Abstract
The transcriptional regulator BrlR is a member of the MerR family of multidrug transport activators that contributes to the high-level drug tolerance of Pseudomonas aeruginosa biofilms. While MerR regulators are known to activate both the expression of multidrug efflux pump genes and their own transcription upon inducer-binding, little is known about BrlR activation. We demonstrate using promoter reporter strains, in vivo and in vitro DNA-binding assays combined with 5’RACE, that BrlR binds to its own promoter, likely via a MerR-like palindromic sequence. Unlike known MerR multidrug transport activators, BrlR and brlR expression are not activated by multidrug transporter substrates. Instead, BrlR-DNA binding was enhanced by the secondary messenger c-di-GMP. In addition to enhanced BrlR-DNA binding, c-di-GMP levels contributed to PbrlR promoter activity in initial attached cells with elevated c-di-GMP levels correlating with increased expression of brlR. While not harboring amino acid motifs resembling previously defined c-di-GMP-binding domains, BrlR was found to bind c-di-GMP in vitro at a ratio of one c-di-GMP per two BrlR. Crosslinking assays confirmed dimer formation to be enhanced in the presence of elevated c-di-GMP levels. Our findings demonstrate BrlR to be an unusual MerR-family member in that BrlR function and expression require the secondary messenger c-di-GMP.
INTRODUCTION
Biofilms are composed of surface-attached microbial communities. A hallmark of biofilms is their profound tolerance to antimicrobial agents (Costerton et al., 1999). Several reasons have been suggested to explain biofilm tolerance including slow growth and starvation-induced growth arrest, increased presence of persister cells that neither grow nor die in the presence of bactericidal antibiotics as well as diffusion limitation of antimicrobials into biofilms (Shah et al., 2006) (Campanac et al., 2002, Picioreanu et al., 2001, Anderl et al., 2000, Lewis, 2001, Stewart & Costerton, 2001, Stewart, 1996, Drenkard, 2003, Nguyen et al., 2011). Recent reports further suggest that bacteria within these microbial communities are physiologically distinct from planktonic bacteria, expressing specific protective factors such as multidrug (MDR) efflux pumps and stress response regulons (Anwar et al., 1992, Sauer & Camper, 2001, Sauer et al., 2002, Stoodley et al., 2002, Donlan & Costerton, 2002, Gilbert et al., 2002, Mah & O’Toole, 2001, Mah et al., 2003, Stewart & Costerton, 2001). In addition, quorum sensing (QS) is required for the formation of the biofilm architecture (Davies et al., 1998), and has been shown to play a role in resistance (Bjarnsholt et al., 2005). Biofilm and planktonic cells further differ in the level of the secondary messenger molecule c-di-GMP, with high concentrations of this molecule correlating with an aggregative or sessile lifestyle (e.g. biofilm formation), while its absence favors motility (e.g. twitching, swarming) and the free swimming lifestyle (D’Argenio & Miller, 2004). While c-di-GMP has not been directly linked to resistance, c-di-GMP contributes to the production of extracellular polysaccharides which has been suggested to offer protection from environmental factors, antimicrobials, and the immune response, likely by limiting diffusion or sequestration (Merritt et al., 2007, Romling et al., 2005, Irie et al., 2012, Mishra et al., 2012). Moreover, low c-di-GMP levels have recently been linked to the expression of proteins required for antimicrobial peptide resistance in Pseudomonas aeruginosa (Chua et al., 2013). The findings indicated that biofilms themselves are not simply a diffusion barrier to these antibiotics, but rather that bacteria within these microbial communities employ distinct mechanisms to resist the action of antimicrobial agents. This was further supported by the finding of the transcriptional regulator BrlR (Biofilm resistance locus Regulator) contributing to the high-level tolerance of biofilms formed by the human pathogen P. aeruginosa (Liao & Sauer, 2012). In particular, BrlR was found to contribute to the tolerance of P. aeruginosa biofilms to hydrogen peroxide and five different classes of antibiotics by affecting MIC and recalcitrance of biofilms to killing by bactericidal antimicrobial agents (Liao & Sauer, 2012), suggesting BrlR may play a role in the activation of genes involved in resistance to antimicrobial agents. The mechanism by which BrlR, a MerR-like regulator, confers tolerance relies on BrlR binding promoter regions and activating at least two multidrug efflux pumps encoded by mexAB-oprM and mexEF-oprN (Liao et al., 2013).
Activation of multidrug transporter genes is a common feature of members of the MerR family of multidrug efflux pump gene activators. However, while MerR activators induce expression of multidrug transporter genes, their induction requires binding of a specific ‘coactivator’ molecule, typically the transporter substrate (Heldwein & Brennan, 2001, Zheleznova et al., 1999, Newberry & Brennan, 2004). For instance, BltR is induced by binding of rhodamine to the C-terminal domain, resulting in activation of transcription of multidrug transporters, TipA is activated by binding thiostrepton, and BmrR by binding both rhodamine and tetraphenylphosphonium (Ahmed et al., 1994, Neyfakh, 2001, Vazquez-Laslop et al., 2000, Brown et al., 2003, Baranova et al., 1999, Godsey et al., 2002, Zheleznova et al., 1999, Holmes et al., 1993). Inducers for the global activator MtaN are not known (Godsey et al., 2001, Godsey et al., 2002). Expression of the Bacillus subtilis multidrug transporter Bmr was shown to be inducible at the level of transcription by two of its diverse substrates, rhodamine 6G and thiostrepton, and this induction is mediated by the regulatory protein BmrR encoded immediately downstream of the bmr gene. Structural analysis of BmrR indicated inducer binding to the C-terminal domain of BmrR (Heldwein & Brennan, 2001, Zheleznova et al., 1999). In addition to activating gene expression of multidrug efflux pump genes, members of this MerR family activate their own transcription upon inducer binding (Summers, 1992, Holmes et al., 1993). Activation of transcription coincides with MerR regulators binding to palindromic promoters in which the spacing between the -35 and -10 elements recognized by the sigma factor is greater than the optimal 17+/-1 bp, usually 19 bp (Heldwein & Brennan, 2001, Godsey et al., 2001, Newberry & Brennan, 2004). Binding to suboptimal 19-bp spacers requires DNA distortion, a DNA-binding mechanism that is common to the entire MerR family (Newberry & Brennan, 2004). Despite MerR family members sharing homologous DNA binding domains and recognizing palindromic promoters with suboptimal spacing, the MerR-DNA binding sites are diverse (Brown et al., 2003).
BrlR harbors two domains, an N-terminal Helix-Turn-Helix DNA binding HTH_BmrR domain, and a C-terminal modulation or “coactivator” GyrI-like binding domain (Liao & Sauer, 2012), (Fig. S1). While BrlR shares similarities to MerR activators including domain organization, sequence homology and activation of multidrug transporter genes (Liao & Sauer, 2012, Liao et al., 2013), little is known about the BrlR-DNA binding sites, the inducer(s) to which BrlR respond in order to activate BrlR-dependent drug tolerance in biofilms, nor its mechanism of activation contributing to the biofilm-specific expression of brlR (Liao & Sauer, 2012, Liao et al., 2013). To begin elucidating the mechanism by which BrlR is activated and brlR is expressed in a biofilm-specific manner, we first mapped the brlR promoter region and investigated the DNA binding properties of BrlR. Promoter reporter strains were then used to probe for cues activating brlR expression. In contrast to known MerR family members, BrlR and brlR expression is not activated by multidrug transporter substrates. Instead, induction of brlR expression coincides with increased levels of the intracellular signaling molecule c-di-GMP, conditions that have previously been linked to attachment and surface attached growth. We further demonstrate that BrlR binds to a palindromic sequence, with BrlR-DNA binding and brlR promoter reporter activity being enhanced by the secondary messenger c-di-GMP. Consistent with BrlR being a c-di-GMP responsive transcription factor, BrlR was found to bind c-di-GMP.
RESULTS
Mapping of the brlR promoter region
To begin elucidating the mechanism by which BrlR is activated and brlR is expressed in a biofilm-specific manner, we first mapped the approximate promoter region as well as the transcriptional start site of brlR. Using 5’RACE, the transcriptional start site was found to be 23 bp upstream of the brlR open reading frame (Figs. 1A, S2). Our transcriptional start site is located within 2 bp of the start site recently identified by Wurtzel et al. (Wurtzel et al., 2012) using RNA-Seq in conjunction with mapping of the ends of untranslated regions (UTR). The 23 bp long UTR harbors a putative Shine-Dalgarno sequence (Fig. 1A). Moreover, a predicted promoter sequence upstream of the transcriptional start site was identified as a σ70-like -10 box centered approximately 4 bp upstream of the transcriptional start site, followed by a -35 box (Fig. 1A). The predicted promoter sequence is located in the intergenic region of brlR and PA4877. The gene PA4877 encoding a hypothetical protein is located 147 bp upstream of the BrlR ATG start codon and is transcribed in the reverse orientation (Fig. 1B). To locate the brlR promoter region, DNA sequences located 120, 250, and 500 bp upstream from the BrlR start codon (Fig. 1B) were PCR amplified and cloned into the pMiniCTX-lacZ vector, yielding three transcriptional promoter fusions (PbrlR-120-lacZ, PbrlR-250-lacZ, PbrlR-500-lacZ). DNA sequences located 23 bp upstream from the BrlR start codon (PbrlR-23-lacZ) were used as control. Under biofilm conditions, no difference in promoter activity was detected for the PbrlR-23-lacZ reporter construct and the vector control (Fig. 1C). However, significantly increased β-galactosidase activities compared to the vector control were detected for the promoter reporter strains harboring DNA located 120, 250, and 500 bp upstream of the brlR start codon (Fig. 1C). In contrast, no difference in β-galactosidase activity was noted under planktonic growth conditions, with β-galactosidase activity levels being comparable to those observed for the vector control (Fig. 1C). These data confirmed previous findings of brlR expression being biofilm-specific (Liao & Sauer, 2012). Moreover, our observations suggested that the brlR promoter was located within 23-120 bp upstream of the BrlR ATG start codon, in the intergenic region between brlR and PA4877 (Fig. 1B). The respective region harbored the putative σ70-like -10 and -35 promoter elements (Fig. 1A).
Figure 1. Mapping of the brlR promoter.
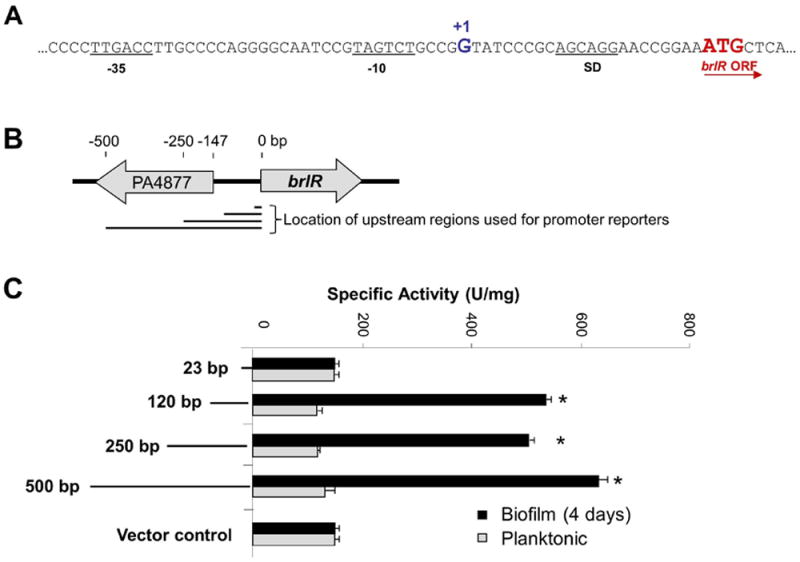
(A) The brlR transcriptional start site (+1) was determined by 5’RACE to be 23 bp upstream of translational start codon (brlR ORF). SD = Shine-Dalgarno sequence. Putative -35 and -10 sequences are underlined. (B) Schematic diagram of brlR position within the P. aeruginosa PAO1 genome. Arrows denote direction of transcription. Numbers above indicate approximate distances while lines below indicate the location and length of the upstream regions used for the construction of promoter reporters. (C) Levels of brlR-lacZ expression were determined by measuring specific β-galactosidase activity in P. aeruginosa PAO1 carrying chromosomal LacZ fusions to various fragments (1-500 bp) upstream of the brlR start codon. β-galactosidase activity was determined under planktonic and biofilm growth conditions. Experiments were carried out at least in triplicate. Error bars indicate standard deviation. *, significantly different from planktonic growth conditions; p < 0.05.
BrlR binds to its own promoter
MerR proteins, including TipA and BmrR, have been shown to bind to their own promoters (Baranova et al., 1999, Newberry & Brennan, 2004, Neyfakh, 2001). To determine whether BrlR acts in a similar manner, we purified BrlR with a C-terminal His6V5-tag (BrlR–His6V5; Fig. 2A) to perform electrophoretic mobility shift assays using promoter fragments spanning the region located 443-543 bp (region A), 223-459 bp (region B), 91-240 bp (region C) and 7-108 bp (PbrlR) upstream of the brlR translational start codon. The tagged BrlR construct has been previously shown to complement a ΔbrlR mutant in vivo (Liao & Sauer, 2012). BrlR did not bind to radiolabeled DNA probes spanning the DNA sequences located 91-543 bp upstream of the brlR start codon (representing regions A-C, Fig. 2B). In contrast, a specific complex was formed when BrlR was added to the radiolabeled DNA probe PbrlR, at a molar ratio of 20:1 (Fig. 2C). The PbrlR sequence is located 7-108 bp upstream of the brlR start codon (Fig. 2C). BrlR-DNA binding was specific as no binding to the labeled non-specific promoter fragment (PpscE-F) was observed, and dependent on both the BrlR protein and brlR promoter DNA concentration. When the molar concentrations of BrlR were increased, binding to PblrR increased. When increasing amounts of unlabeled PbrlR fragment (Fig. 2C) were added to the gel mobility shift assay, decreased BrlR binding to the labeled fragment was observed.
Figure 2. BrlR binds to its own promoter.
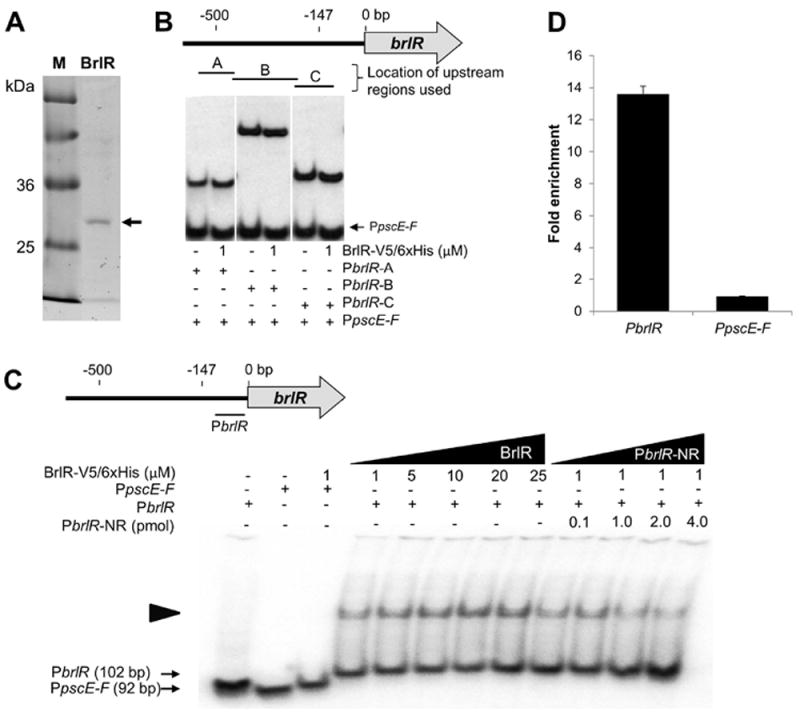
(A) His6V5-Tagged BrlR has an apparent molecular weight of approximately 33 kDa (see arrow). BrlR was purified using Ni-NTA affinity chromatography. M, protein ladder. (B) DNA gel mobility shift assays using purified BrlR and DNA fragments A-C located 443-543, 223-459, and 91-240 bp upstream of the ATG start codon, respectively. The schematic diagram indicates the location and approximate length of the three DNA fragments A-C used in DNA binding assays. A total of 0.1 pmol was used for DNA regions A-C. (C) DNA gel mobility shift assays indicating BrlR binding to a DNA fragment located 7-108 bp upstream of the ATG start codon, demonstrating specificity of BrlR binding. Both the concentration of purified BrlR (1-25 μM BrlR) and the concentration of non-labeled competitor DNA (PbrlR-NR, 0.1-4 pmol) were varied. The PCR amplified DNA fragment PpscE-F (0.1 pmol) was used as a non-specific radiolabeled control. The schematic diagram indicates the location and length of the PbrlR fragment relative to brlR. Experiments were carried out in triplicate and representative images are shown. The schematic diagram indicates the location and length of the PbrlR DNA fragment (7-108 bp) used in DNA binding assays. Numbers above indicate approximate distances to the brlR start codon. (D) Fold enrichment of the promoter sequences of brlR and pscE-F in BrlR-V5/His6 ChIP sample compared to control (ChIP carried out in the presence of untagged BrlR), as determined by qPCR. Experiments were carried out at in triplicate. Error bars indicate standard deviation.
While BrlR binding to the PbrlR promoter appeared to be specific, binding of BrlR was observed at high BrlR:DNA molar ratios. To further confirm BrlR binding to the respective promoter region, we probed for in vivo BrlR-DNA binding using chromatin immunoprecipitation (ChIP) assays and P. aeruginosa biofilms expressing a C-terminally His6V5-tagged BrlR construct (BrlR–His6V5). A P. aeruginosa strain overexpressing the untagged protein was used as control. DNA isolated via anti-V5 antibody immunoprecipitation from PAO1/pJN-brlR and PAO1/pMJT-brlR-His6V5 biofilm samples was subjected to qPCR using primers spanning the promoter region located 7-108 bp upstream of the brlR ATG start codon. On average, the promoter region of brlR was enriched 14-fold, while no enrichment was detected for the non-specific promoter region of PpscE-F (Fig. 2D). The finding suggested BrlR binding to the 7-108 bp region upstream of the BrlR start codon both in vitro and in vivo.
BrlR binds to a palindromic sequence located between the putative -35 and -10-boxes
Binding to palindromic sequences is common among MerR regulators including the MerR regulator BmrR which has been shown using DNA footprint analysis to bind to a palindromic sequence located between the -35 and -10-boxes of the bmrR promoter (Ahmed et al., 1994, Brown et al., 2003). Analysis of the DNA sequence comprising the putative -35 and -10-boxes of the brlR promoter using the software “Palindrome” indicated the presence of a perfect inverted repeat located between the -10 and -35 elements (Fig. 3A). The sequence shared similarity to the symmetrical spacer regions of MerR-like regulated promoter regions including TipA, BmrR, and SoxR as well as the metal-dependent promoters of MerR, CoaR, and CueR (Fig. 3B). The spacer regions have been shown to be the regulator binding sites (Brown et al., 2003). DNA gel mobility shift assays using short DNA sequences lacking either the putative -10 or the -35 promoter elements (prom-35, prom-10, respectively), or a 19bp-long DNA sequence located between the -10 and -35 elements (see “prom-19bp”) demonstrated BrlR-binding to such short DNA sequence (Fig. 3C). Mutating one half of the palindromic sequence by substituting the G’s with T’s impaired BrlR-binding (see “prom-mut1”), suggesting BrlR-binding to occur in-between the putative -35 and -10-boxes in a sequence-specific manner (Fig. 3A, C). Lack of BrlR-binding to a chimeric 19bp long sequence (see “prom-mut2”) composed in part of a portion of the 19 bp spacer region and a DNA sequence located 119-128 bp upstream of the ATG start codon, further supported BrlR-binding to be sequence-specific rather than DNA length-specific (Fig. 3A, C). Interestingly, the 19bp-long DNA sequence is the site of the perfect inverted repeat as determined using the palindrome software analysis tool (Fig. 3A-B).
Figure 3. BrlR binds to a palindromic DNA spacer sequence.
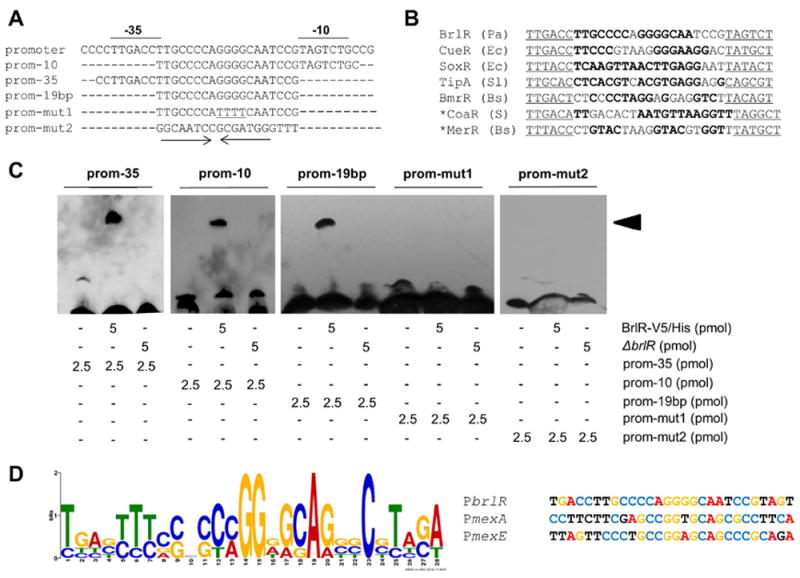
(A) Alignment of BmrR-DNA binding sequence and BrlR-DNA binding sequences used in the DNA gel mobility shift assays. The inverted repeat is highlighted by arrows underneath the alignments. (B) BrlR spacer sequence and spacer sequences found in promoter regions regulated by MerR-like proteins (reviewed in (Brown et al., 2003)). S, Synechocystis sp.; Ec, Escherichia coli; BS, Bacillus subtilis; Pa, Pseudomonas aeruginosa; Sl, Streptomyces lividans. Symmetrical sequences are shown in bold. −35 and −10 sequences are underlined. *, spacer is 20 bp long while all others have 19 bp long spacers (reviewed in (Brown et al., 2003)). (C) DNA gel mobility shift assays indicating binding of BrlR to the putative BrlR-DNA binding sequence. prom, DNA sequence spanning the -10 and -35 promoter elements (underlined); prom-10, DNA sequence lacking the -10 region; prom-35, DNA sequence lacking the -35 region; prom-19 bp, 19 bp spacer region located between the -10 and -35 promoter elements; prom-mut1, 19 bp spacer region located between the -10 and -35 promoter elements in which the G’s on the right side of the palindrome have been substituted with T’s; prom-mut2, 19bp-long sequence in part of a portion of the 19 bp spacer region and a DNA sequence located 119-128 bp upstream of the ATG start codon. A total of 5 pmol purified His6V5-tagged BrlR and 2.5 pmol per each biotinylated DNA sequence was used. BrlR-DNA binding was detected by immunoblot analysis using anti-biotin antibodies. Black arrowheads indicate specific shift. -, no DNA sequences or protein added. Total cell extract obtained from ΔbrlR was used as control. (D) MEME-derived BrlR-DNA binding motif common to BrlR-DNA binding sequence obtained in (A-B) and promoter sequences of the mexAB-oprM and mexEF-oprN operons.
To determine whether the palindromic BrlR-DNA binding sequence shown in Fig. 3A-B is also present in the promoter DNA of mexAB-oprM and mexEF-oprN, which were previously demonstrated to be BrlR-targets (Liao et al., 2013), we made use of MEME to search for common DNA binding motifs (Bailey et al., 2009). One motif common to all sequences was identified as [TC][GTA][ATC][GCT][TC][TC][TC][CGA][CG]X[CG][CT][CA]GG[GAT][GA][CG]A[GA][GCT]C[GCT][TC][ATC][GC][AT], with p-values ranging from 5.3e-9 for the mexAB-oprM promoter region to 4.4e-11 for the mexEF-oprN promoter region (Fig. 3D). The motifs identified by MEME, while not perfect inverted repeats, were found to be located 38-65 bp and 39-66 bp upstream of the transcriptional start sites of mexA and mexE, respectively (Fig. 3D). Moreover, the location of the MEME-derived BrlR-DNA binding motif in the upstream region of the multi-drug efflux pump operons overlapped with the DNA sequences previously used to demonstrate BrlR-binding the promoter DNA of the mexAB-oprM and mexEF-oprN operons (Liao et al., 2013).
brlR expression is enhanced by c-di-GMP in vivo
Our findings suggested BrlR binds to DNA in a manner similar to known MerR family regulators. Considering that MerR multidrug transport activators have been demonstrated to respond to, and to be activated by, the substrates of the multidrug transporters that they are regulating (Ahmed et al., 1994, Neyfakh, 2001, Vazquez-Laslop et al., 2000, Brown et al., 2003, Baranova et al., 1999, Godsey et al., 2002, Zheleznova et al., 1999, Holmes et al., 1993, Heldwein & Brennan, 2001, Summers, 1992), we hypothesized that BrlR may likewise require an inducer. We further hypothesized that such an inducer is not likely to be an antimicrobial agent as brlR expression was found to be biofilm-specific (Liao & Sauer, 2012) (Fig. 1C). To gain a better understanding of the nature of such an inducer, we first determined when brlR expression is induced over the course of initial biofilm formation. This was performed using a gfp promoter reporter and monitoring gfp expression and thus, brlR, by confocal microscopy. No gfp expression was noted in P. aeruginosa cells grown planktonically (not shown) and in cells that were attached via their poles, indicating that brlR expression is not induced in initially or reversibly attached cells (Fig. 4A). The earliest time point at which brlR expression was detected was 6 hr post initial attachment, primarily in cells attached longitudinal to the glass surface. However, only a small percentage of the population expressed gfp. Expression of gfp significantly increased upon continued attachment, with gfp expression, indicative of brlR expression, being maximal following 24 hr of growth (Fig. 4A). brlR expression remained detectable at later biofilm stages (not shown). Under the conditions tested, no gfp expression was detectable when using a vector control (Fig. 4A). Increased brlR expression following 6 hr post-attachment compared to 4 hr post-attachment and vector control was supported by β-galactosidase activity determinations using the PbrlR-120-lacZ promoter reporter strains (Fig. 4D).
Figure 4. brlR expression and BrlR-DNA binding are enhanced by the secondary messenger c-di-GMP.
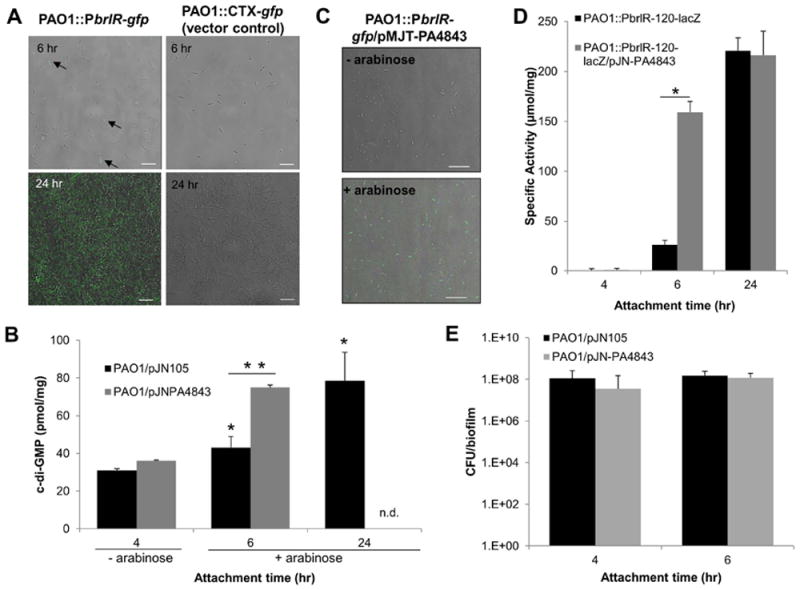
(A) Induction of brlR expression over the course of biofilm formation was monitored by confocal microscopy using a P. aeruginosa PAO1 carrying chromosomal gfp fusions to the upstream region of brlR. Total cells were visualized by brightfield microscopy. Overlays acquired 6 hr and 24 hr post initial attachment are shown. brlR expression was detected as early as 6 hr following initial attachment. Arrows indicate gfp-expressing cells. P. aeruginosa harboring an empty pMini-CTX-gfp vector was used as control. White bar = 10 μm. (B) c-di-GMP levels present in P. aeruginosa harboring an empty pJN105 vector or overexpressing PA4843 (PAO1/pJN-PA4843). The strains were grown attached in tube reactors under flowing conditions for 4, 6, and 24 hr. – arabinose, no arabinose was added to the growth medium. + arabinose, arabinose was added to the growth medium 4 hr post initial attachment to induce PA4843 expression. n.d., not determined. *, significantly different from PAO1/pJN105 at the 4-hr time point; p < 0.05. **, significantly different from PAO1/pJN105 at the 6-hr time point; p < 0.01. (C) Expression of the diguanylate cyclase PA4843 results in enhanced gfp expression indicative of brlR expression, compared to untreated controls. – arabinose, no arabinose was added to the growth medium. + arabinose, arabinose was added to the growth medium 4 hr post initial attachment to induce PA4843 expression. (D) brlR transcriptional reporter activity during surface-attached growth is enhanced upon overexpression of the cyclase PA4843. Strains harboring the brlR-lacZ reporter construct in the absence or presence of the plasmid pJN-PA4843 were grown under continuous flowing conditions in flow cells for 4, 6 hr, and 24 hr. Expression of PA4843 was induced by the addition of arabinose to the growth medium 4 hr post initial attachment. β-galactosidase activity has been corrected by the activity observed for the vector control strain. *, significantly different from PAO1/pJN105 at the 6-hr time point; p < 0.05. (E) Number of viable cells of PAO1/pJN105 and PAO1/pJN-PA4843 attached to glass following 4 and 6 hr post initial attachment under flowing conditions, expressed as CFU/biofilm. All experiments were carried out in triplicate. Error bars indicate standard deviations.
Given the link between initial attachment and induction of brlR expression, we next asked whether factors or conditions associated with surface-attached growth may contribute to the expression of brlR in a biofilm-specific manner. Conditions that have been ascribed to the biofilm mode of growth include slow growth, oxygen and nutrient limitation, presence of oxidative stress, as well as increased viscosity and cell density. However, none of these conditions induced brlR expression (Fig. S3). We likewise excluded a role of multidrug efflux pump substrates tobramycin, norfloxacin, trimethoprim, or a mixture of the three antimicrobial agents as likely inducers of brlR expression (Fig. S4). Biofilms further differ from planktonic cells in the level of the intracellular secondary messenger molecule cyclic di-GMP (c-di-GMP). C-di-GMP has been associated with controlling the transition between a motile and biofilm lifestyle, with high concentrations of this molecule correlating with a sessile lifestyle (e.g. biofilm formation), while its absence favors motility (e.g. twitching, swarming) and the free-swimming lifestyle (Basu Roy et al., 2012, Barraud et al., 2009, D’Argenio & Miller, 2004). We therefore asked whether the secondary messenger c-di-GMP plays a role in brlR expression by determining c-di-GMP levels of cells attached for 4, 6, and 24 hr. Cells attached for 4 hr were characterized by 30±0.95 pmol c-di-GMP per mg biofilm biomass (Fig. 4B). Upon continued attachment, c-di-GMP levels increased significantly to 43 pmol/mg following 6 hr, and 79 pmol c-di-GMP per mg biofilm biomass following 24 hr of attachment under flowing conditions (Fig. 4B). Interestingly, the c-di-GMP levels of cells attached for 4 hr were comparable to levels previously described for planktonic cells while the c-di-GMP level detected in 24 hr-attached cells was comparable to c-di-GMP levels previously described to be present in 4-6 day old biofilms (Basu Roy et al., 2012, Barraud et al., 2009).
The finding suggested induction of brlR expression to correlate with elevated c-di-GMP levels. To further confirm a link between brlR expression and c-di-GMP levels, we introduced the c-di-GMP producing diguanylate cyclase PA4843 under the control of the arabinose inducible PBAD promoter into the PbrlR-gfp promoter reporter to monitor the effect of elevated c-di-GMP levels on brlR expression in P. aeruginosa during attachment. PA4843 was chosen as it was previously demonstrated to be an active diguanylate cyclase (Petrova & Sauer, 2012a). Under the conditions tested, no PbrlR-gfp promoter activity was detected at the 5 hr time point in the absence of arabinose (due to lack of PA4843 expression, PA4843 is under control of PBAD promoter, Fig. 4C). Expression of gfp and thus, expression of brlR, however, was significantly increased upon induction of PA4843 expression via addition of arabinose, as evident by nearly 100% of the attached population expressing gfp (Fig. 4C). β-galactosidase activity assays using a PbrlR-lacZ promoter reporter construct confirmed increased PbrlR promoter activity upon multicopy expression of the cyclase PA4843 (Fig. 4D). The β-galactosidase activity in attached cells overexpressing the cyclase PA4843 at the 6 hr timepoint was similar to the activity detected in the PbrlR-gfp cells attached for 24 hr (Fig. 4D). It is of interest to note that multicopy expression of the cyclase PA4843 correlated with a significant increase in c-di-GMP levels compared to vector control at the 6 hr time point (Fig. 4B). In fact, c-di-GMP levels in attached cells overexpressing cyclase PA4843 were similar to those detected in cells attached for 24 hr (Fig. 4B). However, no significant difference in the number of attached cells was noted upon multicopy expression of the cyclase PA4843 indicating that the difference in brlR expression is likely due to elevated c-di-GMP levels rather than increased biofilm biomass accumulation (Fig. 4E). Moreover, at the 6 hr time point, overexpression of PA4843 correlated with a significant, 7.4±1.3-fold increase in the previously described BrlR-target mexE encoding a component of the MexEF-OprN multidrug efflux pump (Liao et al., 2013), compared to attached cells not overexpressing PA4843. This observation suggested that overexpression of the cyclase PA4843 and subsequent formation of the secondary messenger c-di-GMP enhances brlR promoter reporter activity, and thus, brlR expression as well as expression of BrlR target genes in P. aeruginosa. Moreover, our findings suggested that unlike the known MerR regulators BmrR and TipA, brlR expression is linked to attachment and/or biofilm developmental conditions rather than induced upon exposure to antimicrobial agents.
BrlR is a c-di-GMP responsive transcriptional regulator
To determine whether enhanced brlR expression in the presence of c-di-GMP correlated with enhanced BrlR activity and thus, BrlR-DNA binding, electrophoretic mobility shift assays were performed in the absence or presence of c-di-GMP. While BrlR-DNA binding was detected in the absence of c-di-GMP, addition of c-di-GMP resulted in increased BrlR-DNA binding (Fig. 5A). While c-di-GMP enhanced DNA binding by BrlR, no additional shift was noted (Fig. 5A). To further explore the role of c-di-GMP in BrlR-DNA binding, DNA binding assays using streptavidin magnetic beads were performed in the absence and presence of increasing c-di-GMP concentrations, using the biotinylated PbrlR DNA. BrlR-DNA binding was detected using anti-V5 antibodies. While BrlR-DNA binding was detected in the absence of c-di-GMP, addition of c-di-GMP resulted in increased BrlR-DNA binding in a concentration-dependent manner (Fig. 5B). When increasing amounts of unlabeled PbrlR fragment (Fig. 5C) were added to the streptavidin DNA binding assay, decreased BrlR binding to the labeled fragment was observed. Under the conditions tested, no BrlR binding to the beads alone was detected. Likewise, no DNA binding was observed using cell extracts obtained from P. aeruginosa ΔbrlR (Fig. 5B-C). Enhanced DNA binding was not limited to PbrlR as BrlR-binding to its target promoter PmexE was likewise enhanced by the presence of c-di-GMP (Fig. 5D).
Figure 5. BrlR is a c-di-GMP responsive DNA binding protein.
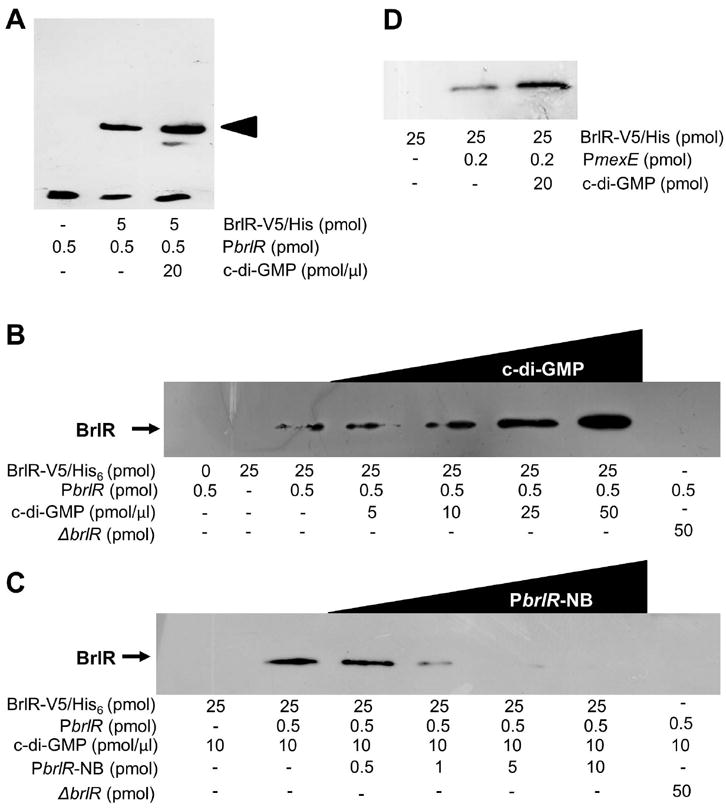
(A) BrlR-DNA gel mobility shift assays using (A) biotinylated PbrlR DNA (0.5 pmol) in the absence and presence c-di-GMP. BrlR-DNA binding was detected by immunoblot analysis using anti-biotin antibodies. Arrowhead denotes shift. (B) Streptavidin BrlR-DNA binding assays using biotinylated PbrlR DNA (0.5 pmol) in the absence and presence of increasing concentrations of c-di-GMP. A total of 25 pmol of purified His6V5-tagged BrlR was used. BrlR-DNA binding was detected by immunoblot analysis using anti-V5 antibodies. (C) BrlR binding to brlR promoter DNA can be outcompeted by increasing concentrations of non-biotinylated PbrlR DNA (PbrlR-NB). (D) BrlR binding to biotinylated PmexE (0.2 pmol) is enhanced by the presence of c-di-GMP as indicated using streptavidin binding assays. Experiments were carried out in triplicate and representative images are shown.
BrlR binds to c-di-GMP in vitro
While lacking predicted secondary structure resembling those of known c-di-GMP binding regions, our findings not only indicated that BrlR-DNA binding was enhanced by c-di-GMP but also that BrlR may be a c-di-GMP responsive transcriptional regulator. To determine whether BrlR binds c-di-GMP in vitro, we made use of a pulldown assay using biotinylated c-di-GMP, with BrlR binding to c-di-GMP being detected by immunoblot analysis using anti-V5 antibodies. V5-tagged BdlA which has been reported to bind heme but not c-di-GMP (Petrova & Sauer, 2012b), as well as the c-di-GMP-responsive transcriptional regulator FleQ that binds c-di-GMP half-maximal at a concentration of 15–25 μM (Hickman & Harwood, 2008) were used as controls. While no c-di-GMP binding was noted for BdlA, FleQ bound c-di-GMP as expected in a concentration-dependent manner (Fig. 6A). Likewise, BrlR bound c-di-GMP in a concentration-dependent manner with c-di-GMP binding detectable at as little as 1.25 μg BrlR (Fig. 6A). c-di-GMP binding to BrlR was characteristic with first order binding kinetics (Fig. 6B). Under the conditions tested, half-maximal binding of c-di-GMP occurred at 1.5 ug BrlR which is equivalent to a concentration of 2.2 μM (Fig. 6B). No c-di-GMP binding was observed to a no-protein control or to cell extracts obtained from P. aeruginosa ΔbrlR (Fig. 6A). BrlR binding of biotinylated c-di-GMP was outcompeted by non-biotinylated c-di-GMP (Fig. 6C) but not by GTP or cyclic AMP (cAMP, Fig. 6C) indicating a specificity of BrlR to c-di-GMP.
Figure 6. BrlR binds c-di-GMP in vitro.
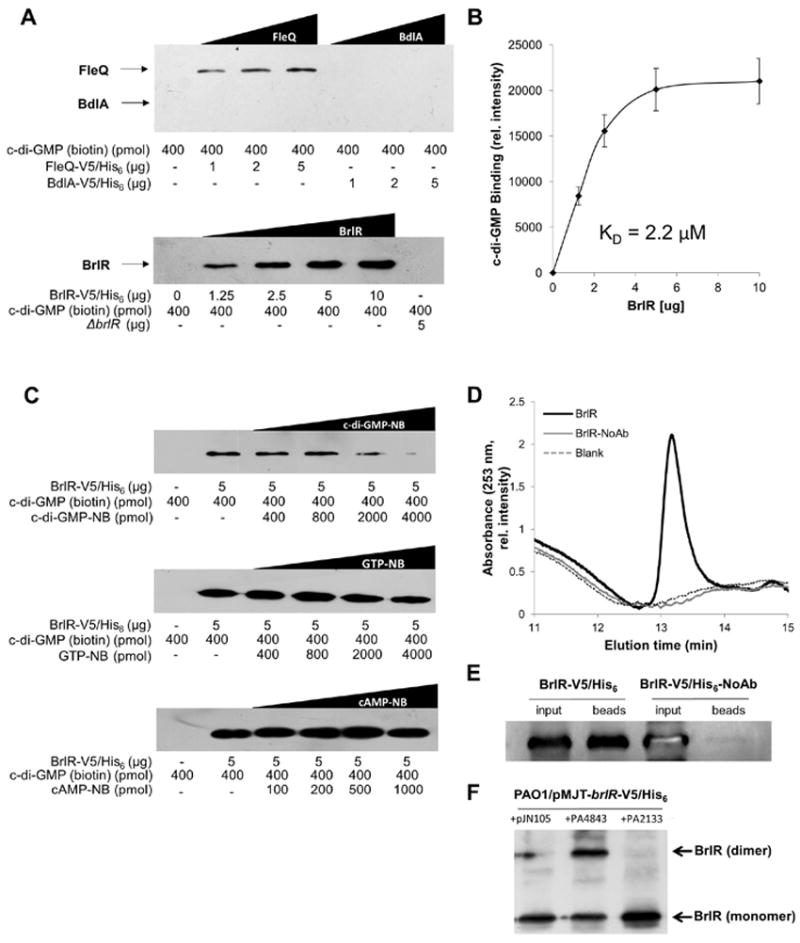
(A) Pulldowns using biotinylated c-di-GMP immobilized to streptavidin magnetic beads demonstrating that BrlR binds c-di-GMP. Increasing concentrations of V5-tagged BrlR were used (1.25-10 μg). A no-protein sample (0 μg) and cell extracts obtained from P. aeruginosa inactivated in brlR (ΔbrlR) were used as controls. BrlR-c-di-GMP binding was detected by immunoblot analysis using anti-V5 antibodies. V5-tagged FleQ and BdlA, having apparent mass of 58.3 and 49.9 kDa, respectively, were used as controls. (B) Concentration-dependent binding of biotinylated c-di-GMP by BrlR. Data are based on relative intensity of bands detectable following probing with anti-biotin antibodies and subsequent analysis using ImageJ. KD, dissociation constant for for c-di-GMP binding to BrlR. (C) BrlR-c-di-GMP binding assays in the presence of increasing concentrations of non-biotinylated c-di-GMP (c-di-GMP-NB), GTP (GTP-NB) and cyclic AMP (cAMP-NB). BrlR-c-di-GMP binding was detected by immunoblot analysis using anti-V5 antibodies. (D) Detection of BrlR-bound c-di-GMP by HPLC analysis. Detection was based on absorbance at 253 nm. C-di-GMP was extracted from BrlR-pulldowns in the absence and presence of anti-V5 antibodies by heat and ethanol extraction. A total of 2 mg of cell extract obtained from P. aeruginosa PAO1/pMJT-brlR-His6V5 was used per pulldown. (E) Immunoblot analysis demonstrating absence/presence of BrlR in pulldown samples prior to (input) and following immunoprecipitation (beads). The respective immunoprecipitation eluates (from beads) were used for c-di-GMP extraction and subsequent HPLC analysis. Immunoblots were probed using anti-V5 antibodies. (F) BrlR oligomerization as determined using immunoblot analysis and crosslinking. Increased dimerization of BrlR is detectable upon multicopy expression of the cyclase PA4843, while only BrlR monomers are detectable upon overexpression of the phosphodiesterase PA2133 as indicated using in vivo crosslinking with dithiobis(succinimidyl propionate) (DSP) and probing with anti-V5 antibodies. Experiments were carried out at least in duplicate and representative data are shown. Error bars represent the standard deviations between replicates
In addition, detection of direct binding of c-di-GMP to BrlR was done using pulldowns, in conjunction with HPLC-based detection of c-di-GMP. To do so, P. aeruginosa-derived BrlR was first immunoprecipitated using anti-V5 antibodies. BrlR-bound c-di-GMP was subsequently extracted from the pulled down protein, and the resulting extracts were subjected to HPLC-based detection of c-di-GMP. Under the conditions tested, extracts obtained from pulldowns using His6V5-tagged BrlR protein in the presence of anti-V5 antibodies were found to contain c-di-GMP (Fig. 6D) based on the identical elution time with commercially available c-di-GMP (Fig. S5). Overall, up to 14 pmol c-di-GMP were detected per 1 μg of BrlR suggesting the presence of 1 pmol c-di-GMP per 2 pmol BrlR. In contrast, no c-di-GMP was detected in pulldown controls lacking anti-V5 antibodies (Fig. 6D). Detection of c-di-GMP by HPLC correlated with the presence of His6V5-tagged BrlR in pulldown eluates as determined by immunoblot analysis (Fig. 6E).
The presence of 1:2 ratio of c-di-GMP:BrlR suggested BrlR dimerization. To confirm dimer formation of BrlR in the presence of c-di-GMP, we made use of in vivo crosslinking of planktonic PAO1/pMJT-brlR-His6V5 either overexpressing the cyclase PA4843 or the phosphodiesterase PA2123. PAO1/pMJT-brlR-His6V5 harboring the empty plasmid pJN105 was used as control. Following cell lysis, the resulting cell extracts were separated by SDS/PAGE, blotted onto PVDF, and probed for BrlR using anti-V5 antibodies. While BrlR in its monomeric form (apparent mass of 33 kDa) was detectable under all conditions tested, a faint band the apparent mass of a BrlR-dimer was detected in of PAO1/pMJT-brlR-His6V5 harboring the empty plasmid pJN105 (Fig. 6F). The band increased in intensity upon overexpression of the cyclase PA4843 but disappeared upon overexpression of the phosphodiesterase PA2123 (Fig. 6F). The finding suggested c-di-GMP to enhance dimerization of BrlR.
DISCUSSION
The MerR family is a group of transcriptional activators with similar N-terminal helix-turn-helix DNA binding regions but diverse C-terminal effector binding regions that are specific to the effector recognized. The signature of the family is amino acid similarity in the first 100 amino acids, including a helix-turn-helix motif followed by a coiled-coil region. The MerR family of transcriptional regulators have further in common that they act on their own promoter to not only regulate their own expression but also induce the expression of their respective regulons. In the present study, we likewise demonstrated that BrlR is capable of binding to its own promoter, likely located within a spacer region between the putative -10 and -35 regions located 5-35 bp upstream of the brlR transcriptional start site. The respective sequence was found to be 19 bp long and characterized by the presence of a perfect inverted repeat, with similar palindromic sequences being present in the previously identified BrlR-dependent mexAB-oprM and mexEF-oprN promoters. The presence of a 19 bp-long spacer region is not uncommon. All MerR family promoter so far identified have an elongated spacer region between the −10 and −35 sequences including all metal-dependent promoters such as the merR, cueR, and coaR promoters (Brown et al., 2003, Ahmed et al., 1994). Moreover, the palindromic spacer region has been shown in many cases to be the regulator binding site (Brown et al., 2003). Likewise, BmrR binds to a dyad symmetrical sequence in the 19 bp spacer region of the bmr promoter and affinity of BmrR for this binding site is increased by addition of the co-activators, rhodamine or tetraphenylphosphonium (TPP) (Newberry & Brennan, 2004, Grkovic et al., 2002). Enhanced DNA binding via a co-activator or inducer is not limited to BmrR as the majority of regulators in the MerR family respond to environmental stimuli, such as oxidative stress, heavy metals or antibiotics (Brown et al., 2003, Ahmed et al., 1994, Heldwein & Brennan, 2001). Similar to other known MerR proteins, DNA binding of BrlR is enhanced by an inducer. However, instead of an environmental stimulus prevalent under biofilm conditions or a multidrug transporter substrate(s) acting as an inducer, here we demonstrate BrlR to be a c-di-GMP responsive regulator, with elevated levels of c-di-GMP not only enhancing BrlR-DNA binding in vitro but also resulting in increased brlR expression for initially attached cells in vivo. Considering that high concentrations of c-di-GMP have been correlated with the sessile lifestyle (e.g. biofilm formation) while its absence or low levels favor motility (e.g. twitching, swarming) and the free living lifestyle (D’Argenio & Miller, 2004), the role of c-di-GMP in BrlR-DNA binding and brlR expression are in agreement with the observed timing of induction of brlR expression upon the transition to surface-associated growth (Liao & Sauer, 2012). Such growth mode dependent expression is unique among members of the MerR family. What makes BrlR stand out even further among members of the MerR family is its c-di-GMP binding capability, with c-di-GMP acting as an inducer. Elevated levels of c-di-GMP were found to correlate with enhanced BrlR dimer formation. While it is unclear whether dimer formation by BrlR is a direct consequence of c-di-GMP binding, other MerR-like regulators have been demonstrated to be dimeric DNA-binding proteins including MerR from B. subtilis and SoxR from E. coli (Helmann et al., 1990, Shewchuk et al., 1989, Watanabe et al., 2008). Moreover, high c-di-GMP levels correlated with increased BrlR binding to PmexE and increased expression of mexE, indicating c-di-GMP also contributes to the expression of BrlR-target genes.
The finding of brlR expression following attachment and BrlR activation being c-di-GMP dependent is in agreement with recent findings of P. aeruginosa biofilms gaining their extraordinary tolerance soon after surface attachment, a stage requiring the action of the two-component hybrid SagSs and with attached cells being characterized by elevated c-di-GMP levels (Petrova & Sauer, 2009, Petrova & Sauer, 2011, Gupta et al., 2013). BrlR has been demonstrated to contribute to Pseudomonas aeruginosa biofilm drug tolerance to antimicrobial agents known as multidrug efflux pumps substrates by activating multidrug-efflux pumps (Liao et al., 2013) but to suppress colistin resistance by repressing of phoPQ expression (Chambers & Sauer, 2013). Our results are in agreement with findings by Chua et al. (Chua et al., 2013) demonstrating that reduced c-di-GMP levels present in dispersed cells correlated with increased production of proteins involved in antimicrobial peptide resistance and increased resistance towards colistin compared to biofilm cells. Together, these observations may explain why colistin, unlike many other antibiotics, kills P. aeruginosa cells in the center rather than the periphery of the biofilm (Pamp et al., 2008)
To our knowledge, no other transcriptional regulator of the MerR family has been described to bind c-di-GMP. Our findings suggest BrlR to be a member of a new and growing class of c-di-GMP binding proteins that are transcriptional regulators and only the second c-di-GMP-responsive transcriptional regulator to be described in P. aeruginosa. C-di-GMP responsive transcriptional regulators include the P. aeruginosa flagella biosynthesis gene activator FleQ (Hickman & Harwood, 2008), the Mycobacterium smegmatis LtmA that broadly regulates the expression of lipid transport and metabolism genes (Li & He, 2012), the Xanthomonas campestis global regulator Clp (Tao et al., 2010), the Burkholderia cenocepacia biofilm formation and virulence regulator Bcam1349 (Fazli et al., 2011), and VpsT responsible for regulating matrix production and motility in Vibrio cholerae (Krasteva et al., 2010). LtmA was found to have a strong binding affinity to c-di-GMP as characterized by a Kd for the specific interaction between LtmA and c-di-GMP of 0.83 μM (Li & He, 2012), FleQ appears to more weakly bind to c-di-GMP as indicated by a Kd of 15-20 μM (Hickman & Harwood, 2008). In comparison, BrlR was characterized by a Kd of 2.2 μM indicating BrlR to bind c-di-GMP stronger than FleQ but weaker than LtmA. The findings suggest BrlR activation and thus, activation of BrlR target genes in P. aeruginosa to occur at lower c-di-GMP levels or at earlier stages of biofilm development than FleQ. While capable of c-di-GMP binding, BrlR, FleQ, and LtmA lack regions that have a predicted secondary structure resembling those of known c-di-GMP binding regions, such as the P. aeruginosa PelD protein, PilZ domains or the I-sites of diguanylate cyclases (Amikam & Galperin, 2006, Chan et al., 2004, Wassmann et al., 2007, De et al., 2008). Likewise, BrlR does not contain any regions with similarity to known c-di-GMP binding regions. However, the in vitro system that we have established for examining BrlR binding to DNA and/or c-di-GMP provides an opportunity to examine in detail the mechanism by which c-di-GMP controls the biochemical activity of a transcriptional regulator as well as reveal alternative determinants of c-di-GMP binding.
Taken together, our results indicate BrlR to be a c-di-GMP responsive transcriptional regulator, with BrlR-DNA binding and brlR expression being enhanced by the secondary messenger c-di-GMP. Considering biofilms having been correlated with elevated levels of c-di-GMP, and that BrlR contributes to the drug tolerance of P. aeruginosa biofilms, with elevated c-di-GMP resulting in increased brlR expression and activation, our findings strongly suggest a contribution of c-di-GMP to biofilm tolerance.
Material and Methods
Bacterial strains, plasmids, media, and culture conditions
All bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strain PAO1 was used as parental strain. All planktonic strains were grown in Lennox Broth (LB, BD Biosciences) in flasks at 220 rpm in the absence or presence of 0.1-1.0% arabinose. Escherichia coli cultures were grown in LB in the absence or presence of 1mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Antibiotics were used at the following concentrations: 50-75 μg/mL gentamicin, 200-250 μg/mL carbenicillin, and 60-75 μg/mL tetracycline for P. aeruginosa; and 20 μg/mL gentamicin, 50 μg/mL ampicillin and 20 μg/mL tetracycline for E. coli.
Table 1.
Strains and plasmids used.
| Strains/Plasmids | Relevant genotype or description | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F- φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk-, mk+) phoA supE44 thi-1 gyrA96 relA1 tonA | Invitrogen Corp. |
| BL21 | F- ompT hsdSB (rB-mB-) gal dcm rne131 (DE3) | Invitrogen Corp |
| Pseudomonas aeruginosa PAO1 | ||
| PAO1 | Wild type strain PAO1 | B.H. Holloway |
| ΔbrlR | PAO1, ΔbrlR (PA4878) | (Liao & Sauer, 2012) |
| Plasmids | ||
| pET101D | Vector for directional cloning and His6V5fusion protein expression, AmpR | Invitrogen Corp |
| pRK2013 | Helper plasmid for triparental mating; mob; tra; KmR | (Figurski & Helinski, 1979) |
| pJN105 | Arabinose-inducible gene expression vector; pBRR-1 MCS; araC-PBAD; GmR | (Newman & Fuqua, 1999) |
| pMJT1 | araC-PBAD cassette of pJN105 cloned into pUCP18, AmpR (CarbR) | (Kaneko et al., 2007) |
| pJN-PA4843 | PA4843 cloned into pJN105; GmR | (Petrova & Sauer, 2012a) |
| pJN-brlR | brlR cloned into pJN105; GmR | (Liao & Sauer, 2012) |
| pJN-bdlA-His6V5 | C-terminal V5/6xHis-tagged bdlA cloned into pJN105 at EcoRI/SpeI | (Petrova & Sauer, 2012b) |
| pMJT-brlR-His6V5 | brlR- His6V5 cloned into pMJT1 | (Liao & Sauer, 2012) |
| pMJT-fleQ-V5 | fleQ-V5 cloned into pMJT1 | This study |
| pMini-CTX-lacZ | attP site-specific integration vector, TetR | (Becher & Schweizer, 2000) |
| pbrlR-23-lacZ | pMini-CTX-lacZ with 1-23 bp upstream of brlR, TetR | This study |
| pbrlR-120-lacZ | pMini-CTX-lacZ with 1-120 bp upstream of brlR, TetR | (Liao & Sauer, 2012) |
| pbrlR-250-lacZ | pMini-CTX-lacZ with 1-250 bp upstream of brlR, TetR | This study |
| pbrlR-500-lacZ | pMini-CTX-lacZ with 1-500 bp upstream of brlR, TetR | This study |
| pMini-CTX-gfp | attP site-specific integration vector, TetR | (Becher & Schweizer, 2000) |
| pbrlR-gfp | pMini-CTX-gfp with 1-750 bp upstream of brlR, TetR | This study |
Strain Construction
Reporter constructs harboring putative promoter sequences of brlR were generated by introducing the respective sequences into pMini-CTX-lacZ or pMini-CTX-gfp. C-terminal V5-tagging of FleQ was accomplished introducing the V5-tag via PCR (using the sequence ggtaagcctatccctaaccctctcctcggtctcgattctacg). The tagged construct was introduced into pMJT-1. The identity of vector inserts was confirmed by sequencing and proper integration of the pMini-CTX vectors confirmed by PCR using Pser-up/-down PCR primers. Plasmids were introduced into P. aeruginosa via conjugation or electroporation. Primers used are listed in Table S1.
Biofilm formation
For reporter studies, biofilms were grown in a continuous flow tube reactor system (1 m long size 13 silicone tubing, Masterflex, Cole Parmer, Inc.) with an inner surface area of 25 cm2 at a flow rate of 0.1 ml/min and in flow cell reactors (BioSurface Technologies) which also allowed for the analysis of biofilm architecture as previously described (Sauer et al., 2002, Sauer et al., 2004, Petrova & Sauer, 2009). Biofilms were grown at 22°C in 20-fold diluted LB medium for up to 4 days. The same growth conditions were used to cultivate biofilms to obtain RNA.
Transcriptional analysis
Microscopic analyses of brlR expression during surface-attached growth were accomplished by allowing the respective reporter strain (PAO1∷PbrlR-gfp) to attach to microscope slides under flowing conditions in flow cells, as previously described (Sauer et al., 2002). Expression of gfp was monitored by confocal scanning laser microscopy using a Leica TCS SP5 confocal microscope (Leica Microsystems, Wetzlar, Germany). To quantitate brlR expression, PbrlR-lacZ fusions were used. β-galactosidase activity of strains harboring the PbrlR-lacZ promoter reporters was determined using the Miller assay (Miller, 1972) with the following modification: instead of using total cells, specific β-galactosidase activity was determined using protein extracts (Petrova & Sauer, 2010, Petrova et al., 2011). Cells were lysed by sonication as previously described (Petrova & Sauer, 2009), and the resulting lysate centrifuged for 2 min at 21,200 × g to pellet unbroken cells and cell debris. Protein determination was done using the modified method of Lowry (Peterson, 1977). A total of 5-10 μg of cell extract was used per assay. An extinction coefficient for o-nitrophenyl-β-galactoside cleavage at 420 nm of 4500 nl/nmol*cm was used.
Transcriptional start site mapping
The transcriptional start site of brlR was determined using Invitrogen’s 5′ RACE system for rapid amplification of cDNA ends (version 2.0) as recommended by the manufacturer (Invitrogen). RNA was isolated from 4-day-old P. aeruginosa wild type biofilms as described above and used as the template. Oligonucleotides used for reverse transcription and PCR are listed in Table S1. Products were separated by agarose gel electrophoresis to assess purity and product size. If necessary, products were excised and gel eluted (Qiagen). The 5′ RACE products were sequenced by the Cornell University Life Sciences Core Laboratories Center (CLC). The sequencing results were interpreted by pairwise alignments of the 5′ RACE product sequence with the P. aeruginosa PAO1 genomic sequence.
Chromatin immunoprecipitation (ChIP) analysis
In order to determine whether BrlR binds to its own promoter region in vivo, 24-hr-old biofilms of P. aeruginosa PAO1/pMJT-brlR-V5/His6, bearing the His6V5-tagged BrlR, were subjected to chromatin immunoprecipitation (ChIP) analysis as previously described (Petrova et al., 2011). P. aeruginosa PAO1 expressing untagged brlR was used as control. Briefly, in vivo DNA-protein crosslinking using 1% formaldehyde for 10 min at 37°C and immunoprecipitation using anti-V5 antibodies (Invitrogen Corp.) were done essentially as previously described (Chiu & Thomas, 2004, Solomon & Varshavsky, 1985, Leech et al., 2008, Liao et al., 2013). Following immunoprecipitation, DNA was liberated by reversing the crosslinking via incubation with 0.5 M NaCl in TE at 65°C for 4 hr. Purified DNA from PAO1/pJN-brlR and PAO1/pMJT-brlR-V5/His6 samples was subjected to qPCR using primers listed in Table S1. Relative transcript quantitation was accomplished using the ep realplex software (Eppendorf AG) by first normalizing transcript abundance (based on Ct value) to mreB followed by determining transcript abundance ratios for PbrlR and PpscE-F using primers listed in Table S1. Melting curve analyses were employed to verify specific single product amplification.
Purification of His-tagged BrlR
For electrophoretic mobility shift assay (EMSA), His6V5-tagged BrlR protein was purified from E. coli or P. aeruginosa. For in vitro c-di-GMP binding assays, His6V5-tagged BrlR was purified from P. aeruginosa. Briefly, supernatants following sonication of planktonic cells, and centrifugation at 21,200 × g were loaded onto nickel-nitrilotriacetic acid (Ni-NTA) affinity resin (Qiagen), washed with buffer, and eluted with an imidazole gradient according to the manufacturer’s instructions for native protein purification. Protein preparations were examined for purity by SDS-PAGE, and fractions containing His6V5-tagged BrlR were pooled and desalted using VivaSpin centrifugal concentrator columns (10 kDa cut-off, Sartorius).
Electrophoretic mobility shift assay (EMSA)
BrlR binding to the region upstream of brlR start codon was performed as described previously (Fuchs et al., 2010, Jones et al., 2010, Liao et al., 2013) using purified His6V5-tagged BrlR. BrlR purification was accomplished by Ni-NTA affinity chromatography (QIAGEN) following the manufacturer’s instructions (see Fig. 3D). The DNA promoter probe was generated by PCR using the indicated oligonucleotides (PbrlR, Table S1) and end-labeled using 10 μCi of [γ-32P]-ATP (GE Healthcare) and 10 U of T4 polynucleotide kinase (New England Biolabs). EMSAs were performed as previously described (Fuchs et al., 2010, Jones et al., 2010). Briefly, probes (0.25 nM each) were incubated in binding buffer (10 mM Tris [pH 7.5], 50 mM KCl, 1 mM EDTA, 1 mM DTT, 5% glycerol and 100 μg/ml bovine serum albumin) containing 50 ng/ml poly (2’-deoxyinosinic-2’-deoxycytidylic acid) [poly(dI-dC)] (Sigma) for 5 min at 25°C. BrlR protein was then added at indicated concentrations for a final reaction volume of 20 μl and incubated for an additional 15 min at 25°C. Samples were subjected to electrophoresis on a 6% polyacrylamide glycine gel (10 mM Tris [pH 7.5], 380 mM glycine, 1 mM EDTA) at 4°C. Imaging and data analyses were performed using a Molecular Imager FX phosphorimager (BioRad) and Imagequant software. Moreover, we made use of biotinylated rather than radiolabeled DNA fragments. To do so, we made use of the LightShift Chemiluminescent EMSA kit (Thermo Scientific) as previously described (Petrova & Sauer, 2010) for determining BrlR-DNA binding to brlR promoter DNA (PbrlR), DNA sequences spanning the 7-108 and the 5-37 bp region located upstream of the transcriptional start site. The respective regions were amplified using primers listed in Table S1. His6V5-tagged BrlR obtained from planktonic cells (5-20 pmol) was used. Samples were separated as described above, blotted onto a Hybond nylon membrane and probed using anti-biotin antibodies. Bands were visualized using the LightShift Chemiluminescent EMSA kit (Thermo Scientific).
Streptavidin magnetic bead DNA binding assay
BrlR binding to the putative brlR promoter DNA was confirmed using the streptavidin magnetic bead DNA binding assay as previously described (Petrova et al., 2011). Briefly, biotinylated target DNA fragment PbrlR was amplified using the primer pair PbrlR-7-108F/R (PbrlR, spanning the 7-108 bp upstream of start codon, Table S1). A total of 0.5 pmol of target DNA was incubated for 30 min at room temperature with cell extract containing 25 pmol of His6V5-tagged BrlR (as indicated by immunoblot analysis using purified His6V5-tagged BrlR) in 25 mM Tris-Cl, pH 8, 5 mM MgCl2, 0.5 mM dithiothreitol, 1 mM EDTA, and 50 ng/uL poly(dI-dC) as nonspecific competitor DNA. For specific competition, non-biotinylated PbrlR target DNA (0-50 pmol) was used. c-di-GMP was added where indicated. Briefly, streptavidin magnetic beads (Thermo Scientific, 100 μg) were used to capture biotinylated DNA. Following three washes, the proteins co-purified with the biotinylated DNA were separated by 10% SDS/PAGE and assessed by immunoblot analysis for the presence of BrlR using anti-V5 antibodies (Invitrogen). An aliquot prior to addition of streptavidin magnetic beads was used to determine total BrlR present in each DNA binding assay (loading control). Bands were visualized by chemiluminescence.
Bioinformatics
Putative BrlR-DNA binding sequences were identified by aligning the upstream DNA region of brlR to the BmrR-DNA binding sequence. The putative BrlR-DNA binding sequences were analyzed for inverted repeats using the palindrome program (http://emboss.bioinformatics.nl/cgi-bin/emboss/palindrome) and default settings.
Detection of and quantitation of BrlR-c-di-GMP binding
To assess the interaction between c-di-GMP and BrlR, the binding was analyzed by pulldown assays using streptavidin magnetic beads (Thermo Scientific, 100 μg) to capture biotinylated c-di-GMP. Following three washes, the beads harboring biotinylated c-di-GMP were incubated with increasing concentration of cell extract (0-10 μg) containing His6V5-tagged BrlR. Cell extracts inactivated in brlR (ΔbrlR) were used as a negative control. Following three washes, proteins co-purified with the biotinylated c-di-GMP were separated by 10% SDS/PAGE and assessed by immunoblot analysis for the presence of BrlR using anti-V5 antibodies (Invitrogen). Bands were visualized by chemiluminescence. Pulldowns were repeated in the presence of non-biotinylated c-di-GMP (0-1000 pmol), GTP (0-1000 pmol), and cAMP (0-1000 pmol).
To directly detect c-di-GMP binding to BrlR and subsequently quantitate bound c-di-GMP, pulldowns were used as previously described (Petrova & Sauer, 2011, Petrova et al., 2011) with some modifications. Briefly, pulldowns were carried out in the presence of 200 pmol c-di-GMP using whole-cell lysates obtained from PAO1/pMJT-brlR-His6V5 grown planktonically to exponential phase and protein A/G agarose beads (Cell Signaling) linked with anti-V5 antibodies (Invitrogen). Controls were done in the absence of anti-V5 antibodies. Following immunoprecipitation using immobilized anti-V5 antibodies, the resulting precipitate was subjected to heat and ethanol precipitation (Morgan et al., 2006) to extract c-di-GMP. C-di-GMP was subsequently extracted in duplicate from pulldowns in the presence and absence of anti-V5 antibodies and quantitated essentially as previously described (Petrova & Sauer, 2011, Basu Roy et al., 2012, Basu Roy et al., 2013). Briefly, c-di-GMP was extracted in duplicate using heat and ethanol precipitation followed by centrifugation. Supernatants were combined, dried using a Speed-Vac and resuspended in 10 mM ammonium acetate. Samples (20 μl) were analyzed using an Agilent 1100 HPLC equipped with an autosampler, degasser, and detector set to 253 nm, and separated using a reverse-phase C18 Targa column (2.1 × 40 mm; 5 μm) at a flow rate of 0.2 ml/min with the following gradient: 0 to 9 min, 1% B; 9 to 14 min, 15% B; 14 to 19 min, 25% B; 19 to 26 min, 90% B; 26 to 40 min, 1% B (buffer A, 10 mM ammonium acetate; buffer B, methanol plus 10 mM ammonium acetate). Commercially available cyclic di-GMP (Biolog) was used as a reference for the identification and quantification of cyclic di-GMP in cell extracts. An aliquot of the pulldown reaction mixture prior to and following immunoprecipitation was retained and subsequently analyzed by immunoblot analysis as described above using anti-V5 antibodies to assess the presence of V5/His-tagged BrlR but the absence of V5/His-tagged BrlR in controls.
Detection of BrlR dimers by protein crosslinking
Dimerization of BrlR was determined via reversible in vivo crosslinking with dithiobis(succinimidyl propionate) (DSP) as previously described (Petrova & Sauer, 2012a). Whole cells of PAO1/pMJT-brlR-His6V5 harboring the empty plasmid pJN105, or PAO1/pMJT-brlR-His6V5 either overexpressing the cyclase PA4843 or the phosphodiesterase PA2123, were used. Briefly, following incubation for 9 min with 2mM DSP (stock solution prepared at 15mM in DMSO), thus treated cells containing V5-tagged BrlR were centrifuged for 1 min at 16000×g, for a total crosslinking time of 10 min. The crosslinking reactions were stopped by resuspending and lysing the cells via sonication directly into TE buffer (10 mM Tris/HCl pH 8.0, 1 mM EDTA, plus 0.3 μg/ml PMSF). Crosslinking was reversed using β-mercaptoethanol (β-ME), while crosslinked samples were treated using a non-reducing SDS-PAGE loading buffer. The resulting cell extracts were separated by SDS/PAGE, and assessed by immunoblot analysis for the presence of V5/His-tagged BrlR using anti-V5-HRP antibodies (0.1 ug/ml, Invitrogen Corp.).
Statistical analysis
A Student’s t-test was performed for pair-wise comparisons of groups, and multivariant analyses were performed using a 1-Way ANOVA followed by a posteriori test using Sigma Stat software.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01 AI080710).
References
- Ahmed M, Borsch CM, Taylor SS, Vazquez-Laslop N, Neyfakh AA. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J Biol Chem. 1994;269:28506–28513. [PubMed] [Google Scholar]
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44:1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar H, Strap JL, Chen K, Costerton JW. Dynamic interactions of biofilms of mucoid Pseudomonas aeruginosa with tobramycin and piperacillin. Antimicrob Agents Chemother. 1992;36:1208–1214. doi: 10.1128/aac.36.6.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova NN, Danchin A, Neyfakh AA. Mta, a global MerR-type regulator of the Bacillus subtilis multidrug-efflux transporters. Mol Microbiol. 1999;31:1549–1559. doi: 10.1046/j.1365-2958.1999.01301.x. [DOI] [PubMed] [Google Scholar]
- Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol. 2009;191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Roy A, Petrova OE, Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol. 2012;194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Roy A, Petrova OE, Sauer K. Extraction and Quantification of Cyclic Di-GMP from Pseudomonas aeruginosa. bio-protocol. 2013 doi: 10.21769/bioprotoc.828. http://www.bio-protocol.org/wenzhang.aspx?id=828. [DOI] [PMC free article] [PubMed]
- Becher A, Schweizer HP. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques. 2000;29:948–952. doi: 10.2144/00295bm04. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JAJ, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Hoiby N, Givskov M. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- Campanac C, Pineau L, Payard A, Baziard-Mouysset G, Roques C. Interactions between Biocide Cationic Agents and Bacterial Biofilms. Antimicrob Agents Chemother. 2002;46:1469–1474. doi: 10.1128/AAC.46.5.1469-1474.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JR, Sauer K. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. Journal of Bacteriology. 2013;195:4678–4688. doi: 10.1128/JB.00834-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc National Acad Sci. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C-M, Thomas CM. Evidence for past integration of IncP-1 plasmids into bacterial chromosomes. FEMS Microbiol Lett. 2004;241:163–169. doi: 10.1016/j.femsle.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Chua SL, Tan SY-Y, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, Tolker-Nielsen T, Yang L, Givskov M. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2013;57:2066–2075. doi: 10.1128/AAC.02499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- D’Argenio DA, Miller SI. Cyclic di-GMP as a bacterial second messenger. Microbiology. 2004;150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 2008;6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 2003;5:1213–1219. doi: 10.1016/j.micinf.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Fazli M, O’Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol. 2011;82:327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs EL, Brutinel ED, Jones AK, Fulcher NB, Urbanowski ML, Yahr TL, Wolfgang MC. The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and - independent mechanisms. J Bacteriol. 2010;192:3553–3564. doi: 10.1128/JB.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P, Maira-Litran T, McBain AJ, Rickard AH, Whyte FW. The physiology and collective recalcitrance of microbial biofilm communities. Adv Microb Physiol. 2002;46:202–256. [PubMed] [Google Scholar]
- Godsey MH, Baranova NN, Neyfakh AA, Brennan RG. Crystal structure of MtaN, a global multidrug transporter gene activator. J Biol Chem. 2001;276:47178–47184. doi: 10.1074/jbc.M105819200. [DOI] [PubMed] [Google Scholar]
- Godsey MH, Zheleznova Heldwein EE, Brennan RG. Structural biology of bacterial multidrug resistance gene regulators. J Biol Chem. 2002;277:40169–40172. doi: 10.1074/jbc.R200018200. [DOI] [PubMed] [Google Scholar]
- Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Marques CNH, Petrova OE, Sauer K. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J Bacteriol. 2013;195:4975–4987. doi: 10.1128/JB.00732-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EEZ, Brennan RG. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature. 2001;409:378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]
- Helmann J, Ballard B, Walsh C. The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science. 1990;247:946–948. doi: 10.1126/science.2305262. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DJ, Caso JL, Thompson CJ. Autogenous transcriptional activation of a thiostrepton-induced gene in Streptomyces lividans. EMBO J. 1993;12:3183–3191. doi: 10.1002/j.1460-2075.1993.tb05987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc National Acad Sci. 2012;109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AK, Fulcher NB, Balzer GJ, Urbanowski ML, Pritchett CL, Schurr MJ, Yahr TL, Wolfgang MC. Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cyclic AMP/Vfr signaling. J Bacteriol. 2010;192:5709–5717. doi: 10.1128/JB.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest. 2007;117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech AJ, Sprinkle A, Wood L, Wozniak DJ, Ohman DE. The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J Bacteriol. 2008;190:581–589. doi: 10.1128/JB.01307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Riddle of Biofilm Resistance. Antimicrob Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, He Z-G. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucl Acids Res. 2012;40:11292–11307. doi: 10.1093/nar/gks923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Sauer K. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol. 2012;194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Schurr MJ, Sauer K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug-efflux pumps in Pseudomonas aeruginosa biofilms. J Bacteriol. 2013;195:3352–3363. doi: 10.1128/JB.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Merritt JH, Brothers KM, Kuchma SL, O’Toole GA. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol. 2007:585–507. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N Y. 1972:352–355. [Google Scholar]
- Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, Wozniak DJ. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cellular Microbiology. 2012;14:95–106. doi: 10.1111/j.1462-5822.2011.01704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry KJ, Brennan RG. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J Biol Chem. 2004;279:20356–20362. doi: 10.1074/jbc.M400960200. [DOI] [PubMed] [Google Scholar]
- Newman JR, Fuqua C. Broad-host-range expression vectors that carry the arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- Neyfakh AA. The ostensible paradox of multidrug recognition. J Mol Microbiol Biotechnol. 2001;3:151–154. [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited Bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol. 2008;68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc National Acad Sci. 2012a;109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol. 2012b;194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Schurr JR, Schurr MJ, Sauer K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol. 2011;81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picioreanu C, van Loosdrecht MCM, Heijnen JJ. Two-dimensional model of biofilm detachment caused by internal stress from liquid flow. Biotechnol Bioeng. 2001;72:205–218. [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Sauer K, Camper AK. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol. 2001;183:6579–6589. doi: 10.1128/JB.183.22.6579-6589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewchuk LM, Helmann JD, Ross W, Park SJ, Summers AO, Walsh CT. Transcriptional switching by the MerR protein: activation and repression mutants implicate distinct DNA and mercury(II) binding domains. Biochemistry. 1989;28:2340–2344. doi: 10.1021/bi00431a053. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother. 1996;40:2517–2522. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. The Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Summers AO. Untwist and shout: a heavy metal-responsive transcriptional regulator. J Bacteriol. 1992;174:3097–3101. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao F, He Y-W, Wu D-H, Swarup S, Zhang L-H. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol. 2010;192:1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Zheleznova EE, Markham PN, Brennan RG, Neyfakh AA. Recognition of multiple drugs by a single protein: a trivial solution of an old paradox. Biochem Soc Trans. 2000;28 [PubMed] [Google Scholar]
- Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. Structure of BeF3-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure. 2007;15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Kita A, Kobayashi K, Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 2012;8:e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheleznova EE, Markham PN, Neyfakh AA, Brennan RG. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell. 1999;96:353–362. doi: 10.1016/s0092-8674(00)80548-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


