Abstract
Sarcocystis spp. are the cyst forming protozoan parasites that prevalent in livestock all around the world. In the presented work, we examined 40 macroscopic and 40 microscopic sarcocysts from Khouzestan and Lorestan provinces, south-western Iran, utilizing PCR–RFLP based on amplification of 18S rRNA gene. Using AvaI, HindII, TaqI and EcoRI restriction enzymes the results represented Sarcocystis gigantea in both macroscopic and microscopic cysts. This result supports the importance of molecular investigations on characterization of Sarcocystis species when we aimed to assess the reliable control and preventive programs.
Keywords: Sarcocystis spp., PCR–RFLP, 18S rRNA, Sheep, Iran
Introduction
Sarcocystosis is a zoonotic disease in domestic animals caused by Sarcocystis spp. (Apicomplexa: Sporozoa), a cyst forming coccidian parasite with obligatory two-host life cycle involving carnivorous as definitive hosts and herbivorous or omnivorous as intermediate hosts. Each intermediate and definitive host may harbour more than one Sarcocystis species (Dubey et al. 1989aa; Dubey and Lindsay 2006).
The parasites are most prevalent in livestock animals including sheep all around the world (Dubey et al. 1989b). Sheep become infected with the parasite via ingesting sporocysts or sometimes sporulated oocysts existed in the food or water (Dubey and Lindsay 2006). Four species of Sarcocystis, known as: Sarcocystis tenella, Sarcocystis arieticanis, S. gigantea and S. medosiformis have been documented and described in the Sheep. Of those Sarcocystis species that infect sheep, Sarcocystis gigantea and S. medusiformis are transmitted by felids and are consider non-pathogenic, whereas microscopic cyst forming species, S. tenella and S. arieticanis, which are transmitted by canids can cause devastating sarcocystosis that may be result in abortion or death depends on the dose of ingested sporocysts and the immune status of the host (Buxton 1998; Dubey et al. 1989b; Dubey and Lindsay 2006).
The conventional tools for species diagnosis of Sarcocystis spp. were based on transmission electron microscopy, structure of the cyst wall in the striated muscles of the intermediate host or information about the lifecycle of the parasite (Jehle et al. 2009). However, because of showing the morphologic variations in these procedures they are not exactly reliable at the species-specific identification. On the other hand, electron microscopy is not a choice for wide and extensive detective studies (McManus and Bowles 1996).
In recent times, various molecular techniques such as PCR and its variants based on sequence changes have been used regarding the sensitivity and rapidity to determine genetic diversity among many parasites, phylogenetic and taxonomic studies and in epidemiological mapping (Erlich et al. 1991; Gonzalez et al. 2006; Maurer 2011).
Currently, PCR-restriction fragment length polymorphism (PCR–RFLP) based on variable regions of the small subunit ribosomal RNA sequences is considered and used widely as a rapid, inexpensive and accurate molecular approach to discriminate different protozoa as well as Sarcocystis spp. (Ellis et al. 1995; Heckeroth and Tenter 1999a; Heckeroth and Tenter 1999b; Motamedi et al. 2011). As sheep are rearing all around Iran and are the strategic animal in livestock industry, so, the aim of this study was to determine the Sarcocystis species of sheep in two economically important provinces, Khouzestan and Lorestan, by PCR–RFLP in basis of amplification of 18S rRNA gene.
Materials and methods
Samples collection
Totally 40 macroscopic cysts (20 from Khouzestan province and 20 from Lorestan province main slaughterhouses) from sheep muscles were collected. Also, at least 50 g from esophagus, diaphragm, intra-costal, abdomen and biceps were sampled for microscopic investigations. The samples were kept at −20 °C until examination. The sampling discontinued when the positive specimens for microscopic cysts reached to 40 specimens (20 from Ahvaz and 20 from Khorammabad).
Digestion method
The method of Dubey et al. (1989a) with some modifications was used for digestion of muscles as described below:
100 g of mixture of collected muscles were minced and digested for 30 min at 37 °C in 100 ml of digestion medium containing 1.3 g of pepsin (Merck), 3.5 ml, and 2.5 g NaCl in 500 ml of distilled water. After digestion, the mixture were centrifuged for 3 min at 3500×g and the sediment was then stained with giemsa and examined by optical microscope at 400× magnification for detecting bradyzoites. After that, the sediments containing bradyzoites washed two times with normal saline and TE solution respectively and preserved at −20 °C until examination.
DNA extraction
DNA was extracted using the genomic DNA extraction Kit (Cinnagen, Iran) according to manufacturer instructions. Next, the extracted DNA was stored at −20 °C until PCR has performed.
PCR amplification and RFLP
Species determination was carried out by PCR–RFLP according to amplification of 18S rRNA gene. PCR protocol and primer selection were adopted according to the previously described by Paikari et al. 2008 with some modification.
Briefly, amplification of the 18S rRNA gene was carried out in 50 μl reaction volumes containing 1 μl of DNA template, 5 pmol of reverse and forward primers, 3.0 mM MgCl2, 5.0 μl 10× PCR Buffer, 200 μM of each dNTP and 2.5 U Taq DNA polymerase. Forward and reverse genus-specific primer sequences used in this study were (Sar-F1): 5′ GCA CTT GAT GAA TTC TGG CA 3′ and (Sar-F2): 5′ CAC CAC CCA TAG AAT CAA G 3′ respectively. The thermal program of PCR was as follows: 94 °C for 5 min, 30 cycles of 94 °C for 2 min, annealing at 57 °C for 30 s, and 72 °C for 2 min, followed by a final extension step at 72 °C for 5 min. To verify the results, 10 μl of each PCR product was electrophoresed in a 1 % agarose gel, stained with safe stain and visualized on a UV transilluminator. The PCR products were identified by size using a 100 bp ladder (Fermentas). The expected PCR product had a length of 609 bp.
To determine the possibility of the cross reaction with related protozoans, Neospora caninum and Toxoplasma gondii, the whole tachyzoites of these two parasites were also analyzed by mentioned primers. The amplified products were analyzed with RFLP using AvaI, HindII, TaqI and EcoRI restriction enzymes. Digested PCR products were resolved on a 1.5 % agarose gel and visualized with safe stain under ultraviolet light.
Results
PCR amplification of T. gondii and N. caninum tachyzoites with mentioned primers revealed no electrophoretic bands (Fig. 1) but, in the same situation our isolates demonstrated a Sarcocystis specific 609 bp band (Figs. 1 and 2).
Fig. 1.
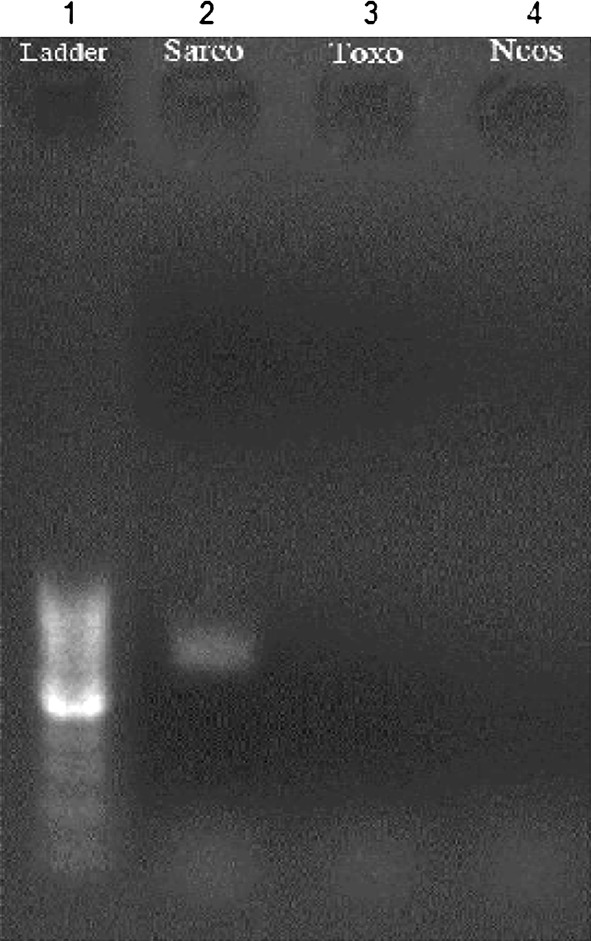
PCR analysis of 18S rRNA gene with T. gondii, N. caninum and Sarcocystis sp. Revealed only 690 bp band for Sarcocystis sp. (1) 100 bp ladder, (2) Sheep Sarcocystis bradyzoite, (3) T. gondii tachyzoite, (4) N. caninum tachyzoite
Fig. 2.
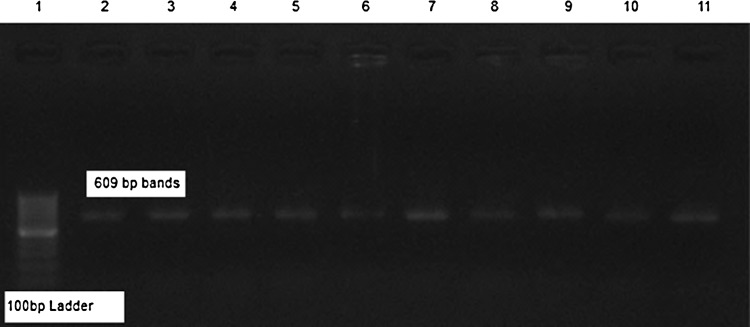
PCR analysis of different isolates of sheep micro and macro sarcocysts (2–11)
Gel electrophoresis of the PCR–RFLP by amplification of 18S rRNA gene from all isolates of micro and macro sarcocysts showed that restriction with TaqI enzyme produced 270 and 339 bp fragments and also, HindII produced 198 and 411 bp fragments (Figs. 3 and 4). These results represent the S. gigantea.
Fig. 3.
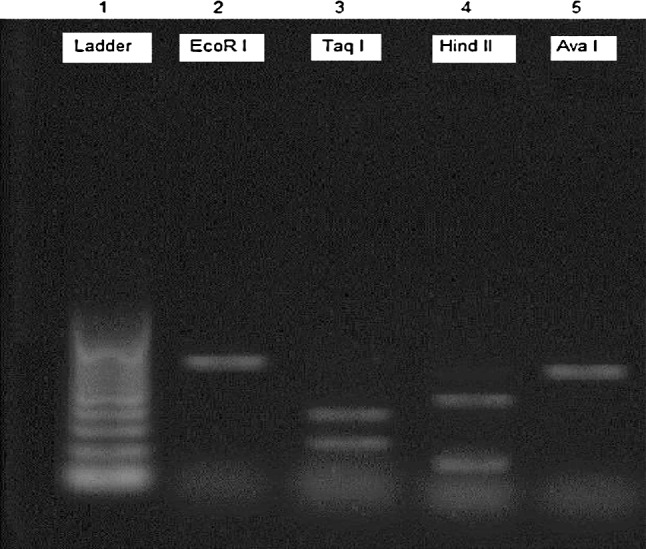
PCR–RFLP analysis of PCR products of the 18S rRNA gene from isolated microcyst of Sarcocystis sp. from examined sheep digested by EcoRI, TaqI, HindII and AvaI restriction enzymes
Fig. 4.
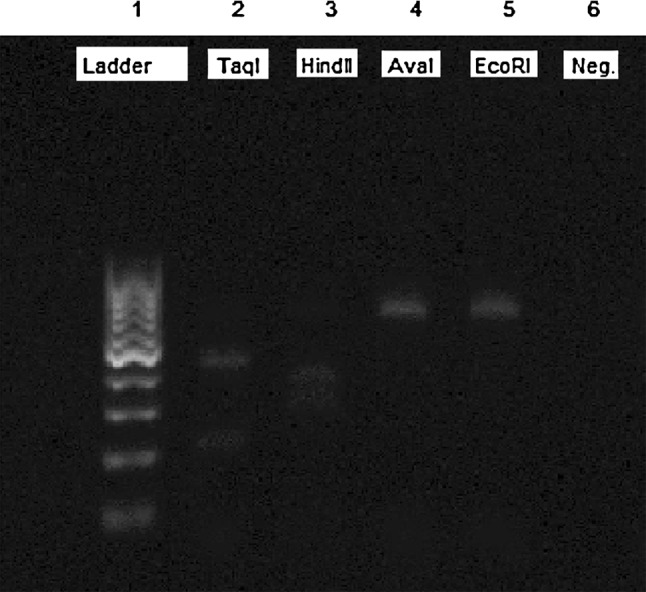
PCR–RFLP analysis of PCR products of the 18S rRNA gene from isolated macrocyst of Sarcocystis sp. from examined sheep digested by EcoRI, TaqI, HindII and AvaI restriction enzymes
Discussion
Thus far, four species of Sarcocystis have detected in sheep including S. tenella, S. arieticanis which transmitted by the felids and S. gigantea and S. medusiformis transmitted by canids (Dubey and Lindsay 2006; Adriana et al. 2008). Among these species S. tenella and S. arieticanis produce microscopic cysts and have pathogenic subsequences as an acute disease presented by abortion, fever, anemia and anorexia in early period of infection and then some chronic disorders maybe develop (Tenter 1995; Pescador et al. 2007). On the other side, macrocyst forming species, S. gigantea and S. medusiformis are considered nonpathogenic, but they can affect the meat quality and marketing and cause economic loss. Sheep maybe infected with these four species simultaneously (Tenter 1995; Dubey and Lindsay 2006).
At yet, various conventional procedures such as trichinoscopy, staining with methylene blue, dob-smear, digestion and histology have been employed for diagnosis of sarcocystosis in meat samples. These methods are genus-specific and just performable on slaughtered carcasses (Williams et al. 1990; Verhasselt et al. 1992; Holmdahl et al. 1993).
On the other hand, some serological assays including enzyme linked immunosorbent assay (ELISA) and indirect fluorescent antibody test (IFAT), based on bradyzoites derived from sarcocysts have been assessed for serological diagnosis of sarcocystosis in sheep as well as some other animals (Uggla and Buxton 1990; Moré et al. 2008; Moré et al. 2010). Since, the bradyzoites of different Sarcocystis spp. have very antigenic similarities and therefore they have considerable cross reactions with other Sarcocystis spp. and other related parasites (Moré et al. 2010), it is obvious that these serological methods have serious limitations for species diagnosis as well as the genus determination.
In recent years, molecular diagnostic techniques have been assessed for specific determination of Sarcocystis spp. (Yang and Zuo 2000; Yang et al. 2001). Among various genomic targets, the highly conserved 18S ribosomal subunit is used widely for species-specific detection of different protozoa as well as Sarcocystis spp. due to presentation of highly variable regions (Yang and Zuo 2000). On the other hand, many authors confirmed the 18S rRNA for firmly species-specific discriminating of sheep sarcocysts (Tenter et al. 1992, 1994; Ellis et al. 1995; Yang et al. 2001). Thus, the aim of our study was to determine the Sarcocystis species of sheep in Khouzestan and Lorestan provinces using PCR–RFLP in basis of amplification of18S rRNA. The sampling was from esophagus, diaphragm, intra-costal, abdomen and leg muscles of sheep. Reported by Buxton (1998) preferred organs for Sarcocystis spp. in the intermediate hosts are heart, diaphragm, and skeletal muscles which the parasite can remain in these sites all long life of the intermediate hosts.
The results of our study revealed that only S. gigantea was distinguished in Khouzestan and Lorestan sheep and multiple infections with Sarcocystis spp. was not present. (Paikari et al. 2008) reported this species for the first time in Qazvin city, Iran but they find also S. arieticanis in their study.
Also, in this work we did not detect any cross reactions with T. gondii and N. caninum. Homan et al. (2000) also reported no cross reaction between T. gondii and Sarcocystis spp. or N. caninum in PCR results.
From the other point of view, since our work demonstrates only the presence of S. gigantea, we can conclude that probably acute clinical sarcocystosis which is related to S. tenella and S. arieticanis is not considerably important in differential diagnosis of sheep disorders such as abortion for example induce by toxoplasmosis in the two different and economically important areas of Iran and because of the definitive hosts of S. gigantean are the cats, so we must concentrate on this animal in control and prevention programs of Sarcocystis infection in sheep. Although, in order to prohibit of probable transmission of other Sarcocystis species access of dogs to sheep herds must be denied (Adriana et al. 2008). These conclusions clearly reveal and support the importance of molecular investigations to identify the etiology of different diseases, especially when these methods are more accurate and economically superior in compare with other methods (Pescador et al. 2007).
Acknowledgments
This study is supported by the Shahid Chamran University for M.Sc thesis. The authors wish to thank the vice-chancellor for research of the Shahid Chamran University for the research.
References
- Adriana T, Mircean V, Blaga R, Bratu CN, Cozma V. Epidemiology and etiology in sheep sarcocystosis. Bull UASVM Vet Med. 2008;65:49–54. [Google Scholar]
- Buxton D. Protozoan infections (Toxoplasma gondii, Neospora caninum and Sarcocystis spp.) in sheep and goats: recent advances. Vet Res. 1998;29:289–310. [PubMed] [Google Scholar]
- Dubey JP, Lindsay DS. Neosporosis, toxoplasmosis, and sarcocystosis in ruminants. Vet Clin Food Anim. 2006;22:645–671. doi: 10.1016/j.cvfa.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Speer CA, Charleston WAG. Ultrastructural differentiation between Sarcocystis of Sarcocystishirsuta and S. hominis. Vet Parasitol. 1989;1989(34):153–157. doi: 10.1016/0304-4017(89)90177-5. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Speer CA, Fayer R. Sarcocystosis of animals and man. Boca Raton: CAC Press Inc; 1989. pp. 105–145. [Google Scholar]
- Ellis JT, Luton K, Baverstock PR, Whitworth G, Tenter AM, Johnson AM. Phylogenetic relationships between Toxoplasma and sarcocystis deduced from a comparison of 18S rDNA sequences. Parasitology. 1995;110:521–528. doi: 10.1017/S0031182000065239. [DOI] [PubMed] [Google Scholar]
- Erlich HA, Gelfand D, Sninsky JJ. Recent advances in the polymerase chain reaction. Science. 1991;252:1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- Gonzalez LM, Villalobos N, Montero E, Morales J, Alamo Sanz R, Muro A, Harrison LJS, Parkhouse RME, Garate T (2006). Differential molecular identification of Taeniid spp. and Sarcocystis spp. cysts isolated from infected pigs and cattle. Vet Parasitol 142:95–101 [DOI] [PubMed]
- Heckeroth AR, Tenter AM. Comparison of immunological and molecular methods for the diagnosis of infections with pathogenic sarcocystis spp in sheep. Tokai J Exp Clin Med. 1999;23:293–302. [PubMed] [Google Scholar]
- Heckeroth AR, Tenter AM. Development and validation of species-specific nested PCR for diagnosis of acute Sarcocystosis in sheep. Int J Parasitol. 1999;29:1331–1349. doi: 10.1016/S0020-7519(99)00111-3. [DOI] [PubMed] [Google Scholar]
- Holmdahl OJ, Mathson JG, Uggla A, Johansson KE. Oligonucleotide probes complementary to variable regions of 18S rRNA from Sarcocystis spp. Mol Cell Probes. 1993;7:481–486. doi: 10.1006/mcpr.1993.1071. [DOI] [PubMed] [Google Scholar]
- Homan WL, Vercammen M, De Braekeleer J, Verschueren H. Identification of a 200–300 fold repetitive 529 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int J Parasitol. 2000;30:69–75. doi: 10.1016/S0020-7519(99)00170-8. [DOI] [PubMed] [Google Scholar]
- Jehle C, Dinkel A, Sander A, Morent M, Romig T, Luc PV, De TV, Thai VV, Mackenstedt U. Diagnosis of Sarcocystis spp. in cattle (Bos taurus) and water buffalo (Bubalus bubalis) in Northern Vietnam. Vet Parasitol. 2009;23:314–320. doi: 10.1016/j.vetpar.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Maurer JJ. Rapid detection and limitations of molecular techniques. Annu Rev Food Sci Technol. 2011;2:259–279. doi: 10.1146/annurev.food.080708.100730. [DOI] [PubMed] [Google Scholar]
- McManus DP, Bowles J. Molecular genetic approaches to parasite identification: their value in diagnostic parasitology and systematics. Int J Parasitol. 1996;26:687–704. doi: 10.1016/0020-7519(96)82612-9. [DOI] [PubMed] [Google Scholar]
- Moré G, Basso W, Bacigalupe D, Venturini MC, Venturini L. Diagnosis of Sarcocystis cruzi, Neospora caninum, and Toxoplasma gondii infections in cattle. Parasitol Res. 2008;102:671–675. doi: 10.1007/s00436-007-0810-6. [DOI] [PubMed] [Google Scholar]
- Moré G, Bacigalupe D, Basso W, Rambeaud M, Venturini MC, Venturini L. Serologic profiles for Sarcocystis sp. and Neospora caninum and productive performance in naturally infected beef calves. Parasitol Res. 2010;106:689–693. doi: 10.1007/s00436-010-1721-5. [DOI] [PubMed] [Google Scholar]
- Motamedi GR, Dalimi A, Nouri A, Aghaeipour K. Ultrastructural and molecular characterization of Sarcocystis isolated from camel (Camelus dromedarius) in Iran. Parasitol Res. 2011;108:949–954. doi: 10.1007/s00436-010-2137-y. [DOI] [PubMed] [Google Scholar]
- Paikari HA, Dalimi AAH, Esmaeilzadeh M, Valizadeh M, Karimi GHR, Motamedi GHR, Abdi Goudarzi M. Identification of sarcocystis species of slaughtered sheep in qazvin. Modares J Med Sci. 2008;11:65–72. [Google Scholar]
- Pescador CA, Corbellini LG, de Oliveira EC, Bandarra PM, Leal JS, Pedroso PMO, Driemeier D. Aborto ovino associado com infecc¸a˜o por Sarcocystis sp. Pesq Vet Bras. 2007;27:393–397. [Google Scholar]
- Tenter AM. Current research on Sarcocystis species of domestic animals. Int J Parasitol. 1995;25:1311–1330. doi: 10.1016/0020-7519(95)00068-D. [DOI] [PubMed] [Google Scholar]
- Tenter AM, Baverstock PR, Johnson AM. Phylogenetic relationships of Sarcocystis species from sheep, goats, cattle and mice based on ribosomal RNA sequences. Int J Parasitol. 1992;22:503–513. doi: 10.1016/0020-7519(92)90151-A. [DOI] [PubMed] [Google Scholar]
- Tenter AM, Luton K, Johnson AM. Species-specific identification of Sarcocystis and Toxoplasma by PCR amplification of small subunit ribosomal RNA gene fragments. Appl Parasitol. 1994;35:173–188. [PubMed] [Google Scholar]
- Uggla A, Buxton D. Immune responses against Toxoplasma and Sarcocystis infections in ruminants: diagnosis and prospects for vaccination. Rev Sci Tech Int Epiz. 1990;9:441–462. doi: 10.20506/rst.9.2.502. [DOI] [PubMed] [Google Scholar]
- Verhasselt P, Voet M, Volckaert G. DNA sequencing by a subcloning-walking strategy using a specific and semi-random primer in the polymerase chain reaction. Int J Parasitol. 1992;28:1053–1060. doi: 10.3109/10425179209030960. [DOI] [PubMed] [Google Scholar]
- Williams JGK, Kubelik AR, Jivak KS, Rafalksi JA, Tingey SV. DNA polymorphisims amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZQ, Zuo YX. The new views of the researchers on cyst forming coccidia species including Sarcocystis by using the molecular biological techniques. Chin J Parasitol Parasit Dis. 2000;18:120–126. [Google Scholar]
- Yang ZQ, Zuo YX, Yao YG, Chen XW, Yang GC, Zhang YP. Analysis of the 18S rRNA genes of Sarcocystis species suggests that the morphologically similar organisms from cattle and water buffalo should be considered the same species. Mol Biochem Parasitol. 2001;115:283–288. doi: 10.1016/S0166-6851(01)00283-3. [DOI] [PubMed] [Google Scholar]


