Abstract
Isobaric tandem mass tags are an attractive alternative to mass difference tags and label free approaches for quantitative proteomics due to the high degree of multiplexing that can be performed with their implementation. A drawback of tandem mass tags are that the co-isolation and co-fragmentation of labeled peptide precursors can result in chimeric MS/MS spectra that can underestimate the fold-change expression of each peptide. Two methods (QuantMode and MS3) have addressed this concern for ion trap and orbitrap instruments, but there is still a need to solve this problem for quadrupole time-of-flight (Q-TOF) instruments. Ion mobility (IM) separations coupled to Q-TOF instruments have the potential to mitigate MS/MS spectra chimeracy since IM-MS has the ability to separate ions based on charge, m/z, and collision cross section (CCS). This work presents results that showcase the power of IM-MS to improve tandem mass tag peptide quantitation accuracy by resolving co-isolated differently charged and same charged peptides prior to MS/MS fragmentation.
Keywords: tandem mass tags, Di-Leu (dimethylated leucine), ion mobility, quantitation, Q-TOF
Introduction
Mass spectrometry (MS)-based proteomics has matured from a technique that could only identify single purified proteins to a technique that can now identify and quantify thousands of proteins at once.[1-4] Many different quantitation strategies are now commonly used in proteomics experiments. There are label-free techniques that measure protein abundance based on peptide ion intensities or spectral counts.[5-7] Mass-difference labeling approaches introduce light and heavy isotopic forms of an isotopic label into peptides to allow binary or tertiary comparisons to be made within the same experiment during MS analysis.[8-12] Isobaric labeling methods allow for the greatest amount of experiment multiplexing enabling quantification measurements of four samples[13, 14], six samples[15-17], eight samples[18], sixteen samples[19, 20], and even fifty-four samples[21] at the same time.
Each of these quantitative approaches has advantages and disadvantages. Label-free methods have the advantage of measuring peptides in their native state, but lack the high throughput of mass-difference and isobaric tagging methods. Mass-difference tags allow for binary or tertiary comparisons, but introduce increased complexity in MS acquisitions that can decrease the confidence and accuracy of quantitation and also limit the sampling depth of proteome. Isobaric tags have the advantage of multiplexing multiple samples together in one run without dramatically affecting MS complexity since quantitation is performed at the MS/MS level. The drawback of this method is that concurrent isolation of multiple precursor ions in the MS scan, termed isobaric interference, can create a chimeric MS/MS spectra leading to an underestimation of protein/peptide fold changes and compression of the quantitative data towards unity.[22-25]
Isobaric interference is one of the most difficult problems facing isobaric tandem mass tag quantification.[24] Simple and effective methods to deal with it have been proposed, such as offline pre-fractionation.[26] However, in exceedingly complex samples, such as those composed of multiple proteomes, pre-fractionation has been shown to make no significant improvement to average ratio accuracy.[27] Savitski et al. have recently characterized two solutions, one involving smaller precursor isolation windows and optimal fragmentation times.[28] Smaller isolation windows can provide a modest improvement to reporter ratios, but may also cause an identification reduction as large as 48.5%.[27] The second method estimates the amount of protein ratio compression using the amount of interference in the precursor scan.[29] While this is a sophisticated approach to the problem, its inherent limitation is that it does not remove interference from a sample. Interference is accounted for and its impact is estimated.
As a comparison to our method, we will focus on two acquisition-method based strategies that have been proposed in the literature. The first solution, named “QuantMode” by the authors, involves using proton-transfer gas phase reactions to reduce the charge state of the target species, separating it from the interfering ion, and performing higher-energy collision-induced dissociation (HCD) on the charge reduced target ion to improve quantification accuracy.[27] The second solution, commonly called the MS3 method, improves quantification accuracy by performing an additional round of tandem MS on a fragment ion in the m/z range 110 – 160 % relative to the precursor ion m/z and then measuring the isobaric tag ratios.[30] Both methods have documented success in the literature, but like all methods possess minor shortcomings. QuantMode works well for different charge precursor ions but can only resolve interference of precursor's with the same charge if the charge-reduced m/z falls outside the precursor isolation window, whereas the triple-stage MS performed by MS3 may produce reporters with greatly reduced intensity leading to decreased quantitative sensitivity.
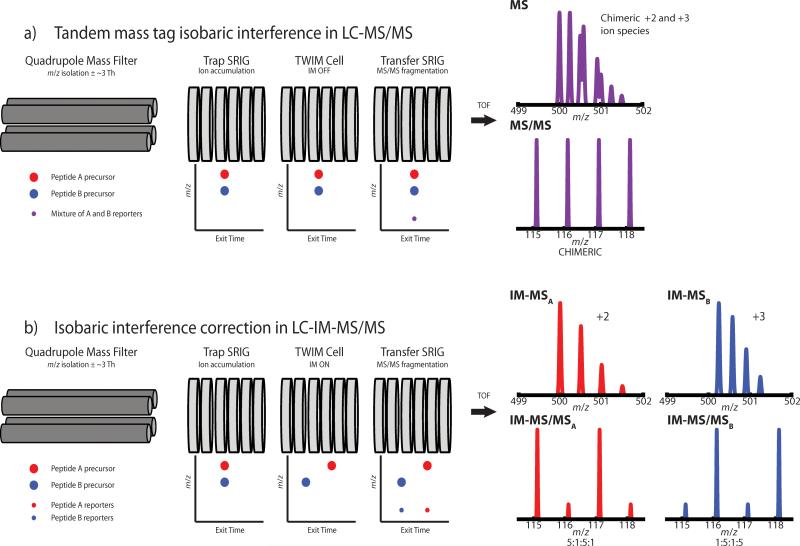
QuantMode and MS3 rely heavily on ion trap and orbitrap mass spectrometers. This leaves an unfilled niche for quadrupole-time-of-flight (QTOF) users wanting to improve isobaric quantitation accuracy. This work investigates the use of traveling wave ion mobility (TWIM)-MS on a QTOF mass spectrometer (SYNAPT G2) to improve peptide/protein quantification. Using ion mobility mass spectrometry (IM-MS), molecular ions can be separated by m/z, charge state, and collisional cross-section (shape and size).[31-34] Scheme 1a illustrates the consequences analyzing two hypothetical co-eluting peptides with similar m/z. Both Peptide A (z = +2) and Peptide B (z = +3) were labeled with N,N-dimethyl leucine (DiLeu) isobaric tags[14] and yield four MS/MS reporters ranging from 115 to 118 Da. Peptide A should display an expression ratio of 5:1:5:1, and Peptide B should display a ratio of 1:5:1:5. However, because their m/z are within a 3 Th isolation window, they are co-isolated and co-fragmented. The resulting chimeric reporter spectrum is the sum of reporter intensities from both precursors. Scheme 1b illustrates how IM separation prior to MS/MS can mitigate the interference. Peptide A and Peptide B can be separated in the IM drift cell and therefore enter the fragmentation cell separately. Each precursor will be individually fragmented and mass analyzed upon exiting the drift cell, thus mitigating the chimeracy observed in Scheme 1a.
Scheme 1.
To illustrate isobaric interference, two co-eluting DiLeu-labeled isobaric peptides, Peptide A (z = +2) and Peptide B (z = +3), are shown being analyzed on a SYNAPT G2 mass spectrometer. Peptide A has an expression ratio of 5:1:5:1 and Peptide B has an expression ratio of 1:5:1:5. (a) If IM separation is disabled, the two peptides will be co-fragmented resulting in an inaccurate reporter spectrum that is not representative of either peptide's true expression ratio. (b) On the other hand, if IM separation is enabled, the precursors will enter the Transfer stack-ring ion guide (SRIG) separately, allowing each peptide to be fragmented and mass analyzed in the TOF individually producing accurate reporter ion expression ratios for each peptide.
The parallel fragmentation of mobility-separated precursors, referred to as time-aligned parallel (TAP) fragmentation, is a vital component to our method.[35-37] It has proven to be a powerful method in proteomics,[38] and has been applied to several large-scale investigations.[39-41] IM-MS has also been used for quantitative analysis involving chemical tags, including isotopic labels[42, 43] and multiplex tags meant to induce mobility differences.[44-46] In 2011, further application to label-based quantification was shown when Waters published a technology briefing detailing the ability of a LC-IM-MS method to separate tandem mass tag (TMT)-labeled bovine serum albumin (BSA) peptides from non-tagged Escherichia coli peptides.[47] The brief report showed evidence that acquisitions not utilizing IM-MS resulted in chimeric MS/MS spectra whereas utilizing IM-MS cleaned up the MS/MS spectra to show purer MS/MS sequence fragments of TMT-labeled BSA peptides and untagged E. coli peptides. An interesting point not made in this report was whether IM-MS has the ability to clean up chimeric MS/MS spectra of two peptides with different isobaric tag ratios. Here, data-dependent analysis (DDA) with and without the use of precursor IM separation is investigated for its ability to correct quantitative inaccuracies caused by isobaric interference of differentially labeled peptides.
Materials and Methods
Chemicals and Materials
Anhydrous acetonitrile, triethylammonium bicarbonate (TEAB), 4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-mthlymorpholinium chloride (DMTMM), Trizma hydrochloride (Tris HCl, ≥ 99.0 %), Iodoacetamide (IAA), acetone (≥ 99.5 %), anhydrous N,N-Dimethylformamide (DMF, ≥ 99.8 %), Triton X-100 (laboratory grade), bovine albumin (BSA, ≥ 96 %), bovine apo-transferrin (≥ 97 %), bovine beta-lactoglobulin (≥ 90 %), horse myoglobin (≥ 90 %), bovine cytochrome C (≥ 95 %) were purchased from Sigma-Aldrich (Saint Louis, MO). Urea, formic acid (Optima LC/MS grade), acetonitrile, and water for LC/MS solvents (Optima LC/MS grade) were purchased from Fisher Scientific (Fair Lawn, NJ). Deionized water (18.2 MΩ·cm) was prepared with a Milli-Q Millipore system (Billerica, MA). DL-dithiothreitol (DTT), trypsin gold (mass spectrometry grade), and rLys-C (mass spectrometry grade) were purchased from Promega (Madison, WI). N-Methylmorpholine was purchased from TCI America (Portland, OR). Sodium dodecyl sulfate (SDS, ≥ 99.8 %) was purchased from US Biological (Marblehead, MA). The BCA protein assay kit and 1X protease and phosphatase inhibitor cocktail were purchased from Thermo Pierce (Rockford, IL). Yeast peptone dextrose (YPD) media was prepared in 1 L deionized water using 10 g yeast extract, 20 g peptone from Becton, Dickinson and Company (Sparks, MD), and 20 g D-(+)-glucose (≥ 99.5 %, Sigma-Aldrich). For solid phase extraction (SPE), Oasis HLB 1cc (10 mg) extraction cartridge were purchased from Waters (Milford, MA) and strong-cation exchange (SCX) spintips were purchased from Protea Biosciences (Morgantown, WV).
Yeast Lysate Protein Preparation
The yeast samples were prepared using a modified Mary Miller protocol[48]. Yeast strain s288c was inoculated with yeast peptone dextrose (YPD) media and shaken for 72 hrs. After shaking, an aliquot of 100 optical density units (ODU) of the yeast culture was transferred in a test tube containing 2% glucose. The yeast sample was concentrated using a Beckman-Coulter J6B centrifuge (Indianapolis, IN) at 2500 × g for 3 min. The media was decanted and the yeast pellet was flash frozen in liquid nitrogen and stored at −80 °C until further use. The yeast pellets were allowed to thaw on ice for 10 min before being resuspended in 1 mL of lysis buffer (50 mM Tris HCl, 0.1 % Triton X-100, 0.5 % SDS, pH = 8.0) and 1X protease and phosphatase inhibitor cocktail. The resuspended yeast was transferred to a new microcentrifuge tube and 200 μL of glass beads were added and vortexed for 2.5 minutes at 4 °C. The combined sample was inverted, and the bottom of the microcentrifuge tube was pierced with a hot 23 gauge needle. The tube was placed atop a 5 mL culture tube and the sample was centrifuged at 2500 × g for three minutes at 4 °C. The collected protein lysate from the pierced microcentrifuge tube was centrifuged at 3000 × g for an additional 5 min and the supernatant was stored at −80 °C until further use.
Because detergents can be detrimental to MS analysis, yeast proteins (~ 2mg per sample) were purified from cell lysis buffer by acetone precipitation. Four parts −20 °C acetone were added to one part yeast protein, the microcentrifuge tube was inverted three times, and protein precipitation was allowed to proceed for 3 hrs at −20 °C. The samples were then centrifuged at 13000 × g for 5 min and the supernatant was removed leaving a precipitated protein pellet. The pellet was reconstituted in 100 μL 8 M urea, 50 mM Tris HCl (pH = 8) and a BCA protein assay was run. The reconstituted yeast protein was divided into 400 μg aliquots for digestion.
Mammalian Protein Mixture Preparation
The five protein equimolar mix was prepared by weighing out 1 mg of BSA, apo-transferrin, beta-lactoglobulin and 0.5 mg of myoglobin, and cytochrome C and resuspending each in 84.5 μL, 75.5 μL, 294 μL, 171.25 μL, and 250 μL 8 M urea, 50 mM Tris HCL (pH = 8), respectively. Eight μL of each reconstituted protein were combined to give approximately 267 μg that was used for digestion.
Protein Digestion
The yeast proteins and the five mammalian protein mix were digested separately using identical protocols. Cysteine residues were reduced by addition of 4 μL 50 mM DTT and incubated for 1 hr at room temperature, then alkylated by addition of 5 μL 85 mM IAA for 20 min in the dark at room temperature. An additional 2 μL aliquot of 50 mM DTT was added to each sample to quench the alkylation reaction. Proteins were digested with rLys-C (1:100 enzyme:protein) for 2 hrs at 37 °C. Prior to trypsin digestion, each digest was diluted with 50 mM Tris HCl (pH = 8) to reduce the urea concentration to ≤ 1 M. Trypsin digestion (1:50 enzyme:protein) was carried out in a 37 °C water bath for 16 hrs. Each digest was acidified to pH = 2 by addition of 10 % formic acid(aq). Each sample was desalted using Oasis HLB, 1cc (10 mg) extraction cartridges following the manufacturer's protocol. The eluted peptides were dried down by vacuum centrifugation using a speedvac (Thermo Scientific, Waltham, MA). Peptides were resuspended in 0.5 M TEAB to a concentration of 5 μg·μL−1.
DiLeu Labeling
DiLeu isobaric tags are isotopically encoded N,N-dimethyl leucines. They were activated by triazine esterification and then attached to primary amines on peptide N-termini and lysine residues. For more details regarding the DiLeu reagent, see Xiang et. al. (2010).[14] Four 50 μg aliquots of yeast protein digest and four 50 μg aliquots of the five protein mix digest were prepared. Each aliquot was labeled with either the 115, 116, 117, or 118 DiLeu label. For each labeling reaction, 1 mg of dried DiLeu label was activated with 50 μL of activation solution (1.86 mg DMTMM, 0.74 μL NMM, 51.5 μL dried DMF) for 1 hr at room temperature, shaking. After 1 hr, 20 μL anhydrous acetonitrile and 25 μL of the appropriate activated DiLeu label were added to each 50 μg protein digest aliquot. Labeling was carried out for 2 hrs at room temperature, shaking. The labeling reaction was quenched by addition of 100 μL deionized water, and the samples were shaken at room temperature for an additional 30 min. The labeled peptide solutions were concentrated to dryness using a speedvac.
Each dried, labeled peptide sample was resuspended in 100 μL of Protea Biosciences SCX resuspension buffer. Alternatively labeled (115, 116, 117, or 118) yeast protein digest samples were aliquoted and combined to give mass ratios of 1:5:1:5 for each respective reporter ion channel. Alternatively labeled five mammalian protein mix digest samples were aliquoted and combined to give mass ratios of 5:1:5:1. Residual labeling chemicals were then removed from each combined sample using SCX spintips following the manufacturer's protocol. The eluate was dried down by speedvac, resuspended in 500 μL in 0.1 % formic acid(aq) (v/v), and desalted using Oasis HLB, 1cc (10 mg) extraction cartridges following the manufacturer's protocol. The eluate was dried down by speedvac and resuspended in 3 % acetonitrile/0.1 % formic acid(aq) (v/v) so that the concentration of each sample was approximately 0.5 μg·μL−1 assuming minimal loss from sample preparation steps. Finally, a mixture of the labeled yeast protein digest and five protein mix digest was prepared by adding one part five protein mix digest to five parts yeast protein digest.
Mass Spectrometry
A Waters SYNAPT G2 QTOF mass spectrometer coupled to a Waters nanoAcquity UPLC was used for nano-liquid chromatography electrospray ionization tandem mass spectrometry (nanoLC-ESI-MS/MS) analysis. Mobile phase A was water in 0.1% FA(aq) (v/v), and mobile phase B was ACN in 0.1% FA(aq) (v/v). For each analysis, approximately 800 ng of DiLeu labeled protein digest was loaded onto a Symmetry C18 nanoAcquity trap column (180 μm × 20 mm, 5 μm) at a flow rate of 5 μL·min−1 of 97% mobile phase A for 3 min. Peptides were separated using a 1.7 μm BEH C18 75 μm × 100 mm column with a 60 min gradient. Mobile phase B was linearly ramped from 9 to 35 % B over 60 min. The flow rate was 300 nL·min−1 and the column temperature was 35 °C. Electrospray emitter tips were prepared in house from 75 μm i.d., 360 μm o.d. capillary tubing (Polymicro Technologies, Phoenix, AZ) using a Sutter P-2000 laser capillary puller (Novator, CA).
Data were acquired in resolution mode using DDA and mobility DDA with a precursor isolation window of ~3 Th. The capillary voltage was set to 2.80 kV, sampling cone voltage to 30 V, extraction cone voltage to 4.0 V, and source temperature to 70 °C. A 1 Hz survey scan was followed by 3 MS/MS scans of the top 3 ions with charge states +2, +3, or +4. Dynamic exclusion was set to 60 seconds. Mobility DDA utilized high purity N2 as the drift gas in the TWIM cell. Pressures in the helium cell and TWIM cell (also called IM-MS cell) were 1.46 × 103 mbar and 3.61 mbar, respectively. Trap DC bias was 48.0 V, IM-MS wave velocity was 1000 ms−1, IMS wave height was 40 V, and IMS wave delay was 450 μs. A lookup table was created and optimized to use specified MS/MS collision energies at specific drift times. The look-up table applies a specific collisional energy to the Transfer cell based on the IM-MS bin. Assuming a rough proportionality between m/z and drift time, collision energies can be tailored to specific m/z. Further details can be found in Figure S1.
Data Analysis
Acquired data were analyzed using MassLynx 4.1 software and DriftScope (version 2.2) software. The LC-MS peak and MS/MS scans of select peptides were manually extracted as 2-D m/z versus drift time data in DriftScope. Reporter ion ratios were then calculated manually in MassLynx. Reporter ion spectra were smoothed once across ± 2 channels. Data were then median centered using a minimum peak width at half height of 4 channels. The centered spectra were based on peak areas. Centroid intensities of the reporter ions were placed into an Excel spreadsheet that calculated the reagent purity-corrected isobaric tag ratios using correction factors and solving four equations simultaneously as outlined previously.[49] A detailed description of the purity-correction calculations can be found on Figure S2.
Mammalian peptides exhibiting isobaric interference were identified by precursor accurate mass matching ( < 20 ppm) to tryptic peptides predicted by Protein Prospector and manual MS/MS annotation of centroid fragment ions ( < 0.05 Th accuracy). The annotated spectra are displayed in Figures S3 – S5. No yeast peptide sequences were identified.
Experimental Plan
The experimental workflow and design is shown in Figure S6. The concept is to create easily identifiable isobaric interference and to maximize its occurrence. The approach is similar to that used by Wegner et al.[27] and Ting et al.[30]. All labeled mammalian peptides should theoretically yield a 5:1:5:1 reporter ratio. By mixing the mammalian peptides with an entire proteome yielding the opposite 1:5:1:5 ratio, the sum of reporter intensities from mutually interfering peptides will trend towards 1:1:1:1. If the reporter ratios extracted at the drift times of interfering parents are closer to 5:1:5:1 and 1:5:1:5 than reporters without IM extraction, it is evidence that IM-MS can partially mitigate isobaric interference. Yeast was selected as the interfering proteome because of its low protein sequence homology to mammals, thus removing some ambiguity when identifying mammalian peptides.
Results
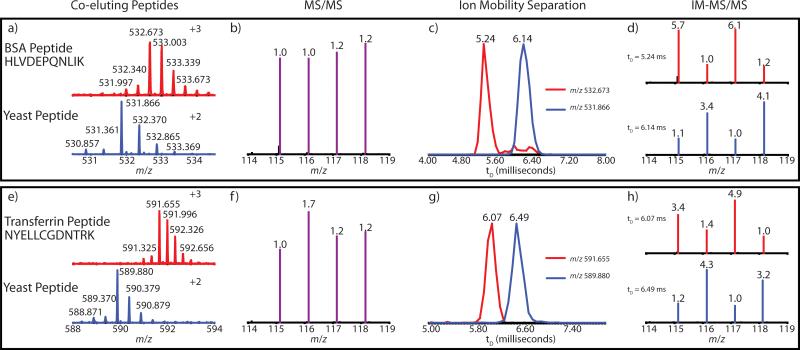
Figure 1 shows the LC-IM-MS/MS analysis of two pairs of co-eluting peptides from mammalian and yeast proteins. Each pair contains peptide ions with different charge states (z = +2, +3). In Figure 1a, the triply charged tryptic BSA peptide HLVDEPQNLIK (m/z 532.673) and doubly charged yeast peptide (m/z 531.866) were co-isolated and co-fragmented to produce the 1.0 : 1.0 : 1.2 : 1.2 chimeric reporter ion spectrum (Figure 1b). Figure 1c shows the extracted ion drift time (tD) distributions for each precursor. The BSA peptide and yeast peptide display apex drift times at 5.24 ms and 6.14 ms, respectively. Each trace was normalized to its own maximum. Figure 1d shows the reporter ratios extracted at the drift times of the BSA and yeast precursors. The ratio at 5.24 ms was 5.7 : 1.0 : 6.1 : 1.2, and the ratio at 6.14 ms was 1.1 : 3.4 : 1.0 : 4.1. Figures 1e - 1h show analogous spectra for the second peptide pair, triply charged transferrin tryptic peptide NYELLCGDNTRK (m/z 591.655) and a doubly charged yeast peptide (m/z 589.880). The initial reporter ratios were 1.0: 1.7: 1.2: 1.2, indicating the possibility of interference. The apex drift time for the transferrin peptide was 6.07 ms and the apex drift time of the yeast peptide was 6.49 ms. The extracted reporter ratios at 6.07 ms and 6.49 ms were 3.4: 1.4: 4.9: 1.0 and 1.2: 4.3: 1.0: 3.2, respectively.
Figure 1.
Example of IM separation improving the quantitative analysis of (a-d) BSA tryptic peptide HLVDEPQNLIK and (e-h) transferrin tryptic peptide NYELLCGDNTRK. In each case, MS/MS without IM separation produced chimeric reporter spectra approaching unity (b, f). After IM separation (c, g), reporter intensities were extracted at drift times of the precursors to produced corrected reporter spectra (d, h). The theoretical ratios for mammalian and yeast peptides were 5:1:5:1 and 1:5:1:5, respectively. IM traces were normalized to their own maximums, and all displayed reporter ratios were obtained from a purity correction algorithm applied to the centroid raw data.
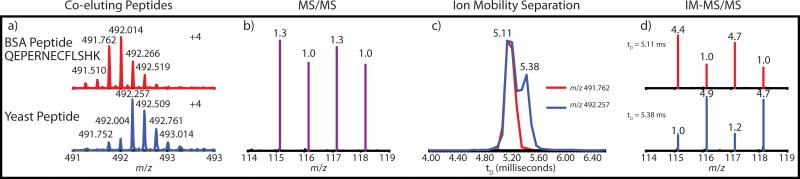
In Figure 2, interfering peptides of the same charge state (z = +4) were analyzed. Tryptic BSA peptide QEPERNECFLSHK (m/z 491.762) and a yeast peptide (m/z 492.257) yield a chimeric reporter ion spectrum with a 1.3: 1.0: 1.3: 1.0 ratio. In Figure 2c, the apex drift time of the BSA peptide was 5.11 ms. The yeast peptide distribution also displays a peak at 5.11 ms, but the true apex was attributed to 5.38 ms. The reporter ratios extracted at 5.11 ms and 5.38 ms were 4.4: 1.0: 4.7: 1.0 and 1.0: 4.9: 1.2: 4.7, respectively.
Figure 2.
IM was used to separate interfering precursors of the same charge (z = +4), tryptic BSA peptide QEPERNECFLSHK and a yeast peptide. MS/MS without IM produced a chimeric reporter spectra approaching unity (b). After IM separation (c), reporter intensities were extracted at drift times of the precursors to produce improved quantitative reporter ion spectra (d). The theoretical ratios for mammalian and yeast peptides were 5:1:5:1 and 1:5:1:5, respectively. IM traces were normalized to their own maximums, and all displayed reporter ratios were obtained from a purity correction algorithm applied to the centroid raw data.
The summaries of the isobaric interference examples, as well as additional considerations involving fold-change, are contained in Table 1. Reporter ion ratios 115/116 and 117/118 were considered independently. Each experimental ratio with IM (IM-MS/MS) and without IM (MS/MS) was compared to the theoretical ratio. The “Log2 Threshold” refers to whether or not an experimental ratio was greater than 1.5 on a Log2 scale. A “yes” or “no” answer is proceeded by the actual ratio on a Log2 scale. A “yes” signifies that IM improved quantitative accuracy. An additional mammalian peptide in Table 1 at m/z 499.316 was clearly experiencing isobaric interference (Figure S7). However, the peptide's mass did not match to any tryptic peptide, single, or double miscleavage for the five proteins within 20 ppm, and MS/MS fragments were not sufficient for full de novo sequencing. The extracted reporter ion ratios unambiguously identify its origin as the mammalian sample, and we hypothesize that the peptide was a fragment from a low abundance protein in the BSA sample (~96% purity) or a peptide that was unintentionally modified during sample preparation.
Table 1.
This table summarizes the examples of isobaric interference mitigation by IM. MS/MS and IM-MS/MS refer to purity-corrected ratios without and with IM separation, respectively. The “Log2 Threshold” refers to whether or not a ratio is greater than 1.5 on a Log2 scale, a common threshold for determining up-regulation.
| Mammalian peptides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Target (m/z, z) | Interferant (m/z, z) | 115/116 Ratio | Log2 Threshold Met? | 117/118 Ratio | Log2 Threshold Met? | ||||||
| Theoretical | MS/MS | IM-MS/MS | MS/MS | IM-MS/MS | Theoretical | MS/MS | IM-MS/MS | MS/MS | IM-MS/MS | ||
| 532.67, 3 | 531.87, 2 | 5.0 | 1.0 | 5.7 | no (0.0) | yes (2.5) | 5.0 | 0.9 | 5.1 | no (−0.2) | yes (2.4) |
| 591.66, 3 | 589.88, 2 | 5.0 | 0.6 | 2.5 | no (−0.7) | no (1.3) | 5.0 | 1.0 | 4.9 | no (0.0) | yes (2.3) |
| 491.76, 4 | 492.26, 4 | 5.0 | 1.3 | 4.4 | no (0.4) | yes (2.1) | 5.0 | 1.3 | 5.7 | no (0.4) | yes (2.5) |
| 499.32, 3 | 499.36, 2 | 5.0 | 0.9 | 3.1 | no (−0.2) | yes (1.6) | 5.0 | 0.8 | 4.3 | no (−0.3) | yes (2.1) |
| Yeast peptides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Target (m/z, z) | Interferant (m/z, z) | 116/115 Ratio | Log2 Threshold Met? | 118/117 Ratio | Log2 Threshold Met? | ||||||
| Theoretical | MS/MS | IM-MS/MS | MS/MS | IM-MS/MS | Theoretical | MS/MS | IM-MS/MS | MS/MS | IM-MS/MS | ||
| 531.87, 2 | 532.67, 3 | 5.0 | 1.0 | 3.3 | no (0.0) | yes (1.7) | 5.0 | 1.0 | 4.2 | no (0.0) | yes (2.1) |
| 589.88, 2 | 591.66, 3 | 5.0 | 1.7 | 3.7 | no (0.8) | yes (1.9) | 5.0 | 1.0 | 3.2 | no (0.0) | yes (1.7) |
| 492.26, 4 | 491.76, 4 | 5.0 | 0.7 | 4.9 | no (−0.5) | yes (2.3) | 5.0 | 0.8 | 4.0 | no (−0.3) | yes (2.0) |
| 499.36, 2 | 499.32, 3 | 5.0 | 1.1 | 3.5 | no (0.1) | yes (1.8) | 5.0 | 1.3 | 5.0 | no (0.4) | yes (2.3) |
Additional instances of isobaric interference were indicated by near-unity reporter ratios produced by other precursors. However, IM separation was unable to mitigate the interference (data not shown).
Discussion
Co-isolation of Interfering Precursors with Different Charge States (z = +2, +3)
The spectra in Figure 1 exemplify the ability of IM to readily separate ions of different charge states. Both pairs of peptides display baseline separation of their extracted drift time distributions and were able to produce IM-MS/MS reporters that were more accurate than the chimeric MS/MS reporters. Even though the yeast peptide sequences were not identified, the pattern of their reporter ratios was strong evidence that they originated from the yeast sample. In Figure 1c, two additional small peaks in the BSA peptide's trace were present at the same drift times as the yeast peptide's peak. It is possible that they represent additional elongated structures of the BSA peptide. Reporters from the elongated BSA peptides could have contributed to the intensity from the yeast reporters and caused the underestimation present in the 6.14 ms reporters. However, similar reporter underestimation was present in both reporter spectra of Figure 1h yet there was no evidence of precursor IM crossover in Figure 1g. Other possibilities for the reporter underestimation could be additional interfering precursors that cannot be resolved in IM, or incomplete labeling by the DiLeu tags.
Co-isolation of Interfering Precursors with the Same Charge States (z = +4)
The examples in Figure 1 demonstrate that IM can separate differently charged interfering precursors, but this is something that can also be readily accomplished by methods described previously by Wegner et al.[27] or Ting et al.[30]. Even the mitigation of differently charged precursors with very similar m/z, such as the unidentified peptide in Figure S7 that differs from its interfering ion by only 40 mTh, would be separated by at least 3 Th after charge reduction. However, precursor IM separation can mitigate interferents of the same charge, which is still possible for the MS3 method but only possible for QuantMode when the difference in m/z is sufficiently large.
The monoisotopic m/z of the quadruply charged BSA and yeast peptides in Figure 2 differ by approximately 0.5 Th. Even if these peptides were charge reduced to doubly charged precursors, the m/z difference would be well within a 3 Th isolation window. After IM separation of these precursors, the largest relative error for the BSA peptide (115:116) was reduced from 74% to 12%, and the largest yeast relative error (118:115) was reduced from 85% to 6%.
It is interesting to note that although Figure 2 arguably presents better reporter correction than Figure 1, the Figure 2 precursors are less resolved by IM separation. The resolution is compounded by the two peaks displayed by the yeast peptide, however we believe the yeast peak at 5.11 ms is a result of the BSA peptide's isotope at m/z 492.266. Decreased IM resolution would be expected for two interfering peaks of the same charge state compared to those with different charges. In a triangular waveform approximation of TW IM-MS, the transit time from the net drift velocity (t) scales proportionally with the cell length (L) and wave speed (s), and inversely with the square of mobility (K) and the net electric field (E):[50]
| (1) |
The Mason-Schamp relationship says that K is directly proportional to ion charge (z) and inversely proportional to CCS (Ω):
| (2) |
Therefore, we can say that t is directly proportional to Ω2 and inversely proportional to z2:
| (3) |
Suppose we call tA the transit time of a doubly charged ion A and tB the transit time of a triply charged ion B of the same mass and CCS as ion A. Using equations 1-3 and assuming all other pertinent variables to be held constant, it can be shown that tB is approximately equal to 4/9 tA. However, if the ions were the same charge and ion A's CCS was 250 Å2, ion B would need a CCS nearly 84 Å2 smaller than ion A to exhibit the same degree of separation. Because of this, we expect that the IM-MS method would have a diminished performance for interferents of the same charge. Figure 2 is only one example of same charge interference and cannot be considered a statistically significant representation, but it does show that IM-MS can perform accurate corrections even when interfering peptides are of the same charge. It is important to note that TW have waveforms more similar to half-sinusoidal than triangular, thus the t/K relationship becomes more quadratic as K increases.[50]
Fold-change Masking
Quantitative proteomic experiments utilizing tandem mass tags are not often concerned with the absolute ratios displayed by reporters. An investigator might not be interested in whether a ratio is 1:5 or 1:8, but rather one might be interested in whether a protein is displaying overall differential regulation. In many instances, ratios are converted to a Log2 scale to represent fold-change. In Table 1, the data was organized and framed relative to fold-change. Ratios were divided up into 115-116 and 117-118 pairs, and a ratio of 1.5 on a Log2 scale (~2.8) was set as a threshold for a peptide to be considered a candidate for up-regulation.
All labeled peptides were mixed to ratios of either 1: 5: 1: 5 or 5: 1: 5: 1 and therefore should surpass the Log2 threshold. However, the fold-change for each peptide in the table was masked by interference and would have been incorrectly passed over for up-regulation candidacy. When the precursors are separated by IM, the ratios meet the Log2 threshold in all examples except for the transferrin miscleavage NYELLCGDNTRK. This result demonstrates that IM-MS is able to correct isobaric interference to a degree that may have a significant impact on real proteomic investigations.
Conclusion and Outlook
Isobaric interference is a very common occurrence in complex samples. In the extreme, Wegner et al. observed only 68% of intensity from the average reporter spectrum came from the intended precursor, and only 3% of MS/MS spectra were obtained from precursor isolations containing less than 1% interference.[27] The results of this investigation demonstrate proof-of-principle that IM separation of tagged precursors prior to MS/MS fragmentation can help mitigate quantitative inaccuracies caused by isobaric interference. IM-MS was able to resolve interference among differentially charged co-isolated peptides, and even among peptides of the same charge state. Contrary to the highlighted example, it is likely that the IM-MS method may have better overall performance with differentially charged interferants compared to pairs with the same charge. The instances of IM separation providing no reporter spectrum correction suggest that, just as is the case with QuantMode and MS3, this method cannot universally solve all interferences.
We would like to emphasize that the IM-MS technique should not be seen as a “competitor” with QuantMode or MS3. Rather, IM-MS should be considered as a potential alternative for Q-TOF instrumentation. The limited examples we presented represent the types of interference that could be encountered, and we selected instances where interference was most obvious. To transform the IM-MS technique into a viable method, significant changes need to be made to the acquisition and analysis software. To the knowledge of the authors, it is not possible to automatically extract specific ion intensities from a single drift time during DDA in MassLynx, DriftScope, or the common search algorithm used for SYNAPT G2 mobility DDA, ProteinLynx Global Server. Consequently, all drift time extractions in this investigation had to be performed manually, making application to large datasets very difficult. In MassLynx, there is an acquisition option to only use MS/MS drift times that were present in the precursor scan. This option is not helpful in this application, because drift times from the interfering peptide would have also been present in the precursor scan. The authors propose that an option allowing only the use of the most abundant isolated peptide's drift time would remedy this situation. Lacking automated analysis to apply the method to large datasets, we cannot explicitly evaluate its utility for large-scale proteomics at this point.
Perhaps the IM-MS method's most imminent need is the applicability to data-independent HDMSE analysis.[39, 51] Mobility DDA's scan cycle commonly operates at 1 to 5 Hz, making it difficult to obtain a large number of peptide IDs and quantified proteins. Additionally, the millisecond-time scale of IM intrinsically requires longer scan rates than analyses without IM. In its current state, precursor IM separation as part of mobility DDA may be appropriate for low-complexity mixtures or targeted analyses. Since HDMSE's performance is largely independent of scan rate, the IM-MS method may be successful with large-scale investigations. The most significant challenge in coupling to HDMSE would be that the lack of quadrupole isolation would mean all co-eluting labeled peptides would be interfering with each other. Future investigations will focus on the use of field-asymmetric ion mobility spectrometry (FAIMS) instruments. Although it is often difficult to predict how the separation of specific ions will compare between FAIMS and electrostatic or TW IM, FAIMS has demonstrated very high resolving powers.[52-55] The SYNAPT G2 is able to achieve drift time resolutions near 25, and based on isobaric interference predictions we made from HDMSE analysis of yeast digest, this would be insufficient for 30.1% of interfering peptide pairs (Table S1). Higher resolution than that of current TW IM instruments may be necessary to minimize apex drift time overlap of co-eluting peptides to make coupling a HDMSE-like data-independent acquisition (DIA) method a possibility. Whether or not this can be achieved by FAIMS must be evaluated empirically.
Supplementary Material
Acknowledgements
The authors would like to thank Tyler Greer and Dustin Frost in the Li Research Group for synthesizing the DiLeu isobaric tags. This work is supported in part by the National Science Foundation grant (CHE-0957784) and the National Institutes of Health grants (1R01DK071801 and 1R56DK071801). L. Li acknowledges an H.I. Romnes Faculty Research Fellowship. R.M.S. acknowledges the NIH-supported Clinical Neuroengineering Training Program Predoctoral Fellowship (T32 EB011434). C.B.L. acknowledges an NIH-supported Chemistry Biology Interface Training Program Predoctoral Fellowship (grant number T32-GM008505) and an NSF Graduate Research Fellowship (DGE-1256259).
References
- 1.Kline KG, Sussman MR. Protein quantitation using isotope-assisted mass spectrometry. Annu. Rev. Biophys. 2010;39:291. doi: 10.1146/annurev.biophys.093008.131339. [DOI] [PubMed] [Google Scholar]
- 2.Ong S-E, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 3.Schulze WX, Usadel B. Quantitation in mass-spectrometry-based proteomics. Annu. Rev. Plant Biol. 2010;61:491. doi: 10.1146/annurev-arplant-042809-112132. [DOI] [PubMed] [Google Scholar]
- 4.de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 5.Lundgren DH, Hwang S-I, Wu L, Han DK. Role of spectral counting in quantitative proteomics. Exp. Rev. Proteomics. 2010;7:39. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- 6.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell Proteomics. 2005;4:1487. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Podwojski K, Eisenacher M, Kohl M, Turewicz M, Meyer HE, Rahnenführer J, Stephan C. Peek a peak: A glance at statistics for quantitative label-free proteomics. Exp. Rev. Proteomics. 2010;7:249. doi: 10.1586/epr.09.107. [DOI] [PubMed] [Google Scholar]
- 8.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009;4:484. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 9.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature. 1999;17:994. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 10.Hsu J-L, Huang S-Y, Chow N-H, Chen S-H. Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 2003;75:6843. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Steen H, Gygi SP. Protein profiling with cleavable isotope-coded affinity tag (cicat) reagents: The yeast salinity stress response. Mol. Cell Proteomics. 2003;2:1198. doi: 10.1074/mcp.M300070-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, silac, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 2002;1:376. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Xiang F, Ye H, Chen R, Fu Q, Li L. N, n-dimethyl leucines as novel isobaric tandem mass tags for quantitative proteomics and peptidomics. Anal. Chem. 2010;82:2817. doi: 10.1021/ac902778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez J-C. Relative quantification of proteins in human cerebrospinal fluids by ms/ms using 6-plex isobaric tags. Anal. Chem. 2008;80:2921. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 16.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AKA, Hamon C. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by ms/ms. Anal. Chem. 2003;75:1895. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Wang Y, Li S. Deuterium isobaric amine-reactive tags for quantitative proteomics. Anal. Chem. 2010;82:7588. doi: 10.1021/ac101306x. [DOI] [PubMed] [Google Scholar]
- 18.Choe L, D'Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. 8 - plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for alzheimer's disease. PROTEOMICS. 2007;7:3651. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dephoure N, Gygi SP. Hyperplexing: A method for higher-order multiplexed quantitative proteomics provides a map of the dynamic response to rapamycin in yeast. Sci. Signal. 2012;5:rs2. doi: 10.1126/scisignal.2002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huttlin EJ, Jedrychowski M, Kuhn K, Dai C, Ting L, McAlister GC, Rad R, Rogers JC, Pike I, Haas W, Gygi SP. Enhanced isobaric labeling enables 18-plexed quantitative exploration of the hsf1-dependent cellular response to multiple proteotoxic stresses at a proteomic scale. Proc. of the 60th Am. Soc. Mass. Spectrom. Conf. 2012 [Google Scholar]
- 21.Everley RA, Kunz RC, Mcallister FE, Kuhn K, Rogers JC, Pike I, Gygi SP. Increasing throughput in kinase inhibition assays: 54-plex quantitation in a single ms run. Proc. of the 60th Am. Soc. Mass. Spectrom. Conf. 2012;ThP05 112 [Google Scholar]
- 22.Bantscheff M, Boesche M, Eberhard D, Matthieson T, Sweetman G, Kuster B. Robust and sensitive itraq quantification on an ltq orbitrap mass spectrometer. Mol. Cell Proteomics. 2008;7:1702. doi: 10.1074/mcp.M800029-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. Addressing accuracy and precision issues in itraq quantitation. Mol. Cell Proteomics. 2010;9:1885. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. Itraq underestimation in simple and complex mixtures: “The good, the bad and the ugly”. J. Proteome Res. 2009;8:5347. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 25.Shirran SL, Botting CH. A comparison of the accuracy of itraq quantification by nlc-esi msms and nlc-maldi msms methods. J. Proteomics. 2010;73:1391. doi: 10.1016/j.jprot.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ow SY, Salim M, Noirel J, Evans C, Wright PC. Minimising itraq ratio compression through understanding lc-ms elution dependence and high-resolution hilic fractionation. Proteomics. 2011;11:2341. doi: 10.1002/pmic.201000752. [DOI] [PubMed] [Google Scholar]
- 27.Wenger CD, Lee MV, Hebert AS, McAlister GC, Phanstiel DH, Westphall MS, Coon JJ. Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat. Meth. 2011;8:933. doi: 10.1038/nmeth.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savitski MM, Sweetman G, Askenazi M, Marto JA, Lang M, Zinn N, Bantscheff M. Delayed fragmentation and optimized isolation width settings for improvement of protein identification and accuracy of isobaric mass tag quantification on orbitrap-type mass spectrometers. Analytical chemistry. 2011;83:8959. doi: 10.1021/ac201760x. [DOI] [PubMed] [Google Scholar]
- 29.Savitski MM, Mathieson T, Zinn N, Sweetman G, Doce C, Becher I, Pachl F, Kuster B, Bantscheff M. Measuring and managing ratio compression for accurate itraq/tmt quantification. Journal of proteome research. 2013;12:3586. doi: 10.1021/pr400098r. [DOI] [PubMed] [Google Scholar]
- 30.Ting L, Rad R, Gygi SP, Haas W. Ms3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nature Methods. 2011;8:937. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLean J, Ruotolo B, Gillig K, Russell D. Ion mobility-mass spectrometry: A new paradigm for proteomics. Int. J. Mass Spectrom. 2005. p. 301.
- 32.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave ims/oa-tof instrument. Int. J. Mass Spectrom. 2007;261:1. [Google Scholar]
- 33.Zhong Y, Hyung S-J, Ruotolo BT. Characterizing the resolution and accuracy of a second-generation traveling-wave ion mobility separator for biomolecular ions. Analyst. 2011;136:3534. doi: 10.1039/c0an00987c. [DOI] [PubMed] [Google Scholar]
- 34.Giles K, Williams JP, Campuzano I. Enhancements in travelling wave ion mobility resolution. Rapid Commun. Mass Spectrom. 2011;25:1559. doi: 10.1002/rcm.5013. [DOI] [PubMed] [Google Scholar]
- 35.Hoaglund-Hyzer CS, Clemmer DE. Ion trap/ion mobility/quadrupole/time-of-flight mass spectrometry for peptide mixture analysis. Analytical chemistry. 2001;73:177. doi: 10.1021/ac0007783. [DOI] [PubMed] [Google Scholar]
- 36.Valentine SJ, Kulchania M, Barnes CAS, Clemmer DE. Multidimensional separations of complex peptide mixtures: A combined high-performance liquid chromatography/ion mobility/time-of-flight mass spectrometry approach. Int J Mass Spectrom. 2001;212:97. [Google Scholar]
- 37.Hoaglund-Hyzer CS, Li J, Clemmer DE. Mobility labeling for parallel cid of ion mixtures. Analytical chemistry. 2000;72:2737. doi: 10.1021/ac0000170. [DOI] [PubMed] [Google Scholar]
- 38.Lee YJ, Hoaglund-Hyzera CS, Srebalus Barnes CA, Hilderbrand AE, Valentine SJ, Clemmer DE. Development of high-throughput liquid chromatography injected ion mobility quadrupole time-of-flight techniques for analysis of complex peptide mixtures. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2002;782:343. doi: 10.1016/s1570-0232(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 39.Myung S, Lee YJ, Moon MH, Taraszka J, Sowell R, Koeniger S, Hilderbrand AE, Valentine SJ, Cherbas L, Cherbas P, Kaufmann TC, Miller DF, Mechref Y, Novotny MV, Ewing MA, Sporleder CR, Clemmer DE. Development of high-sensitivity ion trap ion mobility spectrometry time-of-flight techniques: A high-throughput nano-lc-ims-tof separation of peptides arising from a drosophila protein extract. Analytical chemistry. 2003;75:5137. doi: 10.1021/ac030107f. [DOI] [PubMed] [Google Scholar]
- 40.Taraszka JA, Gao X, Valentine SJ, Sowell RA, Koeniger SL, Miller DF, Kaufman TC, Clemmer DE. Proteome profiling for assessing diversity: Analysis of individual heads of drosophila melanogaster using lc-ion mobility-ms. Journal of proteome research. 2005;4:1238. doi: 10.1021/pr050037o. [DOI] [PubMed] [Google Scholar]
- 41.Valentine SJ, Plasencia MD, Liu X, Krishnan M, Naylor S, Udseth HR, Smith RD, Clemmer DE. Toward plasma proteome profiling with ion mobility-mass spectrometry. Journal of proteome research. 2006;5:2977. doi: 10.1021/pr060232i. [DOI] [PubMed] [Google Scholar]
- 42.Kindy JM, Taraszka JA, Regnier FE, Clemmer DE. Quantifying peptides in isotopically labeled protease digests by ion mobility/time-of-flight mass spectrometry. Analytical chemistry. 2002;74:950. doi: 10.1021/ac010807p. [DOI] [PubMed] [Google Scholar]
- 43.Xun Z, Kaufman TC, Clemmer DE. Stable isotope labeling and label-free proteomics of drosophila parkin null mutants. Journal of proteome research. 2009;8:4500. doi: 10.1021/pr9006238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bohrer BC, Clemmer DE. Biologically-inspired peptide reagents for enhancing ims-ms analysis of carbohydrates. Journal of the American Society for Mass Spectrometry. 2011;22:1602. doi: 10.1007/s13361-011-0168-y. [DOI] [PubMed] [Google Scholar]
- 45.Hilderbrand AE, Myung S, Clemmer DE. Exploring crown ethers as shift reagents for ion mobility spectrometry. Analytical chemistry. 2006;78:6792. doi: 10.1021/ac060439v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerr TJ, Gant-Branum RL, McLean JA. Multiplexed analysis of peptide functionality using lanthanide-based structural shift reagents. Int J Mass Spectrom. 2011;301:28. doi: 10.1016/j.ijms.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hughes C, Vissers H, Langridge J. Resolving chimeric spectra utilizing a data independent mobility acquisition strategy-1. 2011 http://www.google.com/url?sa=t&rct=j&q=chimeric%20spectra&source=web&cd=2&ved=0CCsQFjAB&url=http%3A%2F%2Fwww.18show.cn%2Fbbs%2Fattachment.aspx%3Fattac hmentid%3D3574&ei=BSNDT4uZGsbyggeN9t2YCA&usg=AFQjCNGgjQ5pYy2JCD68igaHOrvy EWMpaw&cad=rja1.
- 48.Miller ME, Cross FR. Distinct subcellular localization patterns contribute to functional specificity of the cln2 and cln3 cyclins of saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:542. doi: 10.1128/mcb.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shadforth IP, Dunkley TPJ, Lilley KS, Bessant C. I-tracker: For quantitative proteomics using itraq. BMC genomics. 2005;6:145. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shvartsburg AA, Smith RD. Fundamentals of traveling wave ion mobility spectrometry. Analytical chemistry. 2008;80:9689. doi: 10.1021/ac8016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li GZ, Vissers JPC, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 52.Shvartsburg AA, Danielson WF, Smith RD. High-resolution differential ion mobility separations using helium-rich gases. Analytical chemistry. 2010;82:2456. doi: 10.1021/ac902852a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shvartsburg AA, Prior DC, Tang K, Smith RD. High-resolution differential ion mobility separations using planar analyzers at elevated dispersion fields. Analytical chemistry. 2010;82:7649. doi: 10.1021/ac101413k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shvartsburg AA, Smith RD. Ultrahigh-resolution differential ion mobility spectrometry using extended separation times. Analytical chemistry. 2011;83:23. doi: 10.1021/ac102689p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shvartsburg AA, Seim TA, Danielson WF, Norheim R, Moore RJ, Anderson GA, Smith RD. High-definition differential ion mobility spectrometry with resolving power up to 500. Journal of the American Society for Mass Spectrometry. 2013;24:109. doi: 10.1007/s13361-012-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.