Abstract
The Nek2 and Plk4 kinases serve as crucial regulators of mitotic processes such as the centrosome duplication cycle and spindle assembly. Deregulation of these processes can trigger chromosome instability and aneuploidy, which are hallmarks of many solid tumors, including breast cancer. Emerging data from the literature illustrated various functions of Nek2 in breast cancer models, with compelling evidence of its prognostic value in breast tumors. The two kinases control distinct steps in the centrosome-centriole cycle and their dysregulation lead to centrosome amplification, marked by the presence of more than two centrosomes within the cell. We found single or composite overexpression of these kinases in breast tumor samples, regardless of subtype, which strongly associated with poor prognosis. Interestingly, in a panel of established cell lines, both kinases are highly expressed in Her2-positive breast cancer cells exhibiting centrosome amplification and trastuzumab resistance. In summary, it appears that Nek2 and Plk4 might synergize to promote breast tumorigenesis and may also be involved in tamoxifen and trastuzumab resistance.
Keywords: Nek2, Plk4, breast cancer, centrosome amplification, chromosome instability, tamoxifen resistance, trastuzumab resistance
2. INTRODUCTION
The Nek2A and Plk4 kinases have been extensively studied with regard to their involvement in diverse mitotic events, including centrosome duplication and separation at the onset of mitosis, spindle assembly, mitotic timing, and cytokinesis. Due to their fluctuating expression and subcellular localization, the two enzymes are important regulators of faithful chromosome segregation with potential further implications in the coordination of cell cycle, cell division, cytoplasmic architecture and tissue polarity. Since these features are frequently deregulated in cancers, a number of studies have attempted to decipher the involvement of these mitotic kinases in human tumors, including breast cancer. These reports provided promising results with potential clinical relevance. Here, we will review the current knowledge on the biological functions of Nek2A (referred to as Nek2), Plk4 and centrosome amplification (CA, defined as the presence of more than two centrosomes within a cell) in breast cancer, introduce preliminary data from our laboratory, propose potential roles for the two kinases in this context, and address future perspectives.
The most traditional role ascribed to Nek2 and Plk4 relates to centrosomes, a pair of non-membranous organelles, each composed of two centrioles surrounded by a protein matrix called pericentriolar material (PCM). The centrosome-centriole duplication cycle ensures proper microtubule organization and establishment of the bipolar spindle pole and is coordinated with the cell cycle, occurring once per mitosis. Consequently, Nek2 and Plk4 are temporally and spatially regulated in a cell-cycle dependent manner. Upon recruitment to centrosomes, Plk4 initiates and catalyzes centriole duplication and elongation throughout the S phase, while Nek2, peaking at G2/S, acts to induce centrosome separation and the formation of the mitotic spindle. Another role of Nek2 addresses its active participation in the spindle assembly checkpoint (SAC), which orchestrates the attachment of spindle microtubules to the kinetochores of chromosomes. The details pertaining to the normal functions of Nek2 and Plk4 are still under active investigation and some of their respective targets, interactions, as well as regulation and localization patterns have been elegantly covered in a series of reviews (1–10). CA can arise as a result of distinct mechanisms that are not mutually exclusive: disengagement of the cell and centrosome duplication cycles, centriole overduplication leading to overproduction of daughter centrioles and centriole accumulation (clustering) caused by impaired cytokinesis and other cell division errors (7, 11–13). CA has been implicated as underlying chromosomal instability, a hallmark of solid tumors, including breast cancer. Herein, we propose that Nek2 and Plk4 kinases mediate CA and breast tumorigenesis.
3. NEK2, PLK4 AND CENTROSOME AMPLIFICATION IN BREAST CANCER
3.1. Nek2 and Plk4 in breast tumors and experimental models
A compelling list of studies addressed the expression and function of Nek2 in breast tumors and experimental models. For example, Nek2 is overexpressed and amplified in breast cancer cell lines and in primary human breast tumors, including ductal carcinoma in situ (DCIS) (14–19). However, the relationship with established biomarkers such as estrogen receptor alpha (ER), progesterone receptor (PR), epidermal growth factor receptor (EGFR), keratin 67 (Ki67) and p53 generated conflicting conclusions. Some studies found no evident links (14, 17), yet positive correlations between elevated Nek2 levels and Ki67 or between Nek2 and Her2-overexpressing or triple negative breast tumors were observed (17, 18).
Work from our group found that concomitant induction of K-RasG12D and c-Myc in mouse mammary glands caused premalignant lesions displaying CA and increased expression of Nek2 at both protein and mRNA level (20). Similarly, simultaneous induction of H-RasG12V and c-Myc in the immortalized non-tumorigenic human mammary epithelial cell line MCF10A significantly increased Nek2 protein expression, while transient knockdown of Nek2 in the same cells reduced CA and DNA synthesis. Collectively, these results suggest that oncogene activation leads to upregulated Nek2 signaling.
More clues about Nek2 were gathered from studies employing gain or loss of function strategies in human mammary epithelial cells. In invasive derivatives of MCF10A cells, basal levels of Nek2 mRNA and protein were higher compared to controls (18). Moreover, Nek2 overexpression caused CA without affecting the cell cycle and its transient knockdown inhibited cell proliferation and caused a G0/G1 arrest, thereby suggesting a role for Nek2 in cell cycle progression (18). Overexpression of Nek2 in HBL100 cells immortalized by the SV40 T antigen induced aneuploidy accompanied by multiple or enlarged nuclei (14). In both MCF-7 and MDA-MB-231 breast cancer cell lines, Nek2 overexpression resulted in augmented numbers of centrosomes and multinucleated cells, whereas its transient silencing decreased cell proliferation and induced aneuploidy (15, 19). Different means of inhibiting Nek2 in various breast cancer cell lines resulted in a wide range of cellular effects responses, differentially exhibited: decreased anchorage-independent growth, migration and colony formation, impaired spindle assembly and chromosome misalignment, aneuploidy, reduced cell proliferation, apoptosis, elevated sensitivity to paclitaxel and doxorubicin, cell cycle defects, abnormal number of centrosomes and binucleation (15, 21–26). Notably, in contrast to breast cancer cells, MCF10A seemed to be insensitive to the treatment with a small molecule inhibitor of Nek2/Hec1 interaction, which destabilized Nek2 protein, alluding to the selectivity of Nek2 inhibition in breast cancer cells (21). The potential dependence of breast cancer cells on Nek2 for their tumorigenic growth was demonstrated in xenograft models, where silencing of Nek2 inhibited the primary and distal tumor growth (15, 19, 21).
Finally, indirect evidence for the role of Nek2 in breast cancer is provided by research of its phosphorylation targets, such as β-catenin, Nlp, Mad2 and Hec1, all of which have been shown to impact aspects of breast carcinogenesis (16, 23, 27–41). Along the same line, a preliminary gene expression microarray from our laboratory indicated that expression of Nek2 and that of its substrate Sgo1 (42) are higher in HCC1954 breast cancer cells compared to MCF10A control.
There is a scarcity of data investigating the putative role of Plk4 in breast cancer. Similarly to the Nek2 results, simultaneous induction of K-RasG12D and c-Myc in murine mammary glands lead to increased expression of Plk4 transcript (20). Our unpublished observations from MCF10A/p53-null cells, previously shown to display chromosome instability, invasiveness and altered therapeutic response (43), indicate significant CA and higher Plk4 protein levels compared to parental cells. There are reports indicating that transient knockdown of Plk4 has anti-proliferative effects in breast cancer cells but not in normal cells and that the response to chemical inhibition of Plk4 is cell-type dependent (44). A recent study found Plk4 overexpression in triple negative breast tumor samples (45) while other groups reported that Plk4 is overexpressed in breast cancer and predicts resistance to therapy (44, 46–49). The contribution of Plk4 to breast cancer biology is also supported by exploration of its interacting proteins. For example, Plk4 binds and phosphorylates Ect2 (50), which is a component of the MammaPrint genetic signature that predicts the recurrence risk of breast cancer (41). Moreover, the aforementioned microarray analysis from our laboratory found that HCC1954 cells overexpress Cep192, demonstrated to recruit Plk4 to the centrosome (51) and Sas-6, a protein that requires Plk4 for its localization to centrioles during mitosis (52).
While this manuscript was under revision, promising anti-proliferative effects were reported from pre-clinical studies using a Plk4-directed agent (53). Based on results from breast cancer cell lines and xenograft mice, applications for phase I trial approvals of the newly developed anti-Plk4 compound CFI-400945 in breast and ovarian cancer patients have been submitted to FDA and Health Canada. This endeavor strongly supports the clinical relevance of Plk4 in breast tumors.
3.2. Centrosome amplification in breast cancer
Owing to their critical roles in regulating the centrosome duplication cycle, the most likely outcome of aberrant Nek2 and Plk4 signaling is CA. Due to the numerous proteins associated with it, a scaffolding role for centrosome was proposed and reviewed with evidence of crosstalk with the cell cycle (54). This organelle represents not only a hub that receives and integrates diverse extra- and intracellular signals and translates them into biological responses. The centrosome is also a direct target susceptible to deregulation, since it provides a signaling platform for the cell cycle machinery, oncogenes, tumor suppressors and transcription factors.
Experimental models of breast cancer demonstrated that induction of CA is linked to various molecular markers of tumor initiation and progression, including ER, p53, Aurora-A, BRCA1 and Ras. For instance, published and ongoing work carried out in our laboratory revealed the involvement of the Her2 and K-RasG12D oncogenes, Cdk4 or E2Fs in generating CA in breast cancer cells and in pre-malignant mammary epithelial lesions (20, 24). Although it initially concluded that in breast tumors CA arises independently of ER and p53 status, the Salisbury group investigated the role of estrogen signaling in CA and chromosome instability (CIN) in rodent models (55). Mouse xenografts using MCF-7 ER-positive cells demonstrated that loss of p53 induces CA and is associated with the development of aggressive tamoxifen-resistant tumors (56). One of the best examples of centrosomal kinases with oncogenic function is Aurora-A, as reflected by its overexpression in breast tumors (57–59). Cell-based and mouse models pointed out that Aurora-A induces CA and tetraploidy in breast cancer through the activation of Akt, BRCA1 and Cdk2 pathways (60–63). In fact, a mechanistic link between estrogen, Aurora kinase A and the development of CA and CIN was proposed in the ACI rat strain with estrogen-induced mammary tumorigenesis (64).
Several breast cancer models suggested a causal connection between CA, CIN, aneuploidy and tumorigenesis. Accordingly, cells with extra centrosomes undergo chromosome missegregation and display tetraploidy (65). Mirroring human cancers, tetraploid mouse mammary epithelial cells devoid of p53 harbor genetic alterations and form tumors in xenograft models (66). The fate of cells with supranumerary centrosomes is not necessarily CIN and aneuploidy. The instability caused by extra centrosomes can often preclude viability. Therefore, cells carrying extra centrosomes may be eliminated via apoptosis, mitotic catastrophe, or replicative senescence (67, 68). However, when viable progeny propagate during tumor development, they can promote the accumulation of multiple centrosomes, allowing pseudobipolar mitosis (11, 12). Therefore, an attractive concept revolves around a microevolution under selective pressure that ultimately results in the generation of cellular subclones containing CA and subsequent genomic instability.
The presence of increased centrosome size in DCIS and the correlation between impaired centrosome function and loss of tissue differentiation (69, 70) imply that CA precedes the acquisition of CIN and aneuploidy (20, 65). A study performed in a large cohort of breast cancer cases and controls examined the relationship between the genetic variation of genes controlling the centrosome and breast cancer risk. The results indicated that haplotypes displaying associations in certain genes rather than single SNPs exhibited a greater predictive value (71). Moreover, genomically unstable and more aggressive aneuploid breast cancers have a greater extent of CA and abnormal mitotic spindles compared to genomically stable aneuploid and diploid tumors (69, 72–74). From a clinical standpoint, the relevance of CIN status resides in its prognostic utility in ER-positive tumors, in which the CIN score is a linear predictor of poor outcome (40, 75). Thus, the poor prognosis associated with CIN may be attributed to the capacity of breast tumors to adapt to environment or stromal pressures, most likely due to genetic heterogeneity (40). In terms of the propensity of particular subtype of breast tumors to develop CA, centrosome anomalies correlate with Her2-overexpression and the absence of ER and PR (73, 76). However, in patients over 50 years old, CA associates with the presence of all of these three receptors (73, 74).
Despite the aforementioned wealth of studies that clearly associate Nek2 and/or Plk4 expression with CA, CIN and aneuploidy in breast cancer, the definitive evidence for a cause-effect relationship is still lacking. Changes in the levels of Nek2 and Plk4 genes, their mislocalization or aberrant activity can potentially compromise the control of the mitotic checkpoint, centrosome duplication and/or mitotic progression and lead to faulty cell decisions such as CA. This event could further induce CIN and aneuploid defects that may ultimately contribute to breast tumorigenesis. In conclusion, identifying the cellular mechanisms underlying CA and its relationship to Nek2 and Plk4, particularly when it occurs in pre-malignant breast lesions, is required in order to establish the contribution of this phenomenon to the breast tumor progression.
4. NEK2 AND PLK4 AS PROGNOSTIC MARKERS
Given their emerging roles in breast cancer biology, we queried publicly available datasets to conduct a retrospective analysis of the expression of Nek2 and Plk4 across human breast tumors. The following breast cancer datasets were downloaded from GEO: GSE2034, GSE3494, GSE6532, GSE4922, GSE11121, GSE7390, GSE2603 and GSE14020. Data were normalized using quantile normalization and then log2 transformed in the Affymetrix Human U133 platform prior to doing the analyses. The datasets were combined, and batch effects were removed using the Bayesian Factor Regression Modeling (BFRM) algorithm to generate the merged database (Yuwanita and Andrechek, unpublished). In addition, a more refined independent analysis was performed in GSE6532 in which only the samples treated with tamoxifen and run on Affymetrix Human U133A were selected (N=190). Since each gene had multiple probes, expression of each probe was categorized into high and low using the median as the cut point. Probe 204641_at for Nek2 and probe 204887_s_at for Plk4 were selected since they had the highest median expression in the resulting combined dataset. A composite score was created by averaging the two probes and further stratified into low (<25th percentile), moderate (25th–75th percentile), and high (>75th percentile) quantiles. Expression of each gene and composite was quantified and compared using ANOVA and the Kruskal-Wallis test and represented in box plots. For survival statistics, eligible GSMs were chosen based on the following reported endpoints: relapse free survival, distant metastasis-free survival and overall survival. Kaplan-Meier survival curves were generated for each outcome comparing the score groups. Log-rank test and Cox proportional hazard regression were used with P-values ≤0.05 considered statistically significant. All statistical analyses were done using SAS Version 9.3 and GraphPad Prism 5.
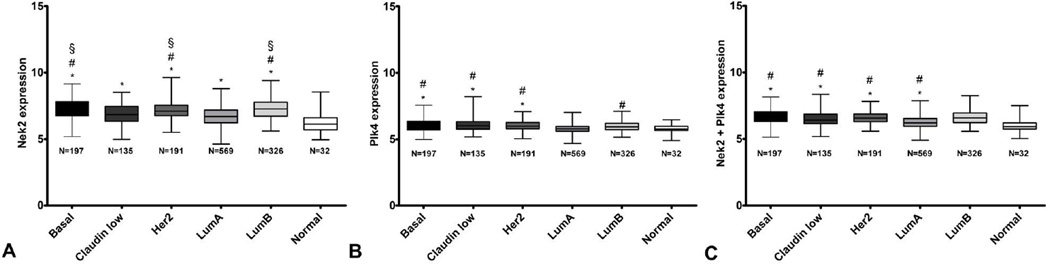
Expression of Nek2 was significantly higher in each of the breast cancer subtypes compared with the normal samples. In fact, with the exception of Her2 or basal vs. luminal B, basal vs. Her2 and claudin low vs. luminal A, all pairwise comparisons were statistically significant (Figure 1A). Expression of Plk4 was significantly higher in each of the basal, claudin low and Her2-amplified subtypes versus normal or luminal A samples. Furthermore, the differential Plk4 expression between luminal A and B subtypes reached statistical significance (Figure 1B). Finally, the composite score of the two genes indicated a combined elevated expression of Nek2 and Plk4 in each of the basal, claudin low, Her2 and luminal B groups versus normal but also compared to the luminal A (Figure 1C).
Figure 1.
Individual and composite Nek2 and Plk4 expression in normal tissue and breast tumor subtypes from the combined datasets. A. Nek2 expression. P ≤ 0.05; * significant compared to normal tissue; # significant compared to luminal A samples; § significant compared to claudin low samples. B. Plk4 expression. P ≤ 0.05; * significant compared to normal tissue; # significant compared to luminal A samples. C. Averaged Nek2+Plk4 expression. P ≤ 0.05; * significant compared to normal tissue; # significant compared to luminal A samples.
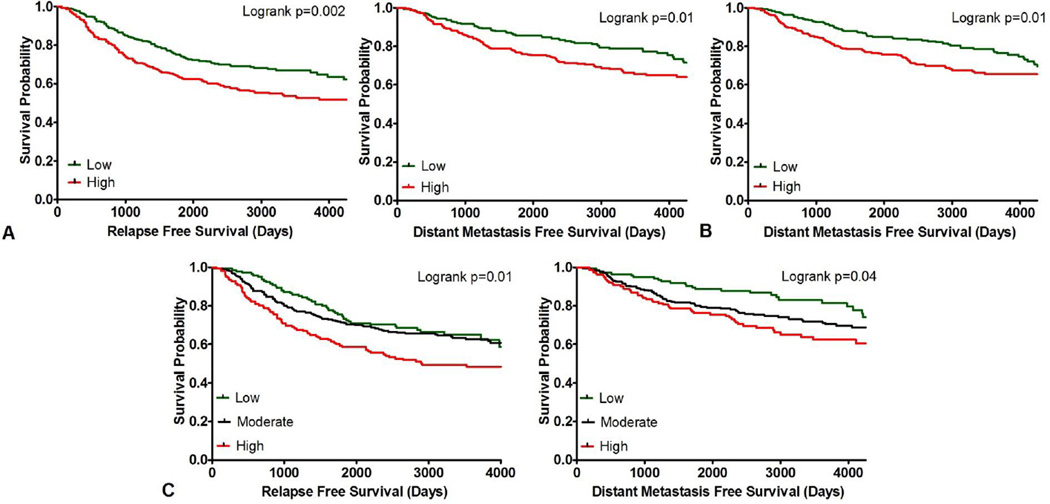
As illustrated in Figure 2A. the relapse free survival of patients with Nek2-overexpressing tumors was significantly worse than that of patients exhibiting low expression, regardless of subtype or treatment (N=603; hazard ratio HR=0.6; 95% confidence interval CI, 0.5 to 0.8; *P=0.002). In addition, Nek2 overexpression showed a significantly poorer prognosis in terms of distant metastasis-free survival compared to Nek2-low tumors (N=594; hazard ratio HR=0.6; 95% confidence interval CI, 0.5 to 0.9; *P=0.01). Plk4-overexpressing tumors were associated with poor distant metastasis-free survival (N=603; hazard ratio HR=0.6; 95% confidence interval CI, 0.5 to 0.9; *P=0.01; Figure 2B). High composite score was associated with significant differences in relapse free survival (N=603; *P=0.01) and distant metastasis-free survival (N=594; *P=0.04; Figure 2C). No significance was found in overall survival for individual or combinatorial overexpression of Nek2 and Plk4. We performed more stringent survival analyses on the subtypes presented in Figure 1, grouped by low/high Nek2 or Plk4 and quantiles of their combined expression for the relapse-free survival, distant metastasis-free survival, and overall survival and found no statistical significance in any of the 45 analyses. This greatly reduced the number of samples and increased the selection bias, therefore we do not show the data as we found them inconclusive.
Figure 2.
Relapse free and distant metastasis-free survival analyses in all breast cancer patients from the combined datasets. A. Kaplan-Meier curves for Nek2 expression in patients grouped by low and high gene expression. B. Kaplan-Meier curve for Plk4 expression in patients grouped by low and high gene expression. C. Kaplan-Meier curves for composite Nek2+Plk4 expression in patients grouped by tertiles.
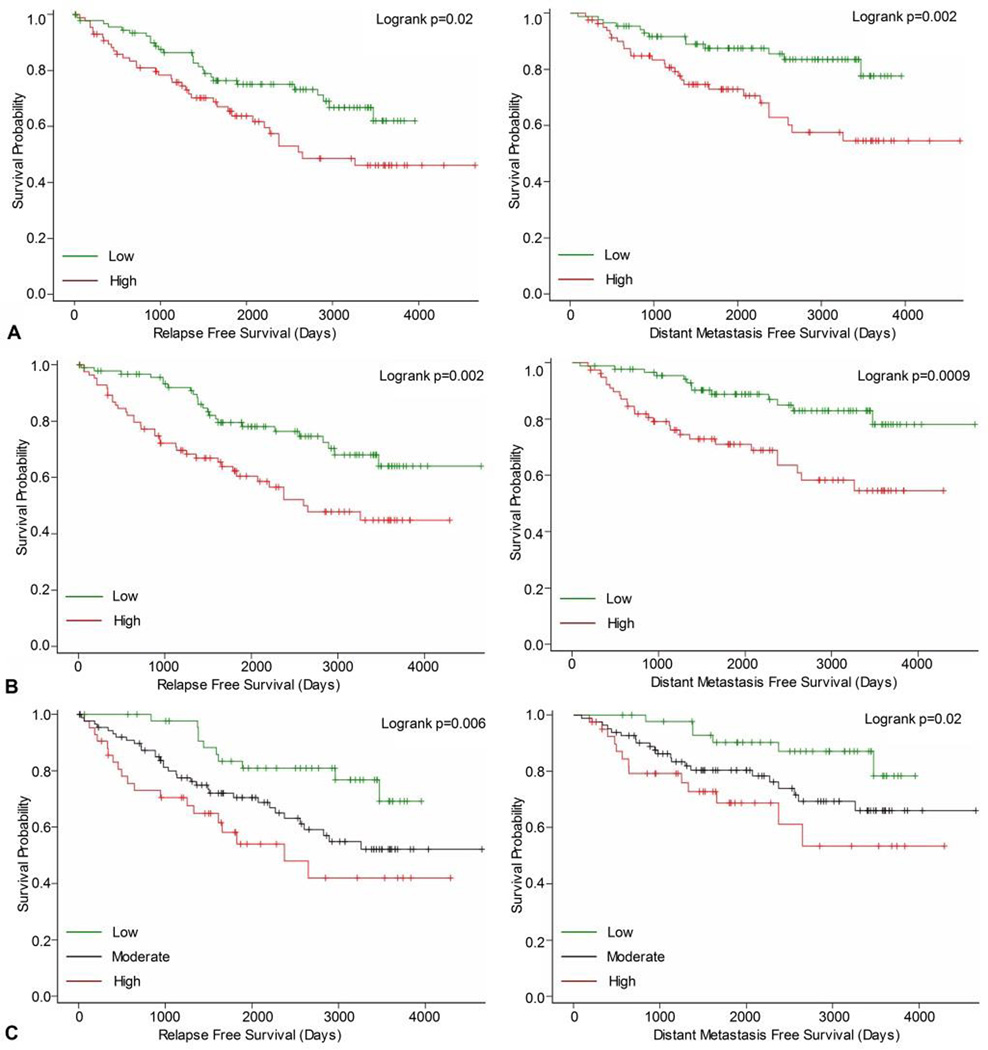
In the independent analysis of GSE6532, Kaplan-Meier curves showed significant differences in relapse free survival and distant metastasis-free survival between high and low expression groups for both Nek2 (Figure 3A) and Plk4 (Figure 3B). Next, a higher composite score was associated with significant differences in relapse free survival and distant metastasis free-survival (Figure 3C). Again, overall survival was not significantly correlated with either single or combined overexpression of Nek2 and Plk4. The significant differences indicating that single and co-overexpression of Nek2 and Plk4 associated with worse outcomes in tamoxifen treated patients than low expressions suggest that these kinases might play a predictive role in endocrine resistance.
Figure 3.
Relapse free and distant metastasis-free in tamoxifen-treated patients from the GSE6532. A. Kaplan-Meier curves for Nek2 expression in patients grouped by low and high gene expression. B. Kaplan-Meier curves for Plk4 expression in patients grouped by low and high gene expression. C. Kaplan-Meier curves for composite Nek2+Plk4 expression in patients grouped by tertiles.
These results are in agreement with previous gene-expression reports addressing the prognostic value of Nek2 and Plk4 in breast tumors. For example, both enzymes are part of a 16-kinase signature present in luminal tumors with poor prognosis and high mitotic index and are highly expressed in basal, luminal Ab, luminal B and Her2-overexpressing subtypes (77). Another study detected Nek2 significantly upregulated in invasive versus benign breast lesions (16). Nek2 was also included in a molecular grade index (MGI) predictive genetic profile that indicated significant difference in distant metastasis-free survival for tamoxifen-treated patients with intermediate and high risk. In this association, the risk classification and prognostic performance of MGI were independent of tumor size, grade, Her2 and PR expression (78, 79). Retrospective analyses of public dataset confirmed that overexpression of Nek2 conferred a poor survival outcome in breast tumors (19, 23). AACR communications mentioned Plk4 overexpression and its predictive relevance to therapy outcome in breast cancer (44, 48, 49).
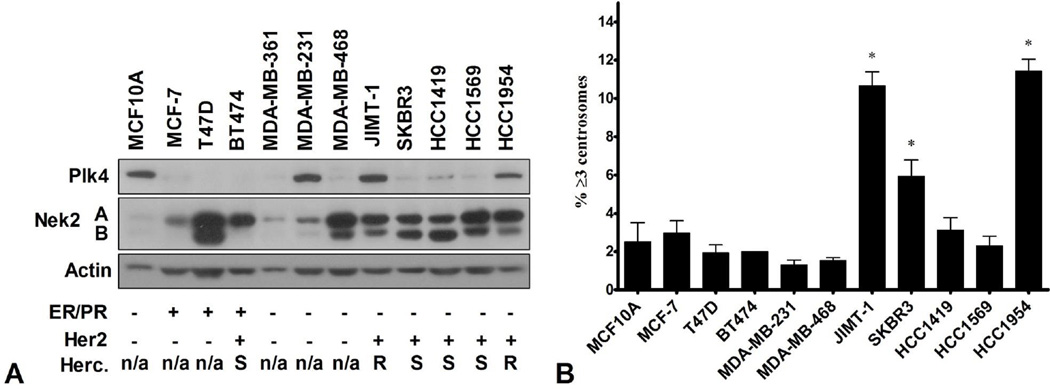
Our finding that some breast cancer cell lines display CA, prompted us to perform a screening of Nek2 and Plk4 protein expression in a panel of asynchronous human mammary epithelial cells of diverse molecular subtypes. HCC1954 and JIMT-1 cell lines emerged as highly expressing both kinases compared to the other cell lines investigated (Figure 4A). As scored by immunofluorescence with antibodies against pericentrin (PCM marker) and quantification of cells with more than two centrosomes, the two cell lines exhibited CA of 11% and 10.7% respectively (Figure 4B). These percentages were significantly higher compared to MCF10A cells (2.5%). In addition, both cell lines are ER-PR-Her2+ and were reported to display primary resistance to trastuzumab (80, 81). The expression patterns observed here suggest that indeed, in some settings these short-live kinases are still under the control of cell-cycle apparatus, while in others subtypes this requirement seems to be eliminated. Consistent with their expression, Nek2 and Plk4 might become hyperactive or synergize very early in breast lesions and translate into functional consequences such as frequent mitotic events and CA.
Figure 4.
Basal levels of Nek2 and Plk4 and centrosome amplification in asynchronous mammary epithelial cell lines. A. Nek2 and Plk4 protein levels determined by immunoblotting. Cells were grouped by ER/PR and Her2 status. Herceptin resistance is listed at n/a, S (sensitive) and R (resistant). B. Centrosome amplification determined by immunofluorescent staining of pericentrin. Cells with ≥ 3 centrosome were expressed as percentage of total number of cells visualized with DAPI. N=3; mean ± SD; P ≤ 0.05; * significant compared to MCF10A cells.
5. NOVEL ROLES FOR NEK2 AND PLK4 IN BREAST CANCER
5.1. Nek2 and Plk4: drivers of breast tumorigenesis
The ability of Nek2 to induce CA in breast cancer cells became evident in MCF10A/DCIS derivatives (18). Data from our group showed that silencing of Nek2 decreased CA present in Her2-positive breast cancer cell lines (24), while a recent report shows that its overexpression in other subtypes of breast cancer cells enhanced CA (25). Interestingly, Nek2 knockdown in MDA-MB-231 triple-negative cells also lead to an increase in centrosome numbers, multipolar mitosis and mitotic catastrophe (19). In light of this evidence, CA seems to be the prevalent outcome of deregulated Nek2, considering its abundance in cells, particularly during the G2/S phase, and its ever-growing range of centrosomal targets and binding partners, including C-Nap1, Nlp and β-catenin. Although there is no direct evidence on the role of Plk4 in promoting CA in breast cancer, experiments conducted in other models of human cancers support the implication of this kinase in CA in tumors. For instance, in osteosarcoma cells, Plk4 associates with centrosome and centrioles throughout the cell cycle, colocalizing with centrin, C-Nap1 and γ-tubulin. Moreover, deregulated Plk4 results in division failure and CA, the induction of which requires the cooperation between Plk4 and Cdk2 (2). Another experiment using osteosarcoma cells revealed that enforced Plk4 expression resulted in extra chromosomes and transient multipolar spindles intermediates in anaphase (65). In prostate cancer cells, CAND1 centrosomal protein controls the stability of Plk4 and synergizes with it to stimulate centriole overduplication (82). It was reported that inhibition of Plk4 in cancer cells blocks centrosome clustering and formation of multipolar spindles (44). In light of the evidence summarized here, it is likely that either kinase could be sufficient to cause CA in mammary epithelial cells. Our preliminary observations from breast cancer cell lines suggest that acquisition of CA might require a synergistic cooperation between Nek2 and Plk4 expressed at high levels. It was demonstrated that Nek2 overexpression causes centriole separation (1) and Plk4 induces centriole reduplication, but the centrioles remain in close configuration (83), thereby Nek2 might be required to resolve the clustered centrioles in Plk4-overexpressing cells, resulting in CA.
A second mean by which Nek2 accomplishes its mitotic functions is through the association with the kinetochore and participation in SAC. Situated at centromeres, the kinetochore is a large protein complex required for spindle attachment, chromosome movement and regulation of the mitotic checkpoint. The mechanism of SAC ensures that cells do not transition to anaphase without the orderly alignment of all sister chromatides to the metaphase plate. Interestingly, given the upregulation of SAC components in aneuploid cancer cells, a SAC-addiction mechanism in tumors was proposed (84). Kinetochore-localized Nek2 is highly active, phosphorylating SAC effectors such as Mad2 and Hec1 (23, 39) and high expression of these substrates has been incorporated in several breast cancer genetic signatures (16, 40, 41). Moreover, overexpression of Mad2 resulted in cells with genomic instability and tumorigenesis in mice (85). Both CA and defective SAC can eventually culminate in chromosome missegregation and aneuploidy. Therefore, it is conceivable that in time, overexpression of Nek2 and/or Plk4 can cause loss of control over proper mitotic division of chromosomes which may in turn facilitate genetic changes favorable to mammary tumor initiation and progression. Alternatively, overexpression of SAC proteins can facilitate centrosome clustering by offering enough time for amplified centrosomes to coalesce to form a megacentrosome. Due to the scaffolding role of the centrosomes and the dynamic relationships between SAC components, Nek2 and/or Plk4 might cooperate with other molecules that promote centrosome clustering, a mechanisms that confers on tumor cells a proliferative advantage.
Besides deregulating mitosis and facilitating the acquisition of genetic instability, Nek2 and Plk4 can directly promote malignant transformation by controlling cell proliferation, viability and motility. Thereby, sustained expression of Nek2 and Plk4 might directly contribute to the progression to aggressive tumor phenotypes, marked by distant metastases.
With regard to cell motility, it was shown that Nek2 knockdown reduced the migratory potential of the highly invasive MDA-MB-231 breast cancer cells (19). The effects of Nek2 inhibition on cell cycle are context-dependent. For example, in MCF10A cells Nek2 silencing lead to a G0/G1 arrest (18). However, in breast cancer cells, the effects of abnormal Nek2 on cell cycle are not uniform. Some subtypes were unaffected by Nek2 knockdown or overexpression while others show significant G2/M arrest and decreased proliferation upon silencing or corresponding increased proliferation with high Nek2 levels (19, 22, 23). Indeed, Nek2 could impact cell cycle progression as it binds to and phosphorylates β-catenin, which in turn induces cyclin D1 expression (86). In myeloma cells, activation of β-catenin/Wnt signaling by Nek2 was in part dependent on Akt (23). Moreover, Nek2 overexpression in these cells repressed the pro-apoptotic genes Bad and PUMA and upregulated the expression of pro-survival genes Bcl-xL and Mcl-1 (23). These results, combined with findings that Nek2 silencing increased the cleavage of caspase-3 in breast cancer cells suggest a possible pro-survival role (19).
Plk4 was proposed to be a regulator of cell proliferation, motility and viability. Thus, in liver cancer models, haploid levels of Plk4 can disrupt Rho-GTPase signaling during cytokinesis, resulting in aneuploidy and tumorigenesis (50). In addition, studies performed in Plk4+/− mouse embryonic fibroblasts suggest that this enzyme promotes activation of Rac1 to induce migration (48). Findings from lung cancer cells support a pro-survival role for Plk4. Downregulation of Plk4 in cells expressing a temperature-sensitive p53 mutant, resulted in apoptosis while its overexpression diminished p53-dependent apoptosis (87). Additional research on lung cancer cells revealed that Plk4 silencing inhibited stress-induced Akt activation and triggered apoptosis. Gradual activation of p53, upon stress conditions, downregulated Plk4 to promote apoptosis and reduce the risk of Plk4-induced CA (88). Finally, pharmacological inhibition of Plk4 blocked proliferation, lead to apoptosis and reduced tumor growth in xenograft models (44).
In order to further unravel their biological relevance in breast cancer, cell culture work shall focus on inducing the gain or loss of Nek2 and/or Plk4 expression in non-tumorigenic and cancer cells of different backgrounds, respectively. This approach can provide preliminary data regarding the direct roles of Nek2 and Plk4 in CA as well as identify cellular and molecular mechanisms by which this event is attained. Next, it would be interesting to explore the collaboration of these two kinases with molecules that promote the clustering of amplified centrosomes. In addition, unbiased phospho-proteomic assays will identify more targets of Nek2 and Plk4 kinases. A likely consequence of abnormal Nek2 and Plk4 expression is alteration in microtubule dynamics that can trigger tissue reorganization during tumorigenesis. Therefore, it is essential to address how the growth of cells deregulated for the two kinases in 3D culture impacts CA, chromosome instability, cell motility and long-term proliferation and survival. With the development of automated techniques to count centrosome markers in tissue samples, the presence of CA in breast cancer could be more accurately assessed. Correlations between the abundance and localization of Nek2 and Plk4, CA status, expression of molecular biomarkers and clinico-pathological features of breast tissue samples are necessary to verify the relevance of Nek2/Plk4-driven CA to malignancy. Furthermore, studies using transgenic mouse models with selective induction of Nek2 and/or Plk4 expression in the mammary gland would help answer additional questions regarding the mechanisms and timeline of CA in vivo. As a final point, inducible knockout models could effectively complete the animal work since, for example, Plk4 knockout mice are not viable (89). In summary, comprehensive approaches including biochemical and pharmacological techniques, gene expression analysis, molecular and cellular assays, as well as translational studies, are required to confirm the proliferative and pro-survival roles of Nek2 and Plk4 and their contribution to promoting CA, CIN and cell motility in breast cancer cells.
5.2. Drug resistance
Whether resulting from gene amplification or loss of transcriptional control, the enhanced levels of Nek2 and Plk4 detected in breast cancer tissues may represent more than predictors of poor outcome. Our retrospective gene expression analysis in tamoxifen-treated ER-positive breast tumors illustrated that single and co-overexpression of Nek2 and Plk4 are significantly correlated with poor relapse free and distant metastasis-free survival. On the other hand, screening of human mammary epithelial cells lines indicated that trastuzumab-insensitive Her2-overexpressing cells exhibit the highest levels of both kinases across all samples. Based on these preliminary results, we propose two roles for Nek2 and Plk4 in breast cancer. First, these kinases hold potential predictive value and could identify tumors prone to respond poorly to tamoxifen or trastuzumab. Secondly, as active components of the centrosomes, Nek2 and Plk4 may contribute to therapy failure by promoting resistance to tamoxifen and trastuzumab. In support of this latter hypothesis is the very idea that tumor cells are able to proliferate with mild genomic instability resulted from defective mitoses and CA. By directly causing CA and chromosome missegregation, Nek2 and Plk4 might be responsible for the propagation of genomically altered breast cancer cells. Such cells harbor genetic changes that under selective pressure can induce dysregulation of pathways implicated in therapy resistance. Consequently, the uptake and metabolism of the therapeutic agents and cellular responses to their inhibitory effects could be greatly influenced.
ER-positive and Her2-amplified breast cancers can become non-responsive to targeted therapies via similar mechanisms involving alteration of cell cycle, mitosis and proliferation. For example, Cdk4 is inhibited during tamoxifen-driven growth arrest (90) and overexpression of cyclin D1 can trigger its activation resulting in tamoxifen resistance (91, 92). Interestingly, exposure of breast cancer cells to the dual Cdk4/6 inhibitor PD-0332991 synergizes with both tamoxifen and trastuzumab treatment (93). Inactivation of Rb can promote tamoxifen resistance (94). Alternatively, ER-positive tumors with low growth factor receptor expression that are initially responsive to tamoxifen can later develop resistance by manifesting elevated levels of Her2 (95). It has become widely acknowledged that in addition to promoting breast tumor growth and survival, activation of the PI3K/Akt signaling pathway is also linked to resistance to tamoxifen (96–99) and trastuzumab (100–102). As summarized above, loss of p53 in MCF-7 ER-positive breast cancer cells leads to CA and is associated with acquisition of heterogeneity for nuclear ER that in turn drives the development of aggressive tamoxifen-resistant tumors (56). Noteworthy, trastuzumab treatment induced repression of several mitotic genes, including Hec1 and Aurora-A (103). Research from our laboratory indicates that Nek2 and Plk4 functions are controlled by E2F and/or Cdk4 in Her2-overexpressing breast cancer cells. In addition, there seems to be a positive feed-back regulation between Nek2 and Cdk4 which acts to mediate CA (24). Two recent publications report interactions between Nek2 or Plk4 and Akt signaling to induce drug resistance and evade apoptosis (23, 88). In addition, ablation of Nek2 in breast cancer cells reduced the levels of phosphorylated Rb (19). It is therefore tempting to speculate that by signaling downstream of the cyclin D1/Cdk4-Rb/E2F axis, Nek2, Plk4 and the cellular events that they trigger might represent biomarkers that can potentially indicate the onset and progression of breast tumors, and/or predict the response to therapeutic agents. Moreover, by interacting with Cdk4, Akt, Rb or other crucial proteins localized at centrosome, Nek2 and Plk4 may play a role in endocrine or trastuzumab resistance.
A preliminary strategy to ascertain the contribution of Nek2 and Plk4 to endocrine and trastuzumab resistance is to answer whether their inhibition can restore drug sensitivity in resistant ER-positive and Her2-overexpressing cell line models. However, their validation as effective biomarkers and therapeutic targets warrants future pre-clinical and translational investigations.
6. PERSPECTIVES AND CLINICAL IMPACT
Over the past decade, the biological relevance of Nek2 and Plk4 went from key mitotic kinases to prognostic markers in breast cancer and potential drivers of tumorigenesis. However, the basic biology of the two kinases still needs to be unraveled in order to better understand their contribution to breast tumorigenesis. Future discoveries will allow the biomedical community to fully exploit Nek2 and Plk4 for the development of novel therapeutic approaches. Both kinases are attractive target drugs since they are involved in specific aspects of microtubule assembly, mitosis and cell division. High throughput screenings to target their kinase activity will address the effects of blocking Nek2 or Plk4 on CA, cell proliferation, survival, motility and drug sensitivity in breast cancer cells. Furthermore, generation of inducible knockout mice is necessary to elucidate the pathophysiological roles of Nek2 and Plk4 as well as the potential side effects of their pharmacological inhibitors.
More importantly, if the effects of Nek2 or Plk4 inhibition on CA can be discretely separated from other effects on the cell cycle per se, the two mitotic kinases could be proposed as targets in a cancer prevention setting. Interfering with Nek2 and/or Plk4 and their cooperating partners that promote centrosome clustering could be detrimental to the tumor progression. Blocking CA would require concentrations of Nek2 or Plk4 inhibitors that leave normal centrosome cycle unaffected and could potentially stall progression and render breast tumors sensitive to therapies, presumably through a suppression of aneuploid populations. Alternatively, if the stoichiometry of Nek2 and Plk4 is essential for the cells, low levels of these kinases can also deregulate the centrosome cycle and/or mitosis, producing populations of breast cancer cells displaying CIN and aneuploidy. In this case, inhibition of Nek2 and Plk4 can ultimately result in aneuploidy and cell death.
ACKNOWLEDGEMENTS
The authors would like to thank Dana Nickleach and Dr. Jeanne Kowalski from the Biostatistics and Bioinformatics Shared Resource, Winship Cancer Institute for the assistance with the GSE6532 analysis and Dr. Eran Andrechek and Mrs. Inez Yuwanita from Michigan State University, for assistance with the combined gene expression database.
Abbreviations
- AACR
American Association for Cancer Research
- CA
centrosome amplification
- CIN
chromosome instability
- DCIS
ductal carcinoma in situ
- EGFR
epidermal growth factor receptor
- ER
estrogen receptor alpha
- GEO
gene expression omnibus
- MGI
molecular grade index
- Nek2
NIMA-related kinase 2
- PCM
pericentriolar material
- Plk4
Polo-like kinase 4
- PR
progesterone receptor
- SAC
spindle assembly checkpoint
Footnotes
This is an, un-copyedited, author manuscript that has been accepted for publication in the Frontiers in Bioscience". Cite this article as appearing in the Journal of Frontiers in Bioscience. Full citation can be found by searching the Frontiers in Bioscience (http://bioscience.org/search/authors/htm/search.htm) following publication and at PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed) following indexing. This article may not be duplicated or reproduced, other than for personal use or within the rule of "Fair Use of Copyrighted Materials" (section 107, Title 17, U.S. Code) without permission of the copyright holder, the Frontiers in Bioscience. From the time of acceptance following peer review, the full final copy edited article of this manuscript will be made available at http://www.bioscience.org/. The Frontiers in Bioscience disclaims any responsibility or liability for errors or omissions in this version of the un-copyedited manuscript or in any version derived from it by the National Institutes of Health or other parties.
REFERENCES
- 1.Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14(7):2876–2889. doi: 10.1091/mbc.E03-02-0108. doi:10.1091/mbc.E03-02-0108 E03-02-0108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7(11):1140–1146. doi: 10.1038/ncb1320. doi:ncb1320 [pii] 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 3.Hayward DG, Fry AM. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006;237(2):155–166. doi: 10.1016/j.canlet.2005.06.017. doi:S0304-3835(05)00561-6 [pii] 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17(5):215–221. doi: 10.1016/j.tcb.2007.03.003. doi:S0962-8924(07)00055-4 [pii] 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Sillibourne JE, Bornens M. Polo-like kinase 4: the odd one out of the family. Cell Div. 2010;5:25. doi: 10.1186/1747-1028-5-25. doi:1747-1028-5-25 [pii] 10.1186/1747-1028-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27(8):307–315. doi: 10.1016/j.tig.2011.05.004. doi:S0168-9525(11)00065-5 [pii] 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korzeniewski N, Hohenfellner M, Duensing S. The centrosome as potential target for cancer therapy and prevention. Expert Opin Ther Targets. 2013;17(1):43–52. doi: 10.1517/14728222.2013.731396. doi:10.1517/14728222.2013.731396. [DOI] [PubMed] [Google Scholar]
- 8.Fry AM, O'Regan L, Sabir SR, Bayliss R. Cell cycle regulation by the NEK family of protein kinases. J Cell Sci. 2012;125(Pt 19):4423–4433. doi: 10.1242/jcs.111195. doi:jcs.111195 [pii] 10.1242/jcs.111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mardin BR, Schiebel E. Breaking the ties that bind: new advances in centrosome biology. J Cell Biol. 2012;197(1):11–18. doi: 10.1083/jcb.201108006. doi:jcb.201108006 [pii] 10.1083/jcb.201108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brownlee CW, Rogers GC. Show me your license, please: deregulation of centriole duplication mechanisms that promote amplification. Cell Mol Life Sci. 2013;70(6):1021–1034. doi: 10.1007/s00018-012-1102-6. doi:10.1007/s00018-012-1102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7(12):911–924. doi: 10.1038/nrc2249. doi:nrc2249 [pii] 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 12.Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci. 2011;7(8):1122–1144. doi: 10.7150/ijbs.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarapore P, Horn HF, Tokuyama Y, Fukasawa K. Direct regulation of the centrosome duplication cycle by the p53-p21Waf1/Cip1 pathway. Oncogene. 2001;20(25):3173–3184. doi: 10.1038/sj.onc.1204424. doi:10.1038/sj.onc.1204424. [DOI] [PubMed] [Google Scholar]
- 14.Hayward DG, Clarke RB, Faragher AJ, Pillai MR, Hagan IM, Fry AM. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004;64(20):7370–7376. doi: 10.1158/0008-5472.CAN-04-0960. doi:64/20/7370 [pii] 10.1158/0008-5472.CAN-04-0960. [DOI] [PubMed] [Google Scholar]
- 15.Tsunoda N, Kokuryo T, Oda K, Senga T, Yokoyama Y, Nagino M, Nimura Y, Hamaguchi M. Nek2 as a novel molecular target for the treatment of breast carcinoma. Cancer Sci. 2009;100(1):111–116. doi: 10.1111/j.1349-7006.2008.01007.x. doi:CAS1007 [pii] 10.1111/j.1349-7006.2008.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bieche I, Vacher S, Lallemand F, Tozlu-Kara S, Bennani H, Beuzelin M, Driouch K, Rouleau E, Lerebours F, Ripoche H, Cizeron-Clairac G, Spyratos F, Lidereau R. Expression analysis of mitotic spindle checkpoint genes in breast carcinoma: role of NDC80/HEC1 in early breast tumorigenicity, and a two-gene signature for aneuploidy. Mol Cancer. 2011;10:23. doi: 10.1186/1476-4598-10-23. doi:1476-4598-10-23 [pii] 10.1186/1476-4598-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Li W, Lv S, Wang Y, Liu Z, Zhang J, Liu T, Niu Y. Abnormal expression of Nek2 and beta-catenin in breast carcinoma: clinicopathological correlations. Histopathology. 2011;59(4):631–642. doi: 10.1111/j.1365-2559.2011.03941.x. doi:10.1111/j.1365-2559.2011.03941.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Li W, Liu N, Zhang F, Liu H, Liu F, Liu J, Zhang T, Niu Y. Nek2A contributes to tumorigenic growth and possibly functions as potential therapeutic target for human breast cancer. J Cell Biochem. 2012;113(6):1904–1914. doi: 10.1002/jcb.24059. doi:10.1002/jcb.24059. [DOI] [PubMed] [Google Scholar]
- 19.Cappello P, Blaser H, Gorrini C, Lin DC, Elia AJ, Wakeham A, Haider S, Boutros PC, Mason JM, Miller NA, Youngson B, Done SJ, Mak TW. Role of Nek2 on centrosome duplication and aneuploidy in breast cancer cells. Oncogene. 2013 doi: 10.1038/onc.2013.183. doi:onc2013183 [pii] 10.1038/onc.2013.183. [DOI] [PubMed] [Google Scholar]
- 20.Zeng X, Shaikh FY, Harrison MK, Adon AM, Trimboli AJ, Carroll KA, Sharma N, Timmers C, Chodosh LA, Leone G, Saavedra HI. The Ras oncogene signals centrosome amplification in mammary epithelial cells through cyclin D1/Cdk4 and Nek2. Oncogene. 2010;29(36):5103–5112. doi: 10.1038/onc.2010.253. doi:onc2010253 [pii] 10.1038/onc.2010.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu G, Qiu XL, Zhou L, Zhu J, Chamberlin R, Lau J, Chen PL, Lee WH. Small molecule targeting the Hec1/Nek2 mitotic pathway suppresses tumor cell growth in culture and in animal. Cancer Res. 2008;68(20):8393–8399. doi: 10.1158/0008-5472.CAN-08-1915. doi:68/20/8393 [pii] 10.1158/0008-5472.CAN-08-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Gollahon L. Nek2-targeted ASO or siRNA pretreatment enhances anticancer drug sensitivity in triplenegative breast cancer cells. Int J Oncol. 2013;42(3):839–847. doi: 10.3892/ijo.2013.1788. doi:10.3892/ijo.2013.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W, Yang Y, Xia J, Wang H, Salama ME, Xiong W, Xu H, Shetty S, Chen T, Zeng Z, Shi L, Zangari M, Miles R, Bearss D, Tricot G, Zhan F. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell. 2013;23(1):48–62. doi: 10.1016/j.ccr.2012.12.001. doi:S1535-6108(12)00514-4 [pii] 10.1016/j.ccr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitner MKH, Saavedra HI. Cdk4 and Nek2 Signal Binucleation and Centrosome Amplification in a Her2+ Breast Cancer Model. PLOS One. 2013 doi: 10.1371/journal.pone.0065971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research. Philadelphia (PA): AACR, Washington, DC; 2013. Coordination of overexpressed NIMA-related kinase 2 and TRF1 results in mitotic abnormalities in breast cancer cells. [Google Scholar]
- 26.Harrison Pitner MK, Saavedra HI. Cdk4 and nek2 signal binucleation and centrosome amplification in a her2+ breast cancer model. PLOS One. 2013;8(6):e65971. doi: 10.1371/journal.pone.0065971. doi:10.1371/journal.pone.0065971 PONE-D-13-04654 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. doi:10.1038/415530a 415530a [pii] [DOI] [PubMed] [Google Scholar]
- 28.Schade B, Lesurf R, Sanguin-Gendreau V, Bui T, Deblois G, O'Toole SA, Millar EK, Zardawi SJ, Lopez-Knowles E, Sutherland RL, Giguere V, Kahn M, Hallett M, Muller WJ. beta-catenin signaling is a critical event in ErbB2-mediated mammary tumor progression. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-3925. doi:0008-5472.CAN-12-3925 [pii] 10.1158/0008-5472.CAN-12-3925. [DOI] [PubMed] [Google Scholar]
- 29.Bahmanyar S, Kaplan DD, Deluca JG, Giddings TH, Jr, O'Toole ET, Winey M, Salmon ED, Casey PJ, Nelson WJ, Barth AI. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008;22(1):91–105. doi: 10.1101/gad.1596308. doi:gad.1596308 [pii] 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97(8):4262–4266. doi: 10.1073/pnas.060025397. doi:10.1073/pnas.060025397 060025397 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howe LR, Brown AM. Wnt signaling and breast cancer. Cancer Biol Ther. 2004;3(1):36–41. doi: 10.4161/cbt.3.1.561. doi:561 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Prosperi JR, Goss KH. A Wnt-ow of opportunity: targeting the Wnt/beta-catenin pathway in breast cancer. Curr Drug Targets. 2010;11(9):1074–1088. doi: 10.2174/138945010792006780. doi:BSP/CDT/E-Pub/00106 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176(6):2911–2920. doi: 10.2353/ajpath.2010.091125. doi:S0002-9440(10)60812-7 [pii] 10.2353/ajpath.2010.091125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rapley J, Baxter JE, Blot J, Wattam SL, Casenghi M, Meraldi P, Nigg EA, Fry AM. Coordinate regulation of the mother centriole component nlp by nek2 and plk1 protein kinases. Mol Cell Biol. 2005;25(4):1309–1324. doi: 10.1128/MCB.25.4.1309-1324.2005. doi:25/4/1309 [pii] 10.1128/MCB.25.4.1309-1324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, Jin S, Song Y, Zhan Q. Cdc2/cyclin B1 regulates centrosomal Nlp proteolysis and subcellular localization. Cancer Biol Ther. 2010;10(9):945–952. doi: 10.4161/cbt.10.9.13368. doi:13368 [pii] 10.4161/cbt.10.9.13368. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Zhan Q. The role of centrosomal Nlp in the control of mitotic progression and tumourigenesis. Br J Cancer. 2011;104(10):1523–1528. doi: 10.1038/bjc.2011.130. doi:bjc2011130 [pii] 10.1038/bjc.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shao S, Liu R, Wang Y, Song Y, Zuo L, Xue L, Lu N, Hou N, Wang M, Yang X, Zhan Q. Centrosomal Nlp is an oncogenic protein that is gene-amplified in human tumors and causes spontaneous tumorigenesis in transgenic mice. J Clin Invest. 2010;120(2):498–507. doi: 10.1172/JCI39447. doi:39447 [pii] 10.1172/JCI39447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King SI, Purdie CA, Bray SE, Quinlan PR, Jordan LB, Thompson AM, Meek DW. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast Cancer Res. 2012;14(2):R40. doi: 10.1186/bcr3136. doi:bcr3136 [pii] 10.1186/bcr3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du J, Cai X, Yao J, Ding X, Wu Q, Pei S, Jiang K, Zhang Y, Wang W, Shi Y, Lai Y, Shen J, Teng M, Huang H, Fei Q, Reddy ES, Zhu J, Jin C, Yao X. The mitotic checkpoint kinase NEK2A regulates kinetochore microtubule attachment stability. Oncogene. 2008;27(29):4107–4114. doi: 10.1038/onc.2008.34. doi:onc200834 [pii] 10.1038/onc.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roylance R, Endesfelder D, Gorman P, Burrell RA, Sander J, Tomlinson I, Hanby AM, Speirs V, Richardson AL, Birkbak NJ, Eklund AC, Downward J, Kschischo M, Szallasi Z, Swanton C. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20(10):2183–2194. doi: 10.1158/1055-9965.EPI-11-0343. doi:1055-9965.EPI-11-0343 [pii] 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. doi:10.1056/NEJMoa021967 347/25/1999 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Fu G, Ding X, Yuan K, Aikhionbare F, Yao J, Cai X, Jiang K, Yao X. Phosphorylation of human Sgo1 by NEK2A is essential for chromosome congression in mitosis. Cell Res. 2007;17(7):608–618. doi: 10.1038/cr.2007.55. doi:cr200755 [pii] 10.1038/cr.2007.55. [DOI] [PubMed] [Google Scholar]
- 43.Weiss MB, Vitolo MI, Mohseni M, Rosen DM, Denmeade SR, Park BH, Weber DJ, Bachman KE. Deletion of p53 in human mammary epithelial cells causes chromosomal instability and altered therapeutic response. Oncogene. 2010;29(33):4715–4724. doi: 10.1038/onc.2010.220. doi:onc2010220 [pii] 10.1038/onc.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mason J, Wei S, Luo X, Nadeem V, Kiarash R, Huang P, Awrey D, Leung G, Beletskaya I, Feher M, Forrest B, Laufer R, Sampson P, Li S-W, Liu Y, Lang Y, Pauls H, Mak TW, Pan JG. Proceedings of the 102nd Annual Meeting of the American Association for Cancer Research. Philadelphia (PA): AACR, Orlando, FL; 2011. Inhibition of Polo-like kinase 4 as an anti-cancer strategy. [Google Scholar]
- 45.Maire V, Nemati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, Gravier E, Marty-Prouvost B, De Koning L, Lang G, Gentien D, Dumont A, Barillot E, Marangoni E, Decaudin D, Roman-Roman S, Pierre A, Cruzalegui F, Depil S, Tucker GC, Dubois T. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013;73(2):813–823. doi: 10.1158/0008-5472.CAN-12-2633. doi:0008-5472.CAN-12-2633 [pii] 10.1158/0008-5472.CAN-12-2633. [DOI] [PubMed] [Google Scholar]
- 46.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115(6):1503–1521. doi: 10.1172/JCI23412. doi:10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berezovska OP, Glinskii AB, Yang Z, Li XM, Hoffman RM, Glinsky GV. Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle. 2006;5(16):1886–1901. doi: 10.4161/cc.5.16.3222. doi:3222 [pii] [DOI] [PubMed] [Google Scholar]
- 48.Brashavitskaya V, Kazazian K, Bagshaw R, Rosario CO, Zih FSW, Haffani Y, Dennis JW, Pawson TJ, Swallow CJ. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research. Philadelphia (PA): AACR, Washington, DC; 2013. RhoGTPase-based regulation of cell motility by Plk4. [Google Scholar]
- 49.Rosario C, Swallow CJ. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research. Philadelphia (PA): AACR, Washington, DC; 2013. Hypoxia induces Plk4 expression and promotes immortalization of proliferating cells. [Google Scholar]
- 50.Rosario CO, Ko MA, Haffani YZ, Gladdy RA, Paderova J, Pollett A, Squire JA, Dennis JW, Swallow CJ. Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci U S A. 2010;107(15):6888–6893. doi: 10.1073/pnas.0910941107. doi:0910941107 [pii] 10.1073/pnas.0910941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sonnen KF, Gabryjonczyk AM, Anselm E, Stierhof YD, Nigg EA. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J Cell Sci. 2013 doi: 10.1242/jcs.129502. doi:jcs.129502 [pii] 10.1242/jcs.129502. [DOI] [PubMed] [Google Scholar]
- 52.Lettman MM, Wong YL, Viscardi V, Niessen S, Chen SH, Shiau AK, Zhou H, Desai A, Oegema K. Direct Binding of SAS-6 to ZYG-1 Recruits SAS-6 to the Mother Centriole for Cartwheel Assembly. Dev Cell. 2013;25(3):284–298. doi: 10.1016/j.devcel.2013.03.011. doi:S1534-5807(13)00160-3 [pii] 10.1016/j.devcel.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laufer R, Forrest B, Li SW, Liu Y, Sampson P, Edwards L, Lang Y, Awrey D, Mao G, Plotnikova O, Leung G, Hodgson R, Beletskaya I, Mason JM, Luo X, Wei X, Yao Y, Feher M, Ban F, Kiarash R, Green E, Mak TW, Pan G, Pauls HW. The Discovery of PLK4 Inhibitors: (E)-3-((1H-indazol-6-yl)methylene)indolin-2-ones as Novel Anti-Proliferative Agents. J Med Chem. 2013 doi: 10.1021/jm400380m. [Epub ahead of print] doi:DOI: 10.1021/jm400380m. [DOI] [PubMed] [Google Scholar]
- 54.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15(6):303–311. doi: 10.1016/j.tcb.2005.04.008. doi:S0962-8924(05)00112-1 [pii] 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 55.D'Assoro AB, Barrett SL, Folk C, Negron VC, Boeneman K, Busby R, Whitehead C, Stivala F, Lingle WL, Salisbury JL. Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res Treat. 2002;75(1):25–34. doi: 10.1023/a:1016550619925. [DOI] [PubMed] [Google Scholar]
- 56.D'Assoro AB, Busby R, Acu ID, Quatraro C, Reinholz MM, Farrugia DJ, Schroeder MA, Allen C, Stivala F, Galanis E, Salisbury JL. Impaired p53 function leads to centrosome amplification, acquired ERalpha phenotypic heterogeneity and distant metastases in breast cancer MCF-7 xenografts. Oncogene. 2008;27(28):3901–3911. doi: 10.1038/onc.2008.18. doi:onc200818 [pii] 10.1038/onc.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20(2):189–193. doi: 10.1038/2496. doi:10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 58.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38(9):1043–1048. doi: 10.1038/ng1861. doi:ng1861 [pii] 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto S, Yamamoto-Ibusuki M, Yamamoto Y, Fujiwara S, Iwase H. A comprehensive analysis of Aurora A; transcript levels are the most reliable in association with proliferation and prognosis in breast cancer. BMC Cancer. 2013;13(1):217. doi: 10.1186/1471-2407-13-217. doi:1471-2407-13-217 [pii] 10.1186/1471-2407-13-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, Deng CX. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25(54):7148–7158. doi: 10.1038/sj.onc.1209707. doi:1209707 [pii] 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 61.Brodie KM, Henderson BR. Characterization of BRCA1 protein targeting, dynamics, and function at the centrosome: a role for the nuclear export signal, CRM1, and Aurora A kinase. J Biol Chem. 2012;287(10):7701–7716. doi: 10.1074/jbc.M111.327296. doi:M111.327296 [pii] 10.1074/jbc.M111.327296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leontovich AA, Salisbury JL, Veroux M, Tallarita T, Billadeau D, McCubrey J, Ingle J, Galanis E, D'Assoro AB. Inhibition of Cdk2 activity decreases Aurora-A kinase centrosomal localization and prevents centrosome amplification in breast cancer cells. Oncol Rep. 2013;29(5):1785–1788. doi: 10.3892/or.2013.2313. doi:10.3892/or.2013.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D'Assoro AB, Liu T, Quatraro C, Amato A, Opyrchal M, Leontovich A, Ikeda Y, Ohmine S, Lingle W, Suman V, Ecsedy J, Iankov I, Di Leonardo A, Ayers-Inglers J, Degnim A, Billadeau D, McCubrey J, Ingle J, Salisbury JL, Galanis E. The mitotic kinase Aurora-A promotes distant metastases by inducing epithelial-to-mesenchymal transition in ERalpha(+) breast cancer cells. Oncogene. 2013 doi: 10.1038/onc.2012.628. doi:onc2012628 [pii] 10.1038/onc.2012.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li JJ, Weroha SJ, Lingle WL, Papa D, Salisbury JL, Li SA. Estrogen mediates Aurora-A overexpression, centrosome amplification, chromosomal instability, and breast cancer in female ACI rats. Proc Natl Acad Sci U S A. 2004;101(52):18123–18128. doi: 10.1073/pnas.0408273101. doi:0408273101 [pii] 10.1073/pnas.0408273101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–282. doi: 10.1038/nature08136. doi:nature08136 [pii] 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437(7061):1043–1047. doi: 10.1038/nature04217. doi:nature04217 [pii] 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 67.Krzywicka-Racka A, Sluder G. Repeated cleavage failure does not establish centrosome amplification in untransformed human cells. J Cell Biol. 2011;194(2):199–207. doi: 10.1083/jcb.201101073. doi:jcb.201101073 [pii] 10.1083/jcb.201101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R. Centrosome amplification causes microcephaly. Nat Cell Biol. 2013 doi: 10.1038/ncb2746. doi:ncb2746 [pii] 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- 69.Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99(4):1978–1983. doi: 10.1073/pnas.032479999. doi:10.1073/pnas.032479999 032479999 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63(6):1398–1404. [PubMed] [Google Scholar]
- 71.Olson JE, Wang X, Pankratz VS, Fredericksen ZS, Vachon CM, Vierkant RA, Cerhan JR, Couch FJ. Centrosome-related genes, genetic variation, and risk of breast cancer. Breast Cancer Res Treat. 2011;125(1):221–228. doi: 10.1007/s10549-010-0950-8. doi:10.1007/s10549-010-0950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kronenwett U, Huwendiek S, Castro J, Ried T, Auer G. Characterisation of breast fine-needle aspiration biopsies by centrosome aberrations and genomic instability. Br J Cancer. 2005;92(2):389–395. doi: 10.1038/sj.bjc.6602246. doi:6602246 [pii] 10.1038/sj.bjc.6602246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneeweiss A, Sinn HP, Ehemann V, Khbeis T, Neben K, Krause U, Ho AD, Bastert G, Kramer A. Centrosomal aberrations in primary invasive breast cancer are associated with nodal status and hormone receptor expression. Int J Cancer. 2003;107(3):346–352. doi: 10.1002/ijc.11408. doi:10.1002/ijc.11408. [DOI] [PubMed] [Google Scholar]
- 74.Guo HQ, Gao M, Ma J, Xiao T, Zhao LL, Gao Y, Pan QJ. Analysis of the cellular centrosome in fine-needle aspirations of the breast. Breast Cancer Res. 2007;9(4):R48. doi: 10.1186/bcr1752. doi:bcr1752 [pii] 10.1186/bcr1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smid M, Hoes M, Sieuwerts AM, Sleijfer S, Zhang Y, Wang Y, Foekens JA, Martens JW. Patterns and incidence of chromosomal instability and their prognostic relevance in breast cancer subtypes. Breast Cancer Res Treat. 2011;128(1):23–30. doi: 10.1007/s10549-010-1026-5. doi:10.1007/s10549-010-1026-5. [DOI] [PubMed] [Google Scholar]
- 76.Montagna C, Andrechek ER, Padilla-Nash H, Muller WJ, Ried T. Centrosome abnormalities, recurring deletions of chromosome 4, and genomic amplification of HER2/neu define mouse mammary gland adenocarcinomas induced by mutant HER2/neu. Oncogene. 2002;21(6):890–898. doi: 10.1038/sj.onc.1205146. doi:10.1038/sj.onc.1205146. [DOI] [PubMed] [Google Scholar]
- 77.Finetti P, Cervera N, Charafe-Jauffret E, Chabannon C, Charpin C, Chaffanet M, Jacquemier J, Viens P, Birnbaum D, Bertucci F. Sixteen-kinase gene expression identifies luminal breast cancers with poor prognosis. Cancer Res. 2008;68(3):767–776. doi: 10.1158/0008-5472.CAN-07-5516. doi:68/3/767 [pii] 10.1158/0008-5472.CAN-07-5516. [DOI] [PubMed] [Google Scholar]
- 78.Jerevall PL, Ma XJ, Li H, Salunga R, Kesty NC, Erlander MG, Sgroi DC, Holmlund B, Skoog L, Fornander T, Nordenskjold B, Stal O. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br J Cancer. 2011;104(11):1762–1769. doi: 10.1038/bjc.2011.145. doi:bjc2011145 [pii] 10.1038/bjc.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Habel LA, Sakoda LC, Achacoso N, Ma XJ, Erlander MG, Sgroi DC, Fehrenbacher L, Greenberg D, Quesenberry CP., Jr HOXB13:IL17BR and molecular grade index and risk of breast cancer death among patients with lymph node-negative invasive disease. Breast Cancer Res. 2013;15(2):R24. doi: 10.1186/bcr3402. doi:bcr3402 [pii] 10.1186/bcr3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tanner M, Kapanen AI, Junttila T, Raheem O, Grenman S, Elo J, Elenius K, Isola J. Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer. Mol Cancer Ther. 2004;3(12):1585–1592. doi:3/12/1585 [pii] [PubMed] [Google Scholar]
- 81.Yao E, Zhou W, Lee-Hoeflich ST, Truong T, Haverty PM, Eastham-Anderson J, Lewin-Koh N, Gunter B, Belvin M, Murray LJ, Friedman LS, Sliwkowski MX, Hoeflich KP. Suppression of HER2/HER3-mediated growth of breast cancer cells with combinations of GDC-0941 PI3K inhibitor, trastuzumab, and pertuzumab. Clin Cancer Res. 2009;15(12):4147–4156. doi: 10.1158/1078-0432.CCR-08-2814. doi:1078-0432.CCR-08-2814 [pii] 10.1158/1078-0432.CCR-08-2814. [DOI] [PubMed] [Google Scholar]
- 82.Korzeniewski N, Hohenfellner M, Duensing S. CAND1 promotes PLK4-mediated centriole overduplication and is frequently disrupted in prostate cancer. Neoplasia. 2012;14(9):799–806. doi: 10.1593/neo.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. doi:S1534-5807(07)00263-8 [pii] 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 84.Colombo R, Caldarelli M, Laura Giorgini ML, Degrassi A, Ciomei M, Pezzetta D, Ballinari D, Montagnoli A, Pesenti E, Donati D, Galvani A. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research. Philadelphia (PA): AACR, Washington, DC; 2013. Targeting aneuploidy with NMS-P153, a tight binder inhibitor of the spindle assembly checkpoint MPS1 (TTK) kinase. [Google Scholar]
- 85.Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11(1):9–23. doi: 10.1016/j.ccr.2006.10.019. doi:S1535-6108(06)00371-0 [pii] 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gehrke I, Gandhirajan RK, Kreuzer KA. Targeting the WNT/beta-catenin/TCF/LEF1 axis in solid and haematological cancers: Multiplicity of therapeutic options. Eur J Cancer. 2009;45(16):2759–2767. doi: 10.1016/j.ejca.2009.08.003. doi:S0959-8049(09)00585-1 [pii] 10.1016/j.ejca.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 87.Li J, Tan M, Li L, Pamarthy D, Lawrence TS, Sun Y. SAK, a new polo-like kinase, is transcriptionally repressed by p53 and induces apoptosis upon RNAi silencing. Neoplasia. 2005;7(4):312–323. doi: 10.1593/neo.04325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakamura T, Saito H, Takekawa M. SAPK pathways and p53 cooperatively regulate PLK4 activity and centrosome integrity under stress. Nat Commun. 2013;4:1775. doi: 10.1038/ncomms2752. doi:ncomms2752 [pii] 10.1038/ncomms2752. [DOI] [PubMed] [Google Scholar]
- 89.Ko MA, Rosario CO, Hudson JW, Kulkarni S, Pollett A, Dennis JW, Swallow CJ. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37(8):883–888. doi: 10.1038/ng1605. doi:ng1605 [pii] 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- 90.Watts CK, Brady A, Sarcevic B, deFazio A, Musgrove EA, Sutherland RL. Antiestrogen inhibition of cell cycle progression in breast cancer cells in associated with inhibition of cyclin-dependent kinase activity and decreased retinoblastoma protein phosphorylation. Mol Endocrinol. 1995;9(12):1804–1813. doi: 10.1210/mend.9.12.8614416. [DOI] [PubMed] [Google Scholar]
- 91.Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Downstream targets of growth factor and oestrogen signalling and endocrine resistance: the potential roles of c-Myc, cyclin D1 and cyclin E. Endocr Relat Cancer. 2005;12(Suppl 1):S47–S59. doi: 10.1677/erc.1.00993. doi:12/Supplement_1/S47 [pii] 10.1677/erc.1.00993. [DOI] [PubMed] [Google Scholar]
- 92.Kilker RL, Planas-Silva MD. Cyclin D1 is necessary for tamoxifen-induced cell cycle progression in human breast cancer cells. Cancer Res. 2006;66(23):11478–11484. doi: 10.1158/0008-5472.CAN-06-1755. doi:66/23/11478 [pii] 10.1158/0008-5472.CAN-06-1755. [DOI] [PubMed] [Google Scholar]
- 93.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, Los G, Slamon DJ. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. doi: 10.1186/bcr2419. doi:bcr2419 [pii] 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bosco EE, Knudsen ES. RB in breast cancer: at the crossroads of tumorigenesis and treatment. Cell Cycle. 2007;6(6):667–671. doi: 10.4161/cc.6.6.3988. doi:3988 [pii] [DOI] [PubMed] [Google Scholar]
- 95.Arpino G, Wiechmann L, Osborne CK, Schiff R. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29(2):217–233. doi: 10.1210/er.2006-0045. doi:er.2006-0045 [pii] 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Frogne T, Jepsen JS, Larsen SS, Fog CK, Brockdorff BL, Lykkesfeldt AE. Antiestrogen-resistant human breast cancer cells require activated protein kinase B/Akt for growth. Endocr Relat Cancer. 2005;12(3):599–614. doi: 10.1677/erc.1.00946. doi:12/3/599 [pii] 10.1677/erc.1.00946. [DOI] [PubMed] [Google Scholar]
- 97.Shin I, Arteaga CL. Expression of active Akt protects against tamoxifen-induced apoptosis in MCF-7 Cells. IUBMB Life. 2006;58(11):664–669. doi: 10.1080/15216540601001681. doi:G41066566U772185 [pii] 10.1080/15216540601001681. [DOI] [PubMed] [Google Scholar]
- 98.Ghayad SE, Vendrell JA, Ben Larbi S, Dumontet C, Bieche I, Cohen PA. Endocrine resistance associated with activated ErbB system in breast cancer cells is reversed by inhibiting MAPK or PI3K/Akt signaling pathways. Int J Cancer. 2010;126(2):545–562. doi: 10.1002/ijc.24750. doi:10.1002/ijc.24750. [DOI] [PubMed] [Google Scholar]
- 99.Ellis MJ, Lin L, Crowder R, Tao Y, Hoog J, Snider J, Davies S, DeSchryver K, Evans DB, Steinseifer J, Bandaru R, Liu W, Gardner H, Semiglazov V, Watson M, Hunt K, Olson J, Baselga J. Phosphatidyl-inositol-3-kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2010;119(2):379–390. doi: 10.1007/s10549-009-0575-y. doi:10.1007/s10549-009-0575-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, Hortobagyi GN, Hung MC, Yu D. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6(2):117–127. doi: 10.1016/j.ccr.2004.06.022. doi:10.1016/j.ccr.2004.06.022 S1535610804002107 [pii] [DOI] [PubMed] [Google Scholar]
- 101.O'Brien NA, Browne BC, Chow L, Wang Y, Ginther C, Arboleda J, Duffy MJ, Crown J, O'Donovan N, Slamon DJ. Activated phosphoinositide 3-kinase/AKT signaling confers resistance to trastuzumab but not lapatinib. Mol Cancer Ther. 2010;9(6):1489–1502. doi: 10.1158/1535-7163.MCT-09-1171. doi:1535-7163.MCT-09-1171 [pii] 10.1158/1535-7163.MCT-09-1171. [DOI] [PubMed] [Google Scholar]
- 102.Baselga J. Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist. 2011;16(Suppl 1):12–19. doi: 10.1634/theoncologist.2011-S1-12. doi:16/suppl_1/12 [pii] 10.1634/theoncologist.2011-S1-12. [DOI] [PubMed] [Google Scholar]
- 103.Le XF, Bedrosian I, Mao W, Murray M, Lu Z, Keyomarsi K, Lee MH, Zhao J, Bast RC., Jr Anti-HER2 antibody trastuzumab inhibits CDK2-mediated NPAT and histone H4 expression via the PI3K pathway. Cell Cycle. 2006;5(15):1654–1661. doi: 10.4161/cc.5.15.3007. doi:3007 [pii] [DOI] [PubMed] [Google Scholar]