Abstract
AIM: To assess the relationship between the P268S, JW1 and N852S polymorphisms and Crohn’s disease (CD) susceptibility in Zhuang patients in Guangxi, China.
METHODS: Intestinal tissues from 102 Zhuang [48 CD and 54 ulcerative colitis (UC)] and 100 Han (50 CD and 50 UC) unrelated patients with inflammatory bowel disease and 72 Zhuang and 78 Han unrelated healthy individuals were collected in the Guangxi Zhuang Autonomous Region from January 2009 to March 2013. Genomic DNA was extracted using the phenol chloroform method. The P268S, JW1 and N852S polymorphisms were amplified using polymerase chain reaction (PCR), detected by restriction fragment length polymorphism (RFLP), and verified by gene sequencing.
RESULTS: Heterozygous mutation of P268S in the NOD2/CARD15 gene was detected in 10 CD cases (six Zhuang and four Han), two Han UC cases, and one Zhuang healthy control, and P268S was strongly associated with the Chinese Zhuang and Han CD populations (P = 0.016 and 0.022, respectively). No homozygous mutant P268S was detected in any of the groups. No significant difference was found in P268S genotype and allele frequencies between UC and control groups (P > 0.05). Patients with CD who carried P268S were likely to be ≤ 40 years of age (P = 0.040), but were not significantly different with regard to race, lesion site, complications, and other clinical features (P > 0.05). Neither JW1 nor N852S polymorphisms of the NOD2/CARD15 gene were found in any of the subjects (P > 0.05).
CONCLUSION: P268S polymorphism may be associated with CD susceptibility in the Zhuang population in the Guangxi Zhuang Autonomous Region, China. In contrast, JW1 and N852S polymorphisms may not be related to CD susceptibility in these patients.
Keywords: Crohn’s disease, NOD2/CARD15, Single nucleotide polymorphisms
Core tip: In this study, P268S, JW1 and N852S polymorphisms of the NOD2/CARD15 gene were genotyped using the PCR-RFLP method and gene sequencing, and the presence of P268S in Guangxi Zhuang Crohn’s disease patients was identified. However, no JW1 or N852S mutants were found in this cohort.
INTRODUCTION
Inflammatory bowel disease (IBD) is a chronic recurrent inflammatory disease of the gastrointestinal tract and includes ulcerative colitis (UC) and Crohn’s disease (CD). In recent years, the incidence of IBD has increased in Western populations with an East-West gradient existing in Europe and is progressively increasing in Asia[1-3]. A recent systematic review revealed that the highest annual incidence of CD was 12.7 per 100000 person-years in Europe, 20.2 per 100000 person-years in North America, and 5.0 per 100000 person-years in Asia and the Middle East[4]. The etiology and pathogenesis of IBD are not completely clear, which involve a complex interaction of factors such as genetics, immunology, environment, and infection[5,6]. Several pathways may be crucial for intestinal homeostasis in IBD, for example barrier function, epithelial restitution, microbial defense, innate immune regulation, adaptive immunity regulation, reactive oxygen species (ROS) generation, and autophagy[7].
Genetic susceptibility to CD shows significant ethnic differences. A recent meta-analysis of multiple genome-wide association studies confirmed that 71 CD susceptibility loci were detected in a European population[8]. However, the majority of these genes could not be verified in the Asian region[9-11]. NOD2/CARD15 was the first verified predisposing gene for CD. Multiple single nucleotide polymorphisms (SNPs) of NOD2/CARD15 were shown to be significantly associated with CD in Caucasian populations[12-14]. Our previous studies confirmed that the R702W, G908R, and L1007fs SNPs of the NOD2 gene were not associated with CD and UC in a Chinese Zhuang population from Guangxi Zhuang Autonomous Region, China[15]. In recent years, some gene mutation sites of NOD2/CARD15 such as P268S, JW1, N852S, D113N, D357A, I363F, and L550V were shown to confer CD susceptibility[16-18]. The P268S SNP of the NOD2/CARD15 gene was also associated with Chinese Han CD susceptibility and its clinical features[19]. The JW1 SNP of the NOD2/CARD15 gene was shown to be associated with CD in Chinese Han, Malay, and Indians in Malaysia[17]. The N852S SNP of the NOD2/CARD15 gene was found to be significantly associated with CD in Ashkenazi Jewish populations[18]. However, there are no data on the correlation between the P268S, JW1, and N852S SNPs of the NOD2/CARD15 gene and the Chinese Zhuang CD population in the Guangxi Zhuang Autonomous Region.
In view of the differences in data regarding the correlation between key regulatory genes and IBD susceptibility, the purpose of the present study was to investigate whether the known gene SNPs (P268S, JW1 and N852S) of the NOD2/CARD15 gene determine susceptibility to CD in the Guangxi Zhuang population from the Guangxi Zhuang Autonomous Region, China. Guangxi has a large Zhuang population in which genetic diseases and genetic SNPs are unique. Therefore, research on the correlation between the P268S, JW1, N852S SNPs of the NOD2/CARD15 gene and CD in Chinese Zhuang patients from the Guangxi Zhuang Autonomous Region is needed.
MATERIALS AND METHODS
Specimen collection
Intestinal tissues from 102 Zhuang (48 CD and 54 UC) and 100 Han (50 CD and 50 UC) unrelated patients with IBD were collected at the Gastroenterology Department, First Affiliated Hospital of Guangxi Medical University, from January 2009 to March 2013. The control group included 72 Zhuang and 78 Han unrelated healthy individuals who did not have liver or gastrointestinal diseases. All patients had a well-established diagnosis of UC or CD based on the modified criteria framed by the World Gastroenterology Organization in 2010[20]. This study was approved by the hospital ethics committee and all the patients or their families provided written informed consent.
DNA extraction
Intestinal mucosa samples were digested using 450 μL of TES buffer (pH = 8.0) which consisted of Tris-HCl, ethylene diamine tetraacetic acid, and sodium chloride, 50 μL sodium dodecyl sulfate (10%), and 5 μL proteinase K (20 g/L) in a 56 °C water bath for 4-6 h. The supernatant was successively extracted by centrifugation at 12000 r/min for 10 min at 4 °C following the sequential addition of equal volumes of phenol, chloroform, and isoamyl alcohol (25:24:1), chloroform, and isoamyl alcohol (24:1). A white floc was precipitated from the final supernatant after the addition of 2.5 volumes of absolute ethanol and repeated aspiration. DNA was extracted from the white floc by centrifugation at 12000 r/min for 5 min at 4 °C after the addition of 75% ethanol. The DNA was dissolved by the addition of 50-120 μL of TE and stored at -20 °C.
Genotyping of P268S, JW1 and N852S
The primer sequences were as published elsewhere[18] and were synthesized by SHENGGONG Biotechnology Co., Ltd., Shanghai, China. PCR reaction mixture contained 2 μL DNA template, 1 μL each of forward and reverse primers (10 μmol/L), 6 μL H2O, and 10 μL of 2 × PCR Master Mix (TIANGEN Biotechnology Co., Ltd., Beijing, China). Reaction conditions consisted of an initial denaturation for 5 min at 94 °C, followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at different temperatures (Table 1) for 45 s, extension at 72 °C for 45 s, and a final extension at 72 °C for 10 min. All of the PCR products were electrophoresed on a 1.5% agarose gel with 1 × Tris-borate-EDTA buffer at 100 V for 30 min and then observed under ultraviolet illumination (Bio-Rad Gel Doc-2000, Hercules, CA, United States).
Table 1.
Polymerase chain reaction primers, annealing temperatures and polymerase chain reaction fragment sizes
| Mutation | Primer | Annealing temperature (°C) | PCR fragment size (bp) |
| P268S | F-TGCCTCTTCTTCTGCCTTCC | 60 | 422 |
| R-AGTAGAGTCCGCACAGAGAG | |||
| JW1 | F-TGCAGTTTTCTTGGGGAGAT | 59 | 220 |
| R-TGTACCTGATCCAGCCCAAT | |||
| N852S | F-CTGTTTGCATGATGGGGGG | 55 | 151 |
| R-CAGCCGTCAGTCAATTTGTAG |
PCR: Polymerase chain reaction.
The PCR products of P268S, JW1, and N852S SNPs of the NOD2/CARD15 gene were digested at 37 °C for 11 h with BamHI, XhoI, and AluI restriction enzymes, respectively (Fermentas, Pittsburgh PA, United States). The digestion reaction contained 5 μL of the PCR product, 2 μL of 10 × buffer, 1 μL of restriction enzyme, and 9 μL of H2O in a total of 17 µL. Following enzymatic digestion, the fragments were separated and visualized using gel electrophoresis (Yito Bio-Instrument Company Ltd., Shanghai, China) (Table 2).
Table 2.
Enzymes and gene polymorphism analysis
| Mutation | Base change | Enzyme |
Restriction fragment size (bp) |
|
| Wild-type | Mutant | |||
| P268S | C→T | BamHI | 422 | Heterozygote 422 + 247 + 175 |
| Homozygote 247 + 175 | ||||
| JW1 | C→T | XhoI | 125 + 95 | Heterozygote 220 + 125 + 95 |
| Homozygote 220 | ||||
| N852S | A→G | AluI | 151 | Heterozygote 151 + 129 + 22 |
| Homozygote 129 + 22 | ||||
The DNA mutative samples which were found by PCR-RFLP were reamplified. The products of each SNP were purified using a PCR purification kit (QIAGEN, Hilden, Germany) and sequenced using ABI 3730XL sequencer (Applied Biosystems, Foster, United States).
Statistical analysis
SPSS version 16.0 software was used for the statistical analysis, while comparisons of genotype and allelic frequencies among the different groups were performed using Fisher’s exact test. The Hardy-Weinberg equilibrium test was used to test the distributions of each mutation genotype frequency. Values of P < 0.05 were considered statistically significant.
RESULTS
PCR
All three SNPs of the NOD2/CARD15 gene were amplified by PCR, and the PCR products were then used for both RFLP analysis and gene sequencing. The target fragment sizes of the P268S, JW1, and N852S mutations were 422 bp (Figure 1A), 220 bp (Figure 1B), and 151 bp (Figure 1C), respectively.
Figure 1.
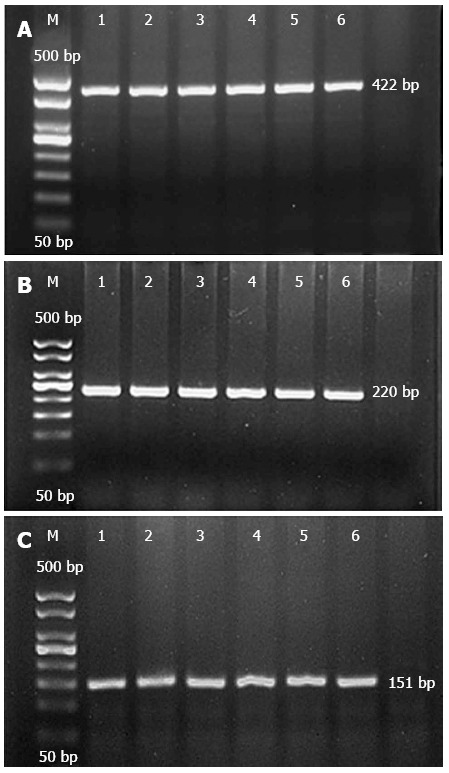
Electrophoresis of P268S, JW1, and N852S PCR products. A: P268S; B: JW1; C: N852S. M: Marker; 1, 2: Ulcerative colitis (UC), Crohn’s disease (CD) of Han; 3, 4: UC, CD of Zhuang; 5, 6: healthy controls.
PCR-RFLP
The PCR products of the P268S, JW1, and N852S mutations were digested using the BamHI, XhoI, and AluI enzymes, respectively. For P268S, a wild-type band of 422 bp was found in the majority of controls, CD patients, and UC patients, while heterozygous mutant bands of 422 bp, 247 bp, and 175 bp were found in six Zhuang CD cases, four Han CD cases, two Han UC cases, and one Zhuang healthy control, however, no homozygous mutants were detected (Figure 2A). For JW1, only wild-type bands of 125 bp and 95 bp were observed in all subjects (Figure 2B). Similarly, just one band of 151 bp was found in wild-type N852S in all subjects (Figure 2C), and no other mutant bands of JW1 or N852S were detected using PCR-RFLP fragment electrophoresis.
Figure 2.
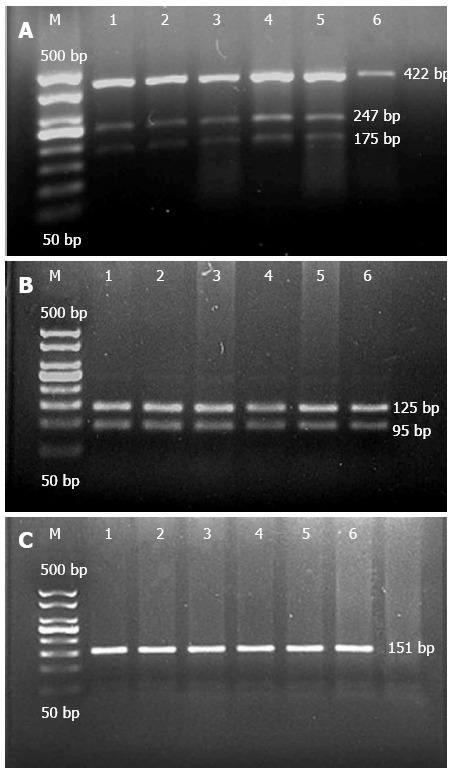
Electrophoresis of P268S, JW1, and N852S digestion products. A: M: Marker; 1-5: Heterozygote of P268S, 6: Wild-type of P268S. B: M: Marker; 1-6: Wild-type of JW1. C: M: Marker; 1-6: Wild-type of N852S.
DNA sequencing
The gene sequencing results of the P268S, JW1 and N852S variants were consistent with those found on PCR-RFLP. For both mutant P268S and JW1, it is a C to T substitution mutation, and for mutant N852S, it is an A to G substitution mutation. In our study, heterozygous (C/T) (Figure 3A) and wild-type (C/C) (Figure 3B) P268S were detected in controls, CD patients, and UC patients, but no homozygous P268S (T/T) was detected. However, only wild-type JW1 (C/C) (Figure 3C) and wild-type N852S (A/A) (Figure 3D) were observed, and no other types (C/T, T/T, A/G, G/G).
Figure 3.
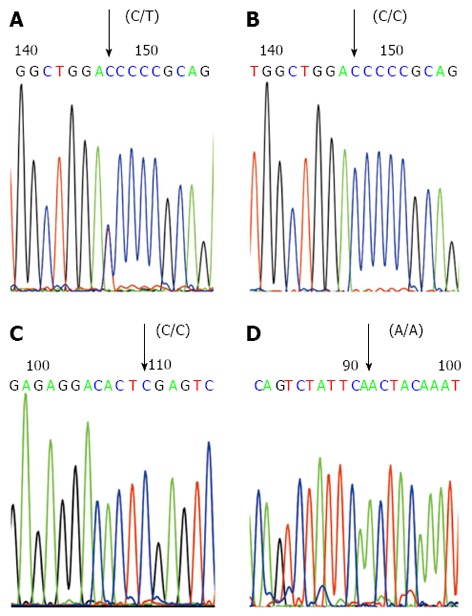
Gene sequencing analysis of P268S, JW1 and N852S polymerase chain reaction products. A: The forward sequencing map of the polymerase chain reaction (PCR) product of heterozygote P268S (C/T); B: The forward sequencing map of the PCR product of wild-type P268S (C/C); C: The forward sequencing map of the PCR product of wild-type JW1 (C/C); D: The forward sequencing map of the PCR product of wild-type N852S (A/A).
Distribution of genotype and allelic frequencies
The distributions of P268S, JW1, and N852S genotypes were in accordance with the Hardy-Weinberg equilibrium test results (P > 0.05). In our cohort, only the P268S heterozygous mutation was found in six (12.5%) of 48 Zhuang CD cases, four (8.0%) of 50 Han CD cases, 0 (0.0%) of 54 Zhuang UC cases, two (4.0%) of 50 Han UC cases, one (1.4%) of 72 Zhuang controls, and zero (0.0%) of 78 Han controls. No P268S homozygous mutations were found. The genotype and allelic frequencies of P268S in the Zhuang and Han populations with CD were significantly higher than those in the control group (aP = 0.016, cP = 0.022 under the genotypic model, and bP = 0.017, dP = 0.022 under the allellic model, respectively); however, the differences between the control group and the UC group were not statistically significant (P > 0.05). The JW1 and N852S genotypes were homozygous wild-type in all three groups of Zhuang and Han. No differences in genotype and allelic frequencies of JW1 and N852S were detected among the groups (P > 0.05) (Tables 3 and 4).
Table 3.
Distribution of genotype and allele frequencies of mutations in Crohn’s disease and ulcerative colitis patients compared with healthy controls in the Guangxi Zhuang population n (%)
| Mutant | Genotype | Allele | Control |
CD |
UC |
||
| P value | P value | ||||||
| P268S | aP1 | NS1 | |||||
| CC | 71 (98.6) | 42 (87.5) | 54 (100.0) | ||||
| CT | 1 (1.4) | 6 (12.5) | 0 (0.0) | ||||
| TT | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| T | 1 (0.7) | 6 (6.2) | cP2 | 0 (0.0) | NS2 | ||
| JW1 | NS1 | NS1 | |||||
| CC | 72 (100.0) | 48 (100.0) | 54 (100.0) | ||||
| CT | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| TT | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| T | 0 (0.0) | 0 (0.0) | NS2 | 0 (0.0) | NS2 | ||
| N852S | NS1 | NS1 | |||||
| AA | 72 (100.0) | 48 (100.0) | 54 (100.0) | ||||
| AG | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| GG | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| G | 0 (0.0) | 0 (0.0) | NS2 | 0 (0.0) | NS2 | ||
Comparisons of genotype frequencies;
Comparisons of allele frequencies.
P < 0.05 vs control using Fisher's exact test;
P < 0.05 vs control using Fisher's exact test. CD: Crohn’s disease; UC: Ulcerative colitis; NS: No significance.
Table 4.
Distribution of genotype and allele frequencies of mutations in Crohn’s disease and ulcerative colitis patients compared with healthy controls in the Guangxi Han population n (%)
| Mutant | Genotype | Allele | Control |
CD |
UC |
||
| P value | P value | ||||||
| P268S | bP1 | NS1 | |||||
| CC | 78 (100.0) | 46 (92.0) | 48 (96.0) | ||||
| CT | 0 (0.0) | 4 (8.0) | 2 (4.0) | ||||
| TT | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| T | 0 (0.0) | 4 (4.0) | dP2 | 2 (2.0) | NS2 | ||
| JW1 | NS1 | NS1 | |||||
| CC | 78 (100.0) | 50 (100.0) | 50 (100.0) | ||||
| CT | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| TT | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| T | 0 (0.0) | 0 (0.0) | NS2 | 0 (0.0) | NS2 | ||
| N852S | NS1 | NS1 | |||||
| AA | 78 (100.0) | 50 (100.0) | 50 (100.0) | ||||
| AG | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| GG | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||||
| G | 0 (0.0) | 0 (0.0) | NS2 | 0 (0.0) | NS2 | ||
Comparisons of genotype frequencies;
Comparisons of allele frequencies.
P < 0.05 vs control using Fisher's exact test;
P < 0.05 vs control using Fisher's exact test. CD: Crohn’s disease; UC: Ulcerative colitis; NS: No significance.
P268S genotype and clinical features of CD
A comparison between CD patients in Guangxi including Zhuang and Han with and without P268S mutations was performed. Eight of the ten patients with CD who carried the P268S mutation were ≤ 40 years of age (eP = 0.040), which suggested that the P268S mutation may be correlated with younger onset of CD in Guangxi patients. However, this mutation was not associated with lesion location, gender, ethnic groups, complications, or lesion severity (P > 0.05) (Table 5).
Table 5.
Clinical characteristics of Crohn’s disease patients in Guangxi with and without P268S mutations
| Phenotype | n | P268S+ | P268S- | P value |
| Age of onset | aP | |||
| ≤ 40 years | 45 | 8 (17.8) | 37 (82.2) | |
| > 40 years | 53 | 2 (3.8) | 51 (96.2) | |
| Location | NS | |||
| Ileum | 58 | 6 (10.3) | 52 (89.7) | |
| Colon/ileocolon | 40 | 4 (10.0) | 36 (90.0) | |
| Gender | NS | |||
| Male | 51 | 7 (13.7) | 44 (86.3) | |
| Female | 47 | 3 (6.4) | 44 (93.6) | |
| Ethnic groups | NS | |||
| Han | 50 | 4 (8.0) | 46 (92.0) | |
| Zhuang | 48 | 6 (12.5) | 42 (87.5) | |
| Comorbidities | NS | |||
| Luminal stenosis | 31 | 4 (12.9) | 27 (87.1) | |
| No luminal stenosis | 67 | 6 (9.0) | 61 (91.0) | |
| Severity | NS | |||
| Severe | 42 | 3 (7.1) | 39 (92.9) | |
| Mild-moderate | 56 | 7 (12.5) | 49 (87.5) |
NS: No significance. P268S+ Mutant P268S; P268S- Wild-type P268S.
P < 0.05 using Fisher's exact test.
DISCUSSION
The NOD2/CARD15 gene is located on chromosome 16q12. The protein that is encoded by the NOD2/CARD15 gene is highly expressed in intestinal mucosal Paneth cells[21]. The NOD2/CARD15 protein has two caspase recruitment domains and includes a nucleotide-binding domain and a leucine-rich repeat (LRR). The LRR may stimulate the secretion of defensin through the identification of bacterial muramyl dipeptide. The level of defensin decreased markedly in patients with CD and gene mutations[22]. LRR may cause a defensive inflammatory reaction by combining bacterial lipopolysaccharide and activating NF-κB[23]. NOD2/CARD15 is the first confirmed predisposing gene for CD, and the R702W, G908R, and L1007fs SNPs of the NOD2/CARD15 gene were found to be significantly associated with CD in Caucasian populations[12-14]. The mutant allele frequencies of these three mutations accounted for approximately 81% of the total CD mutations[24]. Nevertheless, these SNPs were not associated with CD in Japanese, Malaysian, Indian, or Hong Kong, Zhejiang, and Guangxi populations in China, and none of the patients with CD had heterozygous or homozygous variants of R702W, G908R, and L1007fs SNPs[10,11,15,17,25,26]. In addition, the R702W, G908R, L1007fs, P268S, and JW1 SNPs were not correlated with IBD patients in Turkey, instead, the R702W mutation was significantly lower in the IBD group (1.5%) than in the control group (4.8%) (P < 0.05)[27]. Thus, these findings indicate that the NOD2/CARD15 genotype distribution has significant ethnic differences.
This is the first study to report the P268S, JW1, and N852S mutations of the NOD2/CARD15 gene in patients with CD from the Guangxi Zhuang population of China, where the ethnic background is heterogeneous with Han, Zhuang, and other ethnic groups. In this study, the P268S mutation genotype of NOD2/CARD15 was found in some Zhuang and Han patients with CD and was detected only sporadically in healthy individuals and patients with UC in Zhuang and Han. The JW1 and N852S mutations of the NOD2/CARD15 gene were not detected in Guangxi Zhuang or Han patients with IBD.
In recent years, several studies have reported that the P268S mutation of the NOD2/CARD15 gene was found in Ashkenazi Jewish and Irish patients with CD[16,28]. The population-attributable risk of the P268S-JW1 haplotype was 15.1% in Jewish patients with CD[16]. Gasche et al[29] reported that the evolution of P268S occurred in the Middle East and that the mutant was associated with CD in Chinese Tu and Pakistani populations. The P268S SNP of the NOD2/CARD15 gene was also reported to be closely related to CD in Indian patients[17,30]. Similarly, the P268S mutant was confirmed to contribute to CD susceptibility and clinical features in a Han population in Guangdong, China[19]. However, that finding was not in accordance with those of Juyal et al[31], in which the P268S mutant of the NOD2/CARD15 gene was correlated with UC in North India. In our study, we confirmed that the P268S SNP may be involved in the susceptibility of Zhuang or Han patients to CD in Guangxi, China. Our results are in agreement with those from studies on Han and Tu patients with CD from other areas in China[19,29]. However, the P268S homozygous variant was found in Han patients in Guangdong, China, and was not detected in our study population, which may be due to racial heterogeneity or our relatively small sample size. Compared to Europeans (31.2%)[16], we found a lower frequency (12.5%) of mutant P268S in our Zhuang CD patients.
The N852S mutation of the NOD2/CARD15 gene was found to be significantly associated with CD in Ashkenazi Jewish populations[18]; however, since it did not appear as a haploid with R702W, G908R, and L1007fs of the NOD2/CARD15 gene, it is thought to be an independent risk factor for CD[32]. Our results indicated that N852S mutations of the NOD2/CARD15 gene were not detected in Guangxi Zhuang patients with IBD. The JW1 mutant of the NOD2/CARD15 gene was confirmed in Chinese Han in Malaysia[17]. However, we did not find any heterozygous or homozygous mutations of JW1 in the Chinese Zhuang population from the Guangxi Zhuang Autonomous Region. These two novel loci have rarely been reported in China, and further studies are necessary to explore these loci in a larger cohort in China. In summary, the differences in these results may be attributed to the differences in race, geography, environment, and population.
Several studies have proved that the NOD2/CARD15 gene is related to the clinical features of CD including onset location, age, complications, and disease severity[33-35]. It was reported that P268S was related to ileal lesions (P = 0.003), lumen stenosis (P = 0.007), and age ≤ 20 years (P = 0.028) in a Chinese Han population with CD from Guangdong, China[19]. In addition, Chua et al[17] reported that the JW1 mutant tended to correlate with luminal stenosis (P = 0.055) and age < 41 years (P = 0.095) in patients with CD in Malaysia. The results of the present study confirmed that P268S was only related to age ≤ 40 years (P =0.040) in CD patients from the Chinese Zhuang population in the Guangxi Zhuang Autonomous Region. No important relationship was detected between mutant P268S and location, gender, ethnic group, lumen stenosis, and severity of CD. Our results were not in agreement with those studies on Chinese Han patients with CD from Guangdong or patients with CD from Malaysia. This difference may be due to racial heterogeneity, geographic environment, and a relatively small sample size.
In conclusion, this study is the first to demonstrate the relationship between the P268S SNP of the NOD2/CARD15 gene and susceptibility to CD in a Zhuang population from the Guangxi Zhuang Autonomous Region, China. JW1 and N852S SNPs of the NOD2/CARD15 gene were not found in the Zhuang population. Thus, we emphasize that genetic predisposition may be vital in the pathogenesis of IBD. However, the power of this conclusion may be limited by the relatively small sample size in this study. Further studies investigating risk factors and genetic susceptibility to IBD in a larger cohort of patients and in different ethnic groups are needed.
COMMENTS
Background
Inflammatory bowel disease (IBD) is a multifactorial disease with different susceptibility genes in various races. The P268S, JW1, and N852S polymorphisms of NOD2/CARD15 have been confirmed in Crohn’s disease (CD) susceptibility in Chinese Han and Ashkenazi Jewish populations, but there are no reports of a correlation between these three polymorphisms and the Chinese Zhuang CD population in the Guangxi Zhuang Autonomous Region.
Research frontiers
Nucleotide-binding oligomerization domain containing 2/caspase-activation and recruitment domain gene 15 (NOD2/CARD15) is the first confirmed predisposing gene for CD, and the P268S mutation of the NOD2/CARD15 gene was found in Ashkenazi Jewish, Irish, Indian, Pakistani, and Chinese Han and Tu patients with CD, but not in CD in North India. The JW1 SNP of the NOD2/CARD15 gene was shown to be associated with CD in Chinese Han, Malay, and Indians in Malaysia. The N852S mutation of the NOD2/CARD15 gene was only found to be significantly associated with CD in Ashkenazi Jewish populations. The present study assessed whether these known SNPs were associated with IBD in Zhuang patients from Guangxi, China.
Innovations and breakthroughs
The Guangxi Zhuang Autonomous Region of China has the largest Zhuang population, thus genetic diseases and gene polymorphisms are unique. This study is the first to demonstrate the relationship between the P268S, JW1, and N852S polymorphisms of the NOD2/CARD15 gene and susceptibility to CD in the Zhuang population from the Guangxi Zhuang Autonomous Region, China.
Applications
The P268S polymorphism may contribute to CD susceptibility in the Zhuang population in the Guangxi Zhuang Autonomous Region, China. However, JW1 and N852S SNPs may be absent or rare in this population.
Terminology
NOD2/CARD15 is located on chromosome 16q12, and encodes a protein with homology to plant disease resistance-related gene products. Mutant NOD2/CARD15 responds to bacterial muramyl dipeptide and decreases NF-kappaB activation, and these results implicate NOD2/CARD15 in susceptibility to Crohn’s disease. Polymerase chain reaction-restriction fragment length polymorphism is a popular technique used in genetic analysis. It has been used for the detection of intraspecies as well as interspecies variation.
Peer review
This brief paper demonstrates the relationship between the P268S polymorphism in NOD2/CARD15 and Crohn’s disease susceptibility in an ethnic Zhuang population. While the scientific findings in this paper are limited, documentation of genetic variation in CD susceptibility among various ethnic and regional groups is useful.
Footnotes
Supported by Guangxi Graduate Education Innovation Project Fund, No.YCSZ2012035; the Natural Science Foundation of Guangxi Zhuang Autonomous Region, No. 0832009, No. 2012GXNSFAA053143; and Traditional Chinese Medicine Science Fund of Guangxi Zhuang Autonomous Region, China, No. GZPT1238
P- Reviewers: Diehl LJ, Soriano-Ursua M S- Editor: Wen LL L- Editor: Wang TQ E- Editor: Wang CH
References
- 1.Burisch J, Pedersen N, Cukovi X0107-Cavka S, Brinar M, Kaimakliotis I, Duricova D, Shonová O, Vind I, Avnstrøm S, Thorsgaard N, Andersen V, Krabbe S, Dahlerup JF, Salupere R, Nielsen KR, Olsen J, Manninen P, Collin P, Tsianos EV, Katsanos KH, Ladefoged K, Lakatos L, Björnsson E, Ragnarsson G, Bailey Y, Odes S, Schwartz D, Martinato M, Lupinacci G, Milla M, De Padova A, D’Incà R, Beltrami M, Kupcinskas L, Kiudelis G, Turcan S, Tighineanu O, Mihu I, Magro F, Barros LF, Goldis A, Lazar D, Belousova E, Nikulina I, Hernandez V, Martinez-Ares D, Almer S, Zhulina Y, Halfvarson J, Arebi N, Sebastian S, Lakatos PL, Langholz E, Munkholm P; for the EpiCom-group. East-West gradient in the incidence of inflammatory bowel disease in Europe: the ECCO-EpiCom inception cohort. Gut. 2014;63:588–597. doi: 10.1136/gutjnl-2013-304636. [DOI] [PubMed] [Google Scholar]
- 2.Thia KT, Loftus EV, Sandborn WJ, Yang SK. An update on the epidemiology of inflammatory bowel disease in Asia. Am J Gastroenterol. 2008;103:3167–3182. doi: 10.1111/j.1572-0241.2008.02158.x. [DOI] [PubMed] [Google Scholar]
- 3.Zheng JJ, Zhu XS, Huangfu Z, Shi XH, Guo ZR. Prevalence and incidence rates of Crohn’s disease in mainland China: a meta-analysis of 55 years of research. J Dig Dis. 2010;11:161–166. doi: 10.1111/j.1751-2980.2010.00431.x. [DOI] [PubMed] [Google Scholar]
- 4.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42; quiz e30. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 6.Zeng Z, Zhan L, Liao H, Chen L, Lv X. Curcumin improves TNBS-induced colitis in rats by inhibiting IL-27 expression via the TLR4/NF-κB signaling pathway. Planta Med. 2013;79:102–109. doi: 10.1055/s-0032-1328057. [DOI] [PubMed] [Google Scholar]
- 7.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Lin MJ, Zhan LL, Lv XP. Analysis of TLR4 and TLR2 polymorphisms in inflammatory bowel disease in a Guangxi Zhuang population. World J Gastroenterol. 2012;18:6856–6860. doi: 10.3748/wjg.v18.i46.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leong RW, Armuzzi A, Ahmad T, Wong ML, Tse P, Jewell DP, Sung JJ. NOD2/CARD15 gene polymorphisms and Crohn’s disease in the Chinese population. Aliment Pharmacol Ther. 2003;17:1465–1470. doi: 10.1046/j.1365-2036.2003.01607.x. [DOI] [PubMed] [Google Scholar]
- 11.Hirano A, Yamazaki K, Umeno J, Ashikawa K, Aoki M, Matsumoto T, Nakamura S, Ninomiya T, Matsui T, Hirai F, et al. Association study of 71 European Crohn’s disease susceptibility loci in a Japanese population. Inflamm Bowel Dis. 2013;19:526–533. doi: 10.1097/MIB.0b013e31828075e7. [DOI] [PubMed] [Google Scholar]
- 12.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 13.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 14.Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 15.Mei-Jiao Lin, Xiao-Ping Lv, Lan Chen, Ling-ling Zhan. Correlation of R702W,G908R and L1007fs polymorphisms of the NOD2/CARD15 gene with susceptibility to inflammatory bowel disease in Zhuang population in Guangxi, China. Shijie Huaren Xiaohua Zazhi. 2012;20:1210–1215. [Google Scholar]
- 16.Sugimura K, Taylor KD, Lin YC, Hang T, Wang D, Tang YM, Fischel-Ghodsian N, Targan SR, Rotter JI, Yang H. A novel NOD2/CARD15 haplotype conferring risk for Crohn disease in Ashkenazi Jews. Am J Hum Genet. 2003;72:509–518. doi: 10.1086/367848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chua KH, Hilmi I, Ng CC, Eng TL, Palaniappan S, Lee WS, Goh KL. Identification of NOD2/CARD15 mutations in Malaysian patients with Crohn’s disease. J Dig Dis. 2009;10:124–130. doi: 10.1111/j.1751-2980.2009.00374.x. [DOI] [PubMed] [Google Scholar]
- 18.Tukel T, Shalata A, Present D, Rachmilewitz D, Mayer L, Grant D, Risch N, Desnick RJ. Crohn disease: frequency and nature of CARD15 mutations in Ashkenazi and Sephardi/Oriental Jewish families. Am J Hum Genet. 2004;74:623–636. doi: 10.1086/382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv C, Yang X, Zhang Y, Zhao X, Chen Z, Long J, Zhang Y, Zhong C, Zhi J, Yao G, et al. Confirmation of three inflammatory bowel disease susceptibility loci in a Chinese cohort. Int J Colorectal Dis. 2012;27:1465–1472. doi: 10.1007/s00384-012-1450-6. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein CN, Fried M, Krabshuis JH, Cohen H, Eliakim R, Fedail S, Gearry R, Goh KL, Hamid S, Khan AG, et al. World Gastroenterology Organization Practice Guidelines for the diagnosis and management of IBD in 2010. Inflamm Bowel Dis. 2010;16:112–124. doi: 10.1002/ibd.21048. [DOI] [PubMed] [Google Scholar]
- 21.Lala S, Ogura Y, Osborne C, Hor SY, Bromfield A, Davies S, Ogunbiyi O, Nuñez G, Keshav S. Crohn’s disease and the NOD2 gene: a role for paneth cells. Gastroenterology. 2003;125:47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 22.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schäffeler E, Schlee M, Herrlinger KR, Stallmach A, Noack F, Fritz P, et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salucci V, Rimoldi M, Penati C, Sampietro GM, van Duist MM, Matteoli G, Saibeni S, Vecchi M, Ardizzone S, Porro GB, et al. Monocyte-derived dendritic cells from Crohn patients show differential NOD2/CARD15-dependent immune responses to bacteria. Inflamm Bowel Dis. 2008;14:812–818. doi: 10.1002/ibd.20390. [DOI] [PubMed] [Google Scholar]
- 24.Lesage S, Zouali H, Cézard JP, Colombel JF, Belaiche J, Almer S, Tysk C, O’Morain C, Gassull M, Binder V, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahurkar S, Banerjee R, Rani VS, Thakur N, Rao GV, Reddy DN, Chandak GR. Common variants in NOD2 and IL23R are not associated with inflammatory bowel disease in Indians. J Gastroenterol Hepatol. 2011;26:694–699. doi: 10.1111/j.1440-1746.2010.06533.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang ZW, Ji F, Teng WJ, Yuan XG, Ye XM. Risk factors and gene polymorphisms of inflammatory bowel disease in population of Zhejiang, China. World J Gastroenterol. 2011;17:118–122. doi: 10.3748/wjg.v17.i1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ince AT, Hatirnaz O, Ovünç O, Ozbek U. 1007fs, G908R, R702W mutations and P268S, IVS8+158 polymorphisms of the CARD15 gene in Turkish inflammatory bowel disease patients and their relationship with disease-related surgery. Dig Dis Sci. 2008;53:1683–1692. doi: 10.1007/s10620-007-0054-4. [DOI] [PubMed] [Google Scholar]
- 28.Arnott ID, Nimmo ER, Drummond HE, Fennell J, Smith BR, MacKinlay E, Morecroft J, Anderson N, Kelleher D, O’Sullivan M, et al. NOD2/CARD15, TLR4 and CD14 mutations in Scottish and Irish Crohn’s disease patients: evidence for genetic heterogeneity within Europe? Genes Immun. 2004;5:417–425. doi: 10.1038/sj.gene.6364111. [DOI] [PubMed] [Google Scholar]
- 29.Gasche C, Nemeth M, Grundtner P, Willheim-Polli C, Ferenci P, Schwarzenbacher R. Evolution of Crohn’s disease-associated Nod2 mutations. Immunogenetics. 2008;60:115–120. doi: 10.1007/s00251-008-0274-6. [DOI] [PubMed] [Google Scholar]
- 30.Ng SC, Tsoi KK, Kamm MA, Xia B, Wu J, Chan FK, Sung JJ. Genetics of inflammatory bowel disease in Asia: systematic review and meta-analysis. Inflamm Bowel Dis. 2012;18:1164–1176. doi: 10.1002/ibd.21845. [DOI] [PubMed] [Google Scholar]
- 31.Juyal G, Amre D, Midha V, Sood A, Seidman E, Thelma BK. Evidence of allelic heterogeneity for associations between the NOD2/CARD15 gene and ulcerative colitis among North Indians. Aliment Pharmacol Ther. 2007;26:1325–1332. doi: 10.1111/j.1365-2036.2007.03524.x. [DOI] [PubMed] [Google Scholar]
- 32.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung C, Colombel JF, Lemann M, Beaugerie L, Allez M, Cosnes J, Vernier-Massouille G, Gornet JM, Gendre JP, Cezard JP, et al. Genotype/phenotype analyses for 53 Crohn’s disease associated genetic polymorphisms. PLoS One. 2012;7:e52223. doi: 10.1371/journal.pone.0052223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vind I, Vieira A, Hougs L, Tavares L, Riis L, Andersen PS, Locht H, Freitas J, Monteiro E, Christensen IJ, et al. NOD2/CARD15 gene polymorphisms in Crohn’s disease: a genotype-phenotype analysis in Danish and Portuguese patients and controls. Digestion. 2005;72:156–163. doi: 10.1159/000088371. [DOI] [PubMed] [Google Scholar]
- 35.Weiss B, Shamir R, Bujanover Y, Waterman M, Hartman C, Fradkin A, Berkowitz D, Weintraub I, Eliakim R, Karban A. NOD2/CARD15 mutation analysis and genotype-phenotype correlation in Jewish pediatric patients compared with adults with Crohn’s disease. J Pediatr. 2004;145:208–212. doi: 10.1016/j.jpeds.2004.05.024. [DOI] [PubMed] [Google Scholar]


