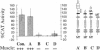Abstract
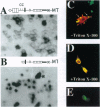
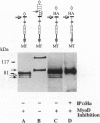
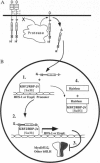
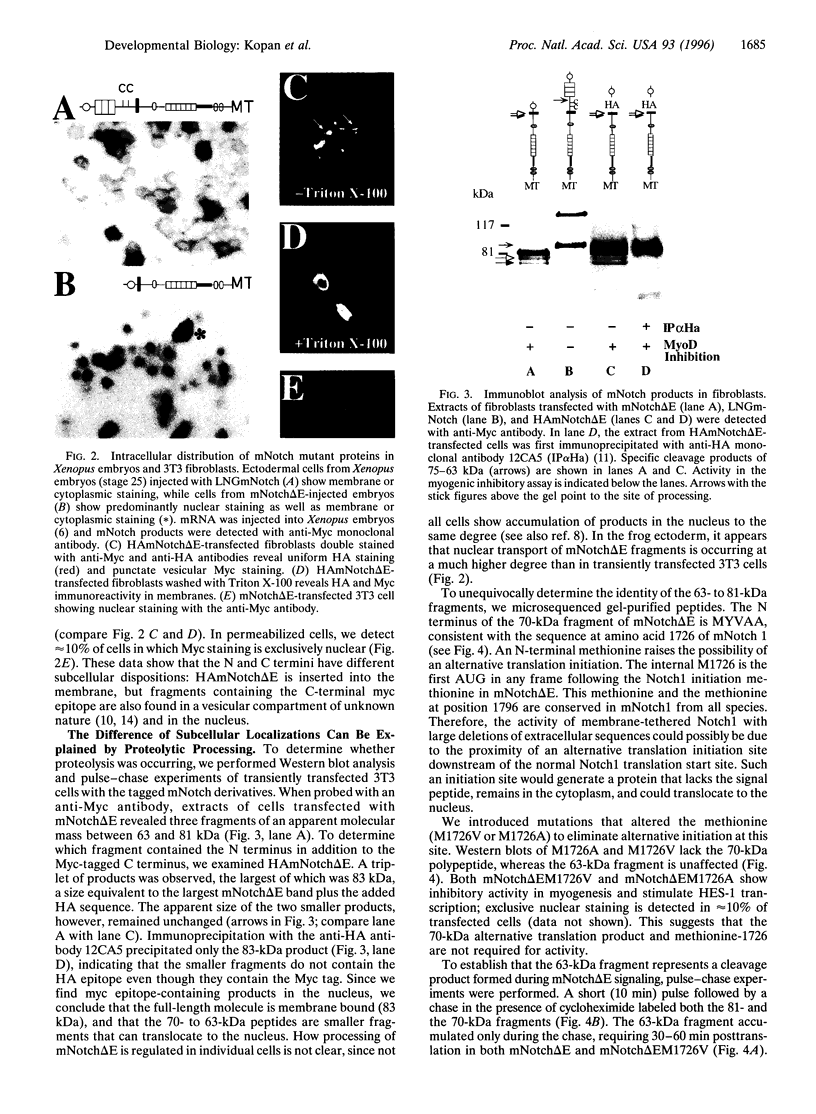
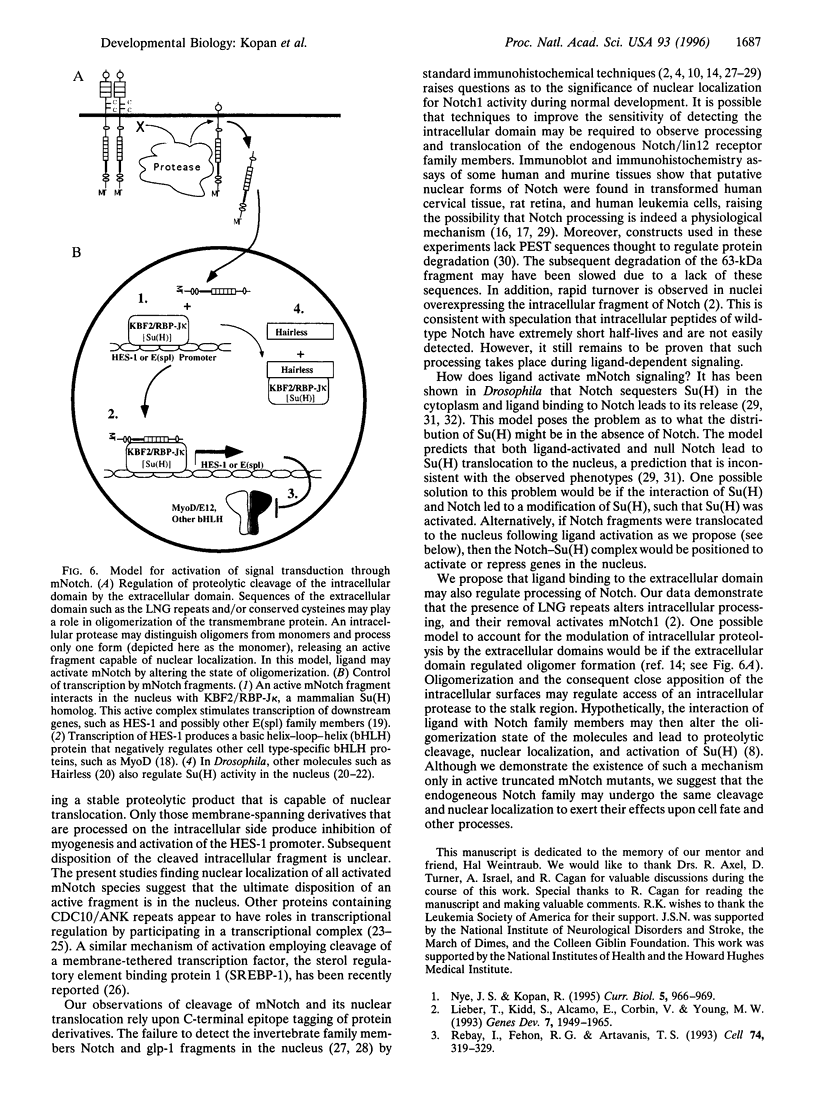
Previous studies imply that the intracellular domain of Notch1 must translocate to the nucleus for its activity. In this study, we demonstrate that a mNotch1 mutant protein that lacks its extracellular domain but retains its membrane-spanning region becomes proteolytically processed on its intracellular surface and, as a result, the activated intracellular domain (mNotchIC) is released and can move to the nucleus. Proteolytic cleavage at an intracellular site is blocked by protease inhibitors. Intracellular cleavage is not seen in cells transfected with an inactive variant, which includes the extracellular lin-Notch-glp repeats. Collectively, the studies presented here support the model that mNotch1 is proteolytically processed and the cleavage product is translocated to the nucleus for mNotch1 signal transduction.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artavanis-Tsakonas S., Matsuno K., Fortini M. E. Notch signaling. Science. 1995 Apr 14;268(5208):225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Aster J., Pear W., Hasserjian R., Erba H., Davi F., Luo B., Scott M., Baltimore D., Sklar J. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harb Symp Quant Biol. 1994;59:125–136. doi: 10.1101/sqb.1994.059.01.016. [DOI] [PubMed] [Google Scholar]
- Brou C., Logeat F., Lecourtois M., Vandekerckhove J., Kourilsky P., Schweisguth F., Israël A. Inhibition of the DNA-binding activity of Drosophila suppressor of hairless and of its human homolog, KBF2/RBP-J kappa, by direct protein-protein interaction with Drosophila hairless. Genes Dev. 1994 Oct 15;8(20):2491–2503. doi: 10.1101/gad.8.20.2491. [DOI] [PubMed] [Google Scholar]
- Coffman C. R., Skoglund P., Harris W. A., Kintner C. R. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell. 1993 May 21;73(4):659–671. doi: 10.1016/0092-8674(93)90247-n. [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Troemel E. R., Evans T. C., Kimble J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development. 1994 Oct;120(10):2901–2911. doi: 10.1242/dev.120.10.2901. [DOI] [PubMed] [Google Scholar]
- Del Amo F. F., Smith D. E., Swiatek P. J., Gendron-Maguire M., Greenspan R. J., McMahon A. P., Gridley T. Expression pattern of Motch, a mouse homolog of Drosophila Notch, suggests an important role in early postimplantation mouse development. Development. 1992 Jul;115(3):737–744. doi: 10.1242/dev.115.3.737. [DOI] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985 Dec;5(12):3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon R. G., Johansen K., Rebay I., Artavanis-Tsakonas S. Complex cellular and subcellular regulation of notch expression during embryonic and imaginal development of Drosophila: implications for notch function. J Cell Biol. 1991 May;113(3):657–669. doi: 10.1083/jcb.113.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini M. E., Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994 Oct 21;79(2):273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Fortini M. E., Rebay I., Caron L. A., Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993 Oct 7;365(6446):555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995 Sep 28;377(6547):355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jennings B., Preiss A., Delidakis C., Bray S. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994 Dec;120(12):3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- Kidd S., Baylies M. K., Gasic G. P., Young M. W. Structure and distribution of the Notch protein in developing Drosophila. Genes Dev. 1989 Aug;3(8):1113–1129. doi: 10.1101/gad.3.8.1113. [DOI] [PubMed] [Google Scholar]
- Kopan R., Nye J. S., Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994 Sep;120(9):2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- LaMarco K., Thompson C. C., Byers B. P., Walton E. M., McKnight S. L. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991 Aug 16;253(5021):789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Lieber T., Kidd S., Alcamo E., Corbin V., Young M. W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993 Oct;7(10):1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- Matsuno K., Diederich R. J., Go M. J., Blaumueller C. M., Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995 Aug;121(8):2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- Nye J. S., Kopan R., Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994 Sep;120(9):2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- Nye J. S., Kopan R. Developmental signaling. Vertebrate ligands for Notch. Curr Biol. 1995 Sep 1;5(9):966–969. doi: 10.1016/s0960-9822(95)00189-8. [DOI] [PubMed] [Google Scholar]
- Pati U. K. Novel vectors for expression of cDNA encoding epitope-tagged proteins in mammalian cells. Gene. 1992 May 15;114(2):285–288. doi: 10.1016/0378-1119(92)90589-h. [DOI] [PubMed] [Google Scholar]
- Rebay I., Fehon R. G., Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993 Jul 30;74(2):319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M. Regulation of enzyme levels by proteolysis: the role of pest regions. Adv Enzyme Regul. 1988;27:135–151. doi: 10.1016/0065-2571(88)90014-3. [DOI] [PubMed] [Google Scholar]
- Roehl H., Kimble J. Control of cell fate in C. elegans by a GLP-1 peptide consisting primarily of ankyrin repeats. Nature. 1993 Aug 12;364(6438):632–635. doi: 10.1038/364632a0. [DOI] [PubMed] [Google Scholar]
- Sasai Y., Kageyama R., Tagawa Y., Shigemoto R., Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992 Dec;6(12B):2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Schweisguth F., Posakony J. W. Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development. 1994 Jun;120(6):1433–1441. doi: 10.1242/dev.120.6.1433. [DOI] [PubMed] [Google Scholar]
- Schweisguth F. Suppressor of Hairless is required for signal reception during lateral inhibition in the Drosophila pupal notum. Development. 1995 Jun;121(6):1875–1884. doi: 10.1242/dev.121.6.1875. [DOI] [PubMed] [Google Scholar]
- Struhl G., Fitzgerald K., Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993 Jul 30;74(2):331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Brown T. A., McKnight S. L. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991 Aug 16;253(5021):762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Turner D. L., Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994 Jun 15;8(12):1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Wang X., Sato R., Brown M. S., Hua X., Goldstein J. L. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994 Apr 8;77(1):53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Sawada J., Yano K., Yamaguchi K., Goto M., Handa H. cDNA cloning of transcription factor E4TF1 subunits with Ets and notch motifs. Mol Cell Biol. 1993 Mar;13(3):1385–1391. doi: 10.1128/mcb.13.3.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagouras P., Stifani S., Blaumueller C. M., Carcangiu M. L., Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci U S A. 1995 Jul 3;92(14):6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]