Significance
Asthma is a chronic inflammatory disorder that is notoriously difficult to diagnose, characterize, and properly treat with tests that are available to clinicians today. Therefore, clinicians would benefit from additional tools that objectively characterize asthma in patients. In this work, we describe a handheld microfluidic device that discriminates asthma from allergic rhinitis patients based on neutrophil function—an inflammatory cell that has been implicated in the pathogenesis of asthma. The device can sort neutrophils from a drop of whole blood within minutes, and was used in a clinical setting to characterize asthmatic and nonasthmatic patients. This technology provides a new tool for clinicians to characterize asthma based on cellular function.
Keywords: diagnostics, microfluidics, KOALA, passive pumping
Abstract
Asthma is a chronic inflammatory disorder that affects more than 300 million people worldwide. Asthma management would benefit from additional tools that establish biomarkers to identify phenotypes of asthma. We present a microfluidic solution that discriminates asthma from allergic rhinitis based on a patient’s neutrophil chemotactic function. The handheld diagnostic device sorts neutrophils from whole blood within 5 min, and generates a gradient of chemoattractant in the microchannels by placing a lid with chemoattractant onto the base of the device. This technology was used in a clinical setting to assay 34 asthmatic (n = 23) and nonasthmatic, allergic rhinitis (n = 11) patients to establish domains for asthma diagnosis based on neutrophil chemotaxis. We determined that neutrophils from asthmatic patients migrate significantly more slowly toward the chemoattractant compared with nonasthmatic patients (P = 0.002). Analysis of the receiver operator characteristics of the patient data revealed that using a chemotaxis velocity of 1.55 μm/min for asthma yields a diagnostic sensitivity and specificity of 96% and 73%, respectively. This study identifies neutrophil chemotaxis velocity as a potential biomarker for asthma, and we demonstrate a microfluidic technology that was used in a clinical setting to perform these measurements.
Asthma is a chronic inflammatory disorder of the lungs that is associated with airway hyperresponsiveness (AHR) and obstructed airflow (1), affecting more than 300 million people worldwide (2). Over the past 30 y, asthma prevalence has increased significantly in many populations, with some indications that prevalence may be reaching a plateau in the developed world. Significant progress has been made in identifying primary mediators involved in the pathophysiology of asthma. Several cell types, such as T helper cells (TH1/TH2), dendritic cells, mast cells, macrophages, eosinophils, and neutrophils play central roles in the pathology of asthma (4–7). Additionally, various cytokines that regulate the leukocyte trafficking, such as interleukins, IFN-γ, and TNF-α, have been identified and targeted in drug therapies. The recruitment of leukocytes to the lungs, particularly eosinophils and neutrophils, is central to the pathogenesis of asthma. Increased numbers of eosinophils are prominently observed in the lung tissue and bronchoalveolar lavage (BAL) fluid for most asthmatics (5). Neutrophils play a more critical role in severe asthma, where elevated counts of neutrophils are often observed in the BAL fluid (7). An overview of the role of neutrophils in asthma is shown in Fig. 1A. Although significant progress has been made in uncovering mediators in the pathology of asthma, these gains have not yet greatly improved our ability to define clinically relevant phenotypes of asthma in patients.
Fig. 1.
Overview of different diagnostic techniques and the role of neutrophils in the pathology of asthma. (A) Summary of the role of neutrophils in the pathology of asthma, showing neutrophil adhesion and transendothelial migration; chemotaxis mediated by macrophages and T-helper cells; and neutrophilia in the lung tissue that leads to airway remodeling and airflow obstruction. (B) Proposed microfluidic method (more details in Fig. S1) for phenotyping asthma patients by measuring upstream of the asthma pathology with rapid neutrophil sorting on a P-selectin–coated surface (1); neutrophil chemotaxis monitored with high-throughput microscopy and automatically tracked with software (2); and asthma characterization on the basis of chemotaxis outputs (3). (C) Traditional clinical asthma diagnostic methods occur downstream of the asthma pathophysiology by measuring the effect of leukocyte inflammation on airway obstruction, nitric oxide output, or clinical symptoms.
Asthma is diagnosed clinically by physicians, informed by the patient’s medical history, spirometry tests that measure lung function, reversibility of AHR, and several other potential metrics (8). These diagnostic techniques measure the effects of the inflammatory response in the lung by assessing airway constriction, nitric oxide production, and the resulting clinical symptoms. However, all of these diagnostic tests require patient compliance, which can be challenging when diagnosing children or the elderly (9). Additionally, many asthma diagnostic tests partially rely on the patient experiencing clinical symptoms that are variable during or around the visit to the physician. Perhaps these common characteristics of current diagnostic techniques contribute to difficulties in diagnosing asthma, particularly in certain subpopulations. For example, in a recent Canadian study involving ∼500 obese and nonobese subjects, Aaron et al. (10) found that ∼30% of the test subjects had been falsely diagnosed with asthma by physicians. Additionally, it is well established that the elderly are consistently underdiagnosed for asthma (11, 12). Therefore, additional tools are needed to improve the diagnosis of asthma. Furthermore, current asthma assessments do not inform the clinician of disease severity, expected clinical course, and risk of exacerbations.
To improve characterization of asthma in the clinic, we have developed a handheld microfluidic chip that can identify functional measures of asthma from a drop of whole blood. Microfluidic systems have several characteristics that make them well-suited for clinical use, including low sample-volume requirements (13, 14); simple integration with automated fluid handling systems (15); and diffusion-dominant laminar fluidic phenomena that allow for precise control of a cell’s microenvironment (16–18). Indeed, microfluidic-based tools are increasingly being used in clinical research for diagnostic purposes (19–26). Neutrophils have been used to diagnose clinical conditions in human patients based on proteomic and genomic analysis (22) and chemotaxis behavior (23, 27), demonstrating that assays measuring cell function can be used for diagnostics. In this work, we assay the neutrophil chemotactic function in a blind study to identify quantitative domains that can be used to discriminate asthma from nonasthmatic allergic rhinitis. This approach of directly measuring the effector cell in the pathology of asthma differs from traditional diagnostic tests, which measure the variable effect of inflammation on airway constriction (Fig. 1 B and C and Table S1). Importantly, we developed methods to simplify the sample preparation, assay protocol, and data analysis that offer significant time savings over traditional macroscale (28–30) and microscale (18) chemotaxis techniques, allowing for the translation of the technology into the clinic. We analyzed 34 patients, and discovered that neutrophil chemotaxis can be used to discriminate asthma from nonasthmatic, allergic rhinitis patients with sensitivity and specificity of 96% and 73%, respectively. The results of the clinical application of our microfluidic device represent a first step demonstration of how asthma can potentially be diagnosed and managed based on cellular function, rather than largely by clinical observations.
Results
We adapted a microfluidic neutrophil chemotaxis platform that we previously developed (31) to assay the neutrophil function of mildly asthmatic and nonasthmatic patients in a clinical setting. Briefly, the diagnostic chip has two primary components: (i) a functionalized migration channel in the base, where the rapid neutrophil purification and chemotaxis are performed, and (ii) a multifunction lid that houses all of the reagents required for the diagnostic test and mitigates evaporation, which can be detrimental for long-term open microfluidic experiments (32). The purification is accomplished using a P-selectin–coated polystyrene surface, whereby whole blood is pumped into the microchannels, allowing neutrophils to bind to the P-selectin, and other components of the whole blood (e.g., erythrocytes, plasma, etc.) are removed using subsequent laminar flow wash steps (33). In our assay, the blood and reagents are passively pumped through the microchannels (34), enabling all of the fluid-handling steps using only a micropipette and requiring no external pumping systems. Once the purification is complete, a lid containing a hydrogel–chemoattractant mixture (H-CA) is placed onto the base component, contacting the H-CA drop with the migration channel and initiating the chemotaxis experiment (Fig. 1B, Fig. S1, and Dataset S1) (31). Following a time-lapse capture of the neutrophil chemotaxis, the data are automatically tracked and analyzed using custom tracking software, dubbed Je’Xperiment (JEX), eliminating the need for onerous manual cell tracking (Fig. S2; source code available upon request). Three outputs were measured to characterize the neutrophil chemotactic function: the absolute migration speed (independent of the direction the cells move); the chemotactic index (displacement of the cell throughout the time lapse divided by its total path length); and chemotaxis velocity (speed of the cell in the direction of increasing concentration of chemoattractant). The microfluidic chip was used in a clinical setting to assay the neutrophil chemotactic function of mildly asthmatic and nonasthmatic patients to elucidate possible diagnostic domains between the two patient groups.
Characterizing Neutrophil Isolation and Chemotaxis.
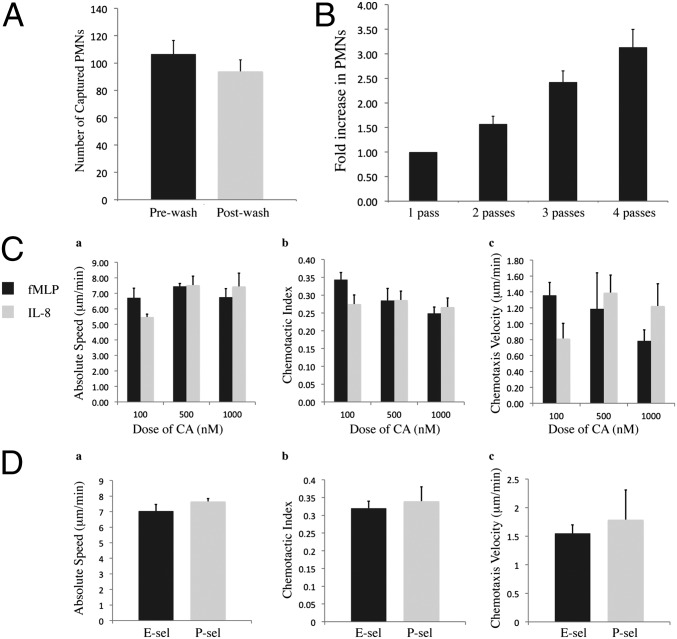
We sought to characterize the neutrophil capture technique for blood samples obtained from the clinic. Using the protein-based neutrophil sorting procedure, we found that ∼89% of neutrophils remained on the surface following subsequent wash steps (Fig. 2A). Furthermore, we found that the surface could be enriched with neutrophils by simply passing whole blood through the microchannels multiple times before the washing procedure (Fig. 2B); this was advantageous if neutrophil capture was particularly low and more neutrophils were required to obtain meaningful chemotaxis data.
Fig. 2.
Characterization of diagnostic chip for performing neutrophil chemotaxis from a drop of blood. (A) Capture of neutrophils, or polymorphonuclear leukocytes (PMNs), from clinical blood samples before and after laminar flow wash steps; capture efficiency was 89% (n = 4). (B) Increase in neutrophil capture on P-selectin–coated microchannels with additional passes of blood across the microchannel substrate (n = 3). (C) Chemoattractant (CA) dose–response for neutrophils obtained from healthy human patients, measuring the absolute migration speed (a), chemotactic index (b), and chemotaxis velocity (c); time-lapse imaging for 120 min (n = 3). Note: no observable neutrophil migration in controls (gel without CA). (D) Neutrophil chemotaxis in a linear gradient of fMLP with a source concentration of 100 nM; time-lapse imaging for 90 min (n = 3). All chemotaxis data tracked and analyzed with JEX. Error bars show SEM.
Following the neutrophil isolation, we characterized the migration of human neutrophils in a linear chemotactic gradient using the potent chemoattractants IL-8 and formyl-methionyl-leucyl-phenylalanine (fMLP; Fig. 2C and Movie S1). For all control experiments (i.e., gel and media, no chemoattractant), neutrophils remained inactivated and there was no observable migration (Movie S1). Additionally, we compared neutrophil chemotaxis for neutrophils on a P-selectin and E-selectin (Fig. 2D), with no significant differences observed in the chemotaxis outputs; this result indicated that either protein coating could be used for the purification and chemotaxis. For the assay of patient samples, we used 100 μg/mL P-selectin for the substrate coating and 100 nM fMLP for the source of chemoattractant, as we observed robust neutrophil capture and chemotaxis using these experimental conditions. To obtain sufficient neutrophil enrichment from clinical samples, blood was passed through the microchannels two times before neutrophil purification.
Measuring Neutrophil Chemotactic Function for Asthmatic and Nonasthmatic Patients.
After we characterized the neutrophil chemotaxis assay using human blood samples, we used the microfluidic device in a clinical setting to analyze the neutrophil function for a cohort of patients that were clinically characterized as asthmatic or nonasthmatic (Table 1). The absolute migration speed, chemotactic index, and chemotaxis velocity for all of the patients are shown in Fig. 3A (see SI Materials and Methods and Movie S2). The absolute migration speed and chemotactic index of the neutrophils did not differ significantly by asthma status (P = 0.33 and P = 0.49, respectively). However, the chemotaxis velocity for asthmatic patients was significantly reduced compared with nonasthmatic patients (P = 0.002). To the authors’ knowledge, this is the first report of reduced neutrophil chemotaxis in mildly asthmatic patients compared with nonasthmatic patients. Furthermore, to eliminate the possibility of medications such as corticosteroids from influencing the chemotaxis results among asthmatic patients (35–40), we measured the chemotaxis velocity of the mildly asthmatic patients that did not report taking these medications to treat their asthma (n = 6; Table S2). The average neutrophil chemotaxis velocity for this group was 1.15 μm/min, in line with the chemotaxis velocity of 1.09 μm/min for all asthmatic patients. Thus, the reduced neutrophil chemotaxis velocity was the manifestation of altered neutrophil physiology for asthmatic patients.
Table 1.
Characteristics of study subjects
| Characteristics | Asthma (n = 23) | No asthma (n = 11) |
| Age* (range), y | 36 (20–52) | 33 (20–48) |
| Men, no. | 13 | 8 |
| Blood EOS count, no. per mm3 | 293 (60) | 242 (73) |
| FEV1 % predicted | 91.2 (18.6) | 104.2 (31.4) |
| FEV1/FVC, ratio | 0.772 (0.161) | 0.802 (0.242) |
| Reversibility, % | 7.66 (0.951) | 3.60 (0.945) |
| FeNO, ppb | 47.0 (9.79) | 24.5 (7.34) |
| Allergic, no. (%) | 22 (95.6) | 10 (90.9) |
| Current asthma symptoms, no. “Yes” | 1 | 0 |
Average value shown with SEM in parentheses, unless otherwise indicated.
Age reported for patient’s baseline characterization.
Fig. 3.
Neutrophil chemotaxis for blood samples obtained from the clinic and comparison with FeNO measurements. (A) No statistically significant difference in neutrophil migration speed (i) and chemotactic index (ii) for asthmatic and nonasthmatic patients; neutrophil chemotaxis velocity (iii) is significantly lower for asthmatic patients (n = 23) compared with nonasthmatic patients (n = 11). *P = 0.002; dark line shows median; boxes show 25th and 75th percentile; endlines show minimum and maximum. (B) Comparison of neutrophil chemotaxis velocity and an emerging clinical diagnostic test (FeNO) showing higher neutrophil chemotaxis velocity for nonasthmatic patients correlating with lower FeNO values. (C) Diagnosis based on multiple chemotaxis outputs, as shown in Fig. 1B; other chemotaxis outputs (chemotactic index and absolute migration speed) do not improve diagnostic performance. All chemotaxis data tracked and analyzed with JEX. Error bars show SEM.
In addition to determining the patients’ neutrophil chemotaxis function, patients were characterized using several other measures commonly used in the clinic, such as the one second force expiratory volume (FEV1) % predicted, the ratio of FEV1 to the forced vital capacity (FEV1/FVC), eosinophil count (EOS#), %Reversibility, and fraction of exhaled nitric oxide (FeNO). Comparisons between our neutrophil chemotaxis outputs and spirometry measurements did not reveal any correlations that could be used to confirm or invalidate our test (Figs. S3–S6). However, the comparison of neutrophil chemotaxis velocity with FeNO measurements—an emerging clinical marker that positively correlates with the number of inflammatory cells present in the lungs—revealed an increase in neutrophil velocity at lower values of FeNO (Fig. 3B); the absolute neutrophil migration speed and chemotactic index did not show similar correlations (Fig. S7). Importantly, patients with increased neutrophil chemotaxis velocity had measures of FeNO below the typical diagnostic cutoff point (20–40 ppb) that clinicians often use to diagnose asthma (41–44).
Determining Chemotaxis Domains for Asthmatic from Nonasthmatic Patients.
Following the identification of the neutrophil chemotaxis velocity as a potential biomarker for asthma, we examined the receiver operator characteristics (ROCs) of the chemotaxis velocity data to determine the optimal domains to discriminate the patient groups (Fig. 4A). We observed that using a chemotaxis velocity of 1.545 μm/min as the threshold for diagnosis of asthma, this biomarker correctly identified 22 of 23 asthma subjects (sensitivity 95.7%, 95% confidence interval 87.0–100.0) and 8 of 11 nonasthma subjects (specificity 72.7%, 95% confidence interval 45.5–100.0). The other chemotaxis measures—chemotactic index and migration speed—had poor sensitivity and specificity due to the nonsignificance in these outputs between asthmatic and nonasthmatic patients (Fig. 3 A and B). Finally, we compared the sensitivity and specificity (45) of our test to other quantitative tests reported in the literature (Fig. 4B and Table S1). We found that our neutrophil chemotaxis diagnostic marker is among the most sensitive measures reported, although not as specific as other diagnostic methods.
Fig. 4.
Performance of microfluidic assay compared with traditional methods. (A) ROC curve for the neutrophil chemotaxis velocity patient data, showing the optimal sensitivity and specificity at a diagnostic cutoff of 1.545 μm/min. (B) Comparison of our method to techniques reported in the literature (see Table S1 for additional details).
Discussion
Asthma is a complex and heterogeneous disease with multiple phenotypes, and accurately diagnosing the severity and/or phenotype of asthma continues to present difficulties for physicians (10–12). In current clinical practice, there is no single test that physicians rely on to diagnose asthma, but rather a series of tools that are both qualitative and quantitative in nature (Fig. 1C). Most quantitative diagnostic tests currently in use measure the physiological effects of inflammatory cells, such as neutrophils and eosinophils, on the lungs, which is in contrast to our direct measurement of neutrophil function (46). One method that has been proposed for defining the different phenotypes of asthma is by the predominant inflammatory cell type involved—eosinophilic (elevated eosinophil counts), neutrophilic (elevated neutrophil counts), and paucigranulocytic (normal neutrophil and eosinophil counts) (4, 5, 47). However, this method of categorizing the patient groups does not take into account the functional status of these inflammatory cells. Mild asthma has been linked most strongly with eosinophils, and neutrophils have been implicated in the pathology of severe asthma. Interestingly, we observed that neutrophils obtained from mild asthmatic patients exhibit impaired chemotactic responses in a linear gradient of chemoattractant compared with neutrophils from nonasthmatic patients (Fig. 3A). To our knowledge, this is the first time reduced neutrophil chemotaxis velocity has been linked to mild asthma.
One possible explanation for the decrease we observed in neutrophil chemotaxis velocity for asthmatic patients is an increase in adhesion between asthmatic neutrophils and the P-selectin substrate coating the microchannels in our diagnostic chip. Dang et al. (48) have reported that leukocytes from allergic-asthmatic patients exhibit higher surface expression of P-selectin glycoprotein ligand-1, which mediates neutrophil binding to P-selectin (49). Therefore, our diagnostic assay may be detecting an increase in neutrophil adhesion to the substrate, resulting in reduced neutrophil chemotaxis velocity toward the chemoattractant. Importantly, this result could not easily be observed by performing a cell count of leukocytes in the peripheral blood or BAL, as has largely been done in past studies (44, 50, 51). We show that our microfluidic chip and automated readout method is sensitive enough to detect these differences in neutrophil function, which can then be used as a basis for classifying asthma. Given this capability of our microfluidic device to measure functional activities of inflammatory cells that correspond to clinical features of disease, we propose that analyses of neutrophils on alternate substrates or the analysis of other inflammatory cells, such as eosinophils, using our method will provide a composite measure for enhanced identification of clinically relevant phenotyping of asthma.
Our proposed method has several features that enabled its translation into a clinical setting. First, a focus on user-friendly operation; transportability; scalable thermoplastic fabrication; and rapid sample processing and data analysis were central to the design of the assay. The fluid handling in our system does not require any external pumps or tubing, and the lid-based method of generating a gradient of chemoattractant enables the robust and repeatable implementation of the technique. Once the sample processing is completed in the clinic, the device can easily be transported to a microscopy facility to conduct the automated imaging and analysis. An important distinction between our approach and other clinical diagnostic methods, such as spirometry or questionnaires, is that our technique does not rely on patient compliance. Our method objectively probes the chemotactic function using neutrophils obtained from a drop of blood, rather than testing clinical symptoms that may or may not be present during the patient’s visit. Indeed, nearly all patients in this study were not experiencing symptoms of asthma during their baseline characterization (Table 1). ROC analysis indicated that using an optimal neutrophil chemotaxis velocity of ∼1.55 μm/min, our method achieved sensitivity and specificity of 96% and 73%, respectively. This performance compares favorably to other diagnostic tests reported in the literature (Fig. 4B and Table S1). Additionally, we observed a correlation between higher neutrophil chemotaxis velocity and lower FeNO values for patients in our study (Fig. 3B), further validating the efficacy of the microfluidic diagnostic test. Note that the sensitivity of our assay is significantly higher than diagnosing asthma based on the spirometry and FeNO measurements of these patients (Fig. 3B and Figs. S3–S6). For example, if a cutoff value of 30 ppb is chosen for diagnosis on the basis of FeNO measurements, 40% (6/15) of asthmatic patients would be characterized as nonasthmatic. These results are a first demonstration of how neutrophil chemotaxis—and more generally, a readout based on effector cell function—can potentially be used to classify asthma, rather than relying solely on clinical symptoms. Furthermore, because our tool provides a quantitative readout, it is interesting to consider the possibility that this tool can also serve as a measure for disease severity. Thus, another potential clinical application is the routine measurement of neutrophil chemotaxis velocity in asthma patients to determine the level of asthma control, or, more importantly, risk for future exacerbations of asthma.
To further validate our method, additional studies are required to determine how much the specificity of our test is influenced by other inflammatory diseases. Chronic obstructive pulmonary disease (COPD) is an example of a disease driven by neutrophil inflammation (52). In general, COPD is difficult to distinguish from severe asthma using existing quantitative clinical tests because the symptoms closely resemble each other; in our study, COPD was an exclusion criterion, so the study subjects were asthmatic, nonasthmatic, allergic, or some combination (Table 1). In practice, our diagnostic technique would likely be used in combination with existing clinical protocols to make the diagnosis, rather than exist as a standalone test. Furthermore, the information-rich readout provided by cell migration analysis allows for the comprehensive study of neutrophil dysfunction that occurs for other inflammatory diseases. For example, we have previously used a similar chemotaxis technology to analyze neutrophils obtained from an arthritic mouse model (53), a disease where neutrophil inflammation is known to regulate dysfunction (54). In this study we observed a ∼10-fold increase in neutrophils from arthritic mice adhering to the substrate, a significant decrease in neutrophil directionality, and no significant difference in chemotaxis velocity compared with wild-type mice (31). Importantly, the neutrophil defect observed in this prior study differs significantly from the asthmatic neutrophil phenotype characterized in this work, suggesting that differences in neutrophil function could be observed to differentiate other inflammatory disorders from asthma. Our method minimizes the training, equipment, and labor requirements necessary to perform these assessments, making the characterization of other inflammatory diseases possible.
Other considerations need to be addressed before widespread clinical use. For example, it has been shown in a modified Boyden chamber that changes in ambient temperature do not significantly alter neutrophil motility; however, deviations in pH can impact motility (55). Other studies have confirmed that in vitro assay conditions can alter neutrophil migration (55–58). Thus, robust operation of the test is sensitive to the proper execution of the protocol, and the design should be optimized based on feedback from more user operation. The method was designed to mitigate user error by using a simplified all-in-one kit design (Dataset S1), and by creating a local cell microenvironment. Furthermore, the use of an “open” microfluidic pumping scheme makes the technology amendable to integration with automation systems (15), which could further reduce variance in the workflow and further standardize the test. However, user operation across multiple clinics would likely reveal changes that could improve the design of the device and protocol. Considerations like these should be addressed in the subsequent development of the technology to vet the method for mainstream clinical practice. In this work, we provide a first demonstration that illustrates the potential of our neutrophil chemotaxis method for the characterization of asthma.
In conclusion, we report a comprehensive microfluidic solution for discriminating asthma from nonasthmatic, allergic rhinitis patients based on the chemotactic function of patients’ neutrophils. The technology is handheld and easily transportable; can purify neutrophils from whole blood obtained from a lancet puncture within 5 min; and reports whether a patient is asthmatic or nonasthmatic with sensitivity of 96% and specificity of 73%. Importantly, the user-friendly design features of the assay enabled the application of the technology in a clinical setting, and we identified a previously unknown correlation between reduced neutrophil chemotaxis velocity and asthmatic patients. This work suggests that neutrophil chemotaxis may be a potential biomarker for asthma, although additional studies are required to investigate whether the specificity is reduced in broader populations.
Materials and Methods
Please refer to SI Materials and Methods for additional details on spirometry measurements; the fabrication and associated methods for the microfluidic assay; the calculation of the chemotaxis outputs; and statistical analysis.
Study Subjects.
Peripheral blood and related clinical data were obtained from normal or asthmatic donors ranging in age from 18 to 55 y. Informed consent was obtained before participation, and the study was approved by the University of Wisconsin Health Sciences Institutional Review Board, protocol nos. H 2008-0096 and 2010-3071.
Clinical Diagnosis of Asthma and Allergy.
Allergic individuals are defined as having a skin test positive to at least one of 12 aeroallergens. Asthmatic individuals are defined as having at least a 6-mo history of asthma based on clinical findings such as cough, wheeze, and shortness of breath. Additionally, asthmatics may be currently taking the following medications; inhaled short-acting B-agonist (as needed and <6 puffs per day during an acute cold), low-dose inhaled corticoid steroids, Advair, and/or a daily nonsteroidal controller medication.
Neutrophil Sorting from Whole Blood and Chemotaxis Assay.
The human subject protocol was approved by the University of Wisconsin Center for Health Sciences Human Subjects Committee. The microfluidic chip was coated with human recombinant P-selectin (R&D Biosystems) at 4 °C for at least 30 min. After obtaining ∼150 µL from the asthma clinic (see SI Materials and Methods), 3 µL of the whole blood was pipetted into a reservoir containing 18 µL of PBS and mixed gently. Neutrophils were captured out of dilute whole blood in preparation of performing the chemotaxis assay by pumping 1 µL of dilute whole blood through each microchannel two times, with each pass 30 s apart. After allowing neutrophils to capture for 4 min, erythrocytes were removed by performing three washes with 3 µL of PBS, alternating the aspiration of PBS–blood mixture between the input and output ports. The PBS was replaced twice with 3 µL of EGM BulletKit media (CC-3124; Lonza) with 20 mM Hepes (25-060-CI; Mediatech). The lid containing the CA and the microchannels were placed in a humidified container at 37 °C for 5 min before placing the lid onto the base and initiating the chemotaxis assay. For all chemotaxis experiments in the microfluidic assay, 100 nM fMLP and 0 nM fMLP (gel only control) were used.
Supplementary Material
Acknowledgments
Support for this work was provided by an Innovation and Economic Development Research Program grant (to E.K.-H.S.), a Morgridge Institute of Research fellowship (to E.B.), National Institutes of Health (NIH) Grant R01 EB010039 (to D.J.B. and E.K.-H.S.) and NIH Program Project Grant HL088584 (to E.A.S., P.S.F., and S.K.M.).
Footnotes
Conflict of interest statement: E.K.-H.S., E.B., and D.J.B. have patent applications pending on technology cited in this work. D.J.B. has ownership in Ratio, Inc., and Bellbrook Labs, LLC. E.K.-H.S., E.B., and D.J.B. have ownership in Salus Discovery, LLC.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324043111/-/DCSupplemental.
References
- 1.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355(21):2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 2.Masoli M, Fabian D, Holt S, Beasley R. Global Initiative for Asthma (GINA) Program The global burden of asthma: Executive summary of the GINA Dissemination Committee report. Allergy. 2004;59(5):469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 3.Nakagome K, Nagata M. Pathogenesis of airway inflammation in bronchial asthma. Auris Nasus Larynx. 2011;38(5):555–563. doi: 10.1016/j.anl.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Wenzel SE. Asthma: Defining of the persistent adult phenotypes. Lancet. 2006;368(9537):804–813. doi: 10.1016/S0140-6736(06)69290-8. [DOI] [PubMed] [Google Scholar]
- 5.Fahy JV. Eosinophilic and neutrophilic inflammation in asthma: Insights from clinical studies. Proc Am Thorac Soc. 2009;6(3):256–259. doi: 10.1513/pats.200808-087RM. [DOI] [PubMed] [Google Scholar]
- 6.Lloyd CM, Hessel EM. Functions of T cells in asthma: More than just T(H)2 cells. Nat Rev Immunol. 2010;10(12):838–848. doi: 10.1038/nri2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wenzel SE, et al. Bronchoscopic evaluation of severe asthma. Persistent inflammation associated with high dose glucocorticoids. Am J Respir Crit Care Med. 1997;156(3 Pt 1):737–743. doi: 10.1164/ajrccm.156.3.9610046. [DOI] [PubMed] [Google Scholar]
- 8.Menzies D, Jackson C, Mistry C, Houston R, Lipworth BJ. Symptoms, spirometry, exhaled nitric oxide, and asthma exacerbations in clinical practice. Ann Allergy Asthma Immunol. 2008;101(3):248–255. doi: 10.1016/S1081-1206(10)60489-9. [DOI] [PubMed] [Google Scholar]
- 9.Dombkowski KJ, Hassan F, Wasilevich EA, Clark SJ. Spirometry use among pediatric primary care physicians. Pediatrics. 2010;126(4):682–687. doi: 10.1542/peds.2010-0362. [DOI] [PubMed] [Google Scholar]
- 10.Aaron SD, et al. Canadian Respiratory Clinical Research Consortium Overdiagnosis of asthma in obese and nonobese adults. CMAJ. 2008;179(11):1121–1131. doi: 10.1503/cmaj.081332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright PL, McClelland RL, Newman AB, Gottlieb DJ, Lebowitz MD. Cardiovascular Health Study Research Group Underdiagnosis and undertreatment of asthma in the elderly. Chest. 1999;116(3):603–613. doi: 10.1378/chest.116.3.603. [DOI] [PubMed] [Google Scholar]
- 12.Urso DL. Asthma in the elderly. Curr Gerontol Geriatr Res. 2009;2009:858415. doi: 10.1155/2009/858415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paguirigan AL, Beebe DJ. Microfluidics meet cell biology: Bridging the gap by validation and application of microscale techniques for cell biological assays. Bioessays. 2008;30(9):811–821. doi: 10.1002/bies.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 15.Puccinelli JP, Su X, Beebe DJ. Automated high-throughput microchannel assays for cell biology: Operational optimization and characterization. JALA Charlottesv Va. 2010;15(1):25–32. doi: 10.1016/j.jala.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Squires T, Quake S. Microfluidics: Fluid physics at the nanoliter scale. Rev Mod Phys. 2005;77:977–1026. [Google Scholar]
- 17.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328(5986):1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Jeon N, et al. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat Biotechnol. 2002;20(8):826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 19.Young EWK, et al. Microscale functional cytomics for studying hematologic cancers. Blood. 2012;119(10):e76–e85. doi: 10.1182/blood-2011-10-384347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai M, et al. In vitro modeling of the microvascular occlusion and thrombosis that occur in hematologic diseases using microfluidic technology. J Clin Invest. 2012;122(1):408–418. doi: 10.1172/JCI58753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chin CD, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011;17(8):1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 22.Kotz KT, et al. Inflammation and the Host Response to Injury Collaborative Research Program Clinical microfluidics for neutrophil genomics and proteomics. Nat Med. 2010;16(9):1042–1047. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler KL, et al. Burn injury reduces neutrophil directional migration speed in microfluidic devices. PLoS ONE. 2010;5(7):e11921. doi: 10.1371/journal.pone.0011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood DK, Soriano A, Mahadevan L, Higgins JM, Bhatia SN. A biophysical indicator of vaso-occlusive risk in sickle cell disease. Sci Transl Med. 2012;4(123):23ra26. doi: 10.1126/scitranslmed.3002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Issadore D, et al. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci Transl Med. 2012;4(141):41ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yager P, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442(7101):412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 27.Berthier E, Surfus J, Verbsky J, Huttenlocher A, Beebe D. An arrayed high-content chemotaxis assay for patient diagnosis. Integr Biol (Camb) 2010;2(11-12):630–638. doi: 10.1039/c0ib00030b. [DOI] [PubMed] [Google Scholar]
- 28.Zigmond SH. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol. 1977;75(2 Pt 1):606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerisch G, Keller HU. Chemotactic reorientation of granulocytes stimulated with micropipettes containing fMet-Leu-Phe. J Cell Sci. 1981;52:1–10. doi: 10.1242/jcs.52.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Sackmann EK, et al. Microfluidic kit-on-a-lid: A versatile platform for neutrophil chemotaxis assays. Blood. 2012;120(14):e45–e53. doi: 10.1182/blood-2012-03-416453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berthier E, Warrick J, Yu H, Beebe DJ. Managing evaporation for more robust microscale assays. Part 1. Volume loss in high throughput assays. Lab Chip. 2008;8(6):852–859. doi: 10.1039/b717422e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agrawal N, Toner M, Irimia D. Neutrophil migration assay from a drop of blood. Lab Chip. 2008;8(12):2054–2061. doi: 10.1039/b813588f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker G, Beebe DJ. A passive pumping method for microfluidic devices. Lab Chip. 2002;2(3):131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 35.Gin W, Kay AB. The effect of corticosteroids on monocyte and neutrophil activation in bronchial asthma. J Allergy Clin Immunol. 1985;76(5):675–682. doi: 10.1016/0091-6749(85)90670-0. [DOI] [PubMed] [Google Scholar]
- 36.Koziol-White CJ, et al. DHEA-S inhibits human neutrophil and human airway smooth muscle migration. Biochim Biophys Acta. 2012;1822(10):1638–1642. doi: 10.1016/j.bbadis.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strandberg K, Blidberg K, Sahlander K, Palmberg L, Larsson K. Effect of formoterol and budesonide on chemokine release, chemokine receptor expression and chemotaxis in human neutrophils. Pulm Pharmacol Ther. 2010;23(4):316–323. doi: 10.1016/j.pupt.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Reid DW, et al. Possible anti-inflammatory effect of salmeterol against interleukin-8 and neutrophil activation in asthma in vivo. Eur Respir J. 2003;21(6):994–999. doi: 10.1183/09031936.03.00109702. [DOI] [PubMed] [Google Scholar]
- 39.Schleimer RP. Glucocorticoids suppress inflammation but spare innate immune responses in airway epithelium. Proc Am Thorac Soc. 2004;1(3):222–230. doi: 10.1513/pats.200402-018MS. [DOI] [PubMed] [Google Scholar]
- 40.Maneechotesuwan K, et al. Formoterol attenuates neutrophilic airway inflammation in asthma. Chest. 2005;128(4):1936–1942. doi: 10.1378/chest.128.4.1936. [DOI] [PubMed] [Google Scholar]
- 41.Schneider A, et al. Diagnosing asthma in general practice with portable exhaled nitric oxide measurement—results of a prospective diagnostic study: FENO < or = 16 ppb better than FENO < or =12 ppb to rule out mild and moderate to severe asthma. Respir Res. 2009;10:15. doi: 10.1186/1465-9921-10-15. and erratum (2009) 10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dupont LJ, Demedts MG, Verleden GM. Prospective evaluation of the validity of exhaled nitric oxide for the diagnosis of asthma. Chest. 2003;123(3):751–756. doi: 10.1378/chest.123.3.751. [DOI] [PubMed] [Google Scholar]
- 43.Miedinger DD, et al. Diagnostic tests for asthma in firefighters. Chest. 2007;131(6):1760–1767. doi: 10.1378/chest.06-2218. [DOI] [PubMed] [Google Scholar]
- 44.Fortuna AM, Feixas T, González M, Casan P. Diagnostic utility of inflammatory biomarkers in asthma: Exhaled nitric oxide and induced sputum eosinophil count. Respir Med. 2007;101(11):2416–2421. doi: 10.1016/j.rmed.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 45.Altman DG, Bland JM. Diagnostic tests. 1: Sensitivity and specificity. BMJ. 1994;308(6943):1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busse WW. What is the best pulmonary diagnostic approach for wheezing patients with normal spirometry? Respir Care. 2012;57(1):39–46, discussion 47–49. doi: 10.4187/respcare.01449. [DOI] [PubMed] [Google Scholar]
- 47.Simpson JLJ, Scott RJR, Boyle MJM, Gibson PGP. Differential proteolytic enzyme activity in eosinophilic and neutrophilic asthma. Am J Respir Crit Care Med. 2005;172(5):559–565. doi: 10.1164/rccm.200503-369OC. [DOI] [PubMed] [Google Scholar]
- 48.Dang BB, Wiehler SS, Patel KDK. Increased PSGL-1 expression on granulocytes from allergic-asthmatic subjects results in enhanced leukocyte recruitment under flow conditions. J Leukoc Biol. 2002;72(4):702–710. [PubMed] [Google Scholar]
- 49.Moore KL, et al. P-selectin glycoprotein ligand-1 mediates rolling of human neutrophils on P-selectin. J Cell Biol. 1995;128(4):661–671. doi: 10.1083/jcb.128.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Samri MT, et al. Variability of sputum inflammatory cells in asthmatic patients receiving corticosteroid therapy: A prospective study using multiple samples. J Allergy Clin Immunol. 2010;125(5):1161–1163. doi: 10.1016/j.jaci.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 51.Shaw DE, et al. Association between neutrophilic airway inflammation and airflow limitation in adults with asthma. Chest. 2007;132(6):1871–1875. doi: 10.1378/chest.07-1047. [DOI] [PubMed] [Google Scholar]
- 52.Barnes PJ. Medicine. Neutrophils find smoke attractive. Science. 2010;330(6000):40–41. doi: 10.1126/science.1196017. [DOI] [PubMed] [Google Scholar]
- 53.Douni E, et al. Transgenic and knockout analyses of the role of TNF in immune regulation and disease pathogenesis. J Inflamm. 1995-1996;47(1-2):27–38. [PubMed] [Google Scholar]
- 54.Németh T, Mócsai A. The role of neutrophils in autoimmune diseases. Immunol Lett. 2012;143(1):9–19. doi: 10.1016/j.imlet.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 55.Phelps P, Stanislaw D. Polymorphonuclear leukocyte motility in vitro. I. Effect of pH, temperature, ethyl alcohol, and caffeine, using a modified Boyden chamber technic. Arthritis Rheum. 1969;12(3):181–188. doi: 10.1002/art.1780120304. [DOI] [PubMed] [Google Scholar]
- 56.Spagnuolo PJ, MacGregor RR. Acute thanol effect on chemotaxis and other components of host defense. J Lab Clin Med. 1975;86(1):24–31. [PubMed] [Google Scholar]
- 57.Li JC, Funahashi H. Effect of blood serum, caffeine and heparin on in vitro phagocytosis of frozen-thawed bull sperm by neutrophils derived from the peripheral blood of cows. Theriogenology. 2010;74(4):691–698. doi: 10.1016/j.theriogenology.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 58.Oakes PW, et al. Neutrophil morphology and migration are affected by substrate elasticity. Blood. 2009;114(7):1387–1395. doi: 10.1182/blood-2008-11-191445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.