Abstract
The rapid determination of nucleic acid sequences is increasing the number of sequences that are available. Inherent in a template or seed alignment is the culmination of structural and functional constraints that are selecting those mutations that are viable during the evolution of the RNA. While we might not understand these structural and functional, template-based alignment programs utilize the patterns of sequence conservation to encapsulate the characteristics of viable RNA sequences that are aligned properly. We have developed a program that utilizes the different dimensions of information in rCAD, a large RNA informatics resource, to establish a profile for each position in an alignment. The most significant include sequence identity and column composition in different phylogenetic taxa. We have compared our methods with a maximum of eight alternative alignment methods on different sets of 16S and 23S rRNA sequences with sequence percent identities ranging from 50% to 100%. The results showed that CRWAlign outperformed the other alignment methods in both speed and accuracy. A web-based alignment server is available at http://www.rna.ccbb.utexas.edu/SAE/2F/CRWAlign.
Keywords: RNA sequence alignment, template-based alignment, comparative analysis, phylogenetic-based alignment
I. Introduction
Darwin's use of comparative analysis was the foundation for his theory on the evolution of biological species [1]. Today the use of comparative analysis is resolving, at the molecular level, the structure, function, and evolution of the cell. Ultra-rapid nucleic acid sequencing methodologies are providing much of the data for this analysis. This monumental increase in data is occurring in parallel with a major paradigm shift in molecular biology. RNA, once considered a molecule only associated with protein synthesis, is now being implicated in all aspects of the cell's basic regulation and metabolism. Many analyses of RNA utilize an alignment of RNA sequences that is composed of similar primary, secondary, and other higher-order structural elements juxtaposed into the same set of columns.
These alignments are analyzed to reveal evolutionary relationships, patterns of structure conservation and variation and secondary and tertiary structures. A few of the discoveries from this analysis are: the identification of Archaea, the third kingdom of life [2], and the phylogenetic relationships for organisms that span the entire tree of life [2]; the accurate determination of RNA secondary and tertiary structures common to sequences from the same RNA family [3, 4], and the use of 16S rRNA to identify the bacteria in microbial ecology environments [5, 6].
De novo RNA sequence alignment programs, such as MAFFT [7], Clustal [8, 9] and SATe [10], generate multiple sequence alignments from sequence information alone. These programs do not utilize any preexisting alignment as a guide to aligning sequences.
Another approach uses a template or seed alignment as reference when aligning a new sequence. Three of the template-based automated multiple RNA sequence aligners available on the web are Silva [11], RDP [12] and Greengenes [13]. Greengenes aligns a new sequence with the Nearest Alignment Space Termination [14] (NAST) algorithm by finding the closest matching seed sequence and performing a pairwise alignment with BLAST [15]. Silva aligns with a dynamic incremental profile sequence aligner called SINA which utilizes a variant of the Needleman-Wunsch algorithm [16]. SINA uses up to 40 related sequences and switches between seed sequences to align sequence regions [11]. With release 10, RDP switched to Infernal [17], a secondary-structure based aligner.
Three of the stand-alone alignment programs available for download are Infernal, HMMER [18] and ssu-align[17]. Infernal builds consensus RNA secondary structure profiles to create new MSAs. Ssu-align is built upon Infernal but adds profile hidden Markov models to the secondary structure profiles. HMMER also builds hidden Markov models, but does not use the consensus secondary structure.
When sequences have minimal identity with one another, de-novo alignment algorithms are unable to determine the correct placement for all of the nucleotides within each sequence into an alignment. In contrast, template-based alignment algorithms utilize information from an existing alignment that has been refined by maximizing the correct juxtaposition of primary and higher-order structural and functional information, and by utilizing the evolutionary relationships of the sequences. Until we are able to capture all of these constraints encrypted in macromolecular sequences into de-novo alignment programs then the addition of new sequences into an existing alignment with template-based alignment algorithms will be more accurate.
We have developed a template-based alignment program called CRWAlign that achieves high accuracy and maintains performance over a large set of sequences. It utilizes the different dimensions of information stored in rCAD to establish a profile for each position in an alignment. The most significant include sequence identity and column composition in different phylogenetic groups. The phylogenetic information in rCAD is obtained from the NCBI Taxonomy database. It can align any RNA molecule for which there is a template alignment and does not require or use the secondary structure of the alignment. Also, the size of the template alignment is limited only by the number of columns and phylogenetic groups in the alignment and the size of available memory. The Gutell lab has used CRWAlign on a 16S bacterial rRNA alignment that is 10,000 columns wide and contains ~140,000 sequences.
The objectives of this paper was to describe CRWAlign and compare it with the most widely-used RNA template-based alignment programs which included three stand-alone programs (ssu-align, Infernal and HMMER) and three web-based aligners (RDP, Silva and GreenGenes). Also, we have included two of the best de novo alignment programs (MAFFT and SATe) in our results. For a rigorous assessment of these programs, we selected the 16S and 23S ribosomal RNAs. These alignments have at least 1400 nucleotides in length and more than 1000 sequences. CRWAlign was more accurate than all other methods on both RNA molecules tested and outperformed the stand-alone methods in execution time.
II. Algorithm
CRWAlign consists of two steps, generation of alignment statistics and aligning sequences. The first step computes alignment statistics of the template sequence for each taxonomic group. The second step is an iterative process to align sequences guided by those statistics. Psuedocode for both phases was not included for lack of space.
A. Alignment Statistics Generation
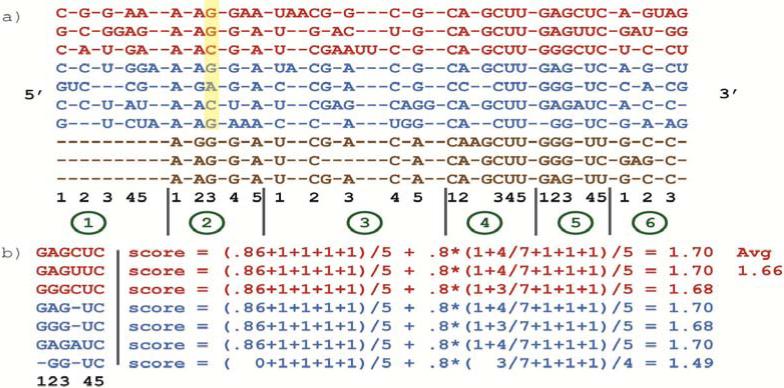
Before a sequence can be aligned, the alignment statistics used by CRWAlign must first be collected from an analysis of the template alignment. A simple example of the statistics generation process involving a ten sequence template alignment is given in Fig. 1.
Figure 1.
Example of the alignment statistics generation process performed by CRWAlign: a) 10 template sequences (red – bacillales, blue – bacilli, brown – bacteria) are used to determine conserved alignment columns; b) computation of the average block 5 score for the bacilli node.
Definition 1: The conservation value of a column i is the ratio of the number of sequences with a nucleotide in column i divided by the total number of sequences in the taxonomic node. If a partial sequence “starts” after or “ends” before column i, it is excluded from the calculation.
The first step is to determine the conservation value for every column calculated at the root taxonomic node (i.e. the entire alignment). Every column with a conservation value greater than 80% is considered a highly-conserved column. Next, the alignment is divided into blocks with each block containing the same number of highly-conserved columns (Fig. 1a). The only block that could have fewer conserved columns is the last block. Note that the last three (partial) sequences in Fig. 1a do not contain the 5’ tail, but they do not affect the conservation value of the columns in block 1. But they are counted in the conservation values for every column after block 1. For the purposes of the example in Fig. 1, the number of conserved columns of each block was set to five, but the best accuracies are seen with the number set between fifteen and twenty. Defining blocks alignment-wide ensures that the statistics for any block covers the same set of columns in every taxonomic node even if the set of conserved columns varies between taxonomic nodes. For example, block 5 in the bacillales (red) node has six conserved columns while it has only five conserved columns in the bacilli or bacteria node (Fig. 1a and 1b).
Alignment statistics are generated for every taxon that contains a minimum number (usually 3-10) of sequences from the seed alignment. In Fig. 1, there are three bacillales (red) sequences, four additional bacilli (blue) sequences and three other bacterial (brown) sequences. The statistics would be generated for the following taxons: bacillales, bacilli, firmicutes (parent of bacilli) and bacteria. There are three sequences in the bacillales node, seven in bacilli and firmicutes (three bacillales and four additional bacilli) and ten sequences in the bacteria node. The statistics generated for each taxon consists of the column conservation values (Def. 1), nucleotide composition of each column (Def. 2), minimum and maximum number of nucleotides in a block and the average score of each block (Def. 3). For each taxon, the complexity of this phase is O(nbs), where n is the number of blocks, b is the length of a block and s is the number of sequences in the taxon.
Definition 2: For any column, there are 4 nucleotide composition values corresponding to A, C, G and U. The value for a given nucleotide is the ratio of the number of sequences with the given nucleotide in that column divided by the number of sequences with any nucleotide in that column. The nucleotide composition value Nip for the ith column and p∈{A,C,G,U}is calculated with:
| (1) |
where SCp(i) is the number of sequences with nucleotide p in column i. For example, for conserved column 3 in block 2 (Fig. 1a, highlighted nucleotides), the nucleotide composition values are A-0%, C-33%, G-67% and U-0% for the bacillales node and A-14%, C-29%, G-57% and U-0% for the bacilli node.
For each alignment block, we compute a score to evaluate how well a given subsequence fit into this block (Def. 3). The numerical measure includes two components: average column conservation values of that subsequence in this alignment block and the average nucleotide composition values of that subsequence.
Definition 3: Given a block in a taxonomic node, let Y be the set of conserved columns. For a given subsequence Si, in this block, let Z be the set of non-gap conserved columns in this subsequence. The score of this subsequence for block b is calculated with:
| (2) |
where Ci is the conservation value of column i and Nj(Sj) is the nucleotide composition value corresponding to the nucleotide at column j in subsequence S. The weight on the second component, 0.8, is determined heuristically. The highest possible score is 1.8.
Fig. 1b contains an example of how an average block score is calculated, specifically block 5 in the bacilli node. The conservation value for conserved columns 2-5 is 1.0 but column 1 has a value of 6/7 = .86 since the last sequence does not contain a nucleotide in that column. The nucleotide composition values are 1.0 for columns 3-5 and also for column 1 since the gap in the last sequence will not affect nucleotide composition, only column conservation. Column 2 has variable nucleotide composition values since it contains 4 As and 3 Gs from the 7 sequences. Note also that the divisor for second term in the score is 4 for the last sequence, but 5 for the others.
B. Alignment Algorithm
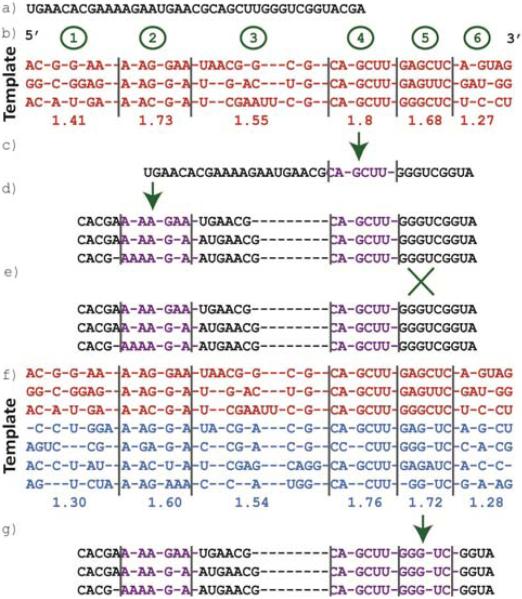
CRWAlign is an iterative alignment algorithm. An example of this process is shown in Fig. 2. The first step is to select the phylogenetic group to align against. If phylogenetic information for the new sequence is available, CRWAlign will use it to select the lowest taxon for which alignment statistics exist. Alternatively, a user may elect to specify theses details. If no such information is available or given, the program will test each taxon at a predefined leaf level, which is 6 in our experiments. The node that aligns the most nucleotides in the first stage will be used from that point on. In the example in Fig. 2, the sequence to be aligned (Fig. 2a) is in the Bacillales taxonomic group. This example uses the same template sequence information as given in Fig. 1, i.e. there are 3 bacillales sequences, 4 additional bacilli sequences and 10 total bacterial sequences. The complexity of the alignment phase is approximately O(nbl), where n is the number of blocks, b is the length of a block and l is the length of the sequence.
Figure 2.
Example of the alignment process performed by CRWAlign: a) unaligned RNA sequences; b) 3 bacillales (red) template sequences and average block scores; c) single match is found for block 5 is aligned; d) 3 matches are found for block 3; e) no matches are found for block 5; f) 7 template sequences (red – bacillales, blue – bacilli) and average block scores; g) a match is found for block 5.
In the first iteration, CRWAlign will only attempt to align the most highly-conserved blocks (i.e. those blocks with an average score greater than 90% of the highest possible score: 1.8 * .9 = 1.62). To ensure the highest accuracy at this stage, the minimum score required for a block to be considered is set quite high (again 1.62), deletions of conserved columns are not allowed and only a limited number of insertions between conserved columns are accepted. This guarantees that any aligned block has a very high probability of being absolutely correct. By first aligning only the most highly-conserved blocks that are closely matched has the effect of reducing the problem of aligning the complete sequence into aligning a set of smaller subsequences. This reduces the complexity of the problem and increases speed and overall accuracy.
In the example, the highly-conserved blocks in the bacillales alignment statistics (Fig. 2b) are 2, 4 and 5 (i.e. only blocks with average scores greater than 1.62). Since block 4 has the highest conservation it is selected first. CRWAlign searches the sequence for a match to the template data. In this case, the matching subsequence must be CAGCUU or something extremely similar to it. The program is able to find a match that scores higher than 1.62 and aligns just those matching nucleotides (Fig. 2c).
CRWAlign will next attempt to align block 2 since it has a higher average score than block 5. Because blocks 2 and 3 have variable lengths, there are multiple possible starts for block 2. In fact, CRWAlign would find 3 different acceptable matches for this block(Fig. 2d). Prematurely choosing the wrong block alignment potentially damages the accuracy of multiple blocks. It solves this problem by carrying multiple potential sequence alignments forward. At each step, the program will attempt to continue the alignment process with each of these acceptable alignments up to a max of the 10 highest-scoring potential alignments. In the examples in Figs. 1 and 2, the block conserved-columns count is set to 5 which will increase the odds that a highly-conserved block will have multiple possible matches in the sequence. By setting this count to a higher number (>10), the highly-conserved blocks will rarely have multiple matches significantly limiting the number of potential sequence alignments CRWAlign must track.
The next block to be aligned is block 5. This block is 3’ of and contiguous to block 4. Therefore, the nucleotides 3’ of the block just aligned must be able to match block 5. In this case, the subsequence GGGUC does not match the template data of G[A/G]GCUC (Fig. 2e). This stage does not allow a conserved column deletion.
After attempting to align all of the most conserved blocks at the lowest taxon possible (i.e. bacillales), CRWAlign does not then move onto less conserved blocks. Instead, it then attempts to align the most conserved blocks in the parent taxonomic node (i.e. bacilli). In this way, the program works its way up the phylogenetic tree. In the bacilli node, there are only 2 conserved blocks (4 and 5) with block average score greater than 1.62. Since block 4 is already aligned, CRWAlign will attempt block 5 and succeed in finding an acceptable match since there are now 5 conserved columns instead of the 6 in the bacillales node (Fig. 2f). Therefore, GGGUC is able to match the block template without requiring a conserved column deletion (Fig. 2g). After this match, there are still 3 potential sequence alignments that will be carried forward.
At this stage in the process, CRWAlign would move up to the parent of bacilli - the firmicutes taxonomic node, but since this node contains no additional template sequences, it is skipped. There are additional template sequences in the bacteria node, but blocks 4 and 5 remain the only highly-conserved blocks. Therefore, this stage of the alignment process is complete. After the completion of any stage, the minimum block score is reduced by 20%. Also, after the first stage, the deletion of a conserved column in a possible alignment is permitted. CRWAlign will take the 3 potential sequence alignments from the previous step (Fig. 2g) and start the process over at the bacillales taxonomic node. The program continues until the entire sequence is aligned.
III. Results
CRWAlign has been evaluated in three different aspects, the accuracy of the alignment results, the running time of the program executions and the scalability for large number of sequences. In addition, CRWAlign has been compared to several existing widely-used automatic alignment programs.
A. Programs Compared
For this study, three template-based programs, ssu-align, Infernal and HMMER (all three available at http://selab.janelia.org/software.html), were tested along with MAFFT (http://mafft.cbrc.jp/alignment/software) and SATe (http://phylo.bio.ku.edu/software/sate/sate.html). Additional testing was done on three web-based aligners, RDP (http://rdp.cme.msu.edu/), Silva (http://www.arbsilva.de) and GreenGenes (http://greengenes.lbl.gov). GreenGenes, RDP and ssu-align are limited to aligning 16S rRNA only while Silva is able to align 23S rRNA in addition to 16S rRNA. The four remaining stand-alone programs, like CRWAlign, are capable of aligning any type of RNA sequence. Infernal, HMMER and CRWAlign are capable of accepting a template alignment as a seed for generating new profiles/statistics. The program ssu-align uses a profile that has already been built and incorporated into the program. In the testing phase, MAFFT-ginsi provided the highest accuracy of the strategies made available by MAFFT. All programs and web sites were run/accessed using the default parameters.
B. Calculating the Accuracy of an Alignment
Randomly selected subsets of the bacterial 16S and 23S rRNA alignments available on the Comparative RNA Web (CRW) Site [19] were used for both test and template sets. There was no overlap between a test and template set, but template sequence alignments were subsets of any larger template alignment (e.g. the 250 bacterial 16S rRNA template alignment was a subset of the 500 bacterial 16S rRNA template alignment). The programs were evaluated through pairwise sequence comparisons. For a pair of sequences i and j, let E be the set of columns containing a nucleotide from either sequence i or j. The pairwise sequence identity for sequences i and j is defined as:
| (3) |
where B is the set of columns containing a nucleotide from both i and j. Pairwise sequence accuracy is defined as:
| (4) |
where S is the set of columns in the test alignment that have an identical stack relative to the correct alignment. For example, if nucleotide 55 (G) of sequence A is stacked with nucleotide 63 (C) of sequence B, then the test alignment must have nucleotide 55 stacked with nucleotide 63 and not with a C nucleotide at any position in sequence B other than nucleotide 63. If a nucleotide from either sequence is stacked with a gap, the test alignment must have the nucleotide stacked with a gap.
C. Accuracy Comparison with other Methods
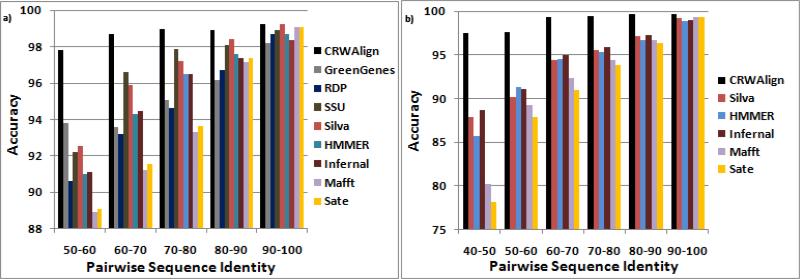
The bacterial 16S rRNA is the only RNA molecule supported by all 9 programs. 1000 bacterial 16S rRNA sequences were aligned by each program and the accuracies based upon ranges of pairwise sequence identity were calculated (Fig. 3a). Each of the 3 programs that accept template alignments (CRWAlign, HMMER and Infernal) were given different template alignments of varying sizes (250, 500, 2000 sequences). CRWAlign, HMMER and Infernal performed best with template alignments of 2000 sequences, 500 sequences and 250 sequences, respectively. The best results for each are used in Fig. 3a.
Figure 3.
Pairwise sequence accuracy results from a) alignment of 1000 bacterial 16S rRNA sequences for 9 alignment programs and b) alignment of 500 bacterial 23S rRNA sequences for 6 alignment programs.
In the 90-100% pairwise sequence identity range, Sate, MAFFT, and Silva essentially equaled the performance of CRWAlign. The other 4 programs had accuracies 0.3% (ssu-align) to 1.0% (GreenGenes) less than CRWAlign. Silva nearly equaled the accuracy of CRWAlign in the 80-90% range, but the gap between CRWAlign and the other 7 programs, including Sate and MAFFT, increased to .6% (ssu-align) to 2.5% (GreenGenes). The difference between CRWAlign and the other programs in the 70-80% sequence identity range varied from .7% (ssu-align) to 4.5% (MAFFT). CRWAlign significantly outperformed every other program in both the 60-70% and 50-60% sequence identity ranges. The de novo programs were able to nearly match the template-based programs in the 80-90% and 90-100% ranges but for lower ranges their accuracies were considerably less than the template-based accuracies.
The web-based aligners, GreenGenes and RDP, and ssu-align did not provide support for 23S rRNA sequences. Fig. 3b contains the results for all of the remaining programs from aligning 500 bacterial 23S rRNA sequences. For HMMER, Infernal and CRWAlign, a 1000 bacterial 23S rRNA template alignment was used. The de novo programs, Sate and MAFFT, nearly equaled the accuracy of CRWAlign in the 90-100% pairwise sequence identity range each scoring only 0.1% less. The other programs had accuracies 0.3% (Silva) to 0.6% (HMMER) less. For the 80-90% range, the gap between CRWAlign and the other programs ranged from 1.3% (Infernal) to 2.0% (Sate) less. This gap continued to grow as the pairwise sequence identity dropped with the 40-50% range having differences ranging from 8.8% (Infernal) to 19.4% (Sate).
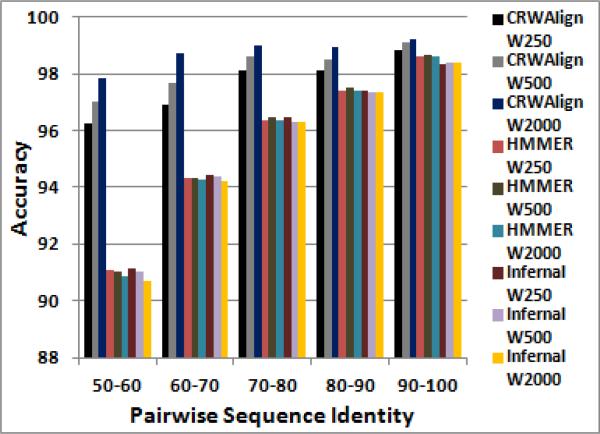
D. Effect of Template Size on Accuracy
To gauge how CRWAlign, HMMER and Infernal dealt with template alignments of differing sizes, each program was given three template alignments containing 250, 500 and 2000 bacterial 16S rRNA sequences as input for aligning 1000 bacterial 16S rRNA sequences (Fig. 4). For both HMMER and Infernal, the overall differences in performance for the 3 template alignment were quite small. In contrast, CRWAlign grew more accurate as the number of sequences in the template alignment grew with large performance gains seen in the 50-60% and 60-70% pairwise sequence identity ranges. But while CRWAlign performed best with the 2000 sequence template alignment, its results using the 250 sequence template alignment were still superior to HMMER and Infernal regardless of template size. In fact, with the 250 sequence template alignment, CRWAlign was still able to outperform overall every other alignment program in this study (Figs. 3a and 4).
Figure 4.
Pairwise sequence accuracy results from alignment of 1000 bacterial 16S rRNA sequences for 3 alignment programs and 3 different template alignments (250, 500 and 2000 sequences).
E. Comparisons of the Run Time of Programs
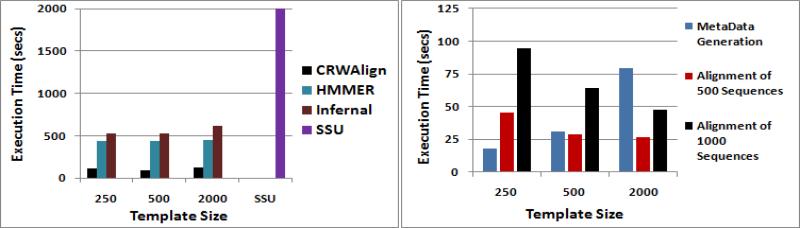
To analyze how fast CRWAlign is able to align RNA sequences, it was compared with the other 3 stand-alone programs, HMMER, Infernal and ssu-align. Fig. 5a shows the wall-clock execution time of each of the programs when aligning 1000 bacterial 16S rRNA sequences when given 3 different template alignments (250, 500 and 1000 sequences). Only 1 data point was collected for ssu-align given that its profile had already been built into the program. Due to platform requirements and software dependencies, CRWAlign has been tested on a Windows platform with an Intel Xeon x7550 @ 2GHz running Windows Server 2008 R2 Enterprise (64-bit). HMMER and Infernal were run on a Linux platform with an Intel Core i7 920 @2.67GHz running Ubuntu (11.10 32-bit). The ssu-align program was run on a Intel Xeon processor 5400 running Solaris 10.0. The results show that CRWAlign is over 15 times faster than ssu-align and on average, 4 and 5 times faster than HMMER and Infernal, respectively (Fig. 5a).
Figure 5.
Execution time for a) aligning 1000 bacterial 16S rRNA sequences for four alignment programs and b) the two phases of the CRWAlign programs when aligning 500 (red) and 1000 (black) bacterial 16S rRNA sequences and using 250, 500 and 2000 template sequences.
F. Scalability of CRWAlign
CRWAlign consists of two phases: 1) statistics generation from a template alignment and 2) aligning unaligned sequences. The total running time of CRWAlign is sensitive to both the number of template sequences as well as the number of sequences aligned. Fig. 5b show the execution time for aligning 500 and 1000 bacterial 16S rRNA sequences with 3 different template alignments (250, 500 and 2000 sequences). The results show that the computational cost of generating alignment statistic is linear to the number of template sequences while the alignment phase has an execution time that increases linearly to the number of sequences to be aligned. Also, as the size of the template alignment grows, CRWAlign is able to align the RNA sequences at a faster rate. This speed increase is most prominent when the template size is small as seen when the template size was increased from 250 to 500 (Fig. 5b). There will be a diminishing return as the size of the template alignment grows. Also, the running time of both phases will grow linearly with the average length of the RNA molecule.
IV. Discussion
The alignment of RNA sequences has in the past required a manual curation stage to create the most accurate alignment. While this has facilitated the creation of very accurate and large alignments at the several of the RNA comparative analysis websites (e.g. rfam, Comparative RNA Web (CRW) Site), this curation is very time-consuming and is unfeasible with the very large number of sequences that are now commonly analyzed. Thus it is imperative that computer programs perform these tasks.
We have developed a template-based alignment algorithm that significantly outperforms in terms of accuracy any existing program. This program utilizes multiple dimensions of data including sequence composition, column conservation and phylogenetic information. This information is stored in rCAD and is used to generate the necessary statistics needed to align new sequences.
The accuracy of CRWAlign was tested on two ribosomal RNA molecules and compared with various template-based and de novo RNA sequence aligners. For both molecules, when the sequence identity range was 90-100%, the competing programs were usually able to nearly match the accuracy of CRWAlign. It is when the sequence identity dropped lower that the other programs were unable to match the accuracy of CRWAlign. CRWAlign was able to maintain a accuracy greater than 97.6% for both RNA molecules in the sequence identity range of 50-60%.
Also, when execution times were compared with the three programs available for download, Infernal, HMMER and ssu-align, our approach ran significantly faster. CRWAlign will also scale to accept large number of sequences in the template alignment and large number of unaligned sequences. Generation of the alignment statistics with templates used in this study showed linear computation time. As the size of the template alignment grows to 10,000 sequences and larger, we expect that the computational cost will increase to linear. As the number of sequences to align grows, the computational cost grows linearly.
The monumental increase in the number of RNA sequences creates new opportunities for the comparative analysis of RNA structure, function, and phylogenetic relationships. These analyses require that these sequences be aligned as accurately and expediently as possible. A web service is available at the CRW Site (http://www.rna.ccbb.utexas.edu/SAE/2F/CRWAlign). This will allow users to obtain a higher quality alignment after uploading unaligned rRNA sequences from any phylogenetic domain.
Acknowledgment
Funding: This project has been funded by grants from Microsoft Research (UTA08-531), National Institutes of Health (GM067317 and GM085337) and Welch Foundation (No. 1427).
Contributor Information
David P. Gardner, Institute for Cellular and Molecular Biology The University of Texas at Austin Austin, TX, USA dpgardn@utexas.edu
Weijia Xu, Texas Advanced Computing Center The University of Texas at Austin Austin, TX, USA xwj@tacc.utexas.edu.
Daniel P. Miranker, Department of Computer Sciences The University of Texas at Austin Austin, TX, USA miranker@cs.utexas.edu
Stuart Ozer, Microsoft Corporation Redmond, WA, USA stuarto@microsoft.com.
Jamie J. Cannone, Center for Computational Biology and Bioinformatics The University of Texas at Austin Austin, TX, USA.
Robin R. Gutell, Center for Computational Biology and Bioinformatics The University of Texas at Austin Austin, TX, USA.
References
- 1.Darwin C. On the Origin of Species London. John Murray; United Kingdom: 1859. [Google Scholar]
- 2.Woese CR. Bacterial evolution. Microbiol. Rev. 1987 Jun;51:221–71. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16-S-like ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 4.Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 2002 Jun;12:301–10. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 5.Robertson CE, Harris JK, Spear JR, Pace NR. Phylogenetic diversity and ecology of environmental Archaea. Curr. Opin. Microbiol. 2005 Dec;8:638–42. doi: 10.1016/j.mib.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Ram JL, Karim AS, Sendler ED, Kato I. Strategy for microbiome analysis using 16S rRNA gene sequence analysis on the Illumina sequencing platform. Syst. Biol. Reprod. Med. 2011 Jun.57:162–170. doi: 10.3109/19396368.2011.555598. [DOI] [PubMed] [Google Scholar]
- 7.Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:212. doi: 10.1186/1471-2105-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73:237–44. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 10.Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T. Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science. 2009 Jun 19;324:1561–4. doi: 10.1126/science.1171243. [DOI] [PubMed] [Google Scholar]
- 11.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005 Jan 1;33(Database Issue):D294–6. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006 Jul;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeSantis TZ, Jr., Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006 Jul 1;34:W394–9. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990 Oct 5;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48:443–53. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 17.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009 May 15;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011 Mar 20;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]