Abstract
Pharmacogenomic testing is viewed as an integral part of precision medicine. To achieve this, we originated The 1200 Patients Project which offers broad, preemptive pharmacogenomic testing to patients at our institution.
We analyzed enrollment, genotype, and encounter-level data from the first year of implementation to assess utility of providing pharmacogenomic results. Results were delivered via a genomic prescribing system (GPS) in the form of traffic lights: green (favorable), yellow (caution), and red (high risk). Additional supporting information was provided as a virtual pharmacogenomic consult, including citation to relevant publications.
Currently, 812 patients have participated, representing 90% of those approached; 608 have been successfully genotyped across a custom array. A total of 268 clinic encounters have occurred at which results were accessible via the GPS. At 86% of visits, physicians accessed the GPS, receiving 367 result signals for medications patients were taking: 57% green lights, 41% yellow lights, and 1.4% red lights. Physician click frequencies to obtain clinical details about alerts varied according to color severity (100% of red were clicked, 72% yellow, 20% green). For 85% of visits, clinical pharmacogenomic information was available for at least one drug the patient was taking, suggesting relevance of the delivered information.
We successfully implemented an individualized health care model of preemptive pharmacogenomic testing, delivering results along with pharmacogenomic decision support. Patient interest was robust, physician adoption of information was high, and results were routinely utilized. Ongoing examination of a larger number of clinic encounters and inclusion of more physicians and patients is warranted.
Keywords: pharmacogenomics, implementation, genomic medicine, precision medicine
INTRODUCTION
Adverse drug reactions are the fifth leading cause of death in the United States [Davies et al., 2007], and thousands of additional patients are prescribed medications from which they derive no benefit; overall efficacy rates for even the most common drugs treating the most prevalent diseases have been estimated to be only about 50% [Spear et al., 2001]. Furthermore, it has been suggested that the health-care system wastes billions of dollars annually on poor prescription drug choices [Altman 2011; Ernst and Grizzle 2001; James et al., 2009] notwithstanding the direct cost of such failures to patients.
The prospect of attaining a future of more informed prescribing practice in our medical system has become today’s possibility through the advance of technology and the ability to rapidly and accurately determine millions of pieces of genetic variation information for large numbers of individuals. This has led to the identification of genetic factors governing response and toxicity for hundreds of drugs [Thorn et al., 2005], with many such associations now replicated, incorporated into drug labels, and encompassed by clinical prescribing guidelines [Relling and Klein 2011]. Indeed, we along with many other groups as well as the United States Government have advocated for increased use of pharmacogenomic testing as a step towards more personalized medicine [Altman et al., 2011; Chan and Ginsburg 2011; Obama 2007; Ratain 2007; Relling et al., 2010].
In response to this challenge to more fully realize the possibilities of personalized or precision medicine, we implemented a project that offers broad, free-of-charge pharmacogenomic testing to patients receiving care at our institution. Entitled “The 1200 Patients Project”, it has the goal of prospectively enrolling and preemptively genotyping (using a panel of variants selected for their pharmacogenomic role) 1200 adults receiving outpatient care [O’Donnell et al., 2012]. Pharmacogenomic results are made available to the physicians caring for enrolled patients through a “Genomic Prescribing System” (GPS) portal [Ratain 2007], which provides instantaneous pharmacogenomic guidance. Our implementation program is designed to simultaneously study this process and model, as determining feasible and effective methods for delivery of pharmacogenomic results is an essential component for determining future sustainability and success of such efforts. Encounter-level data are therefore being collected for the thousands of visits within our project to assess the utility of providing pharmacogenomic results.
Our primary project objective was to determine the feasibility and utility of incorporating preemptive pharmacogenomic testing in routine medical care. Examination of the impact of providing pharmacogenomic results on prescribing decisions and patient outcomes are secondary and future aims that will be assessed. This report describes the initial results from the first year of studying our clinical pharmacogenomics implementation program.
METHODS
Study Design
The overall study conceptualization and schema have been previously described [O’Donnell et al., 2012].
Patient Eligibility
Adult patients receiving care at the University of Chicago were identified for participation by their selected providers, by study research staff, or by expressing self-interest in participating. All patients had to be receiving existing care from one of the designated study physicians who themselves separately signed consent to participate. If an interested patient did not have an existing care relationship with a designated study provider but wanted to participate, study staff assisted the potential patient with arranging a new appointment with a participating study physician. Potential patients were eligible if they were taking, at the time of enrollment, at least one regularly-used prescription medication but not more than six (patients taking >6 medications were ineligible since it is likely that drug-drug interactions, rather than drug-gene interactions, predominate for such individuals; our study was focused on drug-gene interactions); or, if not taking any regularly-used medications at the time of enrollment, the patient was eligible if he/she was ≥65 years or reasonably expected in the opinion of their treating physician to require the use of at least one prescription medication in the next 5 years. Patients were required to have, in the opinion of their treating physician, a life expectancy of ≥3 years. Patients were ineligible if they had previously undergone, or were actively being considered for, liver or kidney transplantation, or if they were unable to provide informed consent to participate.
Genotyping
Patients being genotyped provided a one-time peripheral blood sample for extraction of DNA. Each sample was then genotyped using a MASS-ARRAY/matrix-assisted laser desorption/ionization time-of-flight mass spectrometry method (Sequenom, Inc., San Diego, CA) across (a) a custom-designed panel of selected variants of pharmacogenomic interest and (b) an existing commercial panel consisting of variants typically implicated in drug metabolism (Sequenom ADME panel). All genotyping was performed in a Clinical Laboratory Improvement Amendments (CLIA)-licensed/College of American Pathologists-certified laboratory (Knight Diagnostic Laboratories, Oregon Health & Science University).
Results Delivery via GPS
Genotypes deemed clinically deliverable were translated into drug/variant-specific traffic light-like signals (green, yellow, red, sideways arrow, or dose-calculator) for display in the GPS, as previously described [O’Donnell et al., 2012]. Each traffic signal (or alert) had a corresponding “clinical summary” that was viewable by clicking the alert. Each summary provided a clinical translation of the pharmacogenomic genotype result for that patient, and all summaries were designed to be read in 30 seconds or less to increase likelihood of use during busy clinical care. An example summary was previously published [O’Donnell et al., 2012]. Physician users were not involved in the development of the summaries, but feedback to the research team by users about summaries was welcomed. Results impacting current medications for each patient were summarized and made available to the enrolling provider on a front page within each patient’s chart in the GPS. Results for any newly-prescribed drugs (drugs prescribed anytime after genotyping was performed) were also instantaneously available in the GPS. Separately, providers could also query the GPS (and therefore their patient’s full genotype results) for information for any other drugs, or for potential drugs associated with treatment of various diseases. GPS access was password-restricted to study providers who had signed consent to participate, or their clinical designees. Results provided in the GPS were not available through the medical center’s electronic medical record (EMR). Users were therefore informed that they may wish to reference GPS-provided (CLIA-generated) results in the EMR specifically if they deemed the results clinically important. All individuals with approved GPS access were trained on use of the GPS during a training session prior to study entry, which included mock patients. GPS results were not available to other non-study clinicians in the medical center in this first iteration of our implementation program. Formal dissection of user metrics (time spent in the GPS resource, physician use in the context of clinical workflow) will be undertaken as part of a separate, future analysis.
Analysis
Use of the GPS was studied by analysis of provider-user click logs for outpatient visits from October 2012 (time of first results delivery) through July 2013. Before each clinic day, participating physicians were reminded via email of the given patients on their schedule who were enrolled in the study. Reminders on the day of the clinic visit were also often provided via verbal in-person communication, or via “flagging” of the chart. Providers were simply reminded that a patient was a study participant who had been genotyped, not whether the GPS contained specific pharmacogenomic information for that patient. These reminder systems were in place at the beginning of the study. Study providers were prospectively monitored for whether they accessed the GPS within 72 hours (pre- and/or post) of a clinic visit, and all clicks within the GPS were tracked and categorized. In particular, it was tracked whether physicians “clicked” viewable traffic light signals to receive the associated clinical results summaries.
The study was designed to follow enrolled patient-provider pairs for at least 3 years, as long as the patient-provider relationship exists and unless a party withdraws consent. Because of the potential for bias among our group of early-adopter physicians in this study, we set a high bar for analysis of the primary feasibility endpoint [O’Donnell et al., 2012] – 50% GPS use rate after 3 years for encounters in which medication decisions were being made.
RESULTS
Patient Enrollment
Thus far, 902 patients have been approached for participation in the study, with 815 agreeing to participate (90%). Three (3) consented patients did not meet study eligibility criteria, leaving 812 total patients in the study population to date. Table I shows the baseline characteristics of these patients. The demographics are generally reflective of the larger patient population served by The University of Chicago, although notably, 60% of participants had a college/university-level degree or a post-graduate degree. The most prevalent diseases reflect those not only common in the U.S. population but also representative of enrichment for certain diseases from the clinics of specific subspecialist physician participants in our study. Active enrollment of patients from the clinics of 15 different physicians is currently ongoing toward the initial phase I goal of 1200 patients.
Table 1.
Patient characteristics
| Variable | Frequency (n=812) |
|---|---|
| Male | 354 (44%) |
| Female | 458 (56%) |
| Age in years | |
| < 29 | 38 (5%) |
| 30–49 | 180 (22%) |
| 50–69 | 393 (48%) |
| 70–89 | 199 (25%) |
| > 90 | 2 (0.2%) |
| Race | |
| White | 520 (64%) |
| Black or African American | 228 (28%) |
| Asian | 26 (3%) |
| Native Hawaiian or Other Pacific Islander | 0 |
| American Indian/Alaska Native | 2 (0.2%) |
| More than one race/Other | 13 (2%) |
| Declined to answer/Unknown | 23 (3%) |
| Ethnicity | |
| Not Hispanic or Latino | 304 (37%) |
| Hispanic or Latino | 31 (4%) |
| Not Hispanic | 14 (2%) |
| Declined to answer/Unknown | 463 (57%) |
| Education level | |
| Did not complete high school | 33 (4%) |
| High school graduate/GED | 107 (13%) |
| Attended some college/university | 179 (22%) |
| College/university graduate | 230 (28%) |
| Obtained advanced degree (post-college/university) | 259 (32%) |
| Declined to answer/Unknown | 4 (0.5%) |
| Most common conditions in active patients | |
| Hypertension | 347 (43%) |
| Hyperlipidemia | 221 (27%) |
| Coronary artery disease | 85 (10%) |
| Gastroesophageal reflux disease | 85 (10%) |
| Diabetes mellitus | 80 (10%) |
| Breast cancer | 78 (10%) |
| Crohn’s disease | 60 (7%) |
| Obstructive sleep apnea | 57 (7%) |
| Osteoarthritis | 52 (6%) |
| Osteopenia | 52 (6%) |
| Prostate Cancer | 50 (6%) |
| Anemia | 48 (6%) |
| Asthma | 47 (6%) |
| Hepatitis C | 45 (6%) |
Table II lists the most common prescription medications being taken by the enrolled study population (those listed were being taken by ≥5% of the population; dozens of other medications were being taken by <5% of participants). Of the 17 top drugs, 9 currently have available clinical pharmacogenomic results summaries associated with them within the GPS.
Table 2.
Most common medications in active participants
| Medication | Frequency, n (%) | Genes Reported |
|---|---|---|
| Aspirin | 213 (26%) | LTC4S |
| Hydrochlorothiazide | 169 (21%) | REN, ADD1 |
| Atorvastatin | 138 (17%) | SLCO1B1, GNB3, KIF6 |
| Lisinopril | 118 (15%) | none |
| Amlodipine | 111 (14%) | CYP3A4, CACNA1C |
| Metoprolol | 104 (13%) | ADRB1, GRK4 |
| Levothyroxine | 91 (11%) | none |
| Simvastatin | 69 (8%) | SLCO1B1, ABCB1 |
| Omeprazole | 65 (8%) | CYP2C19 |
| Atenolol | 55 (7%) | LDLR, GNB3, AGT |
| Losartan | 52 (6%) | none |
| Furosemide | 46 (6%) | none |
| Warfarin | 45 (6%) | CYP2C9, VKORC1 |
| Esomeprazole | 44 (5%) | none |
| Metformin | 44 (5%) | none |
| Prednisone | 43 (5%) | none |
| Fluticasone propionate | 40 (5%) | none |
Genotyping Results
To date, genotype results have been generated in a CLIA setting for 608 patients. Assessing compositely all variants that have been interrogated, 94.3% have been successfully genotyped across the entire study population, meaning an interpretable genotype call was assigned. On a per patient basis, the median rate of successful genotyping calls was 96.7% (range: 59.7–99.2%).
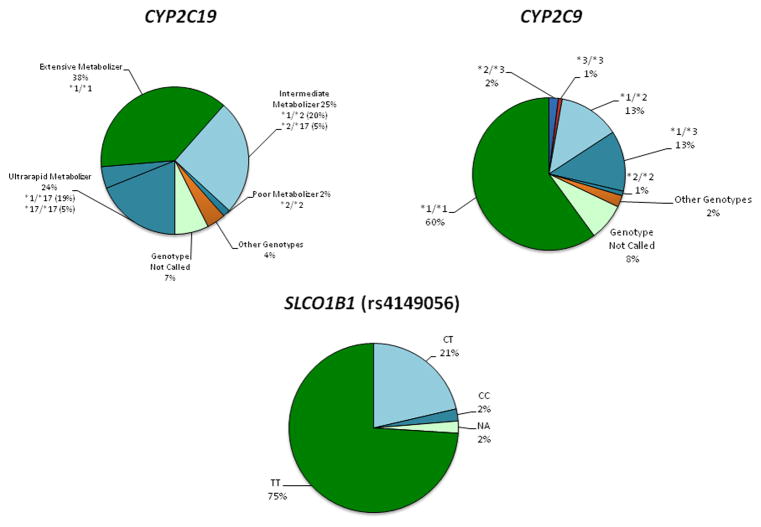
A breakdown of the study population genotype results for several key pharmacogenomic genes being reported in our study is shown in Figure 1.
Figure 1. Genotype results for the study population for several key pharmacogenomic genes being reported.
Rather than being reported in the GPS as shown, each patient’s specific result is translated into a corresponding traffic alert signal color (with a level of evidence designation) with accompanying decision-support information. For some genotypes like those in CYP2C19, this first requires assignment of each genotype into the corresponding enzymatic phenotype group (ultrarapid metabolizer, extensive metabolizer, intermediate metabolizer, poor metabolizer). Genotypes from CYP2C9 are incorporated into a warfarin dosing calculator, with each patient’s results delivered to the physician as a recommended starting dose. NA=genotype not called.
Use of the GPS
The first physician was provided access to his patients’ available pharmacogenomic results in October 2012, and multiple additional physician participants have been “turned live” sequentially since that time (the total number of live physicians is currently n=6; no physicians have dropped-out after being turned live). Once each live physician was actively eligible to receive GPS results for any of their genotyped patients, we prospectively tracked physician use of the GPS at all subsequent study patient visits with that physician. We considered each patient’s visit at which pharmacogenomic results were potentially accessible by their participating physician to be the unit of measure of interest.
A total of 268 such patient encounters occurred during the study period and were analyzed. At 230 of these visits (86%), study physicians accessed the GPS, with no observed increase or decrease in the rate of accession over time.
Importantly, for 85% of the visits for which GPS log-in occurred, there was clinical pharmacogenomic information available for at least one currently prescribed drug.
Relevance of Pharmacogenomic Results Delivered During Outpatient Visits
Examining the visits at which physicians accessed the GPS, the delivered pharmacogenomic information was comprised of 367 pharmacogenomic alerts for medications patients were already taking: 57.2% were green light alerts, 41.4% were yellow light alerts, and 1.4% were red light alerts. Physicians variably clicked delivered alerts to obtain the clinical details about specific pharmacogenomic results, according to color severity: click frequencies were 100% for red lights, 72% for yellow lights, but only 20% for green lights. These results are shown in Figure 2.
Figure 2. Results Delivery via the GPS.
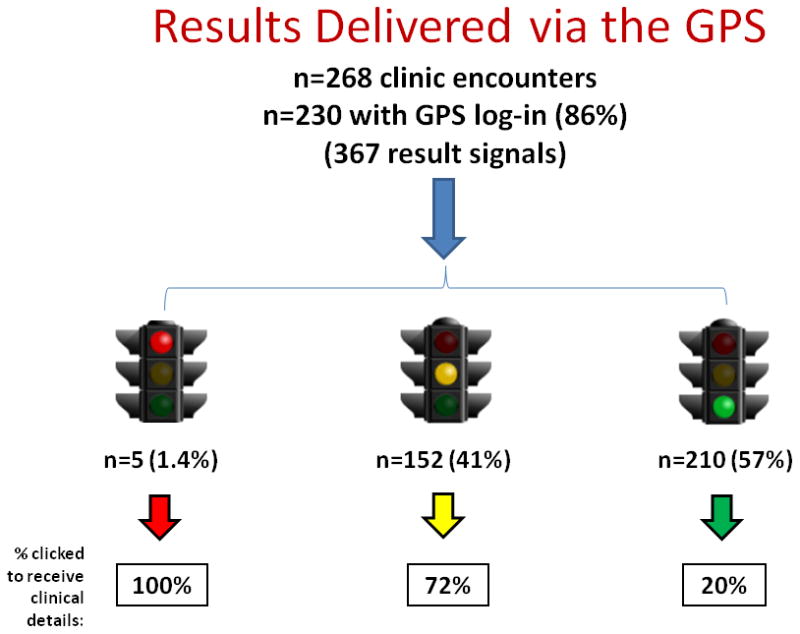
Shown is the rate of physician GPS access since pharmacogenomic results became available in October 2012, the number of alerts delivered by color, and the frequency of results being clicked (by color severity) to obtain clinical consultation summaries that include details supporting each result.
Physician users were asked to report whether provided pharmacogenomic result summaries were “clinically relevant”. First, it should be noted that physician survey completion rates (to answer this question) declined over time: 93% completion rate during the first quarter of evaluable visits, 67% in the second quarter, and 13% and 27% respectively in the third and fourth quarters of reporting. However, the large majority of respondents (30 of 60 physician surveys were returned overall from encounters at which medication changes occurred, including from 5 of the 6 active physician users) said they “agreed strongly” (10%) or “agreed somewhat” (76%), while few said they “disagreed somewhat” (3%) or “disagreed strongly” (10%). Interestingly, 93% of respondents also said they would be “very likely” to enroll other patients to the study after using the GPS.
Continued evaluation of the use, applicability, and utility of this pharmacogenomic results delivery model will occur as the full study population is enrolled and followed over time.
DISCUSSION
Within the Center for Personalized Therapeutics at the University of Chicago, we have created an individualized health care model of preemptive pharmacogenomic testing to permit the widespread implementation and use of pharmacogenomic results during clinical care. Since January 2011, we have prospectively enrolled and preemptively genotyped over 600 adults receiving outpatient care within the Department of Medicine. Patient-specific results have been made available to participating physicians through a created GPS portal, and over 350 pharmacogenomic alerts have been delivered, supporting numerous instances of prescription decisions based upon the results. This manuscript describes the initial results from the project’s first year of implementation. Most importantly, we found that patient interest in being tested was nearly ubiquitous, that physician adoption of the GPS was high, and that delivered results were routinely utilized during office visits. We demonstrated that delivered pharmacogenomic alerts had widespread applicability to our patient population and to the drugs they are routinely prescribed. Initial measures of utility suggest that early-adopter physicians in our project found the availability of pharmacogenomic information to be relevant to their practice, and worthy of repeated use during patient care.
Several aspects of the results generated thus far deserve particular discussion. First, before we could even begin reporting patient genotype results, we found that establishment of a validated, custom-designed genotyping panel to generate accurate genotype calls in a CLIA environment was not trivial. Panel validation required repeat testing of reference samples, analysis, and refinement of the panel over a span of approximately 6 months, and some genotype calls for validated assays nevertheless remain inconsistently reportable, although the overall frequency of missing calls is low. Individual patients having very low overall genotype call rates across multiple variants in the genotyping panels likely represent individual problems with DNA quality, and re-extraction of DNA (without recollection) for these individuals with repeat genotyping is planned. In addition, the results in this manuscript (and in our project) do not yet include clinical reporting of CYP2D6 genotypes because of initial technical hurdles with accurate assignment of haplotypes for this complex gene. These hurdles have recently been overcome and it is expected that we will add reporting of CYP2D6 genotypes in the very near future, a step that will add considerably to the applicability of clinical results within the GPS because CYP2D6 is important for the metabolism of many commonly prescribed medications [De Gregori et al., 2010]. This notwithstanding, the widespread relevance of our existing reportable results (impacting at least one drug for >80% of the analyzed patient visits) is indeed noteworthy in itself. This extensive applicability likely in part reflects our approach favoring broader pharmacogenomic reporting, following a pharmacogenomic ‘non-inferiority’ model [Altman, 2011].
Secondly, it was interesting to note that the breakdown of green, yellow, and red light alerts delivered for the drugs patients were already taking showed a very low prevalence of red light drugs, and a majority of green light drugs. In other words, presumably without specifically knowing it, our patients and physicians generally arrived at genetically-compatible drugs. This may have occurred in part through patient and physician discontinuation of drugs that were causing adverse reactions or non-response [Langley et al., 2005]; it may reflect our study selection bias of enrolling patients taking no more than six drugs; but it also likely reflects the fact that genetic polymorphisms associated with high-risk outcomes would not be expected to be common in human beings or else the impacted drugs would have been unlikely to ever achieve approval or favoritism for clinical use. It was interesting to find that, on exploratory analysis of the genotypes of our 608 tested patients, the lifetime impact on drug prescribing is potentially immense: ~25% of patients have a genotype which would confer a red light signal for at least one of the drugs for which we report results, and over 50% of patients would have a level 1 yellow light alert for at least one drug with results in our system.
Our assessments of initial clinical utility, while encouraging, deserve ongoing study over a larger number of evaluable clinic encounters and including a larger number of physicians. One of our primary goals of the first phase of this project is to identify ways to increase use of the GPS and refine it as a model of pharmacogenomic results delivery. Currently, an expanded number of early-adopter physicians are now enrolling patients in this project, and we anticipate that wider study of the feasibility and utility of results and results delivery will be highly informative to the continued improvement of our project and hopefully also to other pharmacogenomic implementation programs.
It should be noted that none of the results presented relate to instances of drugs being newly-prescribed for a particular patient, and it is this category of prescribing that might ultimately be most relevant for the use of pharmacogenomics. As opposed to deciding whether to discontinue a current medication based on a pharmacogenomic result—an event which might be unlikely if a patient has been doing well on a medication for a long time—new prescribing decisions for treatment of a condition needing attention may benefit greatly from the availability of pharmacogenomic data. Because of the preemptive nature of the pharmacogenomic testing in our project, a dynamic feature of the GPS is that it allows the physician to query their patient’s test results and immediately determine if any results impact drugs they are considering prescribing. Diseases can also be searched, if the physician wants to compare information about multiple drugs that treat a given disease. Our early experience suggests that these features are being under-utilized so far during new prescription decisions by our early-adopter physicians. Analysis of physician-user workflow suggests that this may be occurring because physicians are not using the GPS while in the room with the patient during a visit. Instead, they are checking the GPS ahead of the visit and reviewing pharmacogenomic results on already-prescribed medications in a static fashion, just as they might review other common laboratory results. Additional refinement of the GPS tool, or further user education may be required for the advantages of preemptive genotyping to be fully realized.
In order to continue to provide routine pharmacogenomic results and to be positioned to expand this clinical service to other patients and providers, it will be important to evaluate other high-throughput genotyping options so that marginal costs can be controlled, and turn-around times can be reduced. We are continuing to accrue approximately 30 patients/month to the project, and we anticipate that this number will grow over time as participation is expanded to other clinics, clinical services, or institutions.
Perhaps just as importantly, it will be necessary to continue to study the effects of broad pharmacogenomic results delivery implementation. The value of preemptive testing will ultimately be determined through the evaluation of outcomes measures, including those involving decision-support systems, avoidance of high-risk prescriptions, and decreases in the number of adverse and non-response events.
Acknowledgments
We thank Julianne Faust and Sheena Hussain for their assistance with preparation of this manuscript.
Supported by NIH K23 GM100288-01A1 (PHO), NIH K12 CA139160 (PHO), Bucksbaum Institute Associate Faculty Scholar Pilot Grant (PHO), The Conquer Cancer Foundation of the American Society for Clinical Oncology (MJR), and The William F. O’Connor Foundation (MJR).
Biographies
Peter H. O’Donnell, MD, is an Assistant Professor of Medicine and principal investigator of The 1200 Patients Project at The University of Chicago. He is also Associate Director for Clinical Implementation in The University of Chicago Center for Personalized Therapeutics.
Keith Danahey, MS, is a senior software developer specializing in algorithms and large data sets. After earning a Master of Science in Computer Information Systems, he left algorithmic trading to use his skills for the greater good in the healthcare field.
Michael Jacobs, BS, has worked as a research assistant in the Center for Personalized Therapeutics at The University of Chicago for over a year.
Nisha R. Wadhwa, BS, a member of the University of Chicago Pritzker School of Medicine class of 2016, works in Dr. O’Donnell’s research group in The Center for Personalized Therapeutics at The University of Chicago as part of the Scholarship and Discovery component of her medical education.
Shennin Yuen, BS, worked as a research assistant in The Center for Personalized Therapeutics at The University of Chicago during the time of data collection.
Angela Bush, BS, worked as a research assistant in The Center for Personalized Therapeutics at The University of Chicago during the time of data collection.
Yasmin Sacro, MD, is an Assistant Professor of Medicine at The University of Chicago specializing in primary care.
Matthew J. Sorrentino, MD, is a Professor of Medicine and Associate Director of the Bucksbaum Institute for Clinical Excellence at The University of Chicago. He is a practicing cardiologist, with expertise and research interests in treating hyperlipidemia, hypertension, and coronary artery disease.
Mark Siegler, MD, the Lindy Bergman Distinguished Service Professor of Medicine and Director of the MacLean Center for Clinical Medical Ethics at The University of Chicago, is the Executive Director of the Bucksbaum Institute for Clinical Excellence. He is a practicing physician in internal medicine.
William Harper, MD, is Medical Director of the Program for Personalized Health and Prevention at The University of Chicago. He is a practicing internist with an interest in the optimization of patient care.
Andrea Warrick, BS, is a molecular technologist at the Knight Diagnostic Laboratories of the Oregon Health & Science University.
Soma Das, PhD, is Director of the Clinical Molecular Genetics Laboratory in The University of Chicago’s Department of Human Genetics.
Don Saner, MS, is Director of Clinical and Translational Informatics in the Center for Research Informatics at The University of Chicago.
Christopher L. Corless, MD, PhD, is Executive Director and Chief Medical Officer for Knight Diagnostic Laboratories at The Oregon Health & Science University.
Mark J. Ratain, MD, is the Leon O. Jacobson Professor of Medicine and Director of The Center for Personalized Therapeutics at The University of Chicago. He is also co-PI of the Pharmacogenomics of Anticancer Agents Research (PAAR) Group at The University of Chicago.
Footnotes
Conflict of interest disclosure: M.J.R. is a coinventor holding patents related to pharmacogenetic diagnostics and receives royalties related to UGT1A1 genotyping. No royalties are received from the genotyping performed in this study.
References
- Altman RB. Pharmacogenomics: “noninferiority” is sufficient for initial implementation. Clin Pharmacol Ther. 2011;89(3):348–350. doi: 10.1038/clpt.2010.310. [DOI] [PubMed] [Google Scholar]
- Altman RB, Kroemer HK, McCarty CA, Ratain MJ, Roden D. Pharmacogenomics: will the promise be fulfilled? Nat Rev Genet. 2011;12(1):69–73. doi: 10.1038/nrg2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan IS, Ginsburg GS. Personalized medicine: progress and promise. Annu Rev Genomics Hum Genet. 2011;12:217–244. doi: 10.1146/annurev-genom-082410-101446. [DOI] [PubMed] [Google Scholar]
- Davies EC, Green CF, Mottram DR, Pirmohamed M. Adverse drug reactions in hospitals: a narrative review. Curr Drug Saf. 2007;2(1):79–87. doi: 10.2174/157488607779315507. [DOI] [PubMed] [Google Scholar]
- De Gregori M, Allegri M, De Gregori S, Garbin G, Tinelli C, Regazzi M, Govoni S, Ranzani GN. How and Why to Screen for CYP2D6 Interindividual Variability in Patients Under Pharmacological Treatments. Curr Drug Metab. 2010;11(3):276–282. doi: 10.2174/138920010791196274. [DOI] [PubMed] [Google Scholar]
- Ernst FR, Grizzle AJ. Drug-related morbidity and mortality: updating the cost-of-illness model. J Am Pharm Assoc (Wash) 2001;41(2):192–199. doi: 10.1016/s1086-5802(16)31229-3. [DOI] [PubMed] [Google Scholar]
- James TH, Helms ML, Braund R. Analysis of medications returned to community pharmacies. Ann Pharmacother. 2009;43(10):1631–1635. doi: 10.1345/aph.1M209. [DOI] [PubMed] [Google Scholar]
- Langley C, Marriott J, Mackridge A, Daniszewski R. An analysis of returned medicines in primary care. Pharm World Sci. 2005;27(4):296–299. doi: 10.1007/s11096-005-0354-8. [DOI] [PubMed] [Google Scholar]
- O’Donnell PH, Bush A, Spitz J, Danahey K, Saner D, Das S, Cox NJ, Ratain MJ. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther. 2012;92(4):446–449. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obama B. The genomics and Personalized Medicine Act of 2006. Clin Adv Hematol Oncol. 2007;5(1):39–40. [PubMed] [Google Scholar]
- Ratain MJ. Personalized medicine: building the GPS to take us there. Clin Pharmacol Ther. 2007;81(3):321–322. doi: 10.1038/sj.clpt.6100092. [DOI] [PubMed] [Google Scholar]
- Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89(3):464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, Altman RB, Goetz MP, Evans WE. Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol. 2010;11(6):507–509. doi: 10.1016/S1470-2045(10)70097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7(5):201–204. doi: 10.1016/s1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol Biol. 2005;311:179–191. doi: 10.1385/1-59259-957-5:179. [DOI] [PubMed] [Google Scholar]