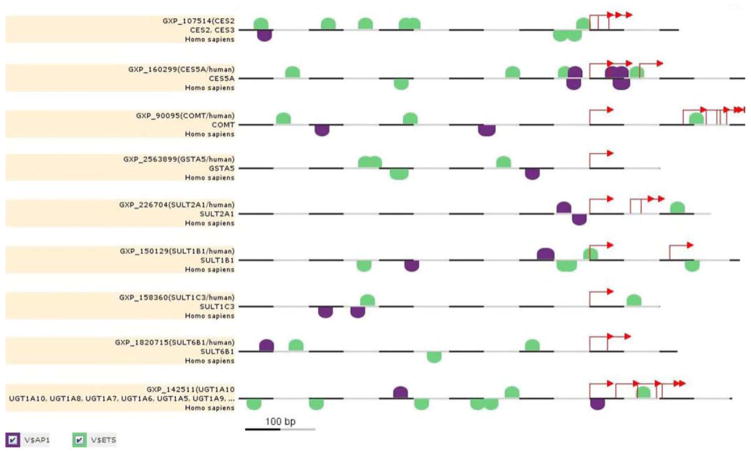
Genomatix was used to assess common transcription factor binding sites 500 bp upstream and 100 bp downstream of the TSS belonging to nine unique BPA-associated loci (all UGT1A functional genes are regulated by a common promoter). Two transcription factor families were common in at least 97% of the promoters of all relevant transcripts. Both activator protein 1 (AP1) and E-twenty-six (ETS) related transcription factor binding sites were identified at least once in the promoter of each locus as seen on the graphical presentation in Figure 1 (depicted in purple and green, respectively). The AP1 family consists of MAF and AP1 related factors such as BACH1/2, MAFA/B/F/G/K, NRL, NFE2L1/2/3 (NRF1/2/3), and NRL. The ETS family consists of a variety of transcription factors containing the Ets DNA binding domain including ELF1/2/4, ETS1/2, ETV1/4/5, SPI1/B/C, among others. The consensus sequence for each family and subfamily along with key transcription factors are listed in Table III. Interestingly, a majority of transcription factor binding sites belonging to the ETS family contain cytosine-guanine (CpG) dinucleotides in the consensus sequence, especially in the ELF, ELK, and ETV subfamilies.
Fig. 1.
Diagram displaying conserved consensus sequence binding sites for AP1 (purple) and ETS (green) transcription factor families at the promoters of BPA-associated candidate genes CES2, CES5A, COMT, GSTA5, SULT2A1, SULT1B1, SULT1C3, SULT6B1, and UGT1A1–10 retrieved from MatInspector. The view includes 500 bp upstream and 100 bp downstream of the transcription start site. Although transcription factor binding sites were identified for multiple transcripts per gene, only one promoter transcript is displayed above.
TABLE III.
Transcription Factors and Consensus Sequences of Common Transcription Factor Binding Sites (TFBS)
| TFBS family | Transcription factor | Genomatix IUPAC sequence |
|---|---|---|
| AP1 (AP1 and MAF related factors) | BACH1 | nsaTGAGtcatgny |
| BACH2 | nrTGAGtcann | |
| MAFA | TGCWgmnynngcn | |
| MAFB | naawntgCTGAcnwarn | |
| MAFK | nwaaawTGCTgactn | |
| NFE2 | hgCTGAgtcay | |
| NFE2L1 (NRF1) | vncGCGCabgcgcvnv | |
| NFE2L2 (NRF2) | nmcCGGAagtgac | |
| NRL | nncTGCTgasn | |
| ETS (Human and murine ETS1 factors) | EHF | nnacCGGAagtnn |
| ELF1 | nnccGGAAgygnn | |
| ELF2 | nnrncaGGAAgnr | |
| ELF3 | naanccGGAArtwnn | |
| ELF4 | nawmccGGAAgtnn | |
| ELF5 | ansmGGAAgtwn | |
| ELK1 | nrccGGAArynn | |
| ELK3 | nnacCGGAagynn | |
| ELK4 | nnacCGGAarynn | |
| ETS1 | vsmGGAAgygn | |
| ETS2 | dacAGGAaryvnkt | |
| ETV1 | nncaGGAAgnn | |
| ETV3 | nnacCGGAartnn | |
| ETV4 | nnacCGGAwrtnn | |
| ETV5 | nnacCGGAwgttn | |
| ETV6 | anccGGAAgtann | |
| GABPA | nnacCGGAagtnn | |
| GABPB1 | nmcCGGAagtgac | |
| SPDEF | nnatcCGGAtgynn | |
| SPI1 | ngnGGAAstn | |
| SPIB | naawgmGGAAgtn | |
| SPIC | naaagmGGAAgtwn |
The table displays individual transcription factors and their consensus sequence motifs for each TFBS family. The sequence motifs follow the standards set by the International Union of Pure and Applied Chemistry (IUPAC), with highly conserved nucleotides in bold. Upper case letters (A, C, G, T) show the most frequent nucleotides occurring at greater than 50% frequency. The degenerate code (R, Y, K, M, S, W) represents two nucleotides that occur at >75% frequency, but individually contribute to <50% frequency. All other frequency distributions are represented by the letter “n”. In particular, several motifs belonging to the ETS family contain cytosine-guanine dinucleotides within their highly conserved sequence.
Methylation Analysis at Candidate Gene Promoters
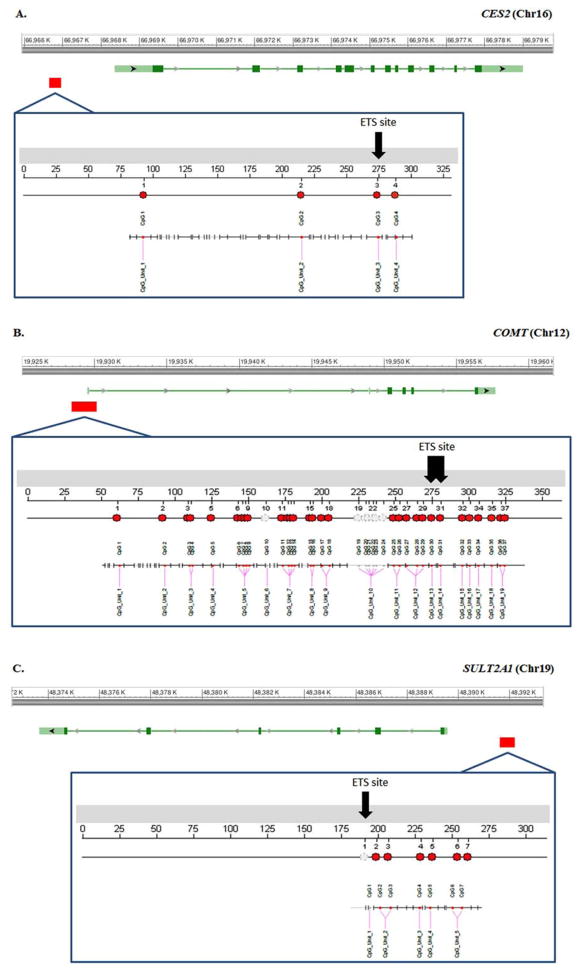
We studied CpG methylation in and surrounding ETS transcription factor binding sites at the promoters of three candidate genes: CES2, COMT, and SULT2A1 (primers in Table IV). These genes were chosen based on their relatively moderate expression levels across fetal specimens irrespective of age, sex, and exposure according to RNA-seq reads. Figure 2 displays the genomic location of the methylation assays with number of CpG sites, CpG units, and ETS transcription factor binding sites. Each assay analyzes multiple CpG units: 4 for CES2 and SULT2A1 and 17 for COMT. Within each assay, we also focused on specific CpG sites located in the ETS consensus sequence: CpG site #3 for CES2, and CpG sites #30 & #31 for COMT. CpG site #1 for SULT2A1 falls within the ETS transcription factor consensus sequences, but was unresolvable with Sequenom.
Fig. 2.
Graphic representation of Sequenom Epityper assays with transcription factor binding sites. A. The CES2 assay examines 4 individual CpG sites located approximately 1,600 to 1,900 bp upstream of the TSS. The CES2 CpG site #3 is found within an ETS transcription factor-binding site. B. The COMT assay covers 30 CpGs sites, although due to technical limitations (e.g. small fragment sizes are not detectable) Sequenom Epi-Typer outputs only 17 CpG units, spanning approximately 250 bp upstream and 100 bp downstream of the TSS. An ETS transcription factor-binding site coincides with CpG sites #30, and #31 on COMT. C. Although CpG methylation on the ETS consensus sequence of SULT2A1 could not be directly assessed (CpG site #1), the assay investigates six individual CpG sites (four CpG units) surrounding the target site, spanning approximately 1,650 to 1,950 bp upstream of the TSS. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The average methylation across all detectable sites in the COMT assay was not significantly associated with tissue BPA after adjusting for age and sex (P =0.108); however, one out of the two CpG sites spanning the putative ETS consensus sequence showed a significant positive trend (P =0.002). An interquartile range (IQR; 10.93 ng/g) increase in total tissue BPA was associated with 0.416% increase in COMT methylation at CpG site #31 (Fig. 2; Table V). Similarly, an IQR (8.28 ng/g) increase in total tissue BPA was associated with a significant 0.821% increase across all six SULT2A1 CpG sites (Fig. 2; Table V; P =0.042). The CES2 assay, however, displayed decreased site-specific methylation at CpG site #3 and average methylation across all sites with increasing BPA concentrations, although neither association was significant (P =0.409 and 0.953, respectively).
TABLE V.
Mixed Effects and Linear Regression Models Comparing Average and Individual CpG (Within ETS binding sites) Methylation With BPA Exposure, Adjusted for Gestational Age and Sex
| Methylation assay | N | Change in % methylation for an IQR change in tissue total BPA (ng/g) | P |
|---|---|---|---|
| CES2 | |||
| Average (4 total CpG; 4 CpG units)a | 50 | −0.008 | 0.953 |
| CpG #3b | 50 | −0.114 | 0.409 |
| COMT | |||
| Average (30 total CpG; 17 CpG units)a | 50 | 0.080 | 0.108 |
| CpG #30b | 50 | −0.010 | 0.851 |
| CpG #31b | 50 | 0.416 | 0.002 |
| SULT2A1 | |||
| Average (6 total CpG; 4 CpG units)a | 40 | 0.821 | 0.042 |
Interquartile Range (IQR) =10.93 ng/g for CES2 and COMT, and 8.28 ng/g for SULT2A1.
Mixed-effects model to account for multiple adjacent CpG sites, adjusted for gestation age and sex.
Linear regression at biologically relevant CpG sites, adjusted for gestational age and sex.
DISCUSSION
The impact of nutritional and environmental exposures on the human xenobiotic metabolism system during early development is not well understood. As previously described, we quantified total BPA concentrations in 50 1st to 2nd trimester fetal liver specimens, ranging from below the limit of quantification to 96.8 ng/g total BPA in tissue [Nahar et al., 2012]. In the current study, we compared gene expression of over 160 XME genes within a subset of fetal liver specimens, dichotomized into low (1.2–2.9 ng/g; n =4) and high (35.4–56.1 ng/g; n =4) total tissue BPA. In total, 14 candidate genes were identified that exhibit significantly reduced expression in fetal liver with higher tissue BPA concentrations. With the exception of the phase I carboxylesterase isoforms, CES2 and CES5A, the majority of BPA-related XME candidate genes belong to the phase II gene family.
Traditionally, the induction of phase I enzymes following drug and xenobiotic exposure is commonly observed, but our finding of BPA-associated repression of predominantly phase II XME expression is novel. Only a handful of studies have previously reported xenobiotic-dependent repression of primarily phase II metabolizing genes. In a 2008 study examining the ability of endocrine disruptors to modify estrogen bioavailability in ER-negative HepG2 cells, 80 μM of BPA or 70 μM of genistein exposure was associated with decreased expression of several phase II metabolism enzymes, including UGT2B7, UGT2B15, and SULT1E1 genes [Hanet et al., 2008]. Of our newly identified exposure-associated XME genes, only the UDP-glucuronosyltransferases have been previously associated with BPA as identified from the Comparative Toxicogenomic Database (http://ctdbase.org/). When MCF7 cells were exposed to 10 nM BPA instead of the control DMSO vehicle, there was a reduction in UGT1A3 mRNA expression [Buterin et al., 2006]. In a second study, BPA was reported to competitively inhibit UGT1A6 and subsequently alter metabolism of serotonin in liver microsomes [Hanioka et al., 2008].
Characterizing XME activity and regulation, especially throughout fetal development, is challenging given the difficulty in acquiring relevant human specimens from sensitive populations. With our unique samples, we were able to compare relative expression for hundreds of genes simultaneously using mRNA-next generation sequencing technology. After compiling all XME related genes, the top and bottom 25% of normalized RNA-seq reads for phase I and phase II metabolizing enzymes were reported for several 1st and 2nd trimester fetal liver tissue in humans, listed in Table II. Detectable protein levels of CYP2C9, CYP2C19, CYP3A4, CYP3A5, and CYP3A7 were previously reported in 1st to 2nd trimester human liver [Hines, 2007]. Specific isoforms for other biotransformation proteins, such as alcohol dehydrogenase (ADH1A), epoxide hydrolase (EPHX1, EPHX2), glutathione S-transferase (GSTA1), and sulfotransferase (SULT1A1) have also been reported in the early fetal liver [Smith et al., 1971; Strange et al., 1989; Omiecinski et al., 1994; Duanmu et al., 2006]. Activities of these phase I and II enzymes vary between gene families and isoforms with relative abundance drastically altering over time until adult levels are reached [Hines and McCarver, 2002; McCarver and Hines, 2002]. Interestingly, a majority of the BPA related candidate genes that were identified exhibited relatively low RNA-seq reads as compared with other phase I and phase II metabolism genes. Thus, the negative association between BPA and specific underdeveloped XME genes may be indicative of repressive effects or delayed maturation of the candidate XME genes to adult levels, potentially altering the response to toxicants later in life.
BPA is well known to interact with nuclear receptors such as estrogen receptor (ERα and β), estrogen related receptor gamma (ERRy), pregnane X receptor (PXR), AhR, androgen receptor, and thyroid receptor [Wetherill et al., 2007; Molina-Molina et al., 2013]. More recently, studies suggest that endocrine active compounds induce rapid responses via nongenomic signal transduction pathways, and disease susceptibility may depend on the coordination between genomic and nongenomic pathways [Watson et al., 2011]. In this study, the co-repression of candidate genes warranted a closer look at common regulators in order to provide insight into novel mechanism of action for BPA that are relevant to the XME pathway. In our bioinformatic investigation of regulatory sites, we identified binding sites for the highly conserved ETS and AP1 related transcription factors. Extracellular signals affecting the MAPK and JNK signaling pathways often converge both upstream and downstream of ETS proteins, which consist of over 25 family members that can hetero-dimerize with other transcription factor families, such as the Fos and Jun domains of AP-1 related proteins [Bassuk and Leiden, 1995; Yordy and Muise-Helmericks, 2000; Kyriakis and Avruch, 2001]. Combinatorial control of specific ETS and AP1 factors, along with other regulators, is likely to influence transcriptional XME specificity during early fetal development, a period in which estriol (E3) predominates and activates nongenomic processes [Watson et al., 2011]. While evidence for estrogen receptor independent mechanisms for BPA is increasing, only a few studies thus far have investigated BPAs influence on ETS and AP1 factor targets. BPA has been reported to stimulate the PU.1 transcription factor, a relative of the SP1A, SP1B, and SP1C regulators, in promyelocytic cells for neutrophilic differentiation [Watanabe et al., 2003]. In human embryonic kidney cells, BPA at μM doses activated Nrf1 and Nrf2 transactivation activity and induced antioxidant response element (ARE) target genes [Chepelev et al., 2013]. Subsequent studies are necessary to investigate specific ETS and AP1 transcription factors and their genomic localization using methods such as chromatin immunoprecipitation coupled with sequencing (CHIP-seq).
Cis-acting regulation at ETS and AP1 related transcription factor binding sites and promoter occupancy, in addition to post-transcriptional modifications, may alter environmental response via signal transduction. In particular, DNA methylation at CpG sites along the consensus sequence motifs, especially for ETS, can inhibit transcription factor binding and transcriptional activity [Hollenhorst et al., 2011]. In our in silico analysis, ETS proteins were prevalent in the candidate gene promoters and many of the ETS consensus sequences contained at least one CpG site. When Hogart et al. characterized epigenome-wide patterns across several different primary murine cells, DNA methylation was highest in hematopoietic stem cells and interestingly, ETS consensus binding motifs were overrepresented within these methylated regions [Hogart et al., 2012]. Given that the fetal liver is the major site of hematopoiesis [Timens and Kamps, 1997], we wanted to examine whether decreased expression along with increased tissue BPA concentrations were associated with increased promoter CpG methylation at ETS binding sequences. Both site-specific methylation at the COMT promoter and average CpG methylation at the SULT2A1 promoter significantly increased with higher BPA concentrations, although the increase in DNA methylation was relatively modest. The less than 1% increase in promoter methylation at the ETS sites of differentially expressed genes suggests that additional regulatory elements, including other epigenetic mechanisms, influence expression. For example, the environmentally sensitive MAPK signaling proteins can associate with chromatin modifying complexes and directly regulate histone modifications [Suganuma and Workman, 2012]. Future work will require further integration of signal transduction pathways with various epigenetic mechanisms, including histone modifications and RNA interference, in response to environmental stress at critical windows of development.
Traditionally toxicological studies employed enzymatic activity assays to understand xenobiotic metabolism in vitro or in animal models; however, analyzing protein activity in the developing fetal liver is far more difficult when access is limited to small amounts of sensitive bio-banked specimens. With the improvement of real-time PCR and utilization of next generation sequencing, hundreds of genes can be reliably profiled simultaneously using small mRNA sample volume and compared with protein activity. In general, mRNA expression is a good proxy for protein level, but gene expression does not always correlate to protein abundance. This discrepancy can be attributed to differences in the rate of protein or mRNA synthesis and turn over, or post-transcriptional regulation [Vogel and Marcotte, 2012]. Hence, examination of target protein activity and post-transcriptional modification of regulatory elements in suitable samples will be necessary in future studies for complete characterization of fetal xenobiotic metabolism. Still, in the case that healthy specimens are available, longitudinal analysis of the development of human XME pathway is challenging given the bioethical concerns for fetal tissue, especially in 3rd trimester. Despite these limitations, gene expression profiling, even at a narrow window of development, can be helpful for exploring early genomic regulation that occur with a changing environment.
While the task of investigating BPAs influence on XMEs may be clearer in animal models in which investigators exercise control over experimental design and model choice, here we utilize human fetal liver samples from an NIH-funded biobank. These samples are derived from individuals who may have been exposed to a wide array of compounds and maternal factors. Thus, a variety of unmeasured but related factors, such as co-exposure to other endocrine active compounds, may confound our results.
In the presence of endogenous hormones and growth factors, phase I and phase II metabolizing gene expression and enzyme function matures until adult baseline levels are attained, either after birth, during childhood, or in adolescence. Therefore, exposure to environmental compounds, especially during early life when the capacity for xenobiotic metabolism is suboptimal, may increase susceptibility to disease in the developing fetus and children [Allegaert et al., 2008]. When XME gene expression was compared in fetal liver exposed in utero to physiologically low and high BPA concentrations, subtle decreases in specific phase I and II metabolizing genes were observed. Although BPA may not immediately dysregulate essential proteins or directly cause disease, the endocrine active compound may stunt XME maturation via signaling pathways and epigenetic mechanisms. Considering the daily and cumulative exposure to a myriad of compounds in humans, the inability to adequately respond to these insults suggests potential consequences for health later in life.
Acknowledgments
Grant sponsor: National Institute of Health; Grant number: ES017524.
Grant sponsor: University of Michigan (UM) National Institute of Environmental Health Science (NIEHS) Core Center; Grant number: P30 ES017885.
Grant sponsor: UM NIEHS Institutional Training Grant; Grant number: T32 ES007062.
The authors acknowledge the University of Washington Birth Defects Research Laboratory (R24HD000836-47) for human tissue sample collection. The authors thank Ellen Pederson and Craig Johnson from the UM Sequencing Core for their assistance with PCR Array and Seque-nom Epityper analysis, Dr. Richard McEachin for bioinformatics consultation, and Dr. Laura Rozek for her supervision of research project and invaluable advice. Authors are also indebted to Dr. Kurunthachalam Kannan and Dr. Chunyang Liao from the Wadsworth Center for BPA analysis in fetal tissue.
Footnotes
AUTHOR CONTRIBUTIONS
D.C.D. and M.S. were involved in project conception and design. M.S.N. and J.H.K. were involved in laboratory processing and data acquisition for PCR Array, RNA-seq, and Sequenom methylation analysis. D.C.D. and M.S.N. helped interpret data and drafted the final manuscript, which was revised and approved by all authors.
References
- Alcorn J, McNamara PJ. Pharmacokinetics in the newborn. Adv Drug Deliv Rev. 2003;55:667–686. doi: 10.1016/s0169-409x(03)00030-9. [DOI] [PubMed] [Google Scholar]
- Allegaert K, van den Anker JN, Naulaers G, de Hoon J. Determinants of drug metabolism in early neonatal life. Curr Clin Pharmacol. 2007;2:23–29. doi: 10.2174/157488407779422294. [DOI] [PubMed] [Google Scholar]
- Allegaert K, Verbesselt R, Naulaers G, van den Anker JN, Rayyan M, Debeer A, de Hoon J. Developmental pharmacology: neonates are not just small adults. Acta Clin Belg. 2008;63:16–24. doi: 10.1179/acb.2008.003. [DOI] [PubMed] [Google Scholar]
- Anderson OS, Nahar MS, Faulk C, Jones TR, Liao C, Kannan K, Weinhouse C, Rozek LS, Dolinoy DC. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ Mol Mutagen. 2012;53:334–342. doi: 10.1002/em.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan B, Henare K, Thorstensen EB, Ponnampalam AP, Mitchell MD. Transfer of bisphenol A across the human placenta. Am J Obstet Gynecol. 2010;202:393, e391–e397. doi: 10.1016/j.ajog.2010.01.025. [DOI] [PubMed] [Google Scholar]
- Bassuk AG, Leiden JM. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010;24:2273–2280. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buterin T, Koch C, Naegeli H. Convergent transcriptional profiles induced by endogenous estrogen and distinct xenoestrogens in breast cancer cells. Carcinogenesis. 2006;27:1567–1578. doi: 10.1093/carcin/bgi339. [DOI] [PubMed] [Google Scholar]
- Caldwell J, Gardner I, Swales N. An introduction to drug disposition: the basic principles of absorption, distribution, metabolism, and excretion. Toxicol Pathol. 1995;23:102–114. doi: 10.1177/019262339502300202. [DOI] [PubMed] [Google Scholar]
- Cao XL, Zhang J, Goodyer CG, Hayward S, Cooke GM, Curran IH. Bisphenol A in human placental and fetal liver tissues collected from Greater Montreal area (Quebec) during 1998–2008. Chemosphere. 2012;89:505–511. doi: 10.1016/j.chemosphere.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chepelev NL, Enikanolaiye MI, Chepelev LL, Almohaisen A, Chen Q, Scoggan KA, Coughlan MC, Cao XL, Jin X, Willmore WG. Bisphenol A activates the Nrf1/2-antioxidant response element pathway in HEK 293 cells. Chem Res Toxicol. 2013;26:498–506. doi: 10.1021/tx400036v. [DOI] [PubMed] [Google Scholar]
- Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu Z, Weckle A, Koukouritaki SB, Hines RN, Falany JL, Falany CN, Kocarek TA, Runge-Morris M. Developmental expression of aryl, estrogen, and hydroxysteroid sulfotransferases in pre-and postnatal human liver. J Pharmacol Exp Ther. 2006;316:1310–1317. doi: 10.1124/jpet.105.093633. [DOI] [PubMed] [Google Scholar]
- Hanet N, Lancon A, Delmas D, Jannin B, Chagnon MC, Cherkaoui-Malki M, Latruffe N, Artur Y, Heydel JM. Effects of endocrine disruptors on genes associated with 17beta-estradiol metabolism and excretion. Steroids. 2008;73:1242–1251. doi: 10.1016/j.steroids.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Hanioka N, Jinno H, Tanaka-Kagawa T, Nishimura T, Ando M. Interaction of bisphenol A with rat hepatic cytochrome P450 enzymes. Chemosphere. 2000;41:973–978. doi: 10.1016/s0045-6535(99)00529-9. [DOI] [PubMed] [Google Scholar]
- Hanioka N, Takeda Y, Tanaka-Kagawa T, Hayashi K, Jinno H, Narimatsu S. Interaction of bisphenol A with human UDP-glucuronosyltransferase 1A6 enzyme. Environ Toxicol. 2008;23:407–412. doi: 10.1002/tox.20345. [DOI] [PubMed] [Google Scholar]
- Hines RN. Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol. 2007;21:169–175. doi: 10.1002/jbt.20179. [DOI] [PubMed] [Google Scholar]
- Hines RN. The ontogeny of drug metabolism enzymes and implications for adverse drug events. Pharmacol Ther. 2008;118:250–267. doi: 10.1016/j.pharmthera.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Hines RN, McCarver DG. The ontogeny of human drug-metabolizing enzymes: phase I oxidative enzymes. J Pharmacol Exp Ther. 2002;300:355–360. doi: 10.1124/jpet.300.2.355. [DOI] [PubMed] [Google Scholar]
- Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogart A, Lichtenberg J, Ajay SS, Anderson S, Margulies EH, Bodine DM. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res. 2012;22:1407–1418. doi: 10.1101/gr.132878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H. Altered drug metabolism during pregnancy: Hormonal regulation of drug-metabolizing enzymes. Expert Opin Drug Metab Toxicol. 2010;6:689–699. doi: 10.1517/17425251003677755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Díaz I, Zafra-Gómez A, Ballesteros O, Navea N, Navalón A, Fernández MF, Olea N, Vílchez JL. Determination of Bisphenol A and its chlorinated derivatives in placental tissue samples by liquid chromatography-tandem mass spectrometry. J Chromatogr B. 2010;878:3363–3369. doi: 10.1016/j.jchromb.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Kennedy M. Hormonal regulation of hepatic drug-metabolizing enzyme activity during adolescence. Clin Pharmacol Ther. 2008;84:662–673. doi: 10.1038/clpt.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Champagne FA. Epigenetic perspective on the developmental effects of bisphenol A. Brain Behav Immun. 2011;25:1084–1093. doi: 10.1016/j.bbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- McCarver DG, Hines RN. The ontogeny of human drug-metabolizing enzymes: Phase II conjugation enzymes and regulatory mechanisms. J Pharmacol Exp Ther. 2002;300:361–366. doi: 10.1124/jpet.300.2.361. [DOI] [PubMed] [Google Scholar]
- Molina-Molina JM, Amaya E, Grimaldi M, Saenz JM, Real M, Fernandez MF, Balaguer P, Olea N. In vitro study on the agonistic and antagonistic activities of bisphenol-S and other bisphenol-A congeners and derivatives via nuclear receptors. Toxicol Appl Pharmacol. 2013;272:127–136. doi: 10.1016/j.taap.2013.05.015. [DOI] [PubMed] [Google Scholar]
- Naciff JM, Khambatta ZS, Reichling TD, Carr GJ, Tiesman JP, Singleton DW, Khan SA, Daston GP. The genomic response of Ishikawa cells to bisphenol A exposure is dose- and time-dependent. Toxicology. 2010;270:137–149. doi: 10.1016/j.tox.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Nahar MS, Liao C, Kannan K, Dolinoy DC. Fetal liver bisphenol A concentrations and biotransformation gene expression reveal variable exposure and altered capacity for metabolism in humans. J Biochem Mol Toxicol. 2012;27:116–123. doi: 10.1002/jbt.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy PJ, Monteiro A, Bhattacharya S, Fraser MJ, Fowler PA. Steroidogenic enzyme expression in the human fetal liver and potential role in the endocrinology of pregnancy. Mol Hum Reprod. 2013;19:177–187. doi: 10.1093/molehr/gas059. [DOI] [PubMed] [Google Scholar]
- Omiecinski CJ, Aicher L, Swenson L. Developmental expression of human microsomal epoxide hydrolase. J Pharmacol Exp Ther. 1994;269:417–423. [PubMed] [Google Scholar]
- Pfeiffer E, Metzler M. Effect of bisphenol A on drug metabolising enzymes in rat hepatic microsomes and precision-cut rat liver slices. Arch Toxicol. 2004;78:369–377. doi: 10.1007/s00204-004-0543-6. [DOI] [PubMed] [Google Scholar]
- Rubin BS. Bisphenol A: an endocrine disruptor with widespread exposure and multiple effects. J Steroid Biochem Mol Biol. 2011;127:27–34. doi: 10.1016/j.jsbmb.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Saghir SA, Khan SA, McCoy AT. Ontogeny of mammalian metabolizing enzymes in humans and animals used in toxicological studies. Crit Rev Toxicol. 2012;42:323–357. doi: 10.3109/10408444.2012.674100. [DOI] [PubMed] [Google Scholar]
- Schonfelder G, Wittfoht W, Hopp H, Talsness CE, Paul M, Chahoud I. Parent bisphenol A accumulation in the human maternal-fetal-placental unit. Environ Health Perspect. 2002;110:703–707. doi: 10.1289/ehp.110-1241091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Bolin JM, Ruotti V, Nguyen BK, Thomson JA, Elwell AL, Stewart R. Single read and paired end mRNA-Seq Illumina libraries from 10 nanograms total RNA. J Vis Exp. 2011:e3340. doi: 10.3791/3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Hopkinson DA, Harris H. Developmental changes and polymorphism in human alcohol dehydrogenase. Ann Hum Genet. 1971;34:251–271. doi: 10.1111/j.1469-1809.1971.tb00238.x. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Strange RC, Howie AF, Hume R, Matharoo B, Bell J, Hiley C, Jones P, Beckett GJ. The development expression of alpha-, mu-and pi-class glutathione S-transferases in human liver. Biochim Biophys Acta. 1989;993:186–190. doi: 10.1016/0304-4165(89)90162-1. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. MAP kinases and histone modification. J Mol Cell Biol. 2012;4:348–350. doi: 10.1093/jmcb/mjs043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timens W, Kamps WA. Hemopoiesis in human fetal and embryonic liver. Microsc Res Tech. 1997;39:387–397. doi: 10.1002/(SICI)1097-0029(19971201)39:5<387::AID-JEMT1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ping J, Peng RX, Yue J, Xia XY, Li QX, Kong R, Hong JY. Changes of multiple biotransformation phase I and phase II enzyme activities in human fetal adrenals during fetal development. Acta Pharmacol Sin. 2008;29:231–238. doi: 10.1111/j.1745-7254.2008.00738.x. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Adachi R, Kusui K, Hirayama A, Kasahara T, Suzuki K. Bisphenol A significantly enhances the neutrophilic differentiation of promyelocytic HL-60 cells. Int Immunopharmacol. 2003;3:1601–1608. doi: 10.1016/S1567-5769(03)00182-6. [DOI] [PubMed] [Google Scholar]
- Watson CS, Jeng YJ, Guptarak J. Endocrine disruption via estrogen receptors that participate in nongenomic signaling pathways. J Steroid Biochem Mol Biol. 2011;127:44–50. doi: 10.1016/j.jsbmb.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherill YB, Akingbemi BT, Kanno J, McLachlan JA, Nadal A, Sonnenschein C, Watson CS, Zoeller RT, Belcher SM. In vitro molecular mechanisms of bisphenol A action. Reprod Toxicol. 2007;24:178–198. doi: 10.1016/j.reprotox.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]