Significance
Rapid degradation of newly synthesized proteins, followed by presentation of the resulting peptides by the MHC molecules, serves as an early alert for the immune system during pathogen infection. This study defines the relative contribution to the MHC peptidome of defective ribosome products (DRiPs), which are newly synthesized and rapidly degraded proteins, vs. mature proteins, degraded at the end of their functional lifetimes (retirees). The rates of synthesis of the individual MHC peptides and their source proteins were followed using stable isotope labeling and quantitative proteomics and peptidomics analyses. We conclude that DRiPs are a significant source of MHC peptides. Many of these DRiPs are misassembled surplus subunits of protein complexes and therefore are degraded soon after synthesis.
Keywords: DRiPome, immunopeptidome, dynamic SILAC
Abstract
MHC class I peptides are products of endogenous cellular protein degradation. Their prompt presentation, after rapid degradation of their newly synthesized source proteins, is needed to alert the immune system during pathogen infection. A possible source for such rapidly degrading proteins can be defective ribosome products (DRiPs), which include polypeptides produced as part of the pioneer round of translation, premature translation termination, and proteins failing to fold properly or to assemble into their multisubunit protein complexes. However, the identities and relative contribution to the MHC peptidome of these mature or newly synthesized and rapidly degraded cellular proteins is not well understood. To clarify these issues, we used dynamic stable isotope labeling by amino acids in cell culture to define the relative rates of synthesis of the HLA class I peptidomes and the source proteomes of three cultured human hematopoietic cell lines. Large numbers of HLA class I peptides were observed to be derived from DRiPs, defined here as HLA peptides that shift from their light to heavy isotope forms faster than their source proteins. Specific groups of proteins, such as ribosomal and T-complex protein 1 (TCP-1), contributed a disproportionately large number of DRiPs to the HLA peptidomes. Furthermore, no significant preference was observed for HLA peptides derived from the amino terminal regions of the proteins, suggesting that the contribution of products of premature translation termination was minimal. Thus, the most likely sources of DRiPs-derived HLA peptides are full-sized, misassembled, and surplus subunits of large protein complexes.
Animal survival requires prompt alerting of the adaptive immune system to the eminent danger of pathogen infections. Degradation of just a few of the pathogens’ proteins and rapid presentation of their degradation products as MHC peptides are sufficient to raise the needed alarm (1, 2). Degradation of newly synthesized proteins, well before the end of their functional lifetimes and even before they fully mature, is essential for such rapid immune response. However, the cellular mechanisms and the identity of the proteins providing peptides for this rapid response pathway are still ill defined. Furthermore, the question of whether the pool of presented MHC peptides (the MHC peptidome or immunopeptidome) is mostly produced from normal, long-lived proteins, at the end of their functional lifetimes (3), or alternatively, from rapidly degraded immature proteins (4, 5) is not yet settled (6–8). In cultured cells, upward to a third of the MHC peptidome was suggested to be derived from the latter. These rapidly degraded defective ribosome products (DRiPs) (4, 9) were proposed to be produced by exotic mechanisms, such as downstream initiation (10), transcripts associated with specific microRNAs (11), premature termination of translation (12–14), short protein segments, produced by the pioneer round of translation (15) during the nonsense mediated decay (NMD) quality control of mRNA (16, 17), and even out-of-frame translated segments of proteins (18). MHC peptides derived from newly synthesized proteins were also suggested to be produced by degradation of normal, fully functional polypeptides, which are the excess subunits of large protein complexes (19, 20). In those cases, where all of the subunits are not simultaneously ready for assembly, the excess subunits should be degraded to avoid interference with the wellbeing of the cells (21, 22).
The main pathway of cytoplasmic and nuclear protein degradation and subsequent production of MHC peptides is assumed to be proteasomal proteolysis (23). The resulting peptides are transported into the ER through the transporter associated with antigen processing (TAP), where they are loaded onto the awaiting MHC class I molecules by specialized chaperones (24–26). It is also suggested that some of the MHC peptides are produced by degradation of ER-resident proteins and their signal peptides (27, 28). Another source of MHC class I peptides was suggested to be proteolysis through the autophagosome-lysosome pathway (29), a pathway that was previously assigned with production of peptides to the MHC class II peptidome (24).
The rates of synthesis of cellular proteins can be determined by the use of large-scale dynamic stable isotope labeling by amino acids in cell culture (dynamic SILAC) experiments (30) or by its newer version—pulsed SILAC (22), both of which are modifications of the classical (static) SILAC approach (31). SILAC is mostly used for mass spectrometry determination of the relative amounts of proteins by metabolic incorporation of stable isotope labeled amino acids. Similarly to its use for proteomics, static or dynamic SILAC can be used to study the synthesis rates of MHC peptidomes (32) and investigate effects of inhibitors on these peptidomes (33–35). Performing dynamic SILAC at both the proteomics and MHC peptidomics levels, using the same cultured cells, enables the comparison between the synthesis rates of the MHC peptides and their source proteins. This approach provides a powerful way to distinguish between MHC peptides derived from degradation of DRiPs or from mature proteins (retirees) (6, 32, 35, 36). It should be emphasized that retirees can be short-lived proteins (SLiPs), which are normal proteins that have relatively short lifetimes in the cells. They can also be slow turning-over proteins, such as most cellular proteins, which are degraded only hours or days after their synthesis. MHC peptides derived from retirees are typified with slower synthesis rates (observed as slower shifts between their light and heavy forms) relative to their proteins of origin, as measured during combined dynamic SILAC proteomics and MHC peptidomics analyses. Conversely, MHC peptides derived from DRiPs are typified with faster shifts from their light to heavy forms relative to their source proteins (35).
The results from this large-scale dynamic SILAC analysis of the endogenous cellular HLA peptidomes and proteomes support the conclusion that a significant portion of the HLA peptidome is derived from DRiPs. The data provide the identities of large numbers of both DRiPs and retirees derived HLA peptides. Furthermore, the results support the notion that full-size immature proteins contribute significantly to the HLA peptidomes of the studied cultured human cells. Importantly, we noticed that protein subunits of large complexes, such as the ribosomes and TCP-1 complex, contribute disproportionately to the formation of the DRiPs-derived HLA peptidomes.
Results
Comparing the Dynamics of HLA Class I Peptides to Their Source Proteins.
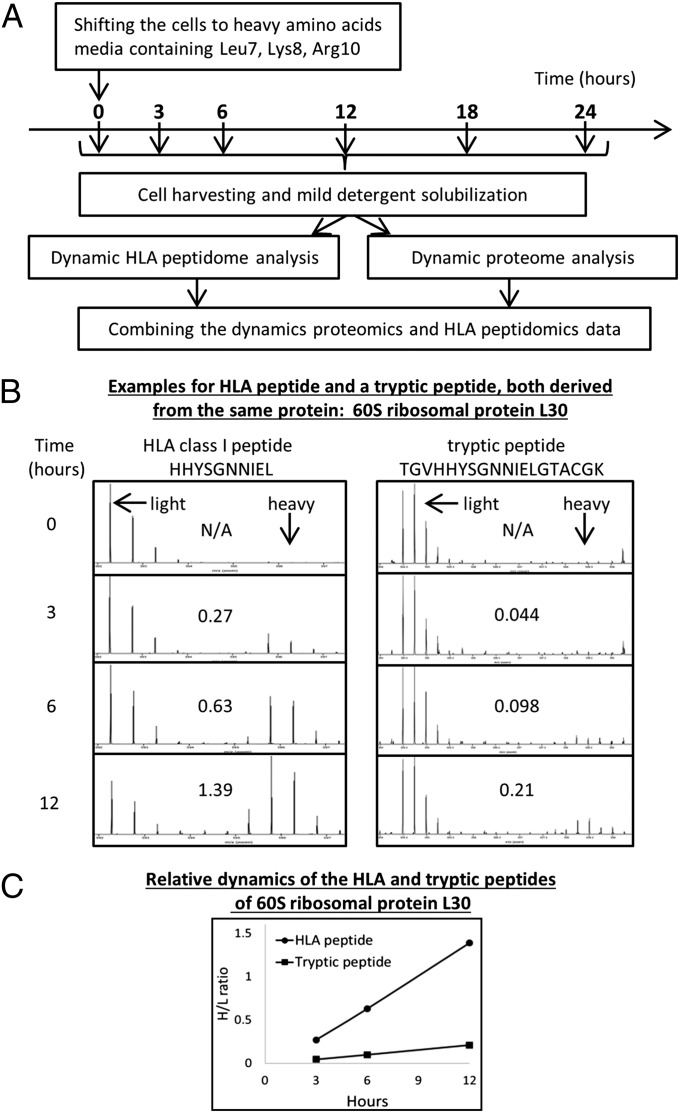
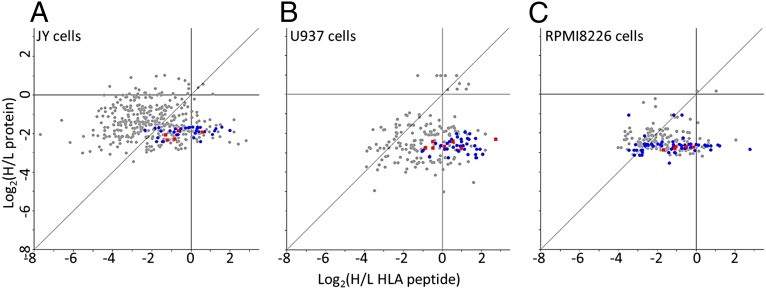
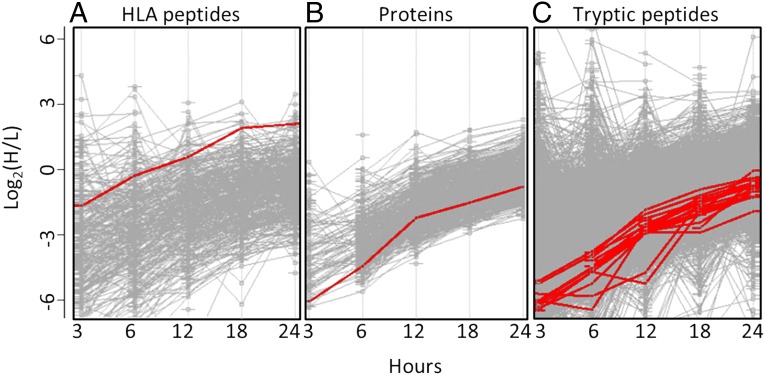
Synthesis rates of the cellular proteins and HLA peptides were determined in parallel from the same human hematopoietic cell cultures. Synthesis rates were followed by the kinetics of incorporation of the three heavy stable-isotope labeled amino acids, lysine (+8), arginine (+10), and leucine (+7), into the cellular proteins and HLA peptides. Leucine was added in addition to the lysine and arginine that are needed for proteomics SILAC (32) because arginine and lysine are not commonly represented within the HLA class I allomorphs expressed in the studied cells. Portions of the cell cultures were taken at 3, 6 12, 18, and 24 h after shifting the cells to the growth media containing the heavy amino acids; the proteins were analyzed after trypsin digestion (the JY cells’ proteins were digested both in solution and in gel slices and the resulting data combined). The HLA class I peptides were analyzed after immunoaffinity purification with the anti-pan HLA-A, -B, and -C monoclonal antibody W6/32. The experimental scheme is displayed in Fig. 1A. An example for measurements of the dynamics of synthesis of an HLA peptide and a tryptic peptide, both derived from the same protein, is shown in Fig. 1B. DRiPs factors are defined as the ratio between the rates of synthesis of the HLA peptides (defined as the heavy to light (H/L) ratios of each HLA peptide at each time point) and of their source proteins (calculated as the median value of H/L ratios of the different tryptic peptides of the protein at each time point). DRiPs-derived HLA peptides shift from their light to heavy forms at the same rate or faster rate than their source proteins, whereas retirees-derived HLA peptides shift slower than their source proteins. Thus, DRiPs factors are ≥1 for DRiPs-derived peptides and <1 for the retirees-derived HLA peptides. The example depicted in Fig. 1 B and C includes the HLA peptide HHYSGNNIEL, which possesses a relatively high DRiPs factor. Its synthesis rate was faster relative to the tryptic peptide TGVHHYSGNNIELGTACGK, derived from the same 60S ribosomal protein L30 (incidentally, this HLA peptide (underlined) is also nested within the displayed tryptic peptide). DRiPs factors of HLA peptide-protein pairs were assumed to be valid only if measurable H/L ratios were observed for the HLA peptides and for at least two tryptic peptides derived from the same protein in at least two shared time points (the example in Fig. 1 B and C meets such criteria). Furthermore, in this study, HLA peptides were defined as derived from relatively certain DRiPs only when they shifted from their light to heavy forms at least 50% (1.5-fold) faster than their source proteins and as derived from certain retirees only if they shifted from the light to heavy forms at 50% slower rates (0.66-fold) relative to their source proteins. Typical examples for datasets of the 12-h time points extracted from the three logarithmically growing cultured cell lines, demonstrating the relative kinetics of the HLA peptides and their source proteins, are shown in Fig. 2. Even though large numbers of HLA peptides and proteins were identified in these analyses, the relative synthesis rates of most of the identified proteins and of the HLA peptides derived from them could not be defined. The reasons for these are the limited overlap between the HLA peptidome and the cellular proteome (32) and the absence of Leu, Arg, and Lys from many of the HLA peptides. The source proteins of many HLA peptides were detected and quantified in the parallel proteomics analyses (statistics in Table 1). The dynamics of synthesis of the proteins and HLA peptides identified and quantified in the JY, RPMI8226, and U937 cell lines at the 12-h time points are displayed in Fig. 2 A–C, respectively. Significant portions of the HLA peptides with definable DRiPs factors had faster dynamics relative to their parental proteins (circles and squares located right to the diagonal lines in Fig. 2), similar to the observation in the breast cancer cell line MCF-7 (35). One possible problem with such calculations, leading to erroneous definition of faster dynamics of HLA peptides relative to their source proteins, can be caused by the larger scatter of the measured H/L ratios of the HLA peptides relative to those of the tryptic peptides of their source proteins. The H/L ratios of the proteins are calculated by using the median value of the H/L ratios of multiple tryptic peptides from each protein. Because these are median values, they are (by definition) less scattered than the single H/L values of each of the HLA peptides. Therefore, we compared the H/L dynamics of the different HLA peptides (Fig. 3A) with those of their source proteins (Fig. 3B) and the individual dynamics of all of the tryptic peptides of these proteins (Fig. 3C). As expected, the H/L ratios of the different tryptic peptides of each protein demonstrated some variance, yet many of the HLA peptides shifted from their light to heavy forms faster than all of the same protein’s tryptic peptides. The entire datasets, with all of the identified proteins and HLA peptides and their dynamics, are listed in Dataset S1 for the JY cells, Dataset S2 for the RPMI8226 cells, and + for the U937 cells.
Fig. 1.
Experimental flowchart depicting the isolation and dynamic SILAC analysis of the proteomes (through its tryptic peptides) and the HLA class I peptidomes of the same cultured cells. The proteins and HLA peptides dynamics were defined by shifting the growth media from light to heavy amino acids (A). Multiple time-point mass spectra of an HLA and a tryptic peptide, both derived from the same protein, illustrating the shift from light to heavy form (calculated H/L ratios are presented in the boxes) (B). Graphical representation of H/L ratios of the same HLA and the tryptic peptides from the 60S ribosomal L30 protein (C).
Fig. 2.
Relative H/L ratios of HLA peptides and of their source proteins at the 12-h time points of the JY (A), U937 (B), and RPMI8226 (C) cells. The different subunits of ribosomes are labeled as blue circles and the subunits of TCP-1 as red squares.
Table 1.
Experimental statistics: The numbers of identified and quantified HLA peptides and proteins observed in the different cell cultures
| Tested cell lines | Identified HLA peptides | Identified proteins | Quantified HLA peptides | Quantified proteins | HLA peptides with DRiPs factor |
| JY | 7,137 | 4,913 | 2,080 | 1,980 | 461 |
| RPMI8226 | 2,359 | 2,116 | 615 | 444 | 145 |
| U937 | 1,651 | 1,821 | 884 | 419 | 228 |
Fig. 3.
Comparative dynamic SILAC data of the proteome and the HLA peptidome of the JY cells: kinetics of HLA peptides (created by Perseus software) (A); protein kinetics, as median of their tryptic peptides kinetics (B); and kinetics of individual tryptic peptides (C). Each gray line indicates the kinetics of one HLA peptide (A), one protein (B) or one tryptic peptide (C). As an example, the bold lines represent the kinetics of an HLA peptide derived from the T-complex protein 1 subunit eta and of the tryptic peptides of the same protein.
Defining the Contribution of the DRiPome to the HLA Peptidome.
A few thousand proteins and HLA peptides were identified in this study and a few hundreds of them could be assigned with DRiPs factors, when relative rates of synthesis could be defined for both proteins and their derived HLA peptides (Table 1 and Datasets S1–S3). Of these, as many as 124 of 461 peptides in the JY cells, 102 of 145 peptides in the RPMI8226 cells, and 116 of 228 peptides in the U937 cells had DRiPs factors >1.5. Similarly, 188, 11, and 18 peptides, respectively, were observed with DRiPs factors <0.66 (Fig. 2 and Table 2). The subset of HLA peptides possessing the high DRiPs factors were derived, to a large extent, from proteins with shared cellular functions. This conclusion was based on analysis with the DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov) (37), using as background the Gene Ontology terms of the proteins with at least two defined DRiPs factors. About a quarter of the proteins with high DRiPs factors were subunits of the ribosomes (enriched with P value of 1.67 × 10−5 in the JY cells). TCP-1 is another protein complex contributing significantly to the high DRiPs factor proteins with three to five (in the different cell lines) of its eight subunits (Fig. 2). The source proteins of the retirees did not seem to originate from any specific molecular pathways of the cells (Table 2 and Datasets S1–S3).
Table 2.
Example for proteins for which DRiPs factors could be defined
| DRiPs |
Retirees |
||||||
| Gene names | DRiPs factor | Gene names | DRiPs factor | Gene names | DRiPs factor | Gene names | DRiPs factor |
| ATP5F1* | 10.51 | RPL10A | 2.36 | PPP2R1A | 0.01 | MAP4 | 0.21 |
| PSMD14 | 8.56 | CLTC | 2.35 | TRIP12 | 0.05 | FASN | 0.21 |
| PSMB8 | 6.39 | CCT8 | 2.32 | PRRC2C | 0.06 | DAZAP1 | 0.22 |
| RPL28 | 6.07 | RPS2 | 2.25 | MKI67 | 0.08 | PCBP2 | 0.23 |
| CCT7 | 5.88 | GRHPR | 2.24 | EIF4G1 | 0.08 | SMARCA2 | 0.23 |
| EIF2S3 | 5.44 | SLC25A3 | 2.23 | MKI67 | 0.09 | PPP2R1A | 0.23 |
| LY75 | 4.95 | PSMB8 | 2.23 | DDX24 | 0.10 | PCBP2 | 0.24 |
| RPN2 | 4.86 | RPL23A | 2.16 | MKI67 | 0.10 | SMARCA2 | 0.24 |
| B2M | 4.81 | CCT4 | 2.14 | DDX21 | 0.11 | TLN1 | 0.25 |
| COX6C | 4.49 | SNRPA | 2.13 | TLN1 | 0.14 | SYMPK | 0.25 |
| ETFB | 4.46 | MFAP1 | 2.12 | CSDE1 | 0.14 | PARP1 | 0.26 |
| ATIC | 4.34 | VARS | 2.10 | SPTBN1 | 0.15 | GTF3C1 | 0.26 |
| RPS7 | 4.24 | ILF2 | 2.09 | LIMA1 | 0.15 | SAP18 | 0.26 |
| PSMD14 | 3.93 | RPL13 | 2.07 | UTP14A | 0.15 | EIF4G1 | 0.27 |
| PHB2 | 3.87 | EFTUD2 | 2.06 | HNRNPL | 0.16 | HNRNPL | 0.28 |
| GRHPR | 3.74 | PSMD14 | 2.00 | LARP1 | 0.16 | TLN1 | 0.28 |
| RPL5 | 3.17 | ILF2 | 2.00 | PRRC2C | 0.16 | RFTN1 | 0.28 |
| RPL14 | 3.15 | RPL6 | 1.96 | MAPRE1 | 0.16 | EIF4A3 | 0.29 |
| HADHA | 3.15 | DDB1 | 1.92 | DDX24 | 0.17 | EIF4G1 | 0.29 |
| RPL10A | 2.99 | RPL13 | 1.87 | RRM1 | 0.17 | RBM4 | 0.30 |
| HIST1H2AB | 2.72 | STT3B | 1.87 | SPTBN1 | 0.18 | RRP15 | 0.30 |
| RPL28 | 2.71 | SF3B3 | 1.86 | DUT | 0.18 | HIST1H1D | 0.31 |
| PSMB8 | 2.68 | RPL7 | 1.84 | PRRC2C | 0.18 | HIST1H1E | 0.32 |
| RPL36 | 2.66 | RPA2 | 1.80 | EIF4G1 | 0.18 | LCP1 | 0.32 |
| NCKAP1L | 2.66 | RPL34 | 1.79 | ATXN2L | 0.18 | PSME3 | 0.32 |
| RPL8 | 2.58 | TCP1 | 1.78 | MKI67 | 0.18 | FASN | 0.33 |
| RPS10 | 2.53 | LYPLA2 | 1.77 | HNRNPK | 0.18 | U2SURP | 0.33 |
| RPL15 | 2.45 | MTA3 | 1.77 | HNRNPUL2 | 0.19 | PTBP1 | 0.34 |
| MS4A1 | 2.44 | NOP14 | 1.75 | PCNA | 0.19 | GORASP2 | 0.34 |
| RPL10A | 2.42 | HLA-DPB1* | 1.74 | ZFR | 0.19 | ADRM1 | 0.34 |
These DRiPs factors were calculated for the 24-h time point in the JY cells. Duplicate names indicate that different DRiPs factors were assigned to separate HLA peptides derived from the same protein. The subunits of ribosomes are bold.
The HLA peptide derived from HLA-DPB1 and ATP5F1 are two examples of peptides originating from known signal sequences. The entire dataset is provided in Dataset S1.
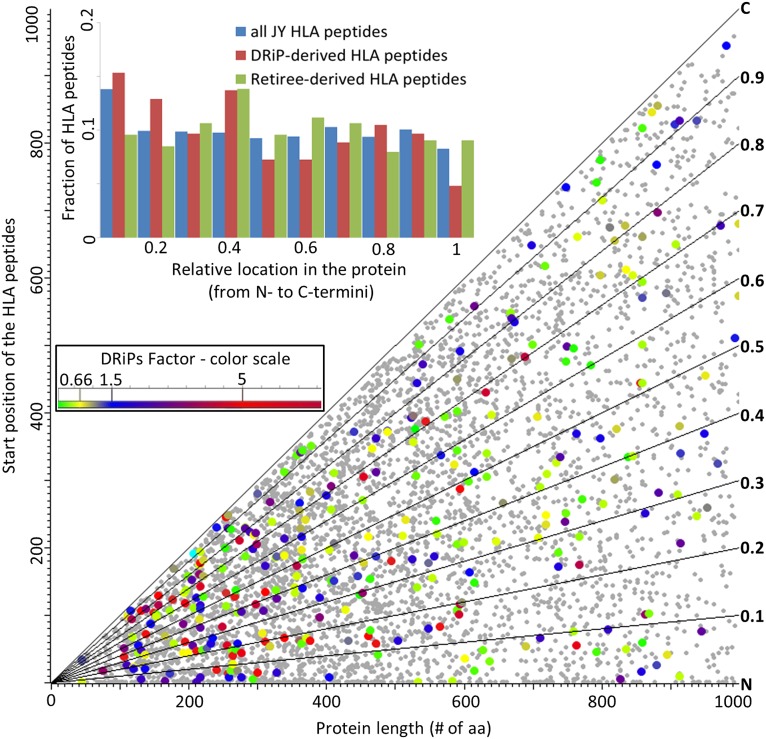
Location of the HLA Peptides Within Their Source Proteins.
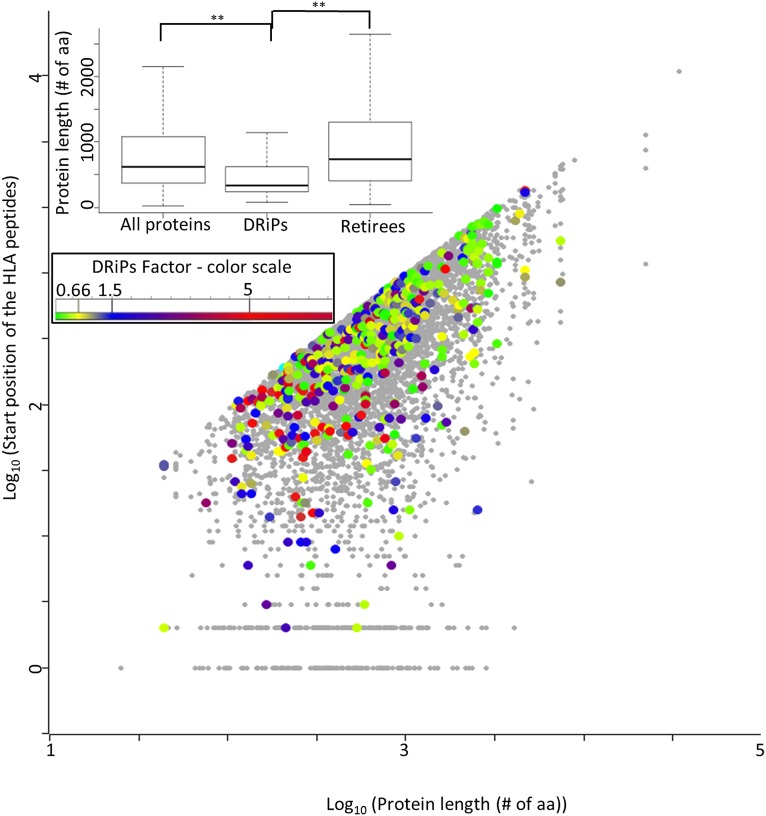
Possible sources of MHC peptides were suggested to be the truncated proteins, possibly created during the pioneer round of translation (15) or due to premature translation termination (12–14). If indeed HLA peptides are largely derived from such truncated and rapidly degraded short segments of the proteins, one expects that significant fractions of the MHC peptides should be derived from regions close to the N termini of their source proteins. Our data do not support a significant contribution of truncated proteins to the DRiPs-derived HLA peptidomes (see Fig. 4 for the JY cell line and Fig. S1 A and B for the other two cell lines). Both the entire HLA peptidomes and their high DRiPs factor subsets were derived from the entire lengths of the proteins. These are displayed as absolute values of the amino acid positions of the HLA peptides within the proteins and as fractional locations within the proteins (Fig. 4). Furthermore, the source proteins of the high DRiPs factor peptides, identified in this study, were mostly derived from relatively short proteins (Fig. 5 and Fig. S2 A and B). The possibility that HLA peptides with high DRiPs factors were largely derived from signal peptides, processed during translocation into the ER, was also evaluated and refuted. The only exceptions in the JY cells for class I peptides derived from DRiPs and from signal peptides were the HLA-DPB1 and ATP5F1 peptides.
Fig. 4.
Relative locations of HLA peptides within the first 1,000 amino acids of their source proteins in the JY cells. The small gray dots represent HLA peptides and their source proteins for which the DRiPs factor was not defined. The colored dots indicate HLA peptides with defined DRiPs factors, with the color indicating the DRiPs factor scale (Inset). Diagonal lines represent the percentile values of the locations of the HLA peptides within their source proteins. The Inset bar graph displays the fractions of high DRiPs factor peptides (derived from the certain DRiPs), low DRiPs factor peptides (derived from the certain retirees), and the entire list of HLA peptides, according to their relative locations within their source proteins.
Fig. 5.
The position of HLA peptides within the entire lengths of their source proteins, in the JY cells. The colors indicate the DRiPs factors of the HLA peptides according to the DRiPs factor scale (Inset). The statistical significance between the protein's length distribution of all proteins, DRiPs, and retiree, were evaluated by the Mann–Whitney test (**P < 0.001).
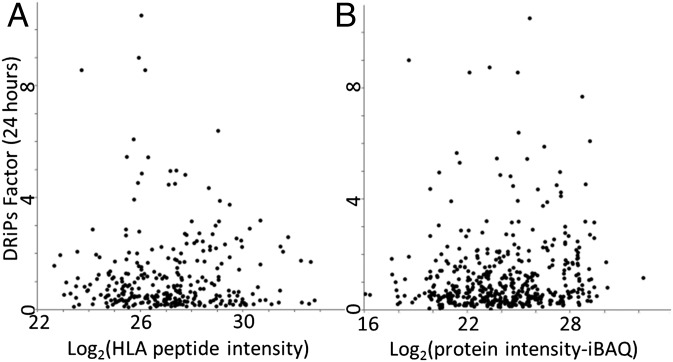
No Correlation Was Observed Between the DRiPs Factors and the Cellular Abundance of the Presented HLA Peptides or Their Source Proteins.
The possibility that the more abundant cellular proteins are translated with higher error rates or folding failures, leading to faster degradation and high DRiPs factor, was not confirmed in this study. No significant correlation was observed between the DRiPs factors and the levels of expression of the different HLA peptides (Fig. 6A) or the levels of expression of the source proteins (Fig. 6B) as calculated by their intensity-based absolute quantification (iBAQ) values (38). For example, the expression levels of the different subunits of the ribosomes or of the TCP-1 were rather similar (in each of these complexes), whereas their DRiPs factors varied significantly (Fig. 2).
Fig. 6.
Correlation between the DRiPs factors and the cellular expression levels of the HLA peptides (A) and of the proteins (B). The cellular levels of the proteins were defined as their iBAQ intensities.
Discussion
Defining the DRiPs Factors of the HLA Peptidome Helps Shed Light on Its Production Pipeline.
An ongoing debate in this field deals with the issue whether the MHC peptidome is mostly derived from degradation of mature proteins (retirees) or immature/DRiPs proteins (3, 7, 24, 39). Clear immunological advantage is gained by early warning on pathogen infection through rapid production of MHC peptides from newly synthesized proteins, followed by their immediate presentation at the cells’ surface (4, 5). However, the identities of the rapidly degraded proteins supplying degradation products to the MHC peptidome were not known thus far. The identities and the DRiPs factors of a few hundred HLA peptides, isolated from cultured human hematopoietic cells, were defined in this study with relatively high certainty. Between 27% and 70% of these HLA peptides have high DRiPs factors, meaning that a significant portion of them are derived from newly synthesized proteins, which are not only rapidly degraded before their full maturation but possibly also channel some of their degradation products directly to the HLA peptidome (40, 41). Large numbers of retirees-derived HLA peptides were also identified in this study, indicating that retirees also contribute significantly to the HLA peptidome (3, 39). Different pathways can lead to higher H/L ratios of HLA peptides relative to their source proteins, as discussed in the Introduction. Such DRiPs can be further divided into the different subtypes (see recent reviews in refs. 6 and 7). Partially or fully unfolded proteins (4, 42, 43), products of premature termination of translation (12–14), downstream initiation (10), products of the pioneer round of translation (15), and out-of-frame translated segments of proteins (18). Another possible source of rapidly degraded proteins are full-sized, surplus subunits of large protein complexes, which are degraded rapidly after being produced in excess of the need for assembly (20, 22, 42). Our data support this last option as a likely source of the rapidly degraded proteins, which supply a significant portion of the MHC peptidome. This conclusion is based on the large contribution of subunits of large complexes, such as the ribosome and TCP-1 subunits, to the high DRiPs factor HLA peptidome. The excess production of such subunits can be caused by limiting production of one or more of the core subunits of the protein complexes (22, 44) or in the case of the ribosomes, by limited availability of some ribosomal subunits (45) or rRNA (Fig. 2 and Table 2). The larger contribution of relatively short proteins to the high DRiPs factor HLA peptidome (Fig. 5) can be explained by the fact that subunits of multisubunits protein complexes are relatively short (46). Furthermore, our data do not support the conclusion that the pioneer round of translation or the premature termination of translation are major sources for the high DRiPs factor HLA peptides or the HLA peptidome in general. One expects products of premature translation termination to be preferentially derived from regions close to the N termini of the proteins (4, 15), which was not observed in this study (Fig. 4). On the other hand, even among the peptides with high DRiPs factors, a few peptides were observed to be derived from proteins known to be homo-multimers (such as ATIC and GRHPR; Table 2). Monomer and homo-multimer proteins cannot be a source for excessively produced subunits and thus are rapidly degraded due to other, yet unknown, defects in their folding or processing. The observation that ribosomal proteins are a significant contributors to the high DRiPs factor HLA peptides is likely unrelated to the suggestion that nuclear translation contributes to the DRiPome through the pioneer round of translation (15). The ribosomes are synthesized in the cytoplasm but assembled in the nucleolus (47) with less than perfect assembly (45). Thus, the detection of newly synthesized ribosomal proteins in the nucleus (45) is expected, because the nucleus is the assembly site of the ribosomes. A very interesting possibility is that HLA peptides are produced in a unique compartment (40), which is associated with the nucleus (48).
Possible Sources for Bias in the Presented Data.
The correlation between the cellular proteome and MHC peptidome is only partial (32, 49). Furthermore, the definition whether HLA peptides are derived from degradation of DRiPs or retirees depends on accurate quantification of the H/L ratios of both the proteins and their derived HLA peptides at multiple time points. Such measurements are less likely to be successful for some proteins that are expressed in the cells mostly as DRiPs with limited parallel expression of their stable counterparts. Such proteins are very rare and do not accumulate in the cells to sufficient levels for detection by mass spectrometry and their transient presence in the cells is only evidenced through the detection of the degradation products as HLA peptides. Therefore, the data presented here are possibly biased against establishing the DRiPs factors of many DRiPs, and the contribution of DRiPs to the HLA peptidome may be even larger than suggested here.
Relevance of the Presence of DRiPs-Derived HLA Peptides to Immunology.
It is not known if cancer cells in the body present more DRiPs-derived HLA peptides than do normal cells. It is possible that the large proportions of MHC peptides derived from DRiPs observed here is a phenomenon unique to cultured cells (after all “life in plastic is not real life”). However, it is known that tumor cells lose significant degree of gene expression control and are thus likely to produce many excess copies of subunits of protein complexes. Such unbalanced cellular protein production, if identified, can be used to design better immunotherapeutics. However, defining the DRiPs factors for HLA peptides in human or animal bodies is likely impossible with the technology described here. The closest attempt to demonstrate the existence of DRiPs in normal cells (to our knowledge) was reported by Schubert et al. (50), who used cultured primary cell lines.
The approach described here includes the analysis of many thousands of HLA peptides and proteins, resulting in definition of the dynamics of >700 pairs of HLA peptides and their source proteins. Such an approach, based on analysis of large sets of endogenous proteins and their derived HLA peptides, is preferred for generalization about the molecular mechanisms of MHC peptides production over assays based on analysis of a transfected, recombinant, single model MHC peptides, which were selected only due the availability of a T-cell clone or an mAb-directed against them. Transfected, recombinant model antigens and their derived MHC peptides may not represent the real cellular events due to their impaired stability (38) and mislocalization (45, 51).
Do Proteins Involved with the Translation Machinery Contribute Much of the DRiPome?
It was suggested before that translational machinery proteins contribute significantly to the MHC peptidome (52–54). However, of the translational machinery components, it was the ribosomal proteins that contributed more significantly to the high DRiPs factor HLA peptidome. Furthermore, when we rechecked our laboratory’s previous data from the MCF-7 breast cancer cells (35) we noticed that also in that study, the ribosomal proteins contributed significantly to the high DRiPs factor HLA peptidome. A reasonable explanation is that ribosomal proteins are relatively abundant and function as large complexes, whose subunits should be synthesized in synchrony with each other. The surplus subunits are likely rapidly degraded and presented as DRiPs-derived MHC peptides.
Are There Dedicated Pipelines for Channeling Large Protein Complexes (Such as Viruses) for Degradation and Display of Their Derived MHC Peptides?
Viruses are large protein complexes (containing also DNA or RNA), and it was already noticed that DNA- and RNA-binding proteins are abundant contributors to the HLA peptidome (52–54). The overtaking of the cellular protein production machinery by the viruses very likely results in less than perfect synchrony in the production of the viral proteins needed for capsid assembly. An interesting possibility is that a dedicated pathway exists for degradation of excess viral proteins (40).
Contribution of Retirees to the HLA Peptidome.
Interestingly, the number of retiree-derived HLA peptides from the less transformed JY cell line was larger than the number of DRiPs-derived peptides, whereas in the more cancerous cell lines (RPMI8226 and U937), there were far less retiree-derived HLA peptides. This finding supports the notion that the relative contribution of DRiPs and mature proteins to the HLA peptidome varies depending on cell types, health state (4), cancerous transformation (11), or inflammation. A possible explanation for such difference in the MHC peptidome’s source proteins is the unbalanced protein synthesis in these more transformed cells, in which larger excess of subunits of protein complexes are produced and degraded.
Conclusions.
DRiPs are mainly derived from surplus, full-sized subunits of protein complexes involved in translational processes. The contribution of DRiPs vs. retiree-derived HLA peptides may vary between cell types and cells' physiological condition. In our opinion, the contribution of various cellular sources to HLA peptidome should better be studied by large-scale studies (55), such as this, rather than by following the dynamics of a single or even a few recombinant and transfected epitopes, selected only because antibodies for their detection are available.
Materials and Methods
Materials.
Bafilomycin was obtained from Santa Cruz Biotechnology, and bortezomib (Velcade) was obtained from Selleckchem. The antibody used was W6/32, a mouse anti–pan-HLA class I (native HLA A, B, and C) produced in mouse ascites fluids.
Cell Culture.
Cultured cells were maintained in DMEM media, supplemented with 10% (vol/vol) FCS for the JY (an EBV-transformed human lymphoblastoid cell line) and the U937 (histolytic lymphoma cell line), and 20% FCS for the RPMI8226 (human multiple myeloma cell line). All cell culture media also contained 2 mM l-glutamine, 1 mM Na-pyruvate, 1% penicillin-streptomycin, 10 mM Hepes buffer (Sigma), and 0.5% of Pluronic F-68 (Sigma) and were grown in a humidified 8% CO2 incubator at 37 °C in shaker flasks (TriForest) rotated on an orbital shaker at 120 rpm.
Dynamic SILAC Labeling with Three Isotopic Labeled Heavy Amino Acids (Leu+7, Arg+10, Lys+8).
At the beginning of each experiment, the growth medium of the cells was changed to DMEM, lacking leucine, lysine, and arginine (Biological Industries) and containing 10% dialyzed FCS (the RPMI8226 cells were grown in 20% FCS). This medium was supplemented with heavy leucine (13C6,15N-Leu), heavy lysine (13C6,15N2-Lys), and arginine (13C6,15N4-Arg) (Cambridge Isotope Labs) at a final concentration of 52, 147.6, and 87.3 mg/L, respectively. Aliquots of cells were taken at the specified times for both proteomics and HLA peptidomics analyses.
Trypsin Digestion of Proteins.
Proteins extracted from ∼1 × 106 cells were dissolved in 8 M urea containing 20 mM DTT and 400 mM ammonium bicarbonate and heated to 60 °C for 30 min. Next, the proteins were carbamidomethylated with 100 mM iodoacetamide at room temperature for 30 min, diluted with three volumes of water, and digested with modified trypsin (Promega) at 37 °C and a 1:50 (wt/wt) enzyme-to-substrate ratio for two cycles of 4 h, respectively. Protein digestion from gel slices was performed by running the proteins in 10% acrylamide gel, staining with Coomassie, and slicing the gel into 5 slices. The proteins in the gel slices were reduced with 2.8 mM DTT at 60 °C for 30 min and carbamidomethylated with 8.8 mM iodoacetamide in 100 mM ammonium bicarbonate at room temperature for 30 min and digested overnight at 37 °C in 10% acetonitrile and 10 mM ammonium bicarbonate with modified trypsin (Promega) at a 1:10 (wt/wt) enzyme-to-substrate ratio. An additional trypsinization was done for 4 h with similar amount of trypsin.
Affinity Purification of the HLA Complexes.
The HLA molecules were purified essentially as in ref. 56 with minor modifications. At each time point, ∼2 × 108 cells were harvested and washed three times with cold PBS by centrifugation and then incubated for 1 h at 4 °C with mild stirring in lysis buffer, containing 1% IGEPAL-CA630, 1 mM EDTA, 1:200 (vol/vol) protease inhibitors mixture, and 1 mM PMSF (Sigma). The cell lysate was spun at 18,000 × g for 30 min, and the supernatant was passed through a column containing the W6/32 antibody covalently bound to AminoLink resin (Pierce), as described in ref. 57. The columns were washed with 10 volumes of 400 mM NaCl and with another 10 volumes of 20 mM Tris⋅HCl, pH 8. The peptides were separated from the heavy subunits of the HLA molecules by elution with 1% trifluoroacetic acid (TFA; Sigma) followed by their concentration and desalting on disposable MicroTip C-18 column (Harvard Apparatus) as in ref. 35. This procedure is a modification of the HLA peptide purification procedure described in ref. 57, as follows. The disposable C-18 columns were washed with 0.1% TFA, and the HLA peptides were eluted with 0.1% TFA and 30% acetonitrile to separate the HLA peptides from the HLA α-chain and the β2-microglobulin. The solvent was evaporated to dryness and the peptides were dissolved in 0.1% TFA and stored until use at −80 °C.
LC-MS/MS Analysis.
The recovered HLA class I peptides and tryptic peptides from the RPMI8226 and U937 cells were analyzed by µLC-MS/MS using a LTQ Orbitrap XL mass spectrometer (Thermo Fisher) coupled to a capillary HPLC (Eksigent), fitted with a C18 trap column, 0.3 × 5 mm (LC-Packings). The HLA class I peptides and tryptic peptides from the JY cells were analyzed with a Q Exactive mass spectrometer (Thermo Fisher) fitted with a capillary UHPLC (EASY-nLC 1000; Thermo Fisher) or Q Exactive Plus mass spectrometer fitted with Ultimate 3000 RSLCnano capillary UHPLC (Thermo Fisher). The peptides were resolved on a homemade capillary column (75-µm ID) packed with Reprosil C18-Aqua (Dr. Maisch GmbH) as in ref. 58 and resolved using 7–40% acetonitrile gradients in the presence of 0.1% formic acid for 2 h for the HLA peptides and the tryptic peptides produced by the “in-gel digests” and 4-h gradients for tryptic peptides of the “in-solution” proteolytic digest.
Orbitrap XL Analyses.
Each full scan (m/z range, 350–2,000) was acquired in the Orbitrap analyzer, followed by MS/MS analyses of the top seven most intense precursor ions using collision-induced disintegration (CID). In the Orbitrap XL, singly, doubly, and triply charged MHC peptides and doubly and triply charged tryptic peptides were selected for fragmentation according to the following criteria: Exclusion time was set to 90 s with lists of containing up to 500 peptides; the automatic gain control (AGC) was set to target value of 107 ions for the full scan MS and the settings for MS/MS were 5 × 105 and 104 ions for Orbitrap and linear ion trap analyzers, respectively. Ion selection threshold was set to 3 × 104 counts and the resolution was set to 7 × 104.
Q Exactive Analyses.
The resolution in full MS mode was set to 7 × 104 at m/z 200, the AGC was set to 3 × 106 with maximum ion time (IT) of 100 ms, and the dynamic exclusion was set to 20 s. Top ten higher-energy collision dissociation fragmentations (HCD) of the same charge states as in the Orbitrap XL were selected from the survey scan of m/z 300–1,800. Tandem mass spectra were acquired starting at m/z 100 with a resolution of 17,500. The target AGC was set to 1 × 105 with a maximum IT of 50 ms, and the normalized collision energy was set to 25 eV.
Data Analysis.
Peptides were identified, and the dynamic SILAC data were quantified using the MaxQuant software tool (59), version 1.3.0.5. Downstream bioinformatics on the MaxQuant output and graphical representation were performed with Perseus, version 1.3.0.4. MaxQuant was used with the Andromeda search engine (60) and the human section of the UniProt/Swiss-Prot database (release 2011_11, 20,257 entries). Peptide precursor and fragment mass tolerances for the LTQ Orbitrap data were set at 6 ppm and 0.5 Da, respectively. For the Q Exactive data, the fragment mass tolerance was set to 20 ppm. Protein amounts were calculated using the iBAQ equation (38) in MaxQuant. The SILAC labels were accepted as variable modification for both tryptic and HLA peptides. For the proteomics analyses, carbamidomethyl cysteine was considered fixed, whereas N-acetylation and oxidation of methionine was considered variable modifications. The minimal peptide length was set to seven amino acid residues, and a maximum of two missed cleavages was allowed for tryptic peptides. The false discovery rate (FDR) was set for tryptic peptides to 0.01 for protein identifications and 0.05 for the MHC peptides. The resulting identified protein tables were filtered to eliminate the identifications derived from the reverse database, as well as from common contaminants.
Supplementary Material
Acknowledgments
The assistance of Ilana Navon in performing the mass spectrometry analyses, the help of Dganit Melamed in data analysis, and the help of Anatoly Meller in bioinformatics definition of the position of the HLA peptides within their source proteins are much appreciated. This research was funded by the Israeli Centers for Research Excellence Program of the Planning and Budgeting Committee and The Israel Science Foundation (Grant 1775/12) and by the Greta Koppel Small Cell Lung Carcinoma Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321902111/-/DCSupplemental.
References
- 1.Esquivel F, Yewdell J, Bennink J. RMA/S cells present endogenously synthesized cytosolic proteins to class I-restricted cytotoxic T lymphocytes. J Exp Med. 1992;175(1):163–168. doi: 10.1084/jem.175.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim Y, Yewdell JW, Sette A, Peters B. Positional bias of MHC class I restricted T-cell epitopes in viral antigens is likely due to a bias in conservation. PLOS Comput Biol. 2013;9(1):e1002884. doi: 10.1371/journal.pcbi.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farfán-Arribas DJ, Stern LJ, Rock KL. Using intein catalysis to probe the origin of major histocompatibility complex class I-presented peptides. Proc Natl Acad Sci USA. 2012;109(42):16998–17003. doi: 10.1073/pnas.1210271109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yewdell JW, Antón LC, Bennink JR. Defective ribosomal products (DRiPs): A major source of antigenic peptides for MHC class I molecules? J Immunol. 1996;157(5):1823–1826. [PubMed] [Google Scholar]
- 5.Reits EA, Vos JC, Grommé M, Neefjes J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature. 2000;404(6779):774–778. doi: 10.1038/35008103. [DOI] [PubMed] [Google Scholar]
- 6.Yewdell JW. DRiPs solidify: Progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 2011;32(11):548–558. doi: 10.1016/j.it.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolan BP, Bennink JR, Yewdell JW. Translating DRiPs: Progress in understanding viral and cellular sources of MHC class I peptide ligands. Cell Mol Life Sci. 2011;68(9):1481–1489. doi: 10.1007/s00018-011-0656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rock KL, Farfán-Arribas DJ, Colbert JD, Goldberg AL. Re-examining class-I presentation and the DRiP hypothesis [published online ahead of print February 21, 2014] Trends Immunol. 2014 doi: 10.1016/j.it.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Princiotta MF, et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18(3):343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 10.Berglund P, Finzi D, Bennink JR, Yewdell JW. Viral alteration of cellular translational machinery increases defective ribosomal products. J Virol. 2007;81(13):7220–7229. doi: 10.1128/JVI.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granados DP, et al. MHC I-associated peptides preferentially derive from transcripts bearing miRNA response elements. Blood. 2012;119(26):e181–e191. doi: 10.1182/blood-2012-02-412593. [DOI] [PubMed] [Google Scholar]
- 12.Schwab SR, Li KC, Kang C, Shastri N. Constitutive display of cryptic translation products by MHC class I molecules. Science. 2003;301(5638):1367–1371. doi: 10.1126/science.1085650. [DOI] [PubMed] [Google Scholar]
- 13.Cardinaud S, Starck SR, Chandra P, Shastri N. The synthesis of truncated polypeptides for immune surveillance and viral evasion. PLoS ONE. 2010;5(1):e8692. doi: 10.1371/journal.pone.0008692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacsina JR, et al. Premature translational termination products are rapidly degraded substrates for MHC class I presentation. PLoS ONE. 2012;7(12):e51968. doi: 10.1371/journal.pone.0051968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apcher S, et al. Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc Natl Acad Sci USA. 2011;108(28):11572–11577. doi: 10.1073/pnas.1104104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: Features and functions. Cell. 2010;142(3):368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106(5):607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 18.Malarkannan S, Horng T, Shih PP, Schwab S, Shastri N. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity. 1999;10(6):681–690. doi: 10.1016/s1074-7613(00)80067-9. [DOI] [PubMed] [Google Scholar]
- 19.Yewdell J. To DRiP or not to DRiP: Generating peptide ligands for MHC class I molecules from biosynthesized proteins. Mol Immunol. 2002;39(3-4):139–146. doi: 10.1016/s0161-5890(02)00097-4. [DOI] [PubMed] [Google Scholar]
- 20.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: Quantitating MHC class I antigen presentation. Nat Rev Immunol. 2003;3(12):952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 21.Yewdell JW, Schubert U, Bennink JR. At the crossroads of cell biology and immunology: DRiPs and other sources of peptide ligands for MHC class I molecules. J Cell Sci. 2001;114(Pt 5):845–851. doi: 10.1242/jcs.114.5.845. [DOI] [PubMed] [Google Scholar]
- 22.Cambridge SB, et al. Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J Proteome Res. 2011;10(12):5275–5284. doi: 10.1021/pr101183k. [DOI] [PubMed] [Google Scholar]
- 23.Gaczynska M, Rock KL, Goldberg AL. Role of proteasomes in antigen presentation. Enzyme Protein. 1993;47(4-6):354–369. doi: 10.1159/000468693. [DOI] [PubMed] [Google Scholar]
- 24.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11(12):823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 25.Sijts EJ, Kloetzel PM. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol Life Sci. 2011;68(9):1491–1502. doi: 10.1007/s00018-011-0657-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henderson RA, et al. HLA-A2.1-associated peptides from a mutant cell line: A second pathway of antigen presentation. Science. 1992;255(5049):1264–1266. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 28.Wei ML, Cresswell P. HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature. 1992;356(6368):443–446. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- 29.Münz C. Antigen processing via autophagy—not only for MHC class II presentation anymore? Curr Opin Immunol. 2010;22(1):89–93. doi: 10.1016/j.coi.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratt JM, et al. Dynamics of protein turnover, a missing dimension in proteomics. Mol Cell Proteomics. 2002;1(8):579–591. doi: 10.1074/mcp.m200046-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 32.Milner E, Barnea E, Beer I, Admon A. The turnover kinetics of major histocompatibility complex peptides of human cancer cells. Mol Cell Proteomics. 2006;5(2):357–365. doi: 10.1074/mcp.M500241-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.García-Medel N, Sanz-Bravo A, Barnea E, Admon A, López de Castro JA. The origin of proteasome-inhibitor resistant HLA class I peptidomes: A study with HLA-A*68:01. Mol Cell Proteomics. 2012;11(1):011486. doi: 10.1074/mcp.M111.011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcilla M, Cragnolini JJ, López de Castro JA. Proteasome-independent HLA-B27 ligands arise mainly from small basic proteins. Mol Cell Proteomics. 2007;6(5):923–938. doi: 10.1074/mcp.M600302-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Milner E, et al. The effect of proteasome inhibition on the generation of the human leukocyte antigen (HLA) peptidome. Mol Cell Proteomics. 2013;12(7):1853–1864. doi: 10.1074/mcp.M112.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolan BP, et al. Distinct pathways generate peptides from defective ribosomal products for CD8+ T cell immunosurveillance. J Immunol. 2011;186(4):2065–2072. doi: 10.4049/jimmunol.1003096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 39.Colbert JD, Farfán-Arribas DJ, Rock KL. Substrate-induced protein stabilization reveals a predominant contribution from mature proteins to peptides presented on MHC class I. J Immunol. 2013;191(11):5410–5419. doi: 10.4049/jimmunol.1300078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lev A, et al. Compartmentalized MHC class I antigen processing enhances immunosurveillance by circumventing the law of mass action. Proc Natl Acad Sci USA. 2010;107(15):6964–6969. doi: 10.1073/pnas.0910997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szeto J, et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2(3):189–199. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- 42.Eisenlohr LC, Huang L, Golovina TN. Rethinking peptide supply to MHC class I molecules. Nat Rev Immunol. 2007;7(5):403–410. doi: 10.1038/nri2077. [DOI] [PubMed] [Google Scholar]
- 43.Ostankovitch M, Robila V, Engelhard VH. Regulated folding of tyrosinase in the endoplasmic reticulum demonstrates that misfolded full-length proteins are efficient substrates for class I processing and presentation. J Immunol. 2005;174(5):2544–2551. doi: 10.4049/jimmunol.174.5.2544. [DOI] [PubMed] [Google Scholar]
- 44.Asher G, Reuven N, Shaul Y. 20S proteasomes and protein degradation “by default”. Bioessays. 2006;28(8):844–849. doi: 10.1002/bies.20447. [DOI] [PubMed] [Google Scholar]
- 45.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17(9):749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brocchieri L, Karlin S. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 2005;33(10):3390–3400. doi: 10.1093/nar/gki615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cisterna B, Biggiogera M. Ribosome biogenesis: From structure to dynamics. Int Rev Cell Mol Biol. 2010;284:67–111. doi: 10.1016/S1937-6448(10)84002-X. [DOI] [PubMed] [Google Scholar]
- 48.Apcher S, et al. Translation of pre-spliced RNAs in the nuclear compartment generates peptides for the MHC class I pathway. Proc Natl Acad Sci USA. 2013;110(44):17951–17956. doi: 10.1073/pnas.1309956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Verteuil D, Granados DP, Thibault P, Perreault C. Origin and plasticity of MHC I-associated self peptides. Autoimmun Rev. 2012;11(9):627–635. doi: 10.1016/j.autrev.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Schubert U, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404(6779):770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 51.Yewdell JW, Lacsina JR, Rechsteiner MC, Nicchitta CV. Out with the old, in with the new? Comparing methods for measuring protein degradation. Cell Biol Int. 2011;35(5):457–462. doi: 10.1042/CBI20110055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fortier MH, et al. The MHC class I peptide repertoire is molded by the transcriptome. J Exp Med. 2008;205(3):595–610. doi: 10.1084/jem.20071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hickman HD, et al. Toward a definition of self: Proteomic evaluation of the class I peptide repertoire. J Immunol. 2004;172(5):2944–2952. doi: 10.4049/jimmunol.172.5.2944. [DOI] [PubMed] [Google Scholar]
- 54.Perreault C. The origin and role of MHC class I-associated self-peptides. Prog Mol Biol Transl Sci. 2010;92:41–60. doi: 10.1016/S1877-1173(10)92003-6. [DOI] [PubMed] [Google Scholar]
- 55.Admon A, Bassani-Sternberg M. The Human Immunopeptidome Project, a suggestion for yet another postgenome next big thing. Mol Cell Proteomics. 2011;10(10):011833. doi: 10.1074/mcp.O111.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunt DF, et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255(5049):1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 57.Bassani-Sternberg M, et al. Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proc Natl Acad Sci USA. 2010;107(44):18769–18776. doi: 10.1073/pnas.1008501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishihama Y, Rappsilber J, Andersen JS, Mann M. Microcolumns with self-assembled particle frits for proteomics. J Chromatogr A. 2002;979(1-2):233–239. doi: 10.1016/s0021-9673(02)01402-4. [DOI] [PubMed] [Google Scholar]
- 59.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 60.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.