Abstract
Background
The transcriptome complexity in an organism can be achieved by alternative splicing of precursor messenger RNAs. It has been revealed that alternations in mRNA splicing play an important role in a number of diseases including human cancers.
Methods
In this study, we exploited whole transcriptome sequencing data from five lung adenocarcinoma tissues and their matched normal tissues to interrogate intron retention, a less studied alternative splicing form which has profound structural and functional consequence by modifying open reading frame or inserting premature stop codons.
Results
Abundant intron retention events were found in both tumor and normal tissues, and 2,340 and 1,422 genes only contain tumor-specific retentions and normal-specific retentions, respectively. Combined with gene expression analysis, we showed that genes with tumor-specific retentions tend to be over-expressed in tumors, and the abundance of intron retention within genes is negatively related with gene expression, indicating the action of nonsense mediated decay. Further functional analysis demonstrated that genes with tumor-specific retentions include known lung cancer driver genes and are found enriched in pathways important in carcinogenesis.
Conclusions
We hypothesize that intron retentions and consequent nonsense mediated decay may collectively counteract the over-expression of genes promoting cancer development. Identification of genes with tumor-specific retentions may also help develop targeted therapies.
Keywords: Intron retentions, RNA-Seq, Lung adenocarcinoma, Gene expression, Nonsense mediated decay
Background
As one of the leading causes of cancer-related mortality in the world, lung cancer accounts for approximately 12 percent of all cancer incidences and 17.6 percent of cancer deaths [1,2]. Of them, lung adenocarcinoma accounts for more than 500,000 deaths per year worldwide and is the most common subtype of non-small cell lung cancer [3]. Although the underlying mechanism of lung adenocarcinoma is still under investigation, studies showed that recurrent mutations in the epidermal growth factor receptor (EGFR) and the anaplastic lymphoma kinase (ALK) fusions could change the efficacy of treatment for patients with lung adenocarcinoma [4-8]. Genetic modifications in other genes, including targeted mutations in BRAF, AKT1, ERBB2 and PIK3CA, as well as ROS1- and RET-involved fusions, may also affect cancer therapy [9]. In addition, a recent study has found frequent copy number changes in NKX2-1, TERT, PTEN, MDM2, CCND1, and MYC in lung adenocarcinoma [3], highlighting the role of various types of genetic alternations in carcinogenesis.
Alternative splicing in multiple-exon genes is prevalent in eukaryotes and it is actively involved in development, cell differentiation and disease. Approximately 90% of multi-exon human genes have splicing variants in different tissues and cell lines [10,11]. Intron retention, or the maintenance of an intron in a mature mRNA transcript, is a less common type of alternative splicing [12] and can have large functional consequence by introducing premature mutations to the mature transcript. Although the impact of intron retentions has been less acknowledged, a recent report suggests that intron retention is one of the most predominant splice events in three breast cancer subtypes [13], and the retention of intron 4 in the wild-type cholecystokinin type 2 (CCK2) receptor shows elevated expression associated with increased tumor growth in a few cancers [14].
The emergence of high-throughput sequencing technologies in the past few years has provided a new platform to perform large-scale transcriptome profiling at an affordable cost. Based on high-throughput sequencing, RNA-Seq can precisely measure mRNA expression and characterize gene isoforms [15,16], and is commonly used to identify somatic mutations [17,18], differentially expressed genes [19], fusion genes in tumor tissue [20-22], and allele-specific expression [23,24]. Here in the present study, we exploited the rich information in RNA-seq data to investigate the potential role of intron retentions in lung adenocarcinoma. Using tumor and matched normal samples, we systematically identified genes with tumor-specific intron retentions. Further investigation suggests a potential protective role of intron retentions in carcinogenesis through the action of nonsense mediated decay (NMD).
Methods
Transcriptome dataset
Transcriptome sequencing data from five lung adenocarcinoma and their paired adjacent normal tissue specimens [17] were downloaded from European Nucleotide Archive (ENA, http://www.ebi.ac.uk/ena/), using the accession number ERP001058. Reads from five patients (LC1, LC5, LC10, LC11, and LC12) were used in this study, and as described in the original study, all protocols were approved by the Institutional Review Board of Seoul National University Hospital (Approval # C-1111-102-387) and Seoul St. Mary’s Hospital (Approval # KC11TISI0678). 101-bp paired-end reads were generated by Illumina Hiseq 2000 sequencer for each sample.
Exon-intron junction data
To extract exon-intron junction sequences, human exon information was first downloaded from Ensembl database (release 69) [25]. To assign exon-intron junction unambiguously, intersecting exons were excluded, resulting in 164,500 non-overlapping exons. Then exon-intron junctions were then determined and 101-bp sequences were extended in each direction for future mapping.
Identification of intron retentions
A strategy of two rounds of short read mapping was adopted to identify retention reads. Bowtie2 [26] was first used to align RNA-Seq reads to human cDNA sequences (Ensembl release 69), with seed length as 20-bp. Unmapped reads were then extracted and mapped to exon-intron junctions by Bowtie2, using the same parameter above. Uniquely mapped reads with a minimum quality score of 30 that cover a 20-bp region centered on the exon-intron junction site and have at most two mismatches within this region were defined as retention reads. A common tumor-specific retention (TSR) was defined as an exon-intron junction that was supported by retention reads from at least two tumor samples and no retention read could be found in any normal sample. Normal-specific retention (NSR) was similarly defined. For each candidate TSR, we estimated its relative abundance by recording the number of reads that covered the junction position in the initial alignment, and reads with insertion or deletions were excluded. Then for a gene, the relative abundance of intron retentions was calculated as:
where Ri is the retention abundance for gene i.
Identification of differentially expressed genes (DEGs)
Gene expression was first calculated by using the RSEM program [27], which effectively uses ambiguously-mapping reads to estimate expression abundance. Next, EdgeR package was used to normalize the data by trimmed mean of M values (TMM) and identify differentially expressed genes [28]. Genes at low expression level (≤ 1 transcript per million reads, TPM) were excluded and DEGs were defined as genes with a p-value < 0.05 after Benjamini-Hochberg adjustment [29].
Identification of tumor-specific variants (TSVs)
Variants in tumor samples were first identified by SAMtools [30] for each patient, and only variants supported by at least three reads with base quality ≥20 were retained. Positions of those variants were then examined in normal samples to make sure they were also covered by reads from the corresponding normal samples and they were not variable in normal samples.
Functional enrichment and pathway analysis
Gene ontology (GO) [31] information for query genes was assigned using bioconductor (http://www.bioconductor.org) package “org.Hs.eg.db”. Enrichment tests were performed by assuming a hypergeometric distribution using “topGO” package [32]. KEGG (Kyoto Encyclopedia of Genes and Genomes) database [33] was used to retrieve pathway annotation information, and Fisher’s exact test was performed to evaluate the enrichment of a pathway. Multiple test correction was conducted using Benjamini-Hochberg method.
Results
Summary statistics of human intron retentions
The data used in this study were whole transcriptome sequencing from tumor and adjacent normal tissues of five patients with lung adenocarcinoma, containing approximately 665 million short reads produced by Illumina HiSeq2000 sequencer, about 67 million per sample (Additional file 1: Table S1). Using Bowtie 2 aligner, about 647 million reads (~97%) can be mapped to human cDNAs. The remaining 18 million unmapped reads were further aligned to exon-intron junctions to identify potential intron retention events. On average, 67,466 and 63,297 retention events were found in each tumor and normal sample, respectively, with ~36,865 retentions in common (Additional file 1: Table S1).
Genes with intron retentions
We next sought to identify genes with tumor-specific intron retentions (TSRs). A TSR is an exon-intron junction that has been covered by retention reads in at least two tumor samples but none of normal samples. As a result, 4,099 TSRs were found by applying above criteria (Table 1), compared to 2,437 normal-specific intron retentions (NSRs). These tissue-specific intron retentions were further mapped to genes, resulting in 2,983 genes with TSRs and 1,991 genes with NSRs, respectively. Of these genes, 500 have both TSRs and NSRs, suggesting they may present different intron retention patterns in tumor and normal samples. For the remaining 2,483 genes only containing TSRs (tumor-specific retention genes, TSRGs, Additional file 2: Table S2), it is possible that some genes may not be expressed in the normal samples and thus lead to biased result. To exclude this possibility, we assessed the expression pattern of TSRGs and retained 2,340 genes that were expressed (>1 TPM) in all tumors and normal controls under investigation. Among them, 576 genes have more than one TSR. Comparatively, 1,422 expressed normal-specific retention genes (NSRGs) were found, and 220 have more than one NSR (Table 1).
Table 1.
Summary statistics for intron retentions in normal and tumor samples
| Normal | Tumor | |
|---|---|---|
| Group-specific retention (GSR)a |
2437 |
4099 |
| Genes with GSRb |
1991 |
2983 |
| Group-specific genesc |
1491 |
2483 |
| Group-specific genes (TPM > 1)d |
1422 |
2340 |
| Group-specific genes (TPM > 1 and GSR > 1)e | 220 | 576 |
aIntron retentions only found in one group (TSR or NSR).
bGenes with TSR or NSR.
cGenes with TSR but not NSR or genes with NSR but not TSR.
dGroup-specific genes that are expressed more than one transcript per million reads (TPM).
eGroup-specific genes that are expressed more than one transcript per million reads (TPM) and have more than one GSR.
Characterization of retained Introns
It has been proposed that in vertebrates, shorter introns have a higher chance to be retained [34,35]. In order to examine whether it is the case here, we compared the size distribution of retained introns and non-retained ones (Figure 1), and found that tumor-specific retained introns are significantly shorter than non-retained introns (1,293 bp versus 1,483 bp, median size, P-value = 4.8 × 10−7, Wilconxon ranksum test). However, no such pattern was found for normal-specific retained introns (1,570 bp versus 1,483 bp, P-value = 0.81). We also found that the position of retained introns are not evenly distributed in the transcripts, and the last introns are most likely to be retained in both tumor and normal samples, but no obvious pattern can be found regarding the forward order of introns (Figure 1).
Figure 1.
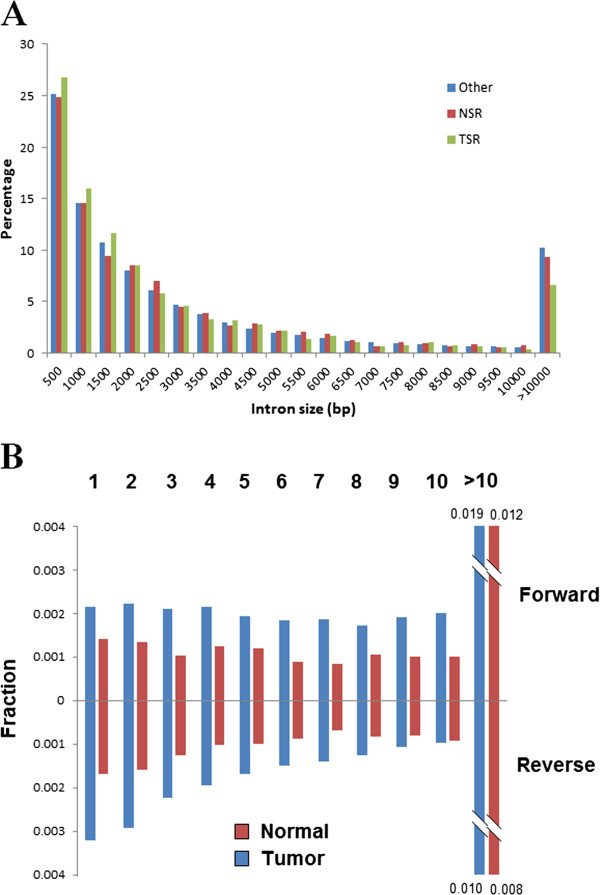
Genomic features of retained introns. A. The size distribution of introns in different classes. The bin size is 500 bp, and introns larger than 10,000 bp were included in one bin. The distribution showed tumor-specific retained introns are larger than other introns (see text). TSR: tumor-specific retained introns; NSR: normal-specific retained introns; Other: non-retained introns. B. The percentage of retained introns in different orders. Forward order is showed above the X-axis, and reverse order is showed under the X-axis. The histogram for introns after ten was truncated to fit the plot. Tumor and normal samples are labeled by blue and darkred.
Gene expression abundance and intron retention abundance
Another question of interest is the relationship between intron retentions and the expression level of genes. To investigate it, we first used RSEM program to estimate gene expression and EdgeR package in bioconductor to identify genes that were differentially expressed between tumors and normal samples. In total, 6,060 differentially expressed genes (DEGs) were identified with a P-value < 0.05 (after Benjamini-Hochberg correction). Of them, 856 (~14%) genes were TSRGs (Figure 2), which were significantly enriched (P-value = 1.2 × 10−9, Fisher’s exact test, FET). Additionally, when only considering 576 TSRGs with multiple retentions, we found a more prominent overrepresentation of DEGs in this set (261 DEGs, P-value = 3.4 × 10−13, FET). Interestingly, a majority of TSRGs (659 of 856, 77%) are up-regulated in tumor samples, which is again highly unexpected (P-value < 2.2 × 10−16, binomial test). This skewness is more substantial in TSRGs with multiple retentions (239 of 261, 92%). We also studied the relationship within tumor samples and a majority of genes (1474 of 2340, 63%) showed a positive correlation between the intron retention count and expression level, which is significantly deviated from the null expectation (50%, P-value < 2.2 × 10−16, binomial test) and is consistent with previous observation [35].
Figure 2.

Overrepresentation of genes with tumor-specific retention (TSR) in differentially expressed genes (DEGs) between tumor and normal samples. A: Venn diagram of DEG and genes with TSR. B: pie chart of the expression pattern in tumor and normal tissues for genes with TSR. A majority of DEGs are up-regulated in tumors. Non-DEG: genes without differentially expression in tumor and normal tissues; up-DEG: differentially expressed genes up-regulated in tumor tissues; down-DEG: differentially expressed genes up-regulated in normal tissues.
One possible explanation for the over-representation of intron retentions in tumors is the inhibition of nonsense mediated decay (NMD), which degrades transcripts with pre-mature codons [36] and is reported to be inhibited in tumor microenvironment [37]. Therefore we investigated the expression pattern of 136 genes involved in NMD process (Additional file 3: Table S3). Among them, only three genes (CTIF, FAU, and RPS28) are significantly down-regulated in tumors, implying NMD may not be inhibited in lung adenocarcinoma and thus cannot explain the large amount of intron retentions in tumors.
It should be noted that the observed correlation could be simply explained by that retentions in over-expressed genes are preferentially identified due to their abundance. Therefore we estimated the intron retention abundance in each TSRG. The mean and median abundance of transcripts with intron retentions for a TSRG in tumor samples are 18% and 5%, consistent with previous observation that intron retentions comprise a minor fraction of splicing forms [10]. Furthermore, we found that up-regulated TSRGs have lower percentage of intron retentions compared with down-regulated genes (4.5% versus 6.7%, median percentage, P-value < 2.2 × 10−16, Wilconxon ranksum test). As transcripts with premature stop codons tend to be degraded by NMD [36], the low level of intron retentions in up-regulated TSRGs and vice versa may suggest the presence of NMD. To validate this, we categorized 2,340 commonly expressed TSRGs as genes with in-frame retention (659) and genes with frame-shift retention (1681), and compared their expression level as well as retention abundance. Genes with in-frame retentions have higher expression than those with frame-shift retentions (1581 versus 1356, mean TPM), but it was not statistically significant (P-value = 0.4561, Wilconxon ranksum test). The retention level in genes with in-frame retentions is significantly higher compared with genes with frame-shift retentions (6.3% versus 4.8%, median percentage, P-value = 4.0 × 10−12, Wilconxon ranksum test), confirming NMD is active in tumor samples.
Functional analysis of TSRGs
To understand the potential functional relevance of those 2,340 TSRGs, we further performed gene ontology analysis. Of 19,520 expressed genes, TSRGs were found to be enriched for 36 GO molecular function terms, including binding terms and kinase activity terms, as well as four cellular component terms (Additional file 4: Table S4). For the subset of TSRGs with multiple TSRs, six biological process terms, 14 molecular function terms and five cellular component terms were found overrepresented (Table 2). Interestingly, several collagen-related terms were found, such as GO0030199 (collagen fibril organization), GO0005583 (fibrillar collagen) and GO0005581 (collagen). As the most abundant proteins in the extracellular matrix, collagen plays an essential role to prevent tumor cells metastasizing to various sites throughout the body [38]. The abundant intron retentions in collagen genes may facilitate their degradation during tumor metastasis.
Table 2.
Enriched gene ontology categories in genes with multiple TSRs
| Category a | Term | Corrected b | Class c |
|---|---|---|---|
| hsa05322 |
Systemic lupus erythematosus |
4.99E-08 |
KEGG |
| GO:0005583 |
Fibrillar collagen |
0.00163 |
GO:CC |
| hsa03010 |
Ribosome |
1.74E-03 |
KEGG |
| GO:0005201 |
Extracellular matrix structural constitu… |
0.006651 |
GO:MF |
| GO:0007411 |
Axon guidance |
0.009976 |
GO:BP |
| GO:0043062 |
Extracellular structure organization |
0.009976 |
GO:BP |
| GO:0030199 |
Collagen fibril organization |
0.009976 |
GO:BP |
| hsa00970 |
Aminoacyl-tRNA biosynthesis |
1.03E-02 |
KEGG |
| hsa04370 |
VEGF signaling pathway |
1.03E-02 |
KEGG |
| GO:0048407 |
Platelet-derived growth factor binding |
0.011402 |
GO:MF |
| GO:0044420 |
Extracellular matrix part |
0.015466 |
GO:CC |
| GO:0005581 |
Collagen |
0.015466 |
GO:CC |
| GO:0030198 |
Extracellular matrix organization |
0.020783 |
GO:BP |
| GO:0006935 |
Chemotaxis |
0.022168 |
GO:BP |
| GO:0042330 |
Taxis |
0.022168 |
GO:BP |
| GO:0031256 |
Leading edge membrane |
0.028215 |
GO:CC |
| hsa04510 |
Focal adhesion |
3.64E-02 |
KEGG |
| GO:0005578 | Proteinaceous extracellular matrix | 0.047652 | GO:CC |
aCategories with more than 1000 genes were removed.
bFisher’s exact test after Benjamini-Hochberg correction.
cGO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; BP: biological process; MF: molecular function; CC: cellular component.
We also conducted pathway analysis for TSRGs and five pathways were overrepresented (Table 2), including the VEGF (vascular endothelial growth factor) signaling pathway (hsa04370). This signal pathway contains several key mediators of angiogenesis and lymphangiogenesis in tumor development [39], and is often found highly expressed in tumors [40]. Enriched intron retentions in these genes, again may activate the mRNA decay mechanism to offset the over-expression.
Investigation of potential cause of intron retentions
One plausible reason for intron retentions is that mutations occurred on the intron splicing sites which change the splicing signal and thus result in an unspliced intron. To explore the prevalence of splicing mutations in tumors, we used SAMtools to identify single nucleotide variants in tumors, and then filtered ones also variable in the matched normal samples. In total, only 27 tumor-specific variants were found to modify the splicing signal (Additional file 5: Table S5). Considering the large number of tumor-specific intron retentions (4,099), it seems that somatic mutations on splicing sites may have a negligible role in causing intron retentions. We also investigated the expression level of several trans-acting splicing activators, including Tra2[41,42] and RNPS1[43], but none shows differential expression between tumors and normal samples.
Intron retentions and tumor genes
By searching the COSMIC database (Catalogue of Somatic Mutations in Cancer, http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/), we found TSRGs include a substantial number of tumor genes, and some are also represented in the Cancer Gene Census [44], which catalogues genes with mutations that have been causally implicated in cancer. Examples include EGFR (epidermal growth factor receptor), KDR (kinase insert domain receptor), ATM (ataxia telangiectasia mutated), and ROS1 (c-ros oncogene 1, receptor tyrosine kinase). Furthermore, three genes were among the top 20 most frequently mutated genes in lung adenocarcinoma: EGFR (34%), ATM (5%) and KDR (5%). TSRG list in this study also targets other genes with a potential role in carcinogenesis, such as MUC16 (mucin 16, cell surface associated), expression of which was found to correlate with clinical outcome in adenocarcinomas [45], as well as RUNX1 (runt-related transcription factor 1), which binds to the core element of many enhancers and promoters and may have various roles in tumors [46,47]. A close investigation further found reads across six exon-intron junctions in MUC16, and the expression of MUC16 is significantly elevated in tumors (p-value = 3.98 × 10−13 after Benjamini-Hochberg correction), but the abundance of intron retention is 3.4%, smaller than 4.5%, the median abundance of up-regulated TSRGs, implying the over-expression of MUC16 in lung adenocarcinoma may be related to the below average intron retention level. Finally, we also prioritized a list of TSRGs which contain multiple frame-shift retentions and were significantly over-expressed in tumor samples (Additional file 6: Table S6). These genes include driver genes such as EGFR, ROS1, and RUNX1, thus functional studies on them should help understand the role of intron retentions in lung tumor development.
Discussion
Recent large-scale efforts from Cancer Genome Atlas Research Network have resulted in lung cancer candidate genes with somatic mutations and copy number alternations [3,48]. However, variations at the mRNA level in these are not fully explored, though the diversity and functionality of tumor-specific transcripts have been highlighted [10,49,50]. Several processes could result in novel mRNA isoforms in tumors, including alterations in promoter usage, exon skipping, and splicing signals, which in consequence changes coding regions and the resulting proteins [51-53]. Thus it is essential to understand the contribution of cancer-related changes emerging at the stage of transcription. The rapid development of sequencing technology makes RNA-Seq a cost-effective way to characterize transcriptome and is therefore frequently used in biomedical studies. Here, we developed a bioinformatics pipeline that explores RNA-Seq data to identify intron retention events, a splicing form of less appreciation but be also important in cancer study [13,54], and further compared their spectrum between lung adenocarcinoma and matched normal tissues. A prevalence of intron retentions was found in carcinoma samples, and over-expressed TSRGs tend to have lower retention abundance compared with under-expressed genes.
One important issue in identifying intron retentions is to distinguish potential contaminations from genomic DNAs or precursor mRNAs during the library preparation process. In order to remove false positive calls caused by contamination, we applied a simple and straightforward filter that requires a candidate intron retention event to be presented in at least two tumor samples and not in any normal sample, or verse visa. If one sample is contaminated and contains false intron retentions, such retentions are not expected to be found in other samples; if multiple samples were contaminated, falsely called intron retentions would be found in both tumor and normal samples, which will also be removed by the filter. However, this filter also removes intron retentions occurred in individual samples, thus the total number of TSRs or NSRs should be even larger than reported here.
The nature of our bioinformatics pipeline determines that it may have limited power in detecting intron retentions in genes with low expression level, partially accounting for the enrichment of intron retentions in over-expressed genes. However, our pipeline also filtered genes with very low expression, the abundance of intron retentions in tumor samples thus cannot be simply explained by the expression bias. Additionally, when focusing on genes with abundant expression, a reverse pattern was demonstrated as the abundance of intron retention is negatively correlated with gene expression, which is likely the result of NMD. Since a substantial proportion of cancer driver genes are over-expressed in tumors, identified intron retentions in those up-regulated genes may suggest a biological role to neutralize over-expression in tumors.
With respect to the mechanism of somatic intron retentions, the most intuitive explanation is that somatic mutations occur at splicing sites and alternate the splicing signal, therefore those splicing sites could not be properly recognized. However, no enrichment of somatic mutations was observed in this dataset (less than 1% of TSRs have somatic mutations in the splicing sites). We also interrogated the expression pattern of several splicing activators, again, no obvious pattern was found. Alternatively, some studies showed that intron retention pattern is different among various tissues [55-57], suggesting other factors, such as cellular environment may also function in promoting the process of intron retention. In addition, the observation of smaller size of retained intron in tumors compared to that in normal samples or non-retained introns is intriguing. Although explanations have been proposed for short retained introns [35], the difference between normal and tumor samples remains unexplained. Future work is therefore necessary to better understand the pattern observed here.
Among genes with tumor-specific retentions, genes with known driver functions in cancer were rediscovered, including EGFR, ROS1, ATM and KDR. Additionally, other growth factor genes were also found with retained introns in tumor samples, such as PDGFRB (platelet-derived growth factor receptor, beta polypeptide), TGFBI (transforming growth factor, beta-induced), EGF (epidermal growth factor), IGF2R (insulin-like growth factor 2 receptor), and ERBB2 (v-erb-b2 erythroblastic leukemia viral oncogene homolog 2), which are also involved in tumor evolution in various studies [58-62]. By detailed investigation, we found intron retentions within these genes all caused frame-shift changes, which tend to invoke NMD. It is well known that cancer driver genes, such as EGFR, are over-expressed or activated by mutations in tumors, further activating downstream pathways associated with cell growth and survival. Therefore intron retentions occurring in these over-expressed or highly mutable driver genes could be protective for the patient by triggering NMD, which in term reduces the expression level or copies of mutable mRNAs. Future validation studies and functional dissections, however, are still critical before we can draw the conclusion.
Conclusions
At the moment of this analysis, only a few studies focus on systematically characterizing the global pattern and contribution of intron retentions in tumorigenesis [63]. Results in this study suggest a potential protective role of intron retentions in lung adenocarcinoma and may benefit further biomarker development. It would also be of interest to investigate the pattern of intron retentions in other cancer types.
Consent
Written informed consent was obtained from patients in the original study and data is released for public use.
Competing interest
The authors declare that they have no competing interest.
Authors’ contribution
QZ and SS conceived the project. QZ, JZ, HJ, HT and JZ carried out the data analysis. QZ and SS drafted the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Summary statistics for each sample.
Genes with tumor-specific retentions and normal-specific retentions.
Genes involved in nonsense mediated decay.
Enriched gene ontology categories for genes with TSRs.
Tumor-specific variants that changes the splicing signal.
List of candidate tumor-specific retention genes for future functional study.
Contributor Information
Qu Zhang, Email: quzhang@post.harvard.edu.
Hua Li, Email: lh@hykgene.com.
Hong Jin, Email: jh@hykgene.com.
Huibiao Tan, Email: thb@hykgene.com.
Jun Zhang, Email: jun_zj10977@163.com.
Sitong Sheng, Email: sst@hykgene.com.
Acknowledgements
We appreciate the helpful comments from two reviewers. QZ thanks Harvard Research Computing group for technical support in computing. This study was funded by the introduction of innovative R&D team program of Guangdong Province (NO. 2009010029), Industry, Education and Academy Cooperation Foundation of Guangdong Province (No.2011A090200114), and Shenzhen Enterprise Engineering Center Project (No. JC201005240008A).
References
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;7(13):1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;7(5):584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansén S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;7(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;7(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;7(11):760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;7(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;7(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;7(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med. 2012;7(3):349–351. doi: 10.1038/nm.2697. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;7(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;7(12):1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;7(5):386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Horvath A, Godbole S, Reddy SD, Mudvari P, Ohshiro K, Cyanam D, Nair S, Fuqua SA, Polyak K, Florea LD, Kumar R. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep. 2013;7:1689. doi: 10.1038/srep01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubi JC. Targeting CCK receptors in human cancers. Curr Top Med Chem. 2007;7(12):1239–1242. doi: 10.2174/156802607780960546. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;7(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;7(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, Lee J, Jung YJ, Kim JO, Yu SB, Kim J, Lee ER, Kang CH, Park IK, Rhee H, Lee SH, Kim JI, Kang JH, Kim YT. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;7(11):2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang J, Jin H, Sheng S. Whole transcriptome sequencing identifies tumor-specific mutations in human oral squamous cell carcinoma. BMC Med Genomics. 2013;7(1):28. doi: 10.1186/1755-8794-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LQ, Cheranova D, Gibson M, Ding S, Heruth DP, Fang D, Ye SQ. RNA-seq reveals novel transcriptome of genes and their isoforms in human pulmonary microvascular endothelial cells treated with thrombin. PLoS One. 2012;7(2):e31229. doi: 10.1371/journal.pone.0031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, Kim YT, Kim JI, Kang JH, Seo JS. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;7(11):2109–19. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y, Kim J, Lee ER, Kang CH, Park IK, Rhee H, Lee SH, Kim JI, Kang JH, Kim YT. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;7(3):375–7. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Ou WB, Marino-Enriquez A, Zhu M, Mayeda M, Wang Y, Guo X, Brunner AL, Amant F, French CA, West RB, McAlpine JN, Gilks CB, Yaffe MB, Prentice LM, McPherson A, Jones SJ, Marra MA, Shah SP, van de Rijn M, Huntsman DG, Dal Cin P, Debiec-Rychter M, Nucci MR, Fletcher JA. 14-3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proc Natl Acad Sci U S A. 2012;7(3):929–934. doi: 10.1073/pnas.1115528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010;7(5992):682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;7(5992):643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, Johnson N, Kähäri AK, Keefe D, Keenan S, Kinsella R, Komorowska M, Koscielny G, Kulesha E, Larsson P, Longden I, McLaren W, Muffato M, Overduin B, Pignatelli M, Pritchard B, Riat HS. et al. Ensembl 2012. Nucleic Acids Res. 2012;7(Database issue):D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;7(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;7:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;7(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;7(1):12. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;7(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;7(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;7(13):1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;7(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M, Berget SM. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994;7(5):3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe NJ, de Souza SJ. Sequence features responsible for intron retention in human. BMC Genomics. 2007;7:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;7(2):89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Wang D, Zavadil J, Martin L, Parisi F, Friedman E, Levy D, Harding H, Ron D, Gardner LB. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol. 2011;7(17):3670–3680. doi: 10.1128/MCB.05704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerenberg PS, Salsas-Escat R, Stultz CM. Collagen–a necessary accomplice in the metastatic process. Cancer Genomics Proteomics. 2007;7(5):319–328. [PubMed] [Google Scholar]
- Waldner MJ, Neurath MF. Targeting the VEGF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;7(1):5–13. doi: 10.1517/14728222.2011.641951. [DOI] [PubMed] [Google Scholar]
- Sia D, Alsinet C, Newell P, Villanueva A. VEGF signaling in cancer treatment. Curr Pharm Des. 2013. [Epub ahead of print] [DOI] [PubMed]
- Tacke R, Tohyama M, Ogawa S, Manley JL. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;7(1):139–148. doi: 10.1016/S0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- Sciabica KS, Hertel KJ. The splicing regulators Tra and Tra2 are unusually potent activators of pre-mRNA splicing. Nucleic Acids Res. 2006;7(22):6612–6620. doi: 10.1093/nar/gkl984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Badolato J, Kobayashi R, Zhang MQ, Gardiner EM, Krainer AR. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 1999;7(16):4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;7(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streppel MM, Vincent A, Mukherjee R, Campbell NR, Chen SH, Konstantopoulos K, Goggins MG, Van Seuningen I, Maitra A, Montgomery EA. Mucin 16 (cancer antigen 125) expression in human tissues and cell lines and correlation with clinical outcome in adenocarcinomas of the pancreas, esophagus, stomach, and colon. Hum Pathol. 2012;7(10):1755–1763. doi: 10.1016/j.humpath.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Ozaki T, Yoshihara Y, Kubo N, Nakagawara A. Runt-related transcription factor 1 (RUNX1) stimulates tumor suppressor p53 protein in response to DNA damage through complex formation and acetylation. J Biol Chem. 2013;7(2):1353–1364. doi: 10.1074/jbc.M112.402594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keita M, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud MC, Sebastianelli A, Trinh XB, Bachvarov D. The RUNX1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle. 2013;7(6):972–986. doi: 10.4161/cc.23963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;7(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, Cyanam D, Mudvari P, Reddy SD, Pakala SB, Nair SS, Florea L, Fuqua SA, Godbole S, Kumar R. Transcriptomic landscape of breast cancers through mRNA sequencing. Sci Rep. 2012;7:264. doi: 10.1038/srep00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;7(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H. et al. The transcriptional landscape of the mammalian genome. Science. 2005;7(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carninci P. Tagging mammalian transcription complexity. Trends Genet. 2006;7(9):501–510. doi: 10.1016/j.tig.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Levy S. Promoting transcriptome diversity. Genome Res. 2007;7(7):965–968. doi: 10.1101/gr.6499807. [DOI] [PubMed] [Google Scholar]
- Masood N, Malik FA, Kayani MA. Unusual intronic variant in GSTP1 in head and neck cancer in Pakistan. Asian Pac J Cancer Prev. 2012;7(4):1683–1686. doi: 10.7314/APJCP.2012.13.4.1683. [DOI] [PubMed] [Google Scholar]
- Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;7(5):757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popielarz M, Cavaloc Y, Mattei MG, Gattoni R, Stevenin J. The gene encoding human splicing factor 9G8. Structure, chromosomal localization, and expression of alternatively processed transcripts. J Biol Chem. 1995;7(30):17830–17835. doi: 10.1074/jbc.270.30.17830. [DOI] [PubMed] [Google Scholar]
- Ledee DR, Chen J, Tonelli LH, Takase H, Gery I, Zelenka PS. Differential expression of splice variants of chemokine CCL27 mRNA in lens, cornea, and retina of the normal mouse eye. Mol Vis. 2004;7:663–667. [PubMed] [Google Scholar]
- Laimer D, Dolznig H, Kollmann K, Vesely PW, Schlederer M, Merkel O, Schiefer AI, Hassler MR, Heider S, Amenitsch L, Thallinger C, Staber PB, Simonitsch-Klupp I, Artaker M, Lagger S, Turner SD, Pileri S, Piccaluga PP, Valent P, Messana K, Landra I, Weichhart T, Knapp S, Shehata M, Todaro M, Sexl V, Höfler G, Piva R, Medico E, Ruggeri BA. et al. PDGFR blockade is a rational and effective therapy for NPM-ALK-driven lymphomas. Nat Med. 2012;7(11):1699–1704. doi: 10.1038/nm.2966. [DOI] [PubMed] [Google Scholar]
- Kim YH, Kwon HJ, Kim DS. Matrix metalloproteinase 9 (MMP-9)-dependent processing of betaig-h3 protein regulates cell migration, invasion, and adhesion. J Biol Chem. 2012;7(46):38957–38969. doi: 10.1074/jbc.M112.357863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial D, McKeown-Longo PJ. Epidermal growth factor (EGF) regulates alpha5beta1 integrin activation state in human cancer cell lines through the p90RSK-dependent phosphorylation of filamin A. J Biol Chem. 2012;7(48):40371–40380. doi: 10.1074/jbc.M112.389577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Mao YQ, Jiang WD, Chen YR, Huang RY, Zhou XB, Wang YF, Shi Z, Wang ZS, Huang RP. Development of IGF signaling antibody arrays for the identification of hepatocellular carcinoma biomarkers. PLoS One. 2012;7(10):e46851. doi: 10.1371/journal.pone.0046851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxnard GR, Binder A, Janne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol. 2013;7(8):1097–1104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W, Chen C, Wang F, Guo X, Lu J, Yang J, Wei M, Tian Z, Guan Y, Tan L, Xu C, Wang L, Gao X, Tian W, Wang J, Yang H, Wang J, Sun Y. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;7(5):806–821. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary statistics for each sample.
Genes with tumor-specific retentions and normal-specific retentions.
Genes involved in nonsense mediated decay.
Enriched gene ontology categories for genes with TSRs.
Tumor-specific variants that changes the splicing signal.
List of candidate tumor-specific retention genes for future functional study.


