Abstract
Nitric oxide (NO) has antimicrobial properties against many pathogens due to its reactivity as an S-nitrosylating agent. It inhibits many of the key enzymes that are involved in the metabolism and virulence of the parasite Entamoeba histolytica through S-nitrosylation of essential cysteine residues. Very little information is available on the mechanism of resistance to NO by pathogens in general and by this parasite in particular. Here, we report that exposure of the parasites to S-nitrosoglutathione (GSNO), an NO donor molecule, strongly reduces their viability and protein synthesis. However, the deleterious effects of NO were significantly reduced in trophozoites overexpressing Ehmeth, the cytosine-5 methyltransferase of the Dnmt2 family. Since these trophozoites also exhibited high levels of tRNAAsp methylation, the high levels suggested that Ehmeth-mediated tRNAAsp methylation is part of the resistance mechanism to NO. We previously reported that enolase, another glycolytic enzyme, binds to Ehmeth and inhibits its activity. We observed that the amount of Ehmeth-enolase complex was significantly reduced in GSNO-treated E. histolytica, which explains the aforementioned increase of tRNA methylation. Specifically, we demonstrated via site-directed mutagenesis that cysteine residues 228 and 229 of Ehmeth are susceptible to S-nitrosylation and are crucial for Ehmeth binding to enolase and for Ehmeth-mediated resistance to NO. These results indicate that Ehmeth has a central role in the response of the parasite to NO, and they contribute to the growing evidence that NO is a regulator of epigenetic mechanisms.
INTRODUCTION
Amoebiasis is a parasitic infection of the human intestine and is caused by the single-celled protozoa Entamoeba histolytica. The disease has a worldwide distribution with substantial morbidity and mortality, and it is one of the three most common causes of death from parasitic disease (1). The clinical spectrum of amoebiasis ranges from asymptomatic infection to colitis, dysentery, or liver abscess. The parasite has two stages in its life cycle: the infective cyst and the invasive trophozoite. In the host, the parasite is exposed to various environmental challenges and is capable of adapting to the demands of its surrounding environment, such as extreme changes in the glucose concentration and the oxidative and nitrosative attacks of the host immune system (2–5).
E. histolytica belongs to the so-called family of “Dnmt2-only” organisms, in that it does not contain any of the canonical DNA methyltransferases (Dnmt1 and Dnmt3). E. histolytica Dnmt2 (Ehmeth) is a weak, but genuine, DNA methyltransferase (6–8), and its ability to catalyze tRNAAsp methylation has been recently demonstrated (9). This dual specificity of Ehmeth for DNA and tRNA has also been proposed for the Dnmt2 homolog in Drosophila melanogaster (10). Although control of gene expression by Ehmeth has been reported (6), this function is apparently not its most important function (11). Since Ehmeth expression fluctuates significantly (2- to 3-fold) between laboratory strains where its expression is barely detectable and strains isolated from patients, these fluctuations suggest that Ehmeth is associated with the parasite's adaptation to its host (reference 8 and unpublished observations).
While the overall biological functions of Dnmt2/Ehmeth are not yet completely understood, recent work has enabled us to view their expressions in terms of the parasite's survival, longevity, and adaptability to metabolic and oxidative stresses. We have recently reported that glucose starvation, with the help of the glycolytic enzyme enolase, regulates the parasite's methylation status (9). Enolase interacts with the catalytic site of Ehmeth and inhibits its methyltransferase activity. Dnmt2 expression has been implicated as a necessary component for maintaining the normal life span in D. melanogaster, and its overexpression induces longevity in fruit flies (12). It has been proposed that the underlying mechanism of extended longevity is an increased resistance to oxidative damage, which has a well-established association with both degenerative diseases and aging (13). Dnmt2 overexpression induces the expression of small heat shock protein (HSP) in D. melanogaster (12) and promotes resistance to H2O2 exposure in E. histolytica (14).
Nitric oxide (NO) is the major cytotoxic molecule that is released by activated macrophages, natural killer cells, and other phagocytic cells for killing E. histolytica trophozoites (15). We have previously reported that NO controls the activity of some of the parasite's virulence factors (16, 17). It has also been recently reported that NO triggers stress responses in E. histolytica and that NO directly inhibits glycolysis and stimulates cysteine synthase activity (18). Evidence is emerging that NO is also a regulator of epigenetic events, because it can modify components of the chromatin remodelling machinery (19, 20). While knowledge on NO as an epigenetic regulator is increasing (20, 21), little is known about the effects of NO on Dnmt activity in general and on Dnmt2 in particular. It is also not known whether the protective effects of Dnmt2 against oxidative stress or heat shock (22) apply to nitrosative stress (14).
In this report, we describe the results of our investigation and describe the underlying molecular mechanisms of increased tolerance to nitrosative stress in E. histolytica trophozoites that overexpress Dnmt2. The findings in this report provide the first evidence of NO-mediated regulation of a Dnmt2 protein.
MATERIALS AND METHODS
Microorganisms.
E. histolytica trophozoites strain HM-1:IMSS were grown under axenic conditions in Diamond's TYI-S-33 medium at 37°C, and trophozoites in the exponential phase of growth were used in all experiments.
Escherichia coli strain BL21(DE3) {F− ompT gal dcm lon hsd SB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 min5])}, a derivative of the E. coli B strain, was used for transformation and protein expression.
DNA constructs.
The pJST4 expression vector and the pJST4-Klp5 vector (23) were kindly provided by A. Lohia, Department of Biochemistry, Bose Institute, India. The pJST4 expression vector enables the expression of the CHH (calmodulin binding domain, hemagglutin [HA], and histidine [His])-tagged protein in E. histolytica; expression of this protein is driven by an actin promoter. The pJST4-Klp5 vector (pcontrol) (23) expresses Klp5, a 99-kDa protein that belongs to the kinesin 5 family (Fig. 1B). This plasmid was used as a control in our previous study in order to exclude the possibility that the CHH tag regulates Ehmeth activity (9), and we used this plasmid for the same purpose in this study. For more details about the construction of pJST4-Ehmeth, see reference 9. The transfection of E. histolytica trophozoites was performed using a previously described protocol (14). Details about the construction of the glutathione S-transferase (GST)–Ehmeth plasmid were previously described in references 8 and 9.
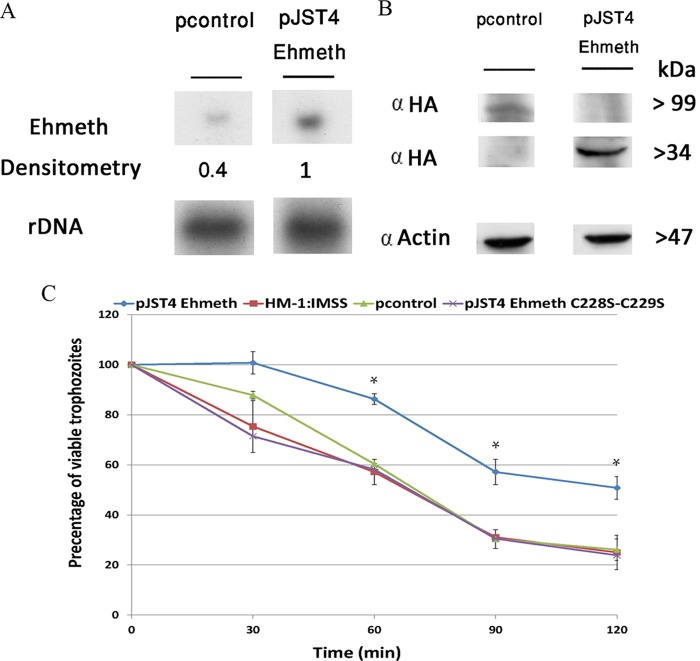
FIG 1.
Overexpression of Ehmeth protects E. histolytica against nitrosative stress. (A) Northern blot analysis was performed using total RNA that was extracted from pJST4-Ehmeth and pcontrol E. histolytica trophozoites. rDNA whose expression was not changed in pJST4-Ehmeth and pcontrol trophozoites were used as controls for RNA loading. The figure displays a representative result from three independent experiments. (B) Western blot analysis was performed on nuclear protein fractions that were prepared from pJST4-Ehmeth and pcontrol E. histolytica trophozoites. The proteins were separated on 12% SDS-PAGE gels and analyzed by Western blotting with an anti-HA (α HA) antibody or an anti-actin antibody. The figure displays a representative result from three independent experiments. (C) The viabilities of wild-type E. histolytica trophozoites from strain HM-1:MSS, E. histolytica trophozoites from a strain that was transfected with a control vector (pcontrol), E. histolytica trophozoites that overexpressed Ehmeth (pJST4-Ehmeth), and E. histolytica trophozoites that overexpressed pJST4-Ehmeth C228S-C229S exposed to 350 μM GSNO for 30, 60, 90, and 120 min were measured. The number of trophozoites at the beginning of each experiment was set at 100%. Bars represent the standard deviations of the means. The means of the different groups for three independent experiments were compared using Student's t test, and statistical significance was set at 5%. The viabilities of the wild-type E. histolytica trophozoites of strain HM-1:MSS, the pcontrol E. histolytica trophozoites, and the pJST4-Ehmeth C228S-C229S E. histolytica trophozoites were not significantly different from each other at any time point, In contrast, the viability of the pJST4-Ehmeth E. histolytica trophozoites was significantly different (P < 0.05) from that of the wild-type E. histolytica trophozoites of strain HM-1:MSS, the pcontrol E. histolytica trophozoites, and the pJST4-Ehmeth C228S-C229S E. histolytica trophozoites after a 60- or 120-min exposure to GSNO.
Site-directed mutagenesis.
The expression of the mutagenic plasmids used for recombinant proteins in E. coli BL21(DE3), namely, Ehmeth C228S-GST, Ehmeth C229S-GST, and Ehmeth C228S-C229S-GST variants, were created by site-directed mutagenesis. Briefly, pairs of complementary mutagenic primers (Ehmeth C228S 5′ and 3′, Ehmeth C229S 5′ and 3′, and Ehmeth C228S-C229S 5′ and 3′ [Table 1]) were used to amplify the entire GST-Ehmeth plasmid with a high-fidelity non-strand-displacing DNA polymerase (PFU DNA polymerase; Promega). The template DNA was eliminated by enzymatic digestion with DpnI, which is specific for methylated DNA, while the mutated plasmid that was generated in vitro was unmethylated and was left undigested. All created mutants were sequenced to ensure the presence of desired mutations and the absence of undesired mutations.
TABLE 1.
Primers used in this study
| Primer name | Sequence | Direction |
|---|---|---|
| Ehmeth C228S 5′ | GATAAAAGGACTTCATGTTTTACTAAGTCA | Sense |
| Ehmeth C228S 3′ | TGACTTAGTAAAACATGAAGTCCTTTTATC | Antisense |
| Ehmeth C229S 5′ | AAAAGGACTTGTTCATTTACTAAGTCATAT | Sense |
| Ehmeth C229S 3′ | ATATGACTTAGTAAATGAACAAGTCCTTTT | Antisense |
| Ehmeth C228S-C229S 5′ | GATAAAAGGACTTCATCATTTACTAAGTCA | Sense |
| Ehmeth C228S-C229S 3′ | TGACTTAGTAAATGATGAAGTCCTTTTATC | Antisense |
| rDNA 5′ | ATGGTGAACAATCATACCTT | Sense |
| rDNA 3′ | TTATCGGATGTGTGAGCCC | Antisense |
| Universal primer | CGCGCGAAGCTTAATACGACTCACTATA | |
| tRNAAsp primer | TGGCGCTTCAACGGGGATT |
For the expression of CHH-tagged Ehmeth C228S-C229S (pJST4 Ehmeth C228S-C229S) in E. histolytica, Ehmeth C228S-C229S was amplified from the plasmid C228S-C229S-GST using the primers Ehmethkpn and EhmethBgl (Table 1) and then cloned in the pJST4 expression vector.
Induction of protein S-nitrosylation.
Trophozoites that were grown in Diamond's TYI-S-33 medium were incubated with freshly prepared S-nitrosoglutathione (GSNO) solution (350 μM; Sigma-Aldrich) for 1 h at 37°C. The trophozoites were then pelleted by centrifugation at 300 × g for 3 min at 4°C, and either total protein extract or total RNA extract was prepared for further analysis.
Northern blotting.
For Northern blotting, total RNA was extracted using the RNA isolation kit TRI reagent (Sigma). RNA (10 μg) was separated on a 1% agarose, 0.3% formaldehyde gel in morpholinepropanesulfonic acid (MOPS) buffer (0.2 M MOPS, 50 mM sodium acetate, 5 mM EDTA; pH 7) and then blotted to Genescreen membranes (NEN Bioproducts, Boston, MA). The RNA was cross-linked to the membrane by UV irradiation (1,200 J/cm2) in a UV Stratalinker apparatus (Stratagene) followed by drying at 80°C for 2 h. The membrane was washed in hybridization buffer (0.5M NaP, 7% SDS, 1 mM EDTA) and then blocked with 100 μg/ml salmon sperm DNA for 1.5 h. Probes were randomly labeled with [α-32P]dCTP by using a random primer DNA labeling mix (Biological Industries, Kibbutz Beit Haemek, Israel) and cleaned on a G-50 column (GE Healthcare). Hybridization with the probes was performed at 60°C overnight. The membrane was then washed several times at 60°C with washing buffer 1 (5% SDS, 40 mM NaP, 1 mM EDTA; pH 7.2), and then with washing buffer 2 (1% SDS, 40 mM NaP, 1 mM EDTA; pH 7.2). The membrane was then exposed to X-ray film (Fujifilm), and the film was developed for detection of the signal.
Western blotting.
For Western blotting, nuclear fractions of pJST4-Ehmeth- or pcontrol-transformed E. histolytica trophozoites were prepared using a previously described protocol (24). Proteins were resolved on 12% SDS-polyacrylamide gel electrophoresis gels and then transferred to nitrocellulose membranes. Blots were then blocked (3% skim milk powder) and probed with 1:500 mouse monoclonal enolase antibody (sc-271384; Santa Cruz Biotechnology), 1:500 rabbit HA antibody (sc-805; Santa Cruz Biotechnology), or 1:2,000 actin mouse monoclonal antibody (ICN691001; MP Biomedicals) for 16 h at 4°C. After incubation with the first antibody, the blots were incubated with a 1:5,000 dilution of a corresponding second antibody for 1 h at room temperature (Jackson ImmunoResearch) and then developed via enhanced chemiluminescence.
Viability assay under NO stress.
E. histolytica trophozoites (1 × 106) were exposed to 350 μM GSNO for 30, 60, 90, and 120 min. At each time point, an aliquot (10 μl) of the culture was stained with eosin (0.1% final concentration), and the number of living trophozoites was counted in a counting chamber under a light microscope.
tRNA bisulfite sequencing.
Total RNA isolation and bisulfite conversions were done using a previously described protocol (25). Bisulfite-treated tRNAs were reverse transcribed by using a tRNA 3′-specific stem-loop primer and amplified with primers that bind only to the deaminated sequences at the 5′ end (Universal primer and Second tRNAAsp primer [Table 1]). Amplicons were subcloned in pGEM-T Easy (Promega) and sequenced (by the Multi-Disciplinary Laboratories Unit, Bruce Rappaport Faculty of Medicine, Technion).
Protein synthesis assay with puromycin.
Trophozoites (2 × 106/ml) that were treated with either 35 μM or 175 μM GSNO for 15 min at 37°C and untreated control trophozoites were incubated with 10 μg/ml puromycin (Sigma) for 20 min. For pretreatment of the trophozoites with cycloheximide (Sigma), the trophozoites were incubated with 100 μg/ml cycloheximide for 5 min before the addition of puromycin. The trophozoites were lysed with 1% Igepal (Sigma) in phosphate-buffered saline (PBS). Puromycin was detected by immunoblotting with a 12D10 clone monoclonal puromycin antibody (Millipore).
Protein synthesis assay with [35S]methionine.
Trophozoites (2 × 106/ml) that were grown in Diamond's TYI-S-33 medium were harvested by low-speed centrifugation at room temperature, washed twice with Dulbecco's modified Eagle medium (DMEM) without serum, and then exposed to 350 μM GSNO for 20 min at 37°C. The trophozoites were washed twice again with DMEM without serum and then incubated with 50 μCi/ml [35S]methionine (Sigma) for 2 h at 37°C. The trophozoites were washed twice with PBS and then lysed using 1% Igepal (Sigma) in PBS. Radiolabeled proteins were precipitated on Whatman filter paper by using trichloroacetic acid and were analyzed by liquid scintillation counting.
Proteolysis of protein samples and mass spectrometry analysis.
Protein samples were resolved by 12% SDS-polyacrylamide gel electrophoresis. The proteins in each gel slice were reduced with 2.8 mM dithiothreitol (DTT; 60°C for 30 min), modified with 8.8 mM iodoacetamide in 100 mM ammonium bicarbonate in the dark at room temperature for 30 min, and digested overnight in 10% acetonitrile and 10 mM ammonium bicabonate with modified trypsin (Promega) at 37°C. The resulting tryptic peptides were resolved by reverse-phase chromatography on 0.075- by 200-mm fused silica capillaries (J&W) packed with Reprosil reverse-phase material (Dr. Maisch GmbH, Germany). The peptides were eluted with linear gradients of 7 to 40% acetonitrile with 0.1% formic acid in water at a flow rate of 0.25 μl/min over 94 min and 95% acetonitrile with 0.1% formic acid in water at a flow rate of 0.25 μl/min over 12 min. Mass spectrometry (MS) was performed by using an ion trap mass spectrometer (Orbitrap; Thermo) in positive mode and repetitively full MS scan followed by collision-induced dissociation (CID) of the seven most dominant ions selected from the first MS scan.
The mass spectrometry data were analyzed using the MaxQuant 1.4.1.2 software (26), which searched the E. histolytica section of the NCBI-nr database with a 1% false-discovery rate, and quantified by label-free analysis using the same software.
Immunoprecipitation assays.
Aliquots of nuclear protein fractions (100 μg) were diluted in 20 mM HEPES (pH 7.5), 150 mM NaCl, 0.1% Triton, 10% glycerol (HNTG buffer; 300 μl final volume) and then incubated with protein G-Sepharose beads (10 μl; Sigma) for 30 min at 4°C. Nonspecific interacting proteins were excluded by centrifugation (3,000 rpm at 4°C for 5 min). The supernatant was incubated with either 1:200 HA antibody or enolase antibody for 2 h at 4°C. Following incubation, protein G-Sepharose beads (20 μl) were added to the samples, which were then incubated for 16 h at 4°C. Immunoprecipitated proteins were collected by centrifugation, washed three times with HNTG buffer, and then resolved by 12% SDS-polyacrylamide gel electrophoresis. The proteins were then transferred to nitrocellulose membranes for Western blot analysis and detected with either a mouse monoclonal enolase antibody (sc-271384; Santa Cruz Biotechnology) or a rabbit six-histidine antibody (sc-803; Santa Cruz Biotechnology).
Expression of recombinant Ehmeth and Ehmeth mutant proteins in E. coli BL21.
E. coli BL-21(DE3) cells that were transfected with the respective vectors (GST-Ehmeth, Ehmeth C228S-GST, Ehmeth C229S-GST, and Ehmeth C228S-C229S-GST) were grown overnight at 37°C in Luria broth (LB) medium that contained 100 μg/ml ampicillin. These precultures were inoculated with a 500 ml of a 1:100 dilution of fresh LB medium that contained 100 μg/ml ampicillin and were further grown for 3 h until the optical density at 600 nm of the medium reached 0.8. These bacteria were induced with 0.5 mM isopropyl-beta-d-thiogalactopyranoside (IPTG) for 16 h at 25°C. At the end of incubation, the induced cells were harvested and lysed in lysis buffer (100 mM KCl, 1 mM DTT, 1 mM phenylmethylsulfonylfluoride, 100 μg/ml lysozyme, and 100 μg/ml leupeptin in PBS). The lysed cells were then sonicated for 5 min with 30-s pulses, with 30 s between each pulsing session. The lysis was completed by adding 1:100 BugBuster protein extraction reagent (Novagen). The lysate was then centrifuged at 2,000 × g for 20 min in order to recover the soluble proteins in the supernatant. GST fusion proteins were purified by affinity purification on glutathione-Sepharose beads. The recombinant proteins were then eluted with glutathione elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM glutathione [Sigma]), and their concentrations were measured by using Bradford's method (27).
Exposure of Ehmeth and mutants proteins to GSNO.
Aliquots (0.04 nmol) of recombinant protein were treated with 5 μM GSNO for 1 h at 37°C in 20 μl of methylation buffer without DTT (100 mM Tris-HCl [pH 7.5], 5% glycerol, 5 mM MgCl2, and 100 mM NaCl). DTT (20 mM) was added to certain protein samples following their exposure to GSNO to revert their S-nitrosylation. Recombinant proteins were resolved by 12% polyacrylamide gel electrophoresis under native conditions and then transferred to nitrocellulose membranes. Membranes were blocked (3% skim milk powder) and then probed with 1:800 rabbit polyclonal S-nitrosocysteine (S-NO-Cys) antibody (N5411; Sigma) for 1 h at room temperature. The membrane were then incubated with 1:5,000 secondary antibody to rabbit IgG (Jackson ImmunoResearch) for 1 h at room temperature and developed by using enhanced chemiluminescence. Ponceau S staining of the blots prior to their blocking was used to control the loading and transfer of the proteins to the membranes.
Molecular docking.
The coordinates for the E. histolytica enolase structure (GenBank accession number XP_649161.1; PDB ID 3QTP) and the E. histolytica methyltransferease EhMeth/Dmnt2 (GenBank accession number XP_655267.2; PDB ID 3QV2) were obtained from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). All nonprotein residues were removed prior to the docking procedure. Subunit B of enolase was considered the receptor, because it was larger (438 residues) than Ehmeth (320 residues), which served as the ligand during docking. The protein-protein docking was done using the Hex 6.3 platform (28) and the shape correlation. Similar clusters of docked proteins were obtained using SwarmDock (29) and ClusPro (30). Docked complexes were visualized, and the images were produced using PyMol (http://www.pymol.org/).
RESULTS
Overexpression of Ehmeth protects E. histolytica against nitrosative stress.
In order to test the hypothesis that Ehmeth is involved in the protection of the parasite against nitrosative stress, we determined the viability of three strains of E. histolytica trophozoites, namely, wild-type strain HM-1:MSS, E. histolytica trophozoites transfected with pcontrol, and E. histolytica trophozoites transfected with pJST4-Ehmeth and that overexpressed Ehmeth, after their exposure to 350 μM GSNO for 120 min. GSNO was selected as the NO donor molecule because it is the main nonprotein S-nitrosothiol (SNO) in human cells and extracellular fluids (31).
The overexpression of Ehmeth as a CHH-tagged protein was confirmed by both Northern and Western blotting (Fig. 1A and B). Figure 1C displays the time-dependent changes in viability of the three types of E. histolytica trophozoites. After 60 min, the viabilities of the wild-type and pcontrol trophozoites were less than that of the pJST4-Ehmeth trophozoites. These differences in viability were exacerbated after 120 min: only 25% of the wild-type E. histolytica trophozoites and pcontrol trophozoites were viable, whereas 50% of the pJST4-Ehmeth trophozoites were viable. These results indicate that Ehmeth contributes toward protecting E. histolytica trophozoites against nitrosative stress.
tRNAAsp methylation and protein synthesis in NO-treated trophozoites.
Eukaryotic protein synthesis is regulated by a variety of tRNA modifications (32). In addition, stress-specific reprogramming of modified ribonucleosides in tRNA is involved in the selective translation of survival proteins (33). We decided to investigate whether Ehmeth is involved in the mechanism of NO resistance in the parasite. For this purpose, we determined the levels of tRNAAsp methylation and protein synthesis in pcontrol trophozoites and pJST4-Ehmeth trophozoites before and after GSNO treatment.
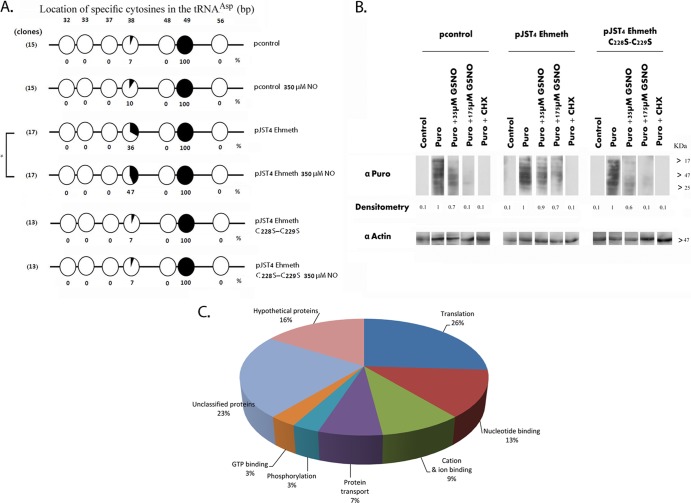
For determining the level of tRNAAsp methylation, we used a recently developed method that is based on bisulfite sequencing of tRNA and enables direct detection of cytosine methylation in tRNA by accurately localizing the methylated cytosines within the sequence (25). The cytosine 38 residue in tRNAAsp is a well-known substrate of Dnmt2 enzymes (25, 34, 35). Amplicons (PCR products) of tRNAAsp were generated from bisulfite-treated total RNA samples that were extracted from E. histolytica strains, and the sequences of several independent amplicons were determined. We observed that cytosine 38 methylation in the pJST4-Ehmeth trophozoites was 5 times greater than that in the pcontrol trophozoites (Fig. 2A). This increase of cysteine 38 methylation in the pJST4-Ehmeth trophozoites that were exposed to 350 μM GSNO for 1 h was even higher (6.7 times) than that in the pcontrol trophozoites (Fig. 2A). There were no differences in the levels of cysteine 38 methylation in the untreated and GSNO-treated pcontrol trophozoites. In contrast, the levels of cysteine 38 methylation in the GSNO-treated pJST4-Ehmeth trophozoites were substantially greater than those in the pJST4-Ehmeth trophozoites that were not exposed to GSNO. We observed 100% methylation of the cytosine 49 residue in the three types of trophozoites, irrespective of whether or not they were exposed to GSNO. These findings suggested that the methylation of the cytosine 49 residue is not catalyzed by Ehmeth (Fig. 2A). We also found complete demethylation of other cytosine residues (32, 33, 37, 48, and 56), and these findings indicated that the bisulfite treatment was efficient (Fig. 2A). Collectively, these results confirm our previous observations about the ability of Ehmeth to methylate tRNAAsp (9) and indicate that Ehmeth is similar to other Dnmt2 proteins in that it methylates the cytosine 38 residue of tRNAAsp. Additionally, our results showed an unexpected positive effect of NO on tRNAAsp methylation in the pJST4-Ehmeth trophozoites.
FIG 2.
Level of tRNAAsp methylation in NO-treated trophozoites. (A) Bisulfite sequencing analysis of tRNAAsp in pcontrol E. histolytica trophozoites, pcontrol E. histolytica trophozoites that were treated with 350 μM GSNO for 1 h, pJST4-Ehmeth trophozoites, pJST4 Ehmeth trophozoites treated with GSNO (350 μM for 1 h), pJST4-Ehmeth C228S-C229S trophozoites, and pJST4-Ehmeth C228S-C229S trophozoites treated with GSNO (350 μM for 1 h). The numbers of clones (sequence reads) are displayed in parentheses on the left side of each row. Black areas in the circles indicate methylated cytosine residues, and white areas indicate unmethylated cytosine residues. The percentage of methylated cytosines is indicated below each circle. The location of specific cytosines in the tRNAAsp is indicated under each row. The levels of C38 tRNAAsp methylation in the untreated pcontrol and GSNO-treated pcontrol trophozoites were not significantly different from each other according to the analysis with Student's t test, for which statistical significance was set at 5%. In contrast, the levels of C38 tRNAAsp methylation in the untreated and GSNO-treated pJST4 Ehmeth trophozoites were significantly different (P < 0.05). (B) Protein synthesis, measured using puromycin-labeled proteins. pcontrol E. histolytica trophozoites, pJST4-Ehmeth E. histolytica trophozoites, and pJST4-Ehmeth C228S-C229S E. histolytica trophozoites were mock treated (control), labeled with 10 μg/ml puromycin (Puro), or treated with 35 μM or 175 μM GSNO for 15 min and then labeled with puromycin for 20 min (Puro + GSNO). Some trophozoites were treated with cycloheximide (CHX; 100 μg/ml) before puromycin labeling (Puro + CHX). The extracts were separated by denaturing electrophoresis and analyzed by Western blotting with a 12D10 clone puromycin antibody. An actin immunoblot is shown as the loading control. The results are representative of two independent experiments. α-Puro, anti-Puro antibody. (C) Functional categories of the upregulated proteins in pJST4-Ehmeth E. histolytica trophozoites and pcontrol E. histolytica trophozoites exposed to 350 μM GSNO for 1 h. The upregulated proteins were classified according to their biological role based on the David Bioinformatics Resources (http://david.abcc.ncifcrf.gov/).
Next, we hypothesized that the hypermethylation of tRNAAsp that we detected in the pJST4-Ehmeth trophozoites promotes protein synthesis. In order to test this hypothesis, we used the surface sensing of translation (SUnSET) technique (36), which uses the antibiotic puromycin (a structural analog of tyrosyl-tRNA) and puromycin antibodies to detect the amount of puromycin that was incorporated into nascent peptide chains.
We observed that pcontrol and pEhmeth trophozoites had comparable rates of protein synthesis under control conditions (Fig. 2B). Treating pcontrol trophozoites with either 35 μM or 175 μM GSNO for 15 min inhibited protein synthesis by 30% and 90%, respectively (Fig. 2B). These inhibitory effects of 35 μM and 175 μM GSNO were less pronounced (10% and 40% inhibition, respectively) in the pJST4-Ehmeth trophozoites (Fig. 2B). Cycloheximide is an inhibitor of protein biosynthesis (37). We also found that the extent of inhibition of protein synthesis by cycloheximide was the same (90%) in the untreated pcontrol and the pJST4-Ehmeth trophozoites. The observation that the inhibitory effect of GSNO in the pJST4-Ehmeth trophozoites was less pronounced than that in the pcontrol trophozoites was confirmed independently by measuring the rate of protein synthesis in these trophozoites by using [35S]methionine (see Fig. S1 in the supplemental material).
We next determined whether Ehmeth overexpression selectively influences the synthesis of proteins that are involved in the resistance to nitrosative stress. For this purpose, we performed quantitative proteomic analysis of pcontrol and pJST4-Ehmeth trophozoites that were exposed or not exposed to 350 μM GSNO for 1 h (see Table S1 in the supplemental material for results of the complete analysis). For the purpose of this study, we decided to focus on proteins that were upregulated in the GSNO-treated pJST4-Ehmeth trophozoites and compare them to those from GSNO-treated pcontrol trophozoites (Fig. 2C). Among these proteins, we identified proteins that are involved in protein translation, such as the 60S and 40S ribosomal proteins and glycyl-tRNA synthetase, protein transport, such as the coatomer beta-subunit and vacuolar sorting protein, and signaling, such as the Rab family GTPases. Interestingly, two proteins that were significantly upregulated were alcohol dehydrogenase 2 (ADH2; 3-fold increase) and the antioxidant peroxiredoxin (4-fold increase). ADH2 is essential for energy metabolism of oxidatively stressed parasites (38), and peroredoxin has been reported to be associated with resistance to nitrosative stress in Leishmania spp. (39).
Collectively, these results strongly suggest that Ehmeth-mediated tRNA methylation has a positive effect on protein synthesis in general and on stress response-related proteins in particular when the parasite is nitrosatively stressed.
The amount of the Ehmeth-enolase inhibitory complex is reduced in NO-treated trophozoites.
Although NO usually inhibits enzymatic activity, it can also activate enzymatic activity, as reported for the E. coli transcription factors OxyR and SoxR (21). Hence, we decided to investigate whether NO also modulates Ehmeth activity. Ehmeth is devoid of any tRNA methyltransferase activity in the absence of DTT (data not shown). On the other hand, the effect of NO on Ehmeth activity could not be determined in the presence of DTT, because DTT reverses the S-nitrosylation of cysteine.
We previously reported that enolase binds to Ehmeth and inhibits its activity (9). We hypothesized that the hypermethylation of tRNAAsp in NO-treated pJST4-Ehmeth trophozoites (Fig. 2A) is due to reduced formation of the enolase-Ehmeth complex. In order to test this hypothesis, we used trophozoites that were transfected with pJST4-Ehmeth plasmids. In the absence of an efficient Ehmeth antibody, we used an HA antibody to immunoprecipitate Ehmeth and visualized it by Western blotting. Using this approach, which had been previously validated (9), we showed that the amount of Ehmeth-enolase complex in the pJST4-Ehmeth trophozoites that were exposed to 350 μM GSNO for 1 h was significantly lower than that found in the untreated pJST4-Ehmeth trophozoites (Fig. 3A). This result can be explained by either a direct effect of NO on the amount of the Ehmeth-enolase complex or by NO limiting the availability of either one or both of the constituents of the complex. According to the results of Western blot analysis, the amount of Ehmeth did not change in the GSNO-treated parasites (Fig. 3B). Collectively, these results indicate that NO can directly influence the amount of the Ehmeth-enolase complex and the methylation status of tRNAAsp.
FIG 3.
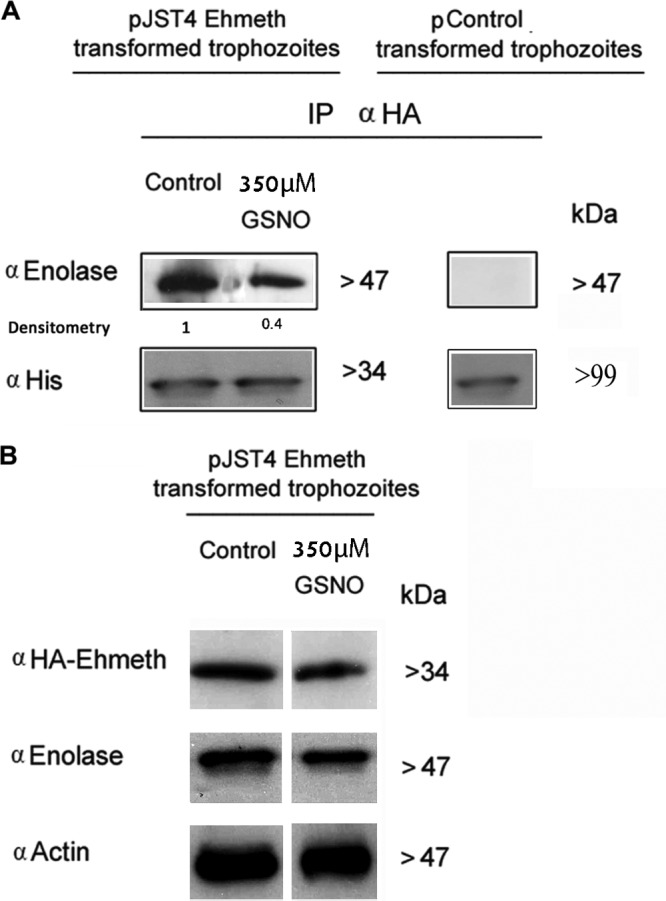
Nitric oxide regulates the amount of Ehmeth-enolase inhibitory complex formed. (A) Ehmeth samples from nuclear lysates of pJST4-Ehmeth and pJST4 Ehmeth E. histolytica trophozoites that were treated with 350 μM GSNO for 1 h was immunoprecipitated (IP) with a monoclonal anti-HA (α HA) antibody. The presence of enolase among the immunoprecipitated proteins was detected by Western blotting with an enolase antibody (left). The presence of CHH-tagged Ehmeth among the immunoprecipitated proteins was detected by Western blotting by using a histidine antibody. The immunoprecipitation experiments were also performed with nuclear lysates of pcontrol E. histolytica trophozoites as a negative control for the expression of CHH-tagged Ehmeth and for the immunoprecipitation of enolase (right). (B) Western blot analysis of nuclear proteins prepared from GSNO-treated pJST4-Ehmeth and pJST4-Ehmeth E. histolytica trophozoites. The proteins were separated on 12% SDS-PAGE gels and analyzed by Western blotting with an HA antibody, an enolase antibody, or an actin antibody. The figure displays a representative result from at least three independent experiments.
Although the crystal structure sizes of Ehmeth and enolase have been determined to be 2.15Å and 1.9Å, respectively (40, 41), the molecular details of the Ehmeth-enolase interaction remains uncharacterized.
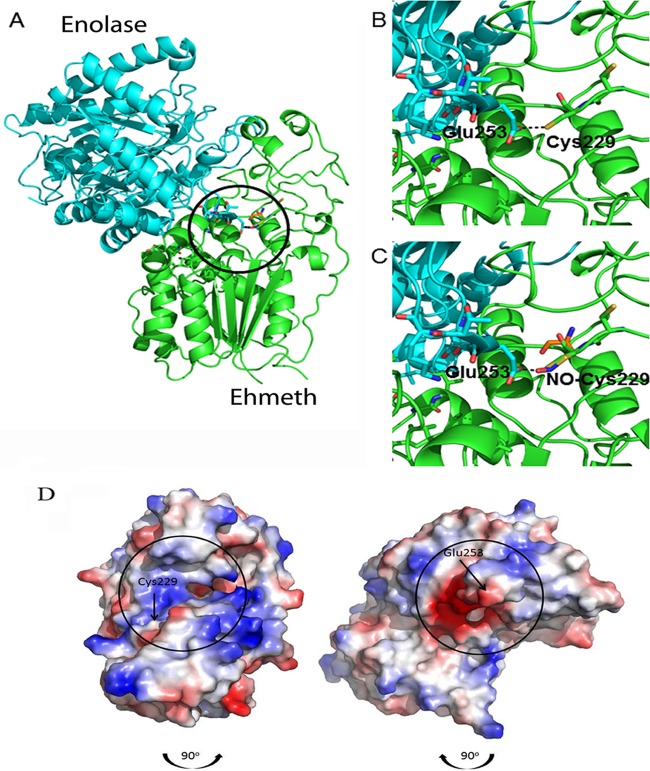
In order to predict which cysteine residues in Ehmeth are involved in the formation of the Ehmeth-Enolase complex and might be potential targets for S-nitrosylation, we performed an in silico docking analysis. Prior to screening for possible complex interfaces, the structures were stripped of nonprotein constituents. The program Hex 6.3 explores the possible energies of protein-protein interactions by using both shape complementarity and electrostatic effects. The top-rated structure, which is displayed in Fig. 4A, showed strong surface complementarity with the loop between residues 250 and 260 of enolase that includes the glutamic acid 253 (Glu253) residue and juts into the aperture that is formed between two globular lobes of the Ehmeth protein. This strong surface complementarity brings the Glu253 residue of enolase into close proximity with the Cys229 residue of Ehmeth. In addition, both the Cys229 residue and its neighbor, the Cys228 residue, of Ehmeth are on a rather unstructured loop, with their SH groups pointing outwards. Hence, the two Cys residues are potentially accessible to NO and to an interacting protein. Figure 4B details the putative interaction between the Glu253 residue of enolase and the Cys229 residue of Ehmeth; the Cys229 residue of Ehmeth was computationally modified by adding NO to the terminal sulfur atom in Fig. 4C.
FIG 4.
Molecular modeling of the putative enolase-Ehmeth complex. The atomic coordinates of E. histolytica enolase and Ehmeth proteins (PDB codes 3QTP and 3QV2, respectively) were docked using the Hex 6.2 platform. (A) The overall best docked structure. (B and C) Close-up images of the interaction between the Glu253 residue of enolase and the Cys229 residue of Ehmeth in the native (B) and NO-modified (C) forms. The complex shown in panel A was disassembled by rotating the enolase 90° in a clockwise direction and the Ehmeth 90° in a counterclockwise direction (the black arrows indicate the direction of complex formation). The interaction interfaces of both proteins are indicated by black circles. (D) Vacuum electrostatic potentials were generated using PyMOL in order to illustrate the charge variance of the E. histolytica enolase and Ehmeth proteins, with red and blue indicating negative and positive surfaces, respectively.
In Fig. 4D, the complex components have been separated and each protein turned 90° in order to reveal the interaction interface. These interfaces have been overlaid with the surface electrostatic potential, which was calculated using the algorithm in PyMol, a molecular visualization system. This analysis clearly showed that a strong negative potential surrounds the Glu253 residue of enolase, while the area that surrounds the Cys229 residue of Ehmeth is strongly positive.
Collectively, the output of the in silico computer-based modeling of the Ehmeth-enolase interaction suggested that the Cys228 and Cys229 residues are accessible to NO and are important sites for the binding of Ehmeth to enolase.
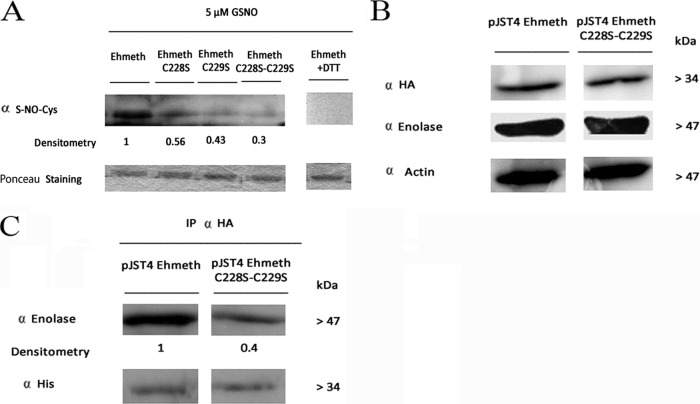
In order to test the accuracy of this model, we created three Ehmeth mutant proteins: Ehmeth228, Ehmeth229, and Ehmeth228-229, in which the Cys228 residue, the Cys229 residue, and the two cysteine residues were, respectively, replaced with a serine. Following the exposure of the wild-type and mutated proteins to 5 μM GSNO for 1 h, their levels of S-nitrosylation were compared by Western blot analysis by using an S-NO-Cys antibody (Fig. 5A). The presence of S-nitrosylated cysteine(s) was detected in the wild-type Ehmeth protein, but the signal was significantly attenuated in the single mutants and even more so in the double mutants. The specificity of the S-NO-Cys antibody was confirmed by the loss of signal in Ehmeth proteins that were treated with DTT immediately after their S-nitrosylation by GSNO (Fig. 5A). Collectively, these results indicated that Cys228 and Cys229 are efficiently S-nitrosylated.
FIG 5.
Role of Ehmeth Cys228 and Cys229 in the formation of the Ehmeth-enolase complex. (A) Western blot analysis of recombinant proteins (Ehmeth, Ehmeth C228S, Ehmeth C229S, and Ehmeth C228S-C229S) that were treated with 5 μM GSNO for 1 h at 37°C. The proteins were resolved on 12% polyacrylamide gels under native conditions, transferred to a nitrocellulose membrane, and then probed with an S-NO-Cys antibody (α S-NO-Cys). Ponceau staining of the membrane prior to its interaction with the S-NO-Cys antibody was used as a loading control. The Ehmeth plus DTT control shows the results with the Ehmeth recombinant protein treated with 5 μM GSNO for 1 h at 37°C followed by incubation with 20 mM DTT for 5 min at 37°C. The figure displays a representative result from at least three independent experiments performed singly. (B) Western blot analysis of nuclear protein fractions prepared from pJST4-Ehmeth and pJST4-Ehmeth C228S-C229S E. histolytica trophozoites performed using an HA antibody, an enolase antibody, or an actin antibody. The figure displays a representative result from at least three independent experiments performed singly. (C) Immunoprecipitation analysis of Ehmeth from pJST4-Ehmeth and pJST4-Ehmeth C228S-C229S E. histolytica trophozoites, performed with an HA antibody. The presence of enolase among the immunoprecipitated proteins was detected by using an enolase antibody. The amounts of Ehmeth and Ehmeth C228S-C229S in the pJST4-Ehmeth and pJST4-Ehmeth C228S-C229S E. histolytica trophozoites were determined by using a histidine antibody.
We next determined the involvement of these cysteines in the formation of the Ehmeth-enolase complex. For this purpose, Ehmeth228-229 was expressed as a CHH-tagged protein in the parasite. According to the results of our Western blot analysis (Fig. 5B), the amounts of Ehmeth and Ehmeth228-229 proteins in the pJST4-Ehmeth and the pJST4-Ehmeth228-229 trophozoites were the same. The amounts of enolase-Ehmeth complex in the pJST4-Ehmeth and the pJST4-Ehmeth228-229 trophozoites were then determined by immunoprecipitation analysis (Fig. 5C). We found that the amount of the Ehmeth-enolase complex in the pJST4-Ehmeth228-229 trophozoites was substantially smaller than that found in the pJST4-Ehmeth trophozoites (Fig. 5C). These results indicated that the Cys228 and Cys229 residues in Ehmeth are involved in the binding of Ehmeth to enolase and strongly suggest that the S-nitrosylation inhibits the formation of the Ehmeth-enolase complex. We next determined the involvement of the Cys228 and Cys229 residues in Ehmeth activity. We found that the levels of tRNAAsp methylation in pcontrol and pJST4-Ehmeth228-229 trophozoites were the same, and this result strongly suggested that these mutations impair the catalytic activity of Ehmeth (Fig. 2A). We also found that untreated and GSNO-treated pcontrol and pJST4-Ehmeth228-229 trophozoites had comparable rates of protein synthesis (Fig. 2B), and this result strongly suggested that the level of protein synthesis correlates with the level of tRNAAsp methylation.
Finally, we compared the protective effect of Ehmeth of nitrosatively stressed pJST4 Ehmeth228-229 trophozoites to that of nitrosatively stressed wild-type, pJST4 pcontrol, and pJST4Ehmeth E. histolytica trophozoites. We found that the protective effect of Ehmeth in nitrosatively stressed trophozoites was lost when the Ehmeth228-229 protein was overexpressed (Fig. 1C).
DISCUSSION
Mammalian defense strategies against pathogens include the production of a chemical arsenal, such as reactive oxygen and nitrogen species. NO plays a major role in this defense process, and NO-induced inhibition of protein synthesis is part of its cytostatic action in mammalian cells. Different mechanisms, such as NO-mediated cleavage of 28S and 18S rRNA (42) and NO-induced phosphorylation of eukaryotic initiation factor 2α (eIF-2α) (43) have been proposed to explain this inhibitory activity. Therefore, one might surmise that any action of the parasite to counteract the deleterious effect of NO on its protein synthesis is an effective means of resistance against nitrosative stress. Our data indicate the existence of a strong correlation between Ehmeth-mediated tRNA methylation and the control of protein synthesis in nitrosatively stressed E. histolytica trophozoites. Most of the recent efforts to explain the role of Dnmt2 in the protection of different organisms from environmental stresses have focused on the organisms' abilities to methylate tRNA (22). Recently, it was proposed that cytosine-5 tRNA methylation in mice promotes tRNA stability and protein synthesis and prevents stress-induced RNase cleavage by angiogenin (44). According to the results of our bioinformatics analysis, the absence of an angiogenin homolog in E. histolytica (data not shown) suggests that this protective mechanism does not exist in E. histolytica, or that a still-undiscovered RNase in E. histolytica has a similar function to that of angiogenin. In contrast, the upregulation of ribosomal proteins 40S and 60S subunits in the GSNO-treated pJST4-Ehmeth trophozoites may be an influential mechanism for maintaining protein synthesis in nitrosatively stressed trophozoites. Our observations are in agreement with the findings of Len and others, indicating that the upregulation of ribosomal proteins 30S and 50S contributes to acid tolerance in Streptococcus mutans (45).
The preservation of protein synthesis as a mechanism of resistance against nitrosative stress is somewhat counterintuitive. The general stratagem in most oxidatively stressed and heat-shocked species, including E. histolytica, typically includes downregulation of protein synthesis (46, 47) in order to stop energy waste and the toxic buildup of damaged or misfolded proteins. In this investigation, we found paradoxical evidence on the maintenance of protein synthesis in GSNO-exposed pJST4-Ehmeth trophozoites. We surmise that the additional protein synthesis is a mechanism of titration/competition against the intracellular accumulation of S-nitrosylated proteins. Essentially, an intracellular protein reservoir is created in order to buffer the effects of nitrosative damage. This notion is well illustrated by the fact that the synthesis and turnover of Fe-S cluster-containing proteins in E. histolytica that were exposed to the NO donor sodium nitroprusside increased in order to overcome the deleterious effects of NO treatment (18).
Our data indicate that expression of specific stress-related proteins is upregulated in GSNO-treated pJST4-Ehmeth trophozoites. This finding is in agreement with those of Santi-Roca and others (18), who reported that the amount of peroxiredoxin transcripts was increased by NO in E. histolytica. It has been reported that ADH2 (48, 49) and peroxiredoxin (50) are associated with the resistance of other organisms to oxidative stress and nitrosative stress (51–53). Accordingly, we posit that these enzymes contribute to the resistance to nitrosative stress of pJST4 Ehmeth trophozoites.
Growing evidence indicates that NO can regulate key epigenetic events, including chromatin remodeling (for a recent review, see reference 21). Our data indicate that S-nitrosylation regulates Dnmt2 activity by inhibiting the formation of an Ehmeth-enolase complex. Since enolase binds to Ehmeth and inhibits its activity (9), this result can be used to explain the significant increase in tRNAAsp methylation that we observed in the GSNO-treated trophozoites. We previously showed that the deletion of the catalytic site in Ehmeth (motif IV) partially suppresses the formation of the Ehmeth-enolase complex, and this finding suggests that other components of Ehmeth are involved in its binding to enolase (9). The results of this investigation shed new light on the formation of this complex and emphasize the importance of the Cys228 and Cys229 residues in Ehmeth in this process. In the future, it will be interesting to challenge and confirm this information for the role of the Glu253 residue of enolase in the formation of the Ehmeth-enolase complex that was obtained in our molecular docking analysis.
In summary, the results of this investigation show that Ehmeth-mediated tRNAAsp methylation is crucial in the protection of E. histolytica against nitrosative stress by maintaining active protein synthesis. Another important finding of this study is that NO influences the amount of Ehmeth-enolase complex and consequently regulates Ehmeth activity. The results of this analysis open the door to many important questions about the regulation of Dnmt activity and the role of NO in Dnmt-protein interactions in other organisms. Finally, the results of this investigation indicate that Ehmeth-mediated tRNAAsp methylation is a potential target for development of drugs to treat amoebiasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Albert Jeltsch and Tomasz Jurkowski of the Institute of Biochemistry, Stuttgart University, Germany, for their constructive comments, the Smoler Proteomics Center at the Technion for their help with the proteomic analysis of the data, and Arieh Bomzon, ConsulWrite, for his editorial assistance in preparing the manuscript.
This study was supported by grants from the Israel Science Foundation and the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Published ahead of print 21 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00031-14.
REFERENCES
- 1.Anonymous. 1997. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28–29 January, 1997. Epidemiol. Bull. 18:13–14 [PubMed] [Google Scholar]
- 2.Baumel-Alterzon S, Weber C, Guillen N, Ankri S. 2013. Identification of dihydropyrimidine dehydrogenase as a virulence factor essential for the survival of Entamoeba histolytica in glucose-poor environments. Cell. Microbiol. 15:130–144. 10.1111/cmi.12036 [DOI] [PubMed] [Google Scholar]
- 3.Tovy A, Hertz R, Siman-Tov R, Syan S, Faust D, Guillen N, Ankri S. 2011. Glucose starvation boosts Entamoeba histolytica virulence. PLoS Negl. Trop. Dis. 5(8):e1247. 10.1371/journal.pntd.0001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mortimer L, Chadee K. 2010. The immunopathogenesis of Entamoeba histolytica. Exp. Parasitol. 126:366–380. 10.1016/j.exppara.2010.03.005 [DOI] [PubMed] [Google Scholar]
- 5.Wolfe AJ. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12–50. 10.1128/MMBR.69.1.12-50.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernes S, Siman-Tov R, Ankri S. 2005. Epigenetic and classical activation of Entamoeba histolytica heat shock protein 100 (EHsp100) expression. FEBS Lett. 579:6395–6402. 10.1016/j.febslet.2005.09.101 [DOI] [PubMed] [Google Scholar]
- 7.Banerjee S, Fisher O, Lohia A, Ankri S. 2005. Entamoeba histolytica DNA methyltransferase (Ehmeth) is a nuclear matrix protein that binds EhMRS2, a DNA that includes a scaffold/matrix attachment region (S/MAR). Mol. Biochem. Parasitol. 139:91–97. 10.1016/j.molbiopara.2004.10.003 [DOI] [PubMed] [Google Scholar]
- 8.Fisher O, Siman-Tov R, Ankri S. 2004. Characterization of cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) in the protozoan parasite Entamoeba histolytica. Nucleic Acids Res. 32:287–297. 10.1093/nar/gkh161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tovy A, Siman Tov R, Gaentzsch R, Helm M, Ankri S. 2010. A new nuclear function of the Entamoeba histolytica glycolytic enzyme enolase: the metabolic regulation of cytosine-5 methyltransferase 2 (Dnmt2) activity. PLoS Pathog. 6(2):e1000775. 10.1371/journal.ppat.1000775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. 2008. Human DNMT2 methylates tRNAAsp molecules using a DNA methyltransferase-like catalytic mechanism. RNA 14:1663–1670. 10.1261/rna.970408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali IK, Ehrenkaufer GM, Hackney JA, Singh U. 2007. Growth of the protozoan parasite Entamoeba histolytica in 5-azacytidine has limited effects on parasite gene expression. BMC Genomics 8:7. 10.1186/1471-2164-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin MJ, Tang LY, Reddy MN, Shen CK. 2005. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J. Biol. Chem. 280:861–864. 10.1074/jbc.C400477200 [DOI] [PubMed] [Google Scholar]
- 13.Zuin A, Castellano-Esteve D, Ayte J, Hidalgo E. 2010. Living on the edge: stress and activation of stress responses promote lifespan extension. Aging (Albany NY) 2:231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher O, Siman-Tov R, Ankri S. 2006. Pleiotropic phenotype in Entamoeba histolytica overexpressing DNA methyltransferase (Ehmeth). Mol. Biochem. Parasitol. 147:48–54. 10.1016/j.molbiopara.2006.01.007 [DOI] [PubMed] [Google Scholar]
- 15.Lin JY, Chadee K. 1992. Macrophage cytotoxicity against Entamoeba histolytica trophozoites is mediated by nitric oxide from L-arginine. J. Immunol. 148:3999–4005 [PubMed] [Google Scholar]
- 16.Siman-Tov R, Ankri S. 2003. Nitric oxide inhibits cysteine proteinases and alcohol dehydrogenase 2 of Entamoeba histolytica. Parasitol. Res. 89:146–149. 10.1007/s00436-002-0716-2 [DOI] [PubMed] [Google Scholar]
- 17.Elnekave K, Siman-Tov R, Ankri S. 2003. Consumption of L-arginine mediated by Entamoeba histolytica L-arginase (EhArg) inhibits amoebicidal activity and nitric oxide production by activated macrophages. Parasite Immunol. 25:597–608. 10.1111/j.0141-9838.2004.00669.x [DOI] [PubMed] [Google Scholar]
- 18.Santi-Rocca J, Smith S, Weber C, Pineda E, Hon CC, Saavedra E, Olivos-Garcia A, Rousseau S, Dillies MA, Coppee JY, Guillen N. 2012. Endoplasmic reticulum stress-sensing mechanism is activated in Entamoeba histolytica upon treatment with nitric oxide. PLoS One 7:e31777. 10.1371/journal.pone.0031777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Illi B, Colussi C, Rosati J, Spallotta F, Nanni S, Farsetti A, Capogrossi MC, Gaetano C. 2011. NO points to epigenetics in vascular development. Cardiovasc. Res. 90:447–456. 10.1093/cvr/cvr056 [DOI] [PubMed] [Google Scholar]
- 20.Watson PM, Riccio A. 2009. Nitric oxide and histone deacetylases: A new relationship between old molecules. Commun. Integr. Biol. 2:11–13. 10.4161/cib.2.1.7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illi B, Colussi C, Grasselli A, Farsetti A, Capogrossi MC, Gaetano C. 2009. NO sparks off chromatin: tales of a multifaceted epigenetic regulator. Pharmacol. Ther. 123:344–352. 10.1016/j.pharmthera.2009.05.003 [DOI] [PubMed] [Google Scholar]
- 22.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24:1590–1595. 10.1101/gad.586710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dastidar PG, Majumder S, Lohia A. 2007. Eh Klp5 is a divergent member of the kinesin 5 family that regulates genome content and microtubular assembly in Entamoeba histolytica. Cell. Microbiol. 9:316–328. 10.1111/j.1462-5822.2006.00788.x [DOI] [PubMed] [Google Scholar]
- 24.Lavi T, Isakov E, Harony H, Fisher O, Siman-Tov R, Ankri S. 2006. Sensing DNA methylation in the protozoan parasite Entamoeba histolytica. Mol. Microbiol. 62:1373–1386. 10.1111/j.1365-2958.2006.05464.x [DOI] [PubMed] [Google Scholar]
- 25.Schaefer M, Pollex T, Hanna K, Lyko F. 2009. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 37:e12. 10.1093/nar/gkn954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 27.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 28.Ritchie DW, Kozakov D, Vajda S. 2008. Accelerating and focusing protein-protein docking correlations using multi-dimensional rotational FFT generating functions. Bioinformatics 24:1865–1873. 10.1093/bioinformatics/btn334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moal IH, Bates PA. 2010. SwarmDock and the use of normal modes in protein-protein docking. Int. J. Mol. Sci. 11:3623–3648. 10.3390/ijms11103623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozakov D, Hall DR, Beglov D, Brenke R, Comeau SR, Shen Y, Li K, Zheng J, Vakili P, Paschalidis I, Vajda S. 2010. Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13–19. Proteins. 78:3124–3130. 10.1002/prot.22835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaston B, Reilly J, Drazen JM, Fackler J, Ramdev P, Arnelle D, Mullins ME, Sugarbaker DJ, Chee C, Singel DJ, et al. 1993. Endogenous nitrogen oxides and bronchodilator S-nitrosothiols in human airways. Proc. Natl. Acad. Sci. U. S. A. 90:10957–10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackman JE, Alfonzo JD. 2013. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip Rev. RNA 4:35–48. 10.1002/wrna.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. 2012. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3:937. 10.1038/ncomms1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311:395–398. 10.1126/science.1120976 [DOI] [PubMed] [Google Scholar]
- 35.Becker M, Muller S, Nellen W, Jurkowski TP, Jeltsch A, Ehrenhofer-Murray AE. 2012. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 40:11648–11658. 10.1093/nar/gks956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6:275–277. 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- 37.Korner A. 1966. Effect of cycloheximide on protein biosynthesis in rat liver. Biochem. J. 101:627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pineda E, Encalada R, Rodriguez-Zavala JS, Olivos-Garcia A, Moreno-Sanchez R, Saavedra E. 2010. Pyruvate:ferredoxin oxidoreductase and bifunctional aldehyde-alcohol dehydrogenase are essential for energy metabolism under oxidative stress in Entamoeba histolytica. FEBS J. 277:3382–3395. 10.1111/j.1742-4658.2010.07743.x [DOI] [PubMed] [Google Scholar]
- 39.Barr SD, Gedamu L. 2003. Role of peroxidoxins in Leishmania chagasi survival. Evidence of an enzymatic defense against nitrosative stress. J. Biol. Chem. 278:10816–10823. 10.1074/jbc.M212990200 [DOI] [PubMed] [Google Scholar]
- 40.Schulz EC, Roth HM, Ankri S, Ficner R. 2012. Structure analysis of Entamoeba histolytica DNMT2 (EhMeth). PLoS One 7:e38728. 10.1371/journal.pone.0038728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulz EC, Tietzel M, Tovy A, Ankri S, Ficner R. 2011. Structure analysis of Entamoeba histolytica enolase. Acta Crystallogr. D Biol. Crystallogr. 67:619–627. 10.1107/S0907444911016544 [DOI] [PubMed] [Google Scholar]
- 42.Cai CQ, Guo H, Schroeder RA, Punzalan C, Kuo PC. 2000. Nitric oxide-dependent ribosomal RNA cleavage is associated with inhibition of ribosomal peptidyl transferase activity in ANA-1 murine macrophages. J. Immunol. 165:3978–3984 [DOI] [PubMed] [Google Scholar]
- 43.Kim YM, Son K, Hong SJ, Green A, Chen JJ, Tzeng E, Hierholzer C, Billiar TR. 1998. Inhibition of protein synthesis by nitric oxide correlates with cytostatic activity: nitric oxide induces phosphorylation of initiation factor eIF-2 alpha. Mol. Med. 4:179–190 [PMC free article] [PubMed] [Google Scholar]
- 44.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. 2012. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19:900–905. 10.1038/nsmb.2357 [DOI] [PubMed] [Google Scholar]
- 45.Len AC, Harty DW, Jacques NA. 2004. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150:1339–1351. 10.1099/mic.0.27008-0 [DOI] [PubMed] [Google Scholar]
- 46.Vogel C, Silva GM, Marcotte EM. 2011. Protein expression regulation under oxidative stress. Mol. Cell. Proteomics 10:M111.009217. 10.1074/mcp.M111.009217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber C, Guigon G, Bouchier C, Frangeul L, Moreira S, Sismeiro O, Gouyette C, Mirelman D, Coppee JY, Guillen N. 2006. Stress by heat shock induces massive down regulation of genes and allows differential allelic expression of the Gal/GaINAc lectin in Entamoeba histolytica. Eukaryot. Cell 5:871–875. 10.1128/EC.5.5.871-875.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espinosa A, Clark D, Stanley SL., Jr 2004. Entamoeba histolytica alcohol dehydrogenase 2 (EhADH2) as a target for anti-amoebic agents. J. Antimicrob. Chemother. 54:56–59. 10.1093/jac/dkh280 [DOI] [PubMed] [Google Scholar]
- 49.Espinosa A, Yan L, Zhang Z, Foster L, Clark D, Li E, Stanley SL., Jr 2001. The bifunctional Entamoeba histolytica alcohol dehydrogenase 2 (EhADH2) protein is necessary for amebic growth and survival and requires an intact C-terminal domain for both alcohol dehydrogenase and acetaldehyde dehydrogenase activity. J. Biol. Chem. 276:20136–20143. 10.1074/jbc.M101349200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akbar MA, Chatterjee NS, Sen P, Debnath A, Pal A, Bera T, Das P. 2004. Genes induced by a high-oxygen environment in Entamoeba histolytica. Mol. Biochem. Parasitol. 133:187–196. 10.1016/j.molbiopara.2003.10.006 [DOI] [PubMed] [Google Scholar]
- 51.Bryk R, Griffin P, Nathan C. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407:211–215. 10.1038/35025109 [DOI] [PubMed] [Google Scholar]
- 52.Echave P, Tamarit J, Cabiscol E, Ros J. 2003. Novel antioxidant role of alcohol dehydrogenase E from Escherichia coli. J. Biol. Chem. 278:30193–30198. 10.1074/jbc.M304351200 [DOI] [PubMed] [Google Scholar]
- 53.Vuorinen K, Ohlmeier S, Lepparanta O, Salmenkivi K, Myllarniemi M, Kinnula VL. 2008. Peroxiredoxin II expression and its association with oxidative stress and cell proliferation in human idiopathic pulmonary fibrosis. J. Histochem. Cytochem. 56:951–959. 10.1369/jhc.2008.951806 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.