Abstract
Background and objective
The advent of next-generation sequencing has significantly facilitated characterization of the oral microbiome. Despite great efforts in streamlining the processes of sequencing and data curation, upstream steps required for amplicon library generation could still influence 16S rRNA gene-based microbial profiles. Among upstream processes, DNA extraction is a critical step that could represent a great source of bias. Accounting for bias introduced by extraction procedures is important when comparing studies that use different methods. Identifying the method that best portrays communities is also desirable. Accordingly, the aim of this study was to evaluate bias introduced by different DNA extraction procedures on oral microbiome profiles.
Design
Four DNA extraction methods were tested on mock communities consisting of seven representative oral bacteria. Additionally, supragingival plaque samples were collected from seven individuals and divided equally to test two commonly used DNA extraction procedures. Amplicon libraries of the 16S rRNA gene were generated and sequenced via 454-pyrosequencing.
Results
Evaluation of mock communities revealed that DNA yield and bacterial species representation varied with DNA extraction methods. Despite producing the lowest yield of DNA, a method that included bead beating was the only protocol capable of detecting all seven species in the mock community. Comparison of the performance of two commonly used methods (crude lysis and a chemical/enzymatic lysis+column-based DNA isolation) on plaque samples showed no effect of extraction protocols on taxa prevalence but global community structure and relative abundance of individual taxa were affected. At the phylum level, the latter method improved the recovery of Actinobacteria, Bacteroidetes, and Spirochaetes over crude lysis.
Conclusion
DNA extraction distorts microbial profiles in simulated and clinical oral samples, reinforcing the importance of careful selection of a DNA extraction protocol to improve species recovery and facilitate data comparison across oral microbiology studies.
Keywords: DNA extraction, bias, oral microbiome
The oral cavity harbors complex microbial communities. Studies using various molecular techniques have helped unravel the diversity of the oral microbiome at different sites, providing new insights on how microbial communities change in relation to oral diseases (1–5). Nevertheless, the use of these culture-independent approaches has limitations related to biases introduced by DNA extraction, polymerase chain reaction (PCR), and sequencing (6–9). The influence of DNA extraction protocols on the characterization of microbial communities has been described as a key issue in studies of the human microbiome of the respiratory tract (10) and colon (11, 12) and in environmental samples (13). Regarding the oral microbiota, the importance of DNA extraction has also been discussed (6, 14); however, to our knowledge, there are no reports that systematically evaluate the effect of DNA extraction on the characterization of oral microbial communities. Thus, the aim of this study was to evaluate the performance of four different DNA extraction protocols on a mock microbial community and to compare the microbial content of supragingival plaque samples after applying two DNA extraction methods commonly used in oral microbiome studies (1, 5, 6, 15, 16).
Methods
Mock community assembly
Mock communities were prepared as previously described (6) by mixing seven representative oral bacteria (Streptococcus oralis 34, Streptococcus mutans NCTC 10449, Lactobacillus casei LR1, Actinomyces oris T14v, Fusobacterium ATCC 10953, Porphyromonas gingivalis ATCC 33277, and Veillonella parvula PK 1910). Bacteria grown in liquid cultures were quantified using a Petroff-Hausser counting chamber, diluted in PBS and mixed in equal proportions to a final concentration for each species of 107 cells µL−1. Cell mixtures (1 µL) were then centrifuged (3,300×g), pellets resuspended in 100 µL of TE buffer (10 mM Tris-HCl pH 7.4, 1 mM EDTA) and stored at −80°C.
Human sampling procedures
Seven subjects participated in this study. Subjects were enrolled at the Dental Clinic of the Faculty of Dentistry at the University of Chile, under an approved protocol. The individuals were systemically healthy, with no antibiotic use in the previous 3 months and were periodontally healthy as determined by no site with probing depths >3 mm and<10% of sites presenting bleeding on probing. A supragingival plaque sample was collected using a sterile curette from the lingual aspect of a first mandibular molar, placed in a vial with 100 µL of TE buffer and stored at −80°C until processing.
DNA extraction
Four DNA extraction methods, described below, were used in this study.
Crude chemical/enzymatic lysis method (C)
Tween 20 (Sigma-Aldrich, St. Louis, MO) at a final concentration of 0.5% v/v and Proteinase K (Promega, Madison, WI) at a final concentration of 0.2 mg mL−1 were added to samples, which were then incubated at 55°C for 2 h and then at 95°C for 5 min. One microliter of this lysate was directly used in PCR reactions. This protocol has been widely used for the characterization of oral microbial communities (1, 15, 16).
Chemical/enzymatic lysis+DNeasy Blood and Tissue kit (Q)
Samples were mixed with lysozyme (final concentration 20 mg mL−1) and incubated at 37°C for 30 min. Buffer AL (from the DNeasy Blood and Tissue kit, Qiagen Valencia, CA) and Proteinase K (to a final concentration of 1.23 mg mL−1) were then added and samples were incubated overnight at 56°C. The following day, samples were incubated for 5 min at 95°C. DNA isolation was then performed using the DNeasy Blood and Tissue kit (Qiagen) in accordance with the manufacturer's instructions and DNA was eluted in MD5 solution (MoBio laboratories, Carlsbad, CA). This protocol has been used by our group for the characterization of the subgingival, mucosal, and salivary microbiomes (5, 6).
Chemical/enzymatic lysis+boiling+DNeasy Blood and Tissue kit (QB)
This method consisted of a modification of the Q protocol, in which an additional step of heating at 100°C for 10 min was introduced immediately after Proteinase K inactivation. Boiling of samples has been previously shown to increase the representation of species that are difficult to lyse, such as Actinomyces (7).
Bead beating+Fast DNA Spin Kit (BB)
Samples were added to a bead beating matrix containing 0.4 g Lysing Matrix B (MP Biomedicals, Santa Ana, CA) supplemented with 1 g of very high density 0.5 mm yttria-stabilized zirconia (95% ZrO2+5% Y2O3) grinding media (Glen Mills Inc., Clifton, NJ). The FastDNA Spin Kit (MP Biomedicals) was then used according to the manufacturer's protocol (for yeast, algae or fungi) with three modifications: (1) decreased Cell Lysis Solution for Yeast (CLS-Y) to 800 µL to allow for sufficient air in the tube for thorough homogenization; (2) three homogenizations at speed 5.0 for 30 sec in the FastPrep-24 Instrument (MP Biomedicals) separated by 5 min on ice to keep samples cool (17), instead of a single homogenization step; (3) the addition of a second wash with Salt Ethanol Wash Solution (SEWS-M) to facilitate removal of lysing solutions, which we found to persist and form precipitates in eluted gDNA if only one wash was performed as in the protocol. We also used a pre-heated incubator (instead of a water bath or heat block) during DNA elution. The lysis solution used with this kit contains detergents and salts only. This protocol was investigated as a possible common one for both bacterial and fungal DNA extraction, since research from our laboratories (unpublished) has demonstrated its effectiveness in releasing DNA from fungi in oral samples.
DNA was isolated from replicate mock communities (independently prepared) using the four methods described above and from equally divided plaque samples using the C and Q methods. All DNA extraction procedures were performed in parallel with a negative control (TE buffer). DNA concentrations were measured using a NanoDrop Instrument (ThermoScientific, Willmington, DE).
Library construction, sequencing, and data processing
Amplicon libraries of 16S rRNA gene V1-V2 hypervariable regions were prepared in triplicate from each extracted sample, combined and sequenced using 454 Titanium chemistry (Roche, 454 Life Sciences, Branford, CT) according to protocols previously described (6). Mothur was used for processing of sequences (18), following the pipeline developed by Schloss et al. (19), as modified by our group (6). Individual sequences were assigned a taxonomy using mothur's version of the Ribosomal Database Project (RDP) classifier (20) and the Human Oral Microbiome Database (HOMD) (21, 22) and RDP database (version 9) as templates, with an 80% bootstrapping cutoff. For mock community analysis, sequences were binned according to their taxonomical assignment. For plaque samples, sequences were clustered into operational taxonomic units (OTUs) (at 97% similarity) and each OTU was assigned a taxonomy, which corresponded to the taxonomic assignment for the majority of sequences within the OTU. When a consensus assignment at the species level was not possible, the highest taxonomical order in which a consensus was reached is reported. In this case, the representative sequence for each OTU (middle sequence) was also compared to the HOMD and its taxonomy reported in parentheses if this comparison resulted in >97% homology to a HOMD Oral Taxon (OT). Sequences were submitted to the Short Reads Archive (SRP038999 for supragingival plaque samples and SRP039007 for mock communities).
Statistical analyses
Differences between the Q and C protocols when applied to supragingival plaque samples were evaluated using paired t-tests for α-diversity metrics and with Analysis of Molecular Variance (AMOVA) (23) for β-diversity comparisons. Differences in the relative abundance of OTUs, genera, and phyla were assessed with Wilcoxon Signed Rank tests with significance thresholds adjusted using the Benjamini-Hochberg false discovery rate method.
Results and discussion
Evaluation of four DNA extraction protocols using a mock community
DNA yield from mock communities varied according to the method used. DNA yield of samples processed with the C protocol could not be determined due to the nature of the product (crude lysate with no DNA purification step). The yield of DNA was 215–295 ng for the Q method, 210–330 ng for the QB method, and 75–95 ng for the BB method. These results are in agreement with those of Yuan et al. (24), who generally observed higher DNA yields from defined species using chemical and enzymatic lysis prior to DNA isolation than when these methods were supplemented with bead beating.
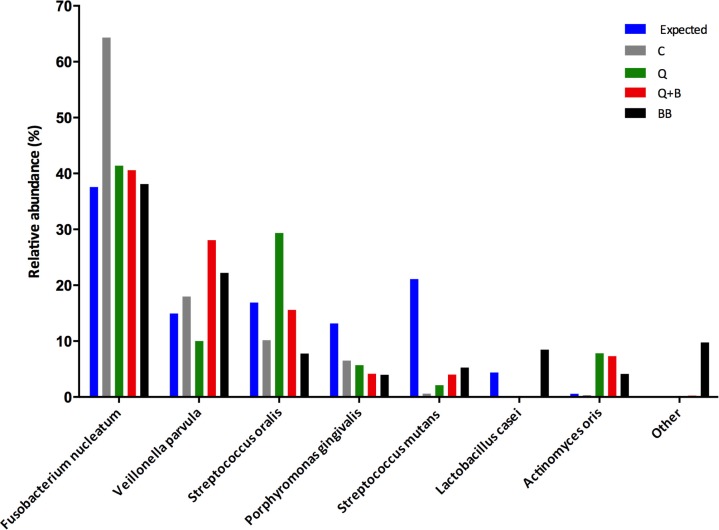
Samples extracted with all methods yielded PCR amplicons, which were sequenced using 454 technology with an average of 5,657±930 sequences per library. As expected, bacterial species representation varied according to DNA extraction procedure. As Fig. 1 shows, an over-representation of F. nucleatum was observed with the C protocol, while the rest of the species, with the exception of V. parvula, were under-represented with this method. The Q method resulted in over-representation of S. oralis, while other Firmicutes such as S. mutans, V. parvula, and L. casei were under-represented, as was P. gingivalis. The addition of a prolonged boiling step to the Q method (Q+B) had the most noticeable effect on V. parvula, which increased after boiling, and S. oralis, which decreased in comparison to Q alone. Importantly, the method that included bead beating (BB) was the only one able to detect all species in the mock community (L. casei only appeared with BB).
Fig 1.
Species representation in a mock community treated with four different DNA extraction methods prior to amplification and sequencing. Graph depicts the expected and obtained relative abundances for seven bacterial species after DNA extraction with a crude chemical/enzymatic lysis protocol (C), chemical/enzymatic lysis +DNeasy Blood and Tissue kit (Q), chemical/enzymatic lysis+boiling+DNeasy Blood and Tissue kit (QB), and bead beating+Fast DNA Spin Kit (BB). The expected relative abundance was calculated taking into account cell input, copy number of 16S RNA molecules for each species and known PCR bias as previously determined by our group (6)
These results are in agreement with other studies, which have observed that species proportions vary according to DNA extraction procedures (10, 12, 24). Use of lysozyme is thought to improve accuracy of community profiles, due to the ability of the enzyme to disrupt peptidoglycans in cell walls, thereby enhancing nucleic acid recovery from Gram-positives. Indeed, inclusion of lysozyme in our study (Q and Q+B methods) generally improved species representation compared to chemical lysis and Proteinase K treatment alone (C), although some Gram-positive species such as S. mutans and L. casei, and also the Gram-negative P. gingivalis, remained greatly under-represented. Use of additional enzymes may therefore be necessary to improve accuracy, as suggested by Yuan et al. (24), who found that a cocktail containing lysozyme, mutanolysin, and staphilysin performed better than lysozyme alone.
Despite low DNA yields, the only method that recovered all species was the BB protocol, which was designed by our group for efficient isolation of fungal DNA from oral samples (25). This method, however, also yielded a high proportion of contaminant sequences, all of them classified as Staphylococcus aureus (see ‘other’ column in Fig. 1). Contamination issues have been previously reported in sequenced mock communities and are attributed to reagent impurity (10). This result highlights the importance of including reagent controls in every run. Nevertheless, the inclusion of a bead beating step seems to be beneficial in improving accuracy in species representation. Our data are again in agreement with Yuan et al. (24), who reported that although the DNA yield of methods that included bead beating tended to be low, this procedure improved accuracy of community portrayals.
Evaluation of the supragingival plaque microbiome using methods Q and C
Seven supragingival plaque samples were split and processed with two DNA extraction methods (C and Q protocols), commonly used in the oral microbiology literature (1, 5, 6, 15, 16). This dataset was comprised of 99,883 sequences (after processing), with an average of 7,135±2,710 sequences per sample. A total of 436 OTUs (defined at 97% homology level) were found across all samples, with a range of 96–193 OTUs per sample. Richness coverage according to CatchAll (26), ranged from 54.8 to 84.4%.
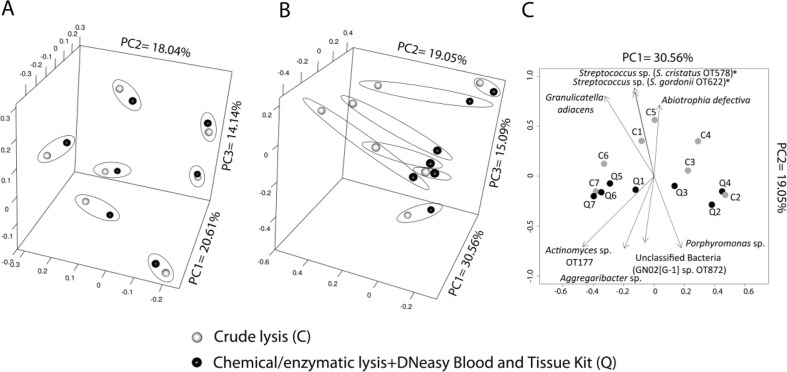
Pairwise comparisons of α-diversity metrics between samples processed with the C and Q methods showed no significant differences (data not shown). When samples were compared based on a β-diversity metric that evaluates distances among communities based on the presence/absence of OTUs (Jaccard metric), DNA extraction was shown not to be an influencing factor on community composition (AMOVA, P=0.968), with samples clustering by subject rather than by method (Fig. 2A). This result agrees with previous findings from studies evaluating the effect of DNA extraction in stool and human colon specimens demonstrating that inter-subject differences are greater than differences introduced by DNA extraction protocols (11, 12). On the contrary, comparison of the distance among samples based on the θYC metric (Fig. 2B), which takes into account species proportions, showed a tendency for clustering according to DNA extraction method, although AMOVA was still not significant (P=0.19). It is evident, however, that a trend existed for sample separation along axis 2, with C samples located towards the positive end. We then determined which taxa showed a significant correlation (Spearman) with axis 2, therefore driving separation of samples along this axis. The most significantly correlated OTUs are depicted in Fig. 2C. Surprisingly, OTUs that drove samples towards the positive end of axis 2 belonged to the Firmicutes and were Gram-positive species. Additionally, OTUs that drove samples towards the negative end of axis 2 included Gram-negative species from the Bacteroidetes and Proteobacteria and expectedly a Gram-positive OTU from the genus Actinomyces.
Fig 2.
Effect of two DNA extraction protocols on global profiles of supragingival plaque communities. Samples from seven subjects were split and processed with the crude chemical/enzymatic lysis (C) protocol (gray circles) or with the chemical/enzymatic lysis+DNeasy Blood and Tissue kit (Q) method (black circles). Principal coordinate analysis plots show distance among samples based on the Jaccard (A) and θYC (B) metrics, which reflect community membership and community structure, respectively. Samples belonging to the same subject were encircled. Panel C depicts a biplot also based on θYC distances with vectors indicating taxa responsible for separation of data points along axis 2. Shown OTUs had a P<0.01 before multiple comparison adjustment. An asterisk indicates OTUs that remained significant after multiple test correction
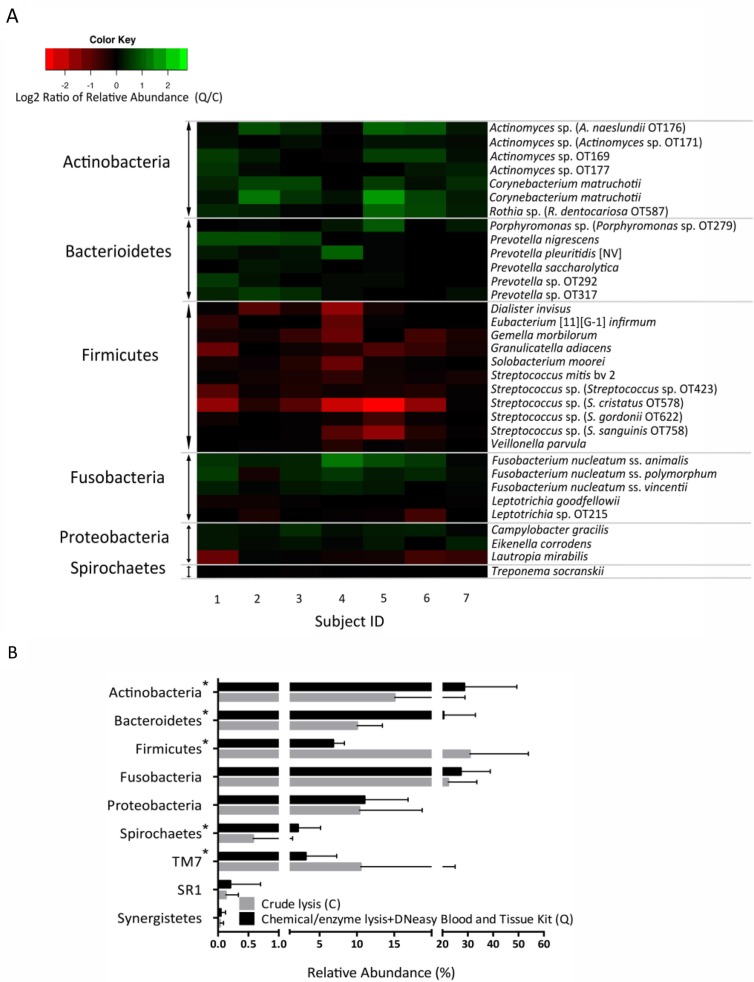
To further evaluate bias introduced by employing the C and Q methods on supragingival plaque microbial profiles, differences in the relative abundance of individual taxa were determined. First, however, we acknowledge that this study has a low sample size (seven subjects), which, in combination with the great inter-subject variability of the human microbiome, could understate differences. With awareness of this limitation, we present data on 33 OTUs that showed a P value of less than 0.05 when their relative abundances in both groups were compared (Fig. 3A). No OTU, however, remained significant after multiple testing adjustment, and therefore these data only indicate trends. In agreement with mock community results, Fig. 3A shows that the Q method improved the recovery of Actinomyces spp. Interestingly, Gram-negative species from the genera Prevotella and Porphyromonas were also better recovered after DNA extraction using the Q protocol. Moreover, in contrast with the over-representation of F. nucleatum in mock communities by the C method, F. nucleatum was seen at greater abundance after Q treatment in the more diverse supragingival samples.
Fig 3.
Effect of two DNA extraction protocols on relative proportions of OTUs and phyla in supragingival plaque. Panel A depicts a heatmap with 33 OTUs showing a P value of less than 0.05 when comparing relative abundances in paired plaque samples extracted via crude lysis (C) or with the chemical/enzymatic lysis+DNeasy Blood and Tissue kit method (Q). OTUs increased in C appear in red, and OTUs increased in Q appear in green. No OTU remained significant after multiple test adjustment. Bar graph in panel B shows relative abundance for all phyla detected. Phyla with a statistically significant differential representation in either C or Q are marked with an asterisk
Figure 3B shows the mean averages for the phyla detected, confirming the results from OTU data in that the Firmicutes were greatly over-represented by the crude lysis method (C), while the Bacteroidetes, Spirochaetes, and Actinobacteria were more efficiently disrupted by the method that included lysozyme treatment and column-based DNA isolation (Q). The cumulative proportions of TM7 also appeared to be higher in the C group. Our results are in agreement with other studies of the gut microbiome reporting Firmicutes as the phylum with the greatest variability according to DNA extraction procedures (11, 12). In the studies cited, however, Firmicutes recovery was improved by utilizing harsher extraction methods, while in our study their relative abundance was decreased with the harsher protocol, which also included lysozyme (Q). This result may be due to increased representation of more difficult to lyse species after using the Q method (e.g. Actinobacteria), rather than an actual change in lysis of Firmicutes, a suggestion that dominant oral Firmicutes (e.g. Mitis-group Streptococcus spp. and Veillonella spp.) may not be as difficult to lyse as Firmicutes of the lower intestinal tract (11, 12). It is also worth noting that a great number of oral Firmicutes species, such as Veillonella, Dialister; and Selenomonas, are Gram-negative and in theory better lysed by Q. These results highlight the importance of understanding bias introduced by DNA extraction protocols in samples specific to the niche of interest.
In conclusion, we have shown that DNA extraction has an impact on microbial profiling, supporting the concern that this step may introduce bias in the portrayal of community structure. Although the use of lysozyme and a column-based extraction procedure improved the recovery of certain species over a crude lysis protocol, additional steps such as bead beating, or inclusion of an enzymatic cocktail as recommended by others (24), may be necessary to maximize accuracy. No protocol, however, is completely unbiased; therefore, consistency among laboratories studying similar sites is required. Our results also raise a cautionary note regarding the possibility that some methods introduce high proportions of contaminant sequences, and therefore appropriate controls should be part of standard operating procedures in every laboratory.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors. This work was supported by grant 5RO1DE02157 from NIDCR-NIH.
References
- 1.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–32. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 3.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simon-Soro A, Pignatelli M, et al. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–85. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–25. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz PI, Dupuy AK, Abusleme L, Reese B, Obergfell C, Choquette L, et al. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol Oral Microbiol. 2012;27:182–201. doi: 10.1111/j.2041-1014.2012.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ, Jr, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72:2837–48. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan JL, Darling AE, Eisen JA. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS One. 2010;5:e10209. doi: 10.1371/journal.pone.0010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahn JH, Kim BY, Song J, Weon HY. Effects of PCR cycle number and DNA polymerase type on the 16S rRNA gene pyrosequencing analysis of bacterial communities. J Microbiol. 2012;50:1071–4. doi: 10.1007/s12275-012-2642-z. [DOI] [PubMed] [Google Scholar]
- 10.Willner D, Daly J, Whiley D, Grimwood K, Wainwright CE, Hugenholtz P. Comparison of DNA extraction methods for microbial community profiling with an application to pediatric bronchoalveolar lavage samples. PLoS One. 2012;7:e34605. doi: 10.1371/journal.pone.0034605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Momozawa Y, Deffontaine V, Louis E, Medrano JF. Characterization of bacteria in biopsies of colon and stools by high throughput sequencing of the V2 region of bacterial 16S rRNA gene in human. PLoS One. 2011;6:e16952. doi: 10.1371/journal.pone.0016952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang C, Ling F, Andersen GL, LeChevallier MW, Liu WT. Evaluation of methods for the extraction of DNA from drinking water distribution system biofilms. Microbes Environ. 2012;27:9–18. doi: 10.1264/jsme2.ME11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazarevic V, Gaia N, Girard M, Francois P, Schrenzel J. Comparison of DNA extraction methods in analysis of salivary bacterial communities. PLoS One. 2013;8:e67699. doi: 10.1371/journal.pone.0067699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schloss PD, Westcott SL. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol. 2011;77:3219–26. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010 doi: 10.1093/database/baq013. baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–91. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA. Redefining the Human Oral Mycobiome with Improved Practices in Amplicon-based Taxonomy: Discovery of Malassezia as a Prominent Commensal. PLoS One. 2014;9(3):e90899. doi: 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunge J. Estimating the number of species with CatchAll. Pac Symp Biocomput. 2011:121–30. doi: 10.1142/9789814335058_0014. [DOI] [PubMed] [Google Scholar]