Significance
Vancomycin is a powerful antibiotic against Gram-positive bacteria that inhibits cell-wall synthesis by binding with high affinity to peptidoglycan precursors. Resistance to vancomycin is due to acquisition of operons encoding, among other enzymes, the zinc-dependent d,d-peptidases VanX, VanY, or VanXY, which catalyze the removal of the drug targets. Structural characterization of VanXY elucidates the molecular basis of their specificity toward vancomycin-susceptible precursors and explains the dual function of VanXY. These studies highlight the striking plasticity of peptidoglycan-modifying enzymes to evolve to antibiotic resistance proteins. They also provide the molecular framework for development of d,d-peptidase inhibitors that may help to curb vancomycin resistance.
Keywords: antibiotic resistance, glycopeptides, enzyme evolution, metallopeptidases, subfamily M15B
Abstract
Vancomycin resistance in Gram-positive bacteria is due to production of cell-wall precursors ending in d-Ala-d-Lac or d-Ala-d-Ser, to which vancomycin exhibits low binding affinities, and to the elimination of the high-affinity precursors ending in d-Ala-d-Ala. Depletion of the susceptible high-affinity precursors is catalyzed by the zinc-dependent d,d-peptidases VanX and VanY acting on dipeptide (d-Ala-d-Ala) or pentapeptide (UDP-MurNac-l-Ala-d-Glu-l-Lys-d-Ala-d-Ala), respectively. Some of the vancomycin resistance operons encode VanXY d,d-carboxypeptidase, which hydrolyzes both di- and pentapeptide. The molecular basis for the diverse specificity of Van d,d-peptidases remains unknown. We present the crystal structures of VanXYC and VanXYG in apo and transition state analog-bound forms and of VanXYC in complex with the d-Ala-d-Ala substrate and d-Ala product. Structural and biochemical analysis identified the molecular determinants of VanXY dual specificity. VanXY residues 110–115 form a mobile cap over the catalytic site, whose flexibility is involved in the switch between di- and pentapeptide hydrolysis. Structure-based alignment of the Van d,d-peptidases showed that VanY enzymes lack this element, which promotes binding of the penta- rather than that of the dipeptide. The structures also highlight the molecular basis for selection of d-Ala–ending precursors over the modified resistance targets. These results illustrate the remarkable adaptability of the d,d-peptidase fold in response to antibiotic pressure via evolution of specific structural elements that confer hydrolytic activity against vancomycin-susceptible peptidoglycan precursors.
The emergence of high-level resistance to vancomycin, a last resort antibiotic against Gram-positive bacteria, in Enterococcus spp. and its spread to methicillin-resistant Staphylococcus aureus is a serious threat to public health (1). Vancomycin acts by binding to the d-alanyl-d-alanine moiety of the uncross-linked N-acetyl-muramyl-l-Ala-d-γ-Glu-l-Lys-d-Ala-d-Ala (pentapeptide[d-Ala]) peptidoglycan precursor blocking the extracellular steps in peptidoglycan synthesis. Resistance is mediated by nine types of operons that replace the d-Ala-d-Ala terminus of peptidoglycan precursors with d-Ala-d-lactate (VanA, -B, -D, and -M types) or d-Ala-d-serine (VanC, -E, -G, -L, and -N types), to which vancomycin exhibits lower binding affinities (2).
A critical step in vancomycin resistance involves depletion of d-Ala–terminating precursors to prevent interaction of vancomycin with its target. This step is facilitated by the d,d-dipeptidase VanX and the d,d-pentapeptidase VanY, which hydrolyze, respectively, d-Ala-d-Ala and the C-terminal d-Ala residue from pentapeptide[d-Ala] (3–6). In the d-Ala-d-Ser form of resistance, a single VanXY enzyme evolved to mediate both d,d-dipeptidase and d,d-pentapeptidase activities (7, 8). Thus, Van d,d-peptidases demonstrate variation in peptidoglycan substrate selectivity that correlates with the specific resistance mechanism. As part of the d-Ala-d-Lac type resistance mechanism, the VanX enzyme shows 105-fold higher catalytic efficiency against d-Ala-d-Ala compared with d-Ala-d-Lac substrates, thus facilitating accumulation of the depsipeptide. However, VanX retains significant activity against d-Ala-d-Ser dipeptide (9). Appropriate to its role in resistance, the VanY enzyme shows carboxypeptidase activity against pentapeptide[d-Ala] but lacks activity against d,d-dipeptide substrates (5, 10). The VanXYC enzyme is selective against resistant dipeptide and pentapeptide peptidoglycan substrates ending in d-Ser (8). Finally, the VanXYG enzyme, first assigned as a dual substrate active “XY” enzyme by sequence similarity (11), was later shown to lack d,d-pentapeptidase activity typical for bona fide VanXY enzymes (12). The molecular determinants responsible for such diversity of substrate specificity among Van d,d-peptidases are unknown, limiting the understanding of their evolution and hampering the development of inhibitors that could be used in combination with glycopeptides.
With the exception of VanYD, which is a penicillin-binding protein (13), all VanX, VanY, and VanXY d,d-peptidases are zinc-dependent enzymes classified into the metallopeptidase clan MD, family M15, according to the MEROPS database (14). Within clan MD, multiple families of enzymes, including M15 representatives, are involved in bacterial cell wall metabolism (15). Three members of this family have been structurally characterized: zinc d-Ala-d-Ala carboxypeptidase from Streptomyces albus (16) (subfamily M15A), bacteriophage l-Ala-d-Glu peptidase PLY500 (17) (subfamily M15C), and VanX (subfamily M15D) from Enterococcus faecium (18). These enzymes display a common core tertiary fold built on a central antiparallel β-sheet arrayed with multiple α-helices on either face of the β-sheet. The fold contains two consensus motifs that form the active site, His-X(6)-Asp and Glu-X(2)-His, with a zinc ion coordinated by the histidine and aspartate residues. In addition, the structure of VanX revealed a small and constricted active site that explains its specificity toward the d-Ala-d-Ala substrate (18). The VanY and VanXY enzymes belonging to subfamily M15B remain structurally uncharacterized.
Given the evolution toward dual substrate specificity of VanXY enzymes, their importance in resistance, and their sequence and functional diversity, we undertook their detailed structural and functional characterization. We determined the crystal structures of VanXYC and VanXYG and performed extensive mutagenesis analysis. Our data led to the identification and characterization of the molecular features responsible for their substrate specificities and demonstrated an exceptional diversification and plasticity within their common metallopeptidase fold in response to drug selective pressure.
Results
Structural Characterization of VanXYC and VanXYG.
Because crystallization of wild-type VanXYC was unsuccessful, after crystallization screening of numerous mutants, we successfully determined the structures of the VanXYC D59S mutant (VanXYCD59S) and wild-type VanXYG to 1.63 Å and 1.40 Å, respectively (SI Materials and Methods and Table S1). The VanXYCD59S structure features two polypeptide chains associated into a dimer (Fig. S1A) whereas VanXYG was monomeric. We confirmed these oligomeric states for VanXYCD59S and VanXYG in solution by size-exclusion chromatography (Fig. S1B).
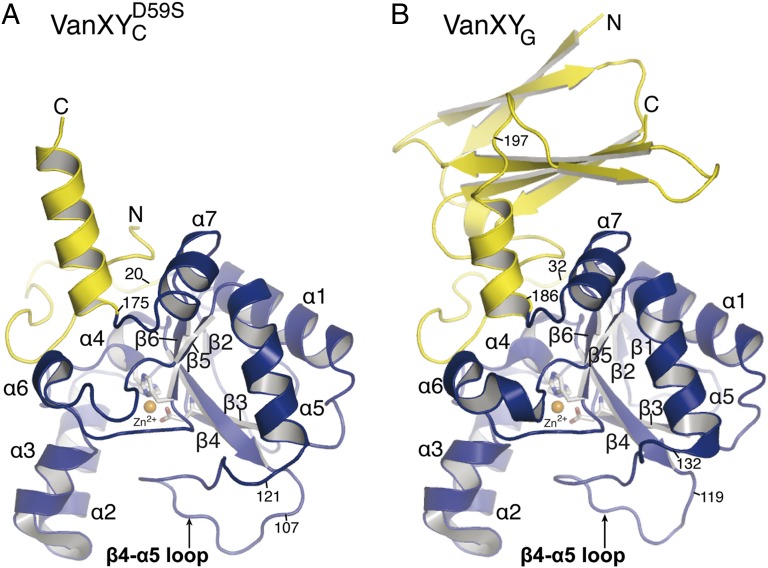
VanXYCD59S and VanXYG protomers contain a common core domain typical of the Zn2+-dependent d,d-peptidase fold (Fig. 1) (15). Spanning residues 21–174 in VanXYCD59S and 33–185 in VanXYG, this domain features a central twisted antiparallel β-sheet (β1–β6) flanked by two pairs of three α-helices (α2, α3, α6, and α1, α5, α7) on the two faces of the β-sheet. The d,d-peptidase folds of VanXYCD59S and VanXYG are essentially identical, superimposing with an rmsd of 0.5 Å over 122 matching Cα atoms. For description of d,d-peptidase domains, the residue numbering of VanXYCD59S will be indicated first and that of VanXYG in parentheses: i.e., His95 (His107).
Fig. 1.
Structures of VanXYCD59S (A) and VanXYG (B). Catalytic domains (VanXYCD59S residues 21–174 and VanXYG residues 33–184) and secondary domains (VanXYCD59S residues 1–20 and 175–188, VanXYG residues 1–32 and 185–250) are in blue and yellow, respectively.
The catalytic center of VanXYCD59S and VanXYG has features consistent with M15 metallopeptidases and their catalytic mechanism (Fig. 2 D and E), including the following: one Zn2+ ion per enzyme chain chelated by histidine residues His95 (His107) and His156 (His167) and an aspartate, Asp102 (Asp114); water molecules closely associated with the Zn2+ ions (hydrogen bond distances of 2.1–2.2 Å), interpreted as the nucleophiles waters in attacking the scissile bond of the substrate (15, 18, 19); the presumed catalytic acid/base, Glu153 (Glu164); and an arginine, Arg62 (Arg74), which stabilizes the negative charge at the tetrahedral center in the transition state (15, 18, 19). Features unique to VanXY include the presence of residues Asp59 (Ser71) and Gln67 (Gln79) that interact on either side of the active site arginine to stabilize its conformation. Both VanXY enzymes possess a prominent loop between β4 and α5 that extends away from the core d,d-peptidase domain and forms a cap over the active site.
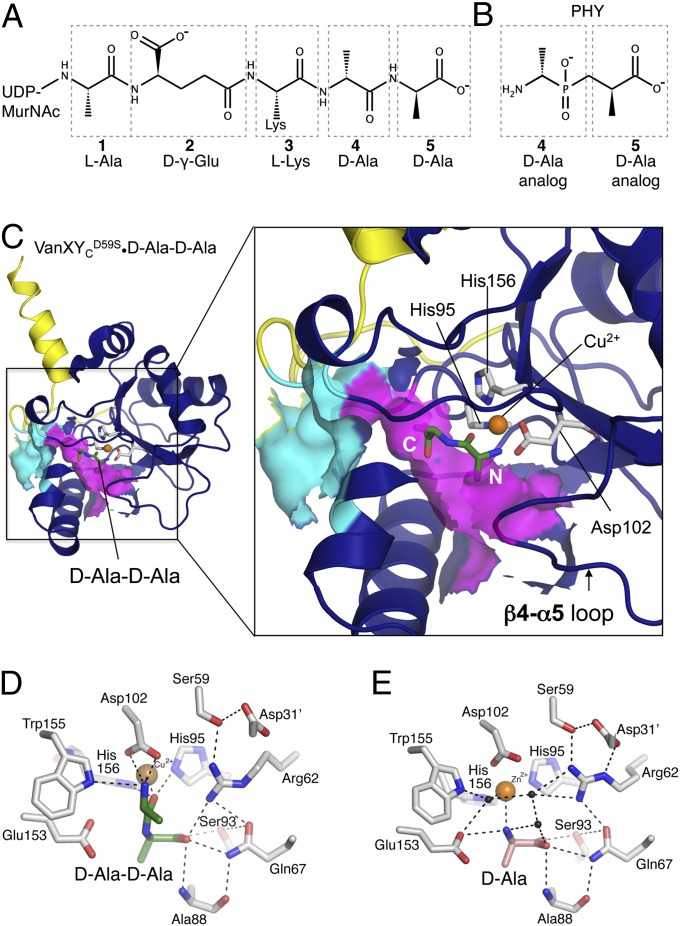
Fig. 2.
Ligand binding site. (A) Chemical structure of pentapeptide[d-Ala], with amino acids numbered from N to C terminus. (B) Chemical structure of PHY, phosphinate d-Ala-d-Ala analog, numbered according to the pentapeptide[d-Ala] it mimics. (C) VanXYCD59S•d-Ala-d-Ala complex. Solvent-exposed surface representation is shown in cyan and purple for putative ligand entry cavity. The N and C termini of d-Ala-d-Ala are indicated in white. (D) Interactions between VanXYCD59S, Zn2+, and d-Ala-d-Ala. (E) Interactions between VanXYCD59S, Cu2+, and d-Ala. Black spheres, water molecules.
According to the VanXYG structure, the N- and C-terminal sequences (residues 1–32 and 186–250) (Fig. 1B) outside the core d,d-peptidase fold form an additional domain not present in VanXYCD59S. This domain contains a four-stranded antiparallel β-sheet sandwiched against a two-stranded antiparallel β-sheet. Similarity searches did not reveal any structural homologs for this domain, yet comparative sequence analysis demonstrated over 50% identity between VanXYG residues 213–250 and corresponding C-terminal regions of VanY (residues 258–294) and VanYM (residues 197–233) but not of VanYB, which lacks this domain (Fig. S2). Furthermore, N-terminal regions of VanXYG (residues 17–35), VanXYC (residues 4–23), and most other M15B subfamily enzymes shared significant sequence similarity (Fig. S2). These observations indicate that selected VanY enzymes may contain an additional domain as observed in VanXYG.
VanXY Interactions with Ligands Reveal the Molecular Details of d,d-Carboxypeptidase Selectivity Essential for Their Adaptability to the d-Ala-d-Ser Resistance Mechanism.
To gain insights into the hydrolytic mechanism and substrate specificity of VanXY enzymes, we determined the crystal structures of VanXYCD59S and VanXYG in complex with the phosphinate transition state analog of d-Ala-d-Ala (PHY, signifying the d,d-diastereoisomer) (Fig. 2 B and C and Fig. S3) to 1.54 Å and 1.36 Å, respectively (Table S1). A mixture of the four diastereoisomers of PHY inhibited VanXYC and VanXYG in a competitive manner with Ki values of 60 and 324 µM, respectively (Fig. S4). We also determined the structure of VanXYCD59S in complex with its substrate d-Ala-d-Ala and product d-Ala (Fig. 2 C–E) to 2.0 and 2.25 Å, respectively. The enzyme•substrate complex structure was captured by replacing the VanXYCD59S active site zinc with copper, which has been shown to inhibit the activity of this enzyme (8) (SI Materials and Methods).
The position of the inhibitor was identical in both structures and similar to that of the d-Ala-d-Ala substrate, with the N termini of both ligands pointed outwards to the opening of the VanXY active site cavity and their C termini located deep in the catalytic site (Fig. 2C and Fig. S3). Due to the racemic mixture for crystallization, PHY and its (l,d)-diastereoisomer were observed in both VanXY complexes (Fig. S5). However, the (l,d)-diastereoisomer will not be discussed further because VanXYC was 384-fold less efficient in hydrolyzing l-Ala-d-Ala than d-Ala-d-Ala and had no activity against the (l, l)- or (d, l)-diastereoisomers (Table S2).The two oxygens of the tetrahedral phosphate of PHY chelated the Zn2+ ion, mimicking the expected state of the tetrahedral hydrolysis intermediate (Fig. S5) (9, 18). Similarly, the peptide oxygen of d-Ala-d-Ala formed an interaction with the Cu2+ ion (Fig. 2D). In line with this observation, the side chain of Arg62 (Arg74) formed a bidentate interaction with the other phosphate oxygen of PHY (Fig. S3), while remaining hydrogen-bonded to Gln67 (Gln79) and Ser59 (Ser71).
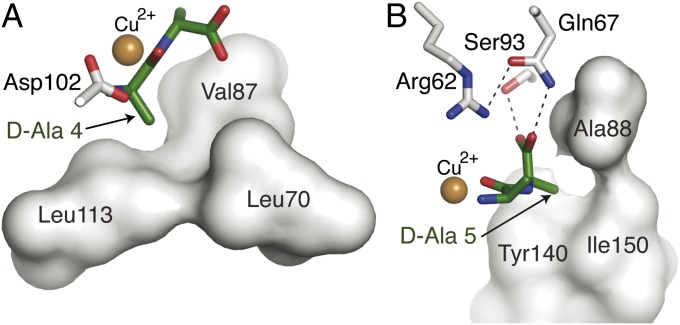
The positions of the two methyl groups of PHY and of d-Ala-d-Ala were defined by hydrophobic pockets in the VanXYC active center and revealed the molecular mechanism for selectivity toward hydrolysis of the d-Ala-d-Ala target. d-Ala-4 was accommodated into a pocket formed by Leu70 (Ile82), Val87 (Val99), and Leu113 (Phe124) (Fig. 3A) and d-Ala-5 by Ala88 (Ala100), Tyr140 (Tyr150), and Ile150 (Ile161) (Fig. 3B). Importantly, modeling any larger sidechain in either position of d-Ala-d-Ala or PHY led to steric clashes with its binding pocket in both VanXY structures, suggesting strict selectivity for alanine at both positions of the substrate. The carboxylates of d-Ala-d-Ala and PHY occupied the deepest part of the active site and were oriented by interactions with Gln67 (Gln79), Arg62 (Arg74), and Ser93 (Ser105) (Figs. 2D and 3B). Gln67 (Gln79) is conserved only in the M15B subfamily (Fig. S2), suggesting that this is a key residue for substrate specificity.
Fig. 3.
d-Ala binding pockets of VanXYC. (A) Interactions between VanXYCD59S and N terminus and d-Ala-4 of d-Ala-d-Ala. (B) Interactions between VanXYCD59S and C terminus and d-Ala-5 of d-Ala-d-Ala. Residues forming hydrophobic interactions with methyl groups of each d-Ala are shown as gray surface.
The position of d-Ala in the VanXYCD59S•product complex (Fig. 2E) corresponded to that of d-Ala-5 of d-Ala-d-Ala or PHY in their respective VanXYCD59S complex structures (Fig. 2D and Fig. S5). The amino group of the d-Ala product interacted with Glu153 of VanXYCD59S, consistent with the proposed role of this residue as a general acid to the leaving amine group of d-Ala after hydrolysis (19). The Zn2+ ion in the VanXYCD59S•d-Ala structure remained closely bound to a water molecule. These observations are in line with the hydrolytic mechanism proposed for the aminopeptidase VanX (18, 20) and suggest a similar mechanism for VanXYC d,d-carboxypeptidase activity.
VanXY d,d-Carboxypeptidase Active Site Is Distinct from That of VanX d,d-Aminopeptidase.
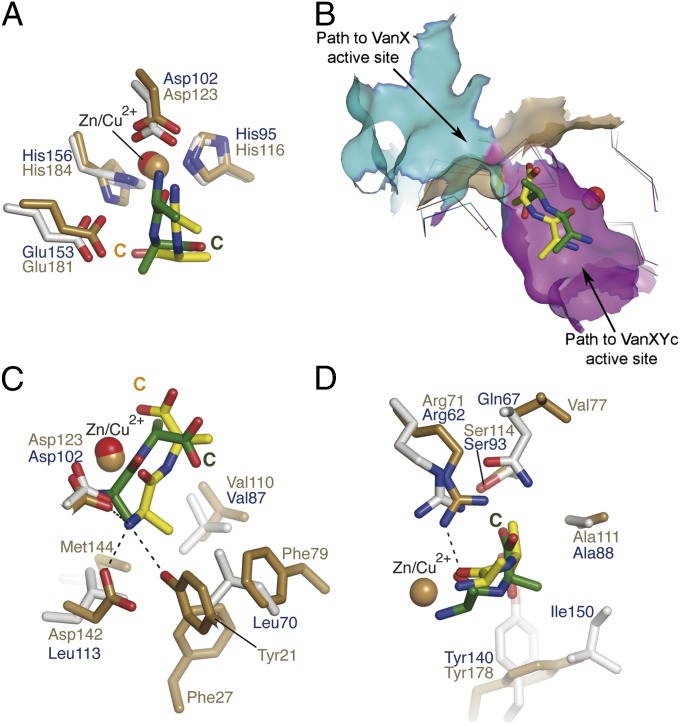
The catalytic domains of VanXY enzymes (VanXYCD59S residues 31–160, VanXYG residues 43–170) superimposed well with that of VanX (residues 39–188) with pairwise rmsd values of 2.3 and 2.4 Å for VanXYCD59S and VanXYG, respectively. The similarity between VanXY and VanX structures extended to the core elements involved in hydrolysis (Fig. 4A), with both enzymes interacting with their respective substrates in the same vicinity of the active center (Fig. 4B). However, comparative analysis of the VanXY and VanX d-Ala-d-Ala binding sites revealed important differences. The VanXY•d-Ala-d-Ala complex displayed two distinctive cavities leading to the active center (Fig. 2C). One of the cavities (cyan in Figs. 2C and 4B) is constricted by Ala88 (Ala100), Tyr140 (Tyr151), and Ile150 (Ile161) and is bounded by the conserved N-terminal region of M15B enzymes (VanXYC residues 4–23, VanXYG residues 17–35). This cavity corresponded roughly to the shallow d-Ala-d-Ala binding pocket in VanX (gold in Fig. 4B). In VanX, the N terminus of d-Ala-d-Ala and the sidechain of Ala-1 were bound into this cavity’s deepest region, with the C terminus of the substrate facing solvent (18). The second cavity (magenta in Figs. 2C and 4B) in the VanXY active center was relatively larger and had no equivalent in VanX (Fig. S6). This cavity accommodated d-Ala-d-Ala or PHY with their C termini and d-Ala-5 bound in the depth of this pocket and their N termini facing toward solvent. Of note, the VanXY active site larger cavity was capped by the prominent β4–α5 loop, suggesting its potential role in mediating substrate interactions. Overall, these observations indicate that, whereas VanXY enzymes retained a cavity that is seen in VanX, the binding of the dipeptide substrate in these enzymes is shifted to a distinct and unique region of the active center.
Fig. 4.
Comparison of d-Ala-d-Ala binding by VanX and VanXY. (A) Ligand binding to VanXYCD59S (gray/blue sticks) and VanX (gold sticks). Green sticks, d-Ala-d-Ala bound to VanXYCD59S; yellow sticks, d-Ala-d-Ala bound to VanX. (B) Superposition of VanXYCD59S•d-Ala-d-Ala (blue and green) and VanX•d-Ala-d-Ala (gold and yellow) complexes. Solvent-exposed surface representation shown for substrate entry cavities of VanXYCD59S, colored cyan and purple, as in Fig. 2C, and gold for VanX. (C) Comparison of interactions between VanXYCD59S and VanX with d-Ala-4 of d-Ala-d-Ala, colored as in A. Hydrogen bonds indicated are between VanX and d-Ala-d-Ala. (D) Detailed view of interactions between VanXYCD59S and VanX with d-Ala-5 of d-Ala-d-Ala, colored as in A. Hydrogen bonds indicated are between VanXYCD59S and d-Ala-d-Ala.
Comparison of the positions of d-Ala-d-Ala in VanX and VanXY revealed that the sidechains of the C-terminal moieties of the ligands do not superimpose: the peptide bond in d-Ala-d-Ala was rotated 180° between the VanXY and VanX complexes (Fig. 4A). Consequently, the methyl group of the C-terminal residue of d-Ala-Ala in the VanX complex occupied a position corresponding to the carboxylate of d-Ala-d-Ala/PHY in the VanXY complexes. Furthermore, the VanX d-Ala-d-Ala binding pocket provided more interactions with the N-terminal d-Ala of the substrate than the equivalent residues of VanXY (Fig. 4C). In contrast, the C termini and d-Ala-5 of d-Ala-d-Ala/PHY interacted more extensively with VanXY than with VanX. Interactions with the reaction product d-Ala also pointed to functional differences between these enzymes. In the case of VanX, the d-Ala product was bound at the same position as the N-terminal alanine of d-Ala-d-Ala whereas, in the VanXYCD59S•product complex, d-Ala occupied a position corresponding to the C-terminal alanine of d-Ala-d-Ala/PHY. Overall, these comparative observations reflect the d,d-carboxypeptidase specificity of VanXY versus the d,d-aminopeptidase specificity of VanX (9, 18).
Structure-Guided Mutagenesis Probe into the Role of VanXY Active Site Residues.
In agreement with previous data (8, 12, 21), hydrolysis assays against d-Ala-d-Ala and pentapeptide[d-Ala] demonstrated that, despite strong structural similarities, VanXYC was 22-fold more efficient than VanXYG against d-Ala-d-Ala and that VanXYC was 13-fold more efficient against d-Ala-d-Ala than against pentapeptide[d-Ala] (Fig. 5C and Table S3). To identify the molecular determinants responsible for these differences, active site residues were mutated in VanXYC and VanXYG (Table S4), and the activity of these variants was measured against the two substrates (Fig. 5C and Table S3).
Fig. 5.
Mutational analysis of putative VanXYC substrate binding surface. (A) Comparison of the conformation of the β4–α5/bisubstrate selectivity loop of VanXYCD59S when bound to d-Ala-d-Ala (loop and ligand colored beige) and PHY (loop and ligand colored green). (B) Surface representation of VanXYCD59S•d-Ala-d-Ala complex. Residues identified through mutagenesis to contact pentapeptide[d-Ala] are colored purple. (C) Catalytic efficiencies of selected VanXYC mutants for hydrolysis of d-Ala-d-Ala and pentapeptide[d-Ala]. For full list of mutants, see Table S3.
Mutation of the catalytic base Glu153 (Glu164) and of the transition state-stabilizing Arg62 (Arg74) severely abrogated VanXY activity, confirming their essentiality (Table S3). We observed a similar effect with mutation of Gln67 (Gln79) to glutamate, suggesting that the amide nitrogen was also critical for VanXY catalysis. Mutation of VanXYC Asp59 to serine resulted in a sixfold lower catalytic efficiency whereas the reciprocal mutation of VanXYG Ser71 to aspartate had an opposite effect, yielding a twofold increase. These results are in line with the structural analysis suggesting that this VanXYC residue coordinates the catalytic arginine and plays an important role in substrate positioning. The fact that the Ser71Asp mutant of VanXYG was not sufficient to match the level of d,d-dipeptidase activity indicates that other active site residues contribute to this difference.
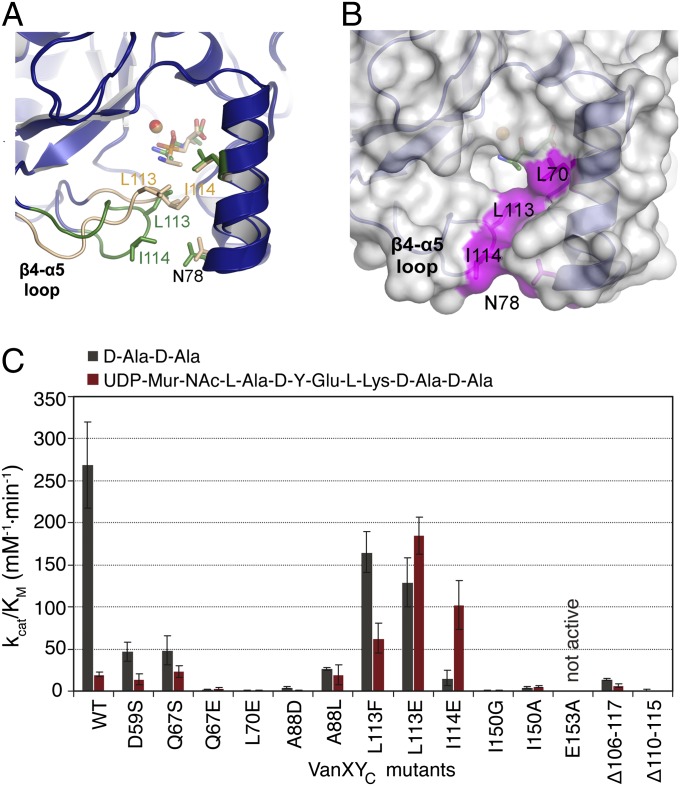
Next, we targeted the binding pocket for the N-terminal alanine of the substrate. The VanXYC Leu70Glu mutation led to a significant drop in activity against both substrates whereas substitution of the corresponding residue Ile82 to alanine in VanXYG resulted in threefold higher catalytic efficiency against d-Ala-d-Ala compared with the wild-type enzyme. This result points to the importance of a hydrophobic sidechain at this position and suggests that the smaller alanine sidechain may provide a catalytic advantage in interaction with substrate’s alanine. The VanXYG mutant Ile125Ala had a drastic decrease in catalytic activity. At the equivalent position, the VanXYC mutant Ile114Glu retained only 12% activity against d-Ala-d-Ala but showed a fivefold higher catalytic efficiency against pentapeptide[d-Ala]. Mutation Leu113Glu in VanXYC reduced d,d-dipeptidase activity of this enzyme by twofold. Notably, both Leu113 (Leu124) and Ile114 (Ile125) residues belong to the β4–α5 loop, which is prominently exposed near the VanXY-specific substrate binding cavity. The drastic changes in activity prompted by these mutations suggest that this structural element may be involved in d,d-dipeptidase activity.
Within the binding pocket for the C-terminal alanine of the substrate, mutation of VanXYC Ala88 to either leucine or aspartate severely compromised the d,d-dipeptidase activity whereas the d,d-pentapeptidase activity was affected by Ala88Asp but not by Ala88Leu substitution (Table S3). Mutation of VanXYC Ile150 resulted in abrogation of both activities and demonstrated its importance for the bisubstrate activity of VanXYC.
Combined mutagenesis and biochemical analysis points to the prominent β4–α5 loop (VanXYCD59S residues 107–120) as a VanXY-specific structural element involved in interactions with the substrates. This loop formed one face of the VanXY-specific cavity proximal to the PHY binding site (Fig. 2C), adopted different conformations according to the bound ligand (Fig. 5A), and possessed significant flexibility, as judged by the corresponding B-factors (Fig. S7). To further test its potential role in VanXY activity, we probed the other residues forming this loop and those interacting with it by mutagenesis. In agreement with our previous results, mutations Asn78Phe, Leu113Glu, or Ile114Glu, resulted in three- to ninefold increases in VanXYC catalytic efficiency and up to threefold greater affinity for pentapeptide[d-Ala] (Fig. 5C and Table S3).
To test whether the rise in activity was due to destabilization of the β4–α5 loop by mutagenesis, we introduced two deletions in this region, VanXYC Δ106–117 and Δ110–115. The deletant derivatives demonstrated, respectively, 20- and 245-fold decreases in d,d-dipeptidase activity (Fig. 5C and Table S3). The VanXYC Δ106–117 variant showed a threefold decrease in d,d-pentapeptidase activity whereas the VanXYC Δ110–115 mutant completely lost activity against this substrate. These results confirmed the key role played by the β4–α5 loop, which we now term the bisubstrate selectivity loop, in the dual activity of VanXY. In the absence of a costructure between VanXYC and pentapeptide[d-Ala], we speculate that the observed increases in catalytic efficiency of VanXY bisubstrate selectivity loop mutants were due to introduction of additional interactions with this substrate.
Discussion
Metallopeptidases are important contributors to recycling and metabolism of peptidoglycan and are essential in glycopeptide resistance. Due to antibiotic selection, enzymes from subfamilies M15D and M15B evolved as resistance-conferring proteins due to their activities in reducing the proportion of peptidoglycan precursors containing the glycopeptide-susceptible terminal d-Ala moiety by hydrolyzing d-Ala-d-Ala and pentapeptide[d-Ala], respectively. The evolution of VanXY, most similar to members of the M15B subfamily yet capable of mediating both dipeptidase and pentapeptidase reactions, is an example of the emergence of bifunctional antibiotic resistance enzymes (22). Bacteria might gain several advantages from this strategy, such as greater flexibility in enzyme activity, greater reaction efficiency, more efficient use of transcription and translation, coupled gene regulation, or ease of movement of mobile genetic resistance elements (22).
We provide insights into the structure, function, and evolution of VanXY d,d-peptidases, revealing the mechanism of dual specificity of this class of enzymes to mediate vancomycin resistance. This adaptation mechanism is centered on a mobile loop, appropriately positioned over the catalytic center and participating in both the d,d-dipeptidase and d,d-pentapeptidase reactions. Such flexible structural elements often mediate substrate selectivity and support diverse enzymatic reactions, including peptide hydrolysis (23, 24). In the case of VanXYC, the bisubstrate selectivity loop may alter its conformation based on the size of the substrate, providing the necessary chemical environment to accommodate its distinct substrates.
Structure-based multiple sequence alignment (Fig. S2) indicated that VanY enzymes generally lack the elements corresponding to the β4-α5/bisubstrate selectivity loop in VanXY and to the d,d-aminopeptidase binding site of VanX. These features are consistent with the observation that the VanY enzymes lack activity against the dipeptide substrates and select for larger peptidoglycan substrates (5, 10). The expected open architecture of the active site of VanY likely provides a suitable binding surface for peptidoglycan fragments larger than dipeptides. These results further highlight acquisition of the bisubstrate selectivity loop as the key adaptation conferring dual substrate specificity to VanXY enzymes.
The data also pointed to VanY rather than VanX enzymes as possible ancestors for bisubstrate-specific VanXY because the “open” active center architecture of VanY enzymes seems more suitable for incorporation of the bisubstrate specificity loop onto a pentapeptide-binding d,d-carboxypeptidase active site. Such a modification would have allowed a VanXY ancestor to graft d,d-dipeptidase activity onto a d,d-pentapeptidase active site already optimized as a carboxypeptidase. An appropriate scaffold could have been an ancestral M15B subfamily enzyme with a role either in peptidoglycan metabolism, such as L,d-carboxypeptidases (25, 26), and/or self-resistance of a glycopeptide producer, such as VanYn (27). This evolutionary model is consistent with sequence analysis and extensive phylogenetic reconstruction (Fig. S8) showing that VanXY are most related to VanY enzymes, and speculation that they would share a substrate binding site less constricted than that of VanX (7). It is also in line with a general trend for antibiotic resistance proteins, where an ancestral “protoresistance” enzyme scaffold with a housekeeping function is recruited for resistance through structural changes that affect its substrate specificity or activity (28). To the best of our knowledge, elucidation of the structural adaptations of the M15 metallopeptidase fold in response to antibiotic pressure is the first such observation for this enzyme family.
Structural analysis also showed that VanXY enzymes are optimized to select for d-Ala at the terminal position of the peptidoglycan substrates. The main catalytic residues of the VanX and VanXY families superimpose well, implying a conserved hydrolysis mechanism. However, the VanXY active site features subtle differences, such as acquisition of the Gln67/Gln79 residue that positions the carboxylate of the substrates. These features would position the unsuitable substrate into the hydrophobic environment of the VanXY d-Ala binding pockets, enabling discrimination against d-Ser at the C-terminal position. Analysis of VanY sequences indicates that these enzymes also contain this key glutamine residue and the general characteristics of this hydrophobic selectivity pocket, suggesting that they also rely on these interactions for d,d-carboxypeptidase activity.
Although our data provided significant insights into the molecular function of VanXY enzymes, several aspects of VanXYG activity remain to be clarified. Specifically, it is unclear why VanXYG lacks d,d-pentapeptidase activity and has relatively poor d,d-dipeptidase activity, even though VanXYG and VanXYC have very similar interactions with PHY.
Another intriguing structural feature is the presence of a secondary domain in VanXYG formed by residues 1–32 and 186–250. The specific role of this domain remains to be determined. Phylogenetic analysis of VanXY enzymes placed VanXYG closest to the VanY enzymes and suggested that the sequence corresponding to the VanXYG secondary domain is partially conserved in these enzymes (Figs. S2 and S8). Given that VanY enzymes are membrane-associated whereas VanXY enzymes are not, one might speculate that the secondary domain might have a role in membrane association. In this scenario, presence of this domain in VanXYG could be a relic of the evolution of VanXY from VanY enzymes.
Finally, the molecular structures of the VanXY and VanY enzymes pave the way for the design of inhibitors that would target vancomycin resistance. Given that the shallow nature of the VanX active site posed challenges in designing potent inhibitors (29), the less constricted pentapeptide[d-Ala] binding sites of VanXY or VanY suggest that these enzymes are more attractive targets for inhibitor development. The presented crystal structures provide the necessary molecular framework for searches for suitable inhibitory compounds.
Our data demonstrate that the d,d-peptidase fold is able to expand its role in vancomycin resistance as a result of specific structural changes in its active site. This study highlights the remarkable adaptability of this enzyme class to accommodate widely different peptidoglycan substrates. This observation underscores the adaptability of microorganisms in response to antibiotic challenge and demonstrates the susceptibility of established antibiotics long thought to be exempt from resistance.
Materials and Methods
VanXY proteins were purified using N-terminal His-tags from Escherichia coli BL21-CodonPlus (DE3)-RIPL cells. Site-directed mutagenesis was performed using the oligonucleotide pairs in Table S1. All crystals were grown using hanging-drop vapor diffusion. Crystals of PHY complexes were obtained by soaking into apoprotein crystals. Crystals of the VanXYCD59S-d-Ala complex were obtained by cocrystallization with acetyl-l-Lys-d-Ala-d-Ala. Crystals of the VanXYCD59S-d-Ala-d-Ala complex were obtained by replacing Zn2+ in apoprotein crystals using phenanthroline, followed by soaking of CuCl2 and d-d-Ala-d-Ala. d,d-dipeptidase and d,d-pentapeptidase activities were determined by the amino acid oxidase-lactate dehydrogenase coupled assay. Full details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank P. E. Reynolds for having incited us to initiate this study. We thank R. Di Leo for technical help, Z. Wawrzak for data collection and interpretation, C. H. Park for coordinates for VanX•d-Ala-d-Ala and VanX•phosphonate analog complexes, D. Mengin-Lecreulx for providing pentapeptide[D-Ala], and M. Anderluh and S. Gobec for providing transition-state analog PHY. This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (Contracts HHSN272200700058C and HHSN272201200026C) and by an unrestricted grant from Reckitt Benckiser.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4F78, 4MUQ–4MUT, and 4OAK).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402259111/-/DCSupplemental.
References
- 1.Arias CA, Murray BE. The rise of the Enterococcus: Beyond vancomycin resistance. Nat Rev Microbiol. 2012;10(4):266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Périchon B, Courvalin P. 2011. Glycopeptide resistance. Antibiotic Discovery and Development, eds Dougherty TJ, Pucci MJ (Springer, Boston), pp 515–542.
- 3.Reynolds PE, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of D-alanyl-D-alanine. Mol Microbiol. 1994;13(6):1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 4.Lessard IA, et al. Homologs of the vancomycin resistance D-Ala-D-Ala dipeptidase VanX in Streptomyces toyocaensis, Escherichia coli and Synechocystis: Attributes of catalytic efficiency, stereoselectivity and regulation with implications for function. Chem Biol. 1998;5(9):489–504. doi: 10.1016/s1074-5521(98)90005-9. [DOI] [PubMed] [Google Scholar]
- 5.Wright GD, Molinas C, Arthur M, Courvalin P, Walsh CT. Characterization of vanY, a DD-carboxypeptidase from vancomycin-resistant Enterococcus faecium BM4147. Antimicrob Agents Chemother. 1992;36(7):1514–1518. doi: 10.1128/aac.36.7.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur M, Depardieu F, Snaith HA, Reynolds PE, Courvalin P. Contribution of VanY D,D-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994;38(9):1899–1903. doi: 10.1128/aac.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds PE, Arias CA, Courvalin P. Gene vanXYC encodes D,D -dipeptidase (VanX) and D,D-carboxypeptidase (VanY) activities in vancomycin-resistant Enterococcus gallinarum BM4174. Mol Microbiol. 1999;34(2):341–349. doi: 10.1046/j.1365-2958.1999.01604.x. [DOI] [PubMed] [Google Scholar]
- 8.Podmore AHB, Reynolds PE. Purification and characterization of VanXY(C), a D,D-dipeptidase/D,D-carboxypeptidase in vancomycin-resistant Enterococcus gallinarum BM4174. Eur J Biochem. 2002;269(11):2740–2746. doi: 10.1046/j.1432-1033.2002.02946.x. [DOI] [PubMed] [Google Scholar]
- 9.Wu Z, Wright GD, Walsh CT. Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1995;34(8):2455–2463. doi: 10.1021/bi00008a008. [DOI] [PubMed] [Google Scholar]
- 10.Arthur M, Depardieu F, Cabanié L, Reynolds P, Courvalin P. Requirement of the VanY and VanX D,D-peptidases for glycopeptide resistance in enterococci. Mol Microbiol. 1998;30(4):819–830. doi: 10.1046/j.1365-2958.1998.01114.x. [DOI] [PubMed] [Google Scholar]
- 11.McKessar SJ, Berry AM, Bell JM, Turnidge JD, Paton JC. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob Agents Chemother. 2000;44(11):3224–3228. doi: 10.1128/aac.44.11.3224-3228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Depardieu F, Bonora MG, Reynolds PE, Courvalin P. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol Microbiol. 2003;50(3):931–948. doi: 10.1046/j.1365-2958.2003.03737.x. [DOI] [PubMed] [Google Scholar]
- 13.Casadewall B, Courvalin P. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J Bacteriol. 1999;181(12):3644–3648. doi: 10.1128/jb.181.12.3644-3648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawlings ND, Barrett AJ, Bateman A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40(Database issue):D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlings ND, Barrett AJ. 2012. Introduction: Metallopeptidases and their clans. Handbook of Proteolytic Enzymes, eds Rawlings ND, Salvesen G (Elsevier, Amsterdam), pp 325–369.
- 16.Charlier P, Wery J-P, Dideberg O, Frère J-M. 2011. Streptomyces albus G D-Ala-D-Ala carboxypeptidase. Encyclopedia of Inorganic and Bioinorganic Chemistry, ed Scott RA (Wiley, Chichester, UK)
- 17.Korndörfer IP, et al. Structural analysis of the L-alanoyl-D-glutamate endopeptidase domain of Listeria bacteriophage endolysin Ply500 reveals a new member of the LAS peptidase family. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 6):644–650. doi: 10.1107/S0907444908007890. [DOI] [PubMed] [Google Scholar]
- 18.Bussiere DE, et al. The structure of VanX reveals a novel amino-dipeptidase involved in mediating transposon-based vancomycin resistance. Mol Cell. 1998;2(1):75–84. doi: 10.1016/s1097-2765(00)80115-x. [DOI] [PubMed] [Google Scholar]
- 19. Auld DS (2012) Catalytic mechanisms for metallopeptidases. Handbook of Proteolytic Enzymes, eds Rawlings ND, Salvesen G (Elsevier, Amsterdam), pp 370–396.
- 20.McCafferty DG, Lessard IA, Walsh CT. Mutational analysis of potential zinc-binding residues in the active site of the enterococcal D-Ala-D-Ala dipeptidase VanX. Biochemistry. 1997;36(34):10498–10505. doi: 10.1021/bi970543u. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds PE, Courvalin P. Vancomycin resistance in enterococci due to synthesis of precursors terminating in D-alanyl-D-serine. Antimicrob Agents Chemother. 2005;49(1):21–25. doi: 10.1128/AAC.49.1.21-25.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Fisher JF, Mobashery S. The bifunctional enzymes of antibiotic resistance. Curr Opin Microbiol. 2009;12(5):505–511. doi: 10.1016/j.mib.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Bruce LA, et al. Hydrogen bond residue positioning in the 599-611 loop of thimet oligopeptidase is required for substrate selection. FEBS J. 2008;275(22):5607–5617. doi: 10.1111/j.1742-4658.2008.06685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malabanan MM, Amyes TL, Richard JP. A role for flexible loops in enzyme catalysis. Curr Opin Struct Biol. 2010;20(6):702–710. doi: 10.1016/j.sbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Courtin P, et al. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an L,D-carboxypeptidase involved in peptidoglycan maturation. J Bacteriol. 2006;188(14):5293–5298. doi: 10.1128/JB.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barendt SM, Sham L-T, Winkler ME. Characterization of mutants deficient in the L,D-carboxypeptidase (DacB) and WalRK (VicRK) regulon, involved in peptidoglycan maturation of Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol. 2011;193(9):2290–2300. doi: 10.1128/JB.01555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binda E, Marcone GL, Pollegioni L, Marinelli F. Characterization of VanYn, a novel D,D-peptidase/D,D-carboxypeptidase involved in glycopeptide antibiotic resistance in Nonomuraea sp. ATCC 39727. FEBS J. 2012;279(17):3203–3213. doi: 10.1111/j.1742-4658.2012.08706.x. [DOI] [PubMed] [Google Scholar]
- 28.Morar M, Wright GD. The genomic enzymology of antibiotic resistance. Annu Rev Genet. 2010;44:25–51. doi: 10.1146/annurev-genet-102209-163517. [DOI] [PubMed] [Google Scholar]
- 29.Crowder MW. Combating vancomycin resistance in bacteria: Targeting the D-ala-D-ala dipeptidase VanX. Infect Disord Drug Targets. 2006;6(2):147–158. doi: 10.2174/187152606784112146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.