Significance
Bile salts are extremely abundant molecules in the mammalian intestine and play important roles in food digestion. Much less well known is the fact that bile salts are also highly antimicrobial and control bacterial growth in the gut. Here, we report the discovery that bile salts affect bacterial growth by causing widespread unfolding and aggregation of cytosolic proteins in bacteria, while simultaneously triggering a prooxidizing shift in the cellular ratio of reduced to oxidized glutathione. Our studies therefore reveal an important and unrealized property of a set of compounds long known to be important in human physiology, demonstrate how bacteria such as Escherichia coli defend themselves against bile salts, and uncover perhaps the first physiological condition that causes disulfide stress in vivo.
Keywords: protein folding, oxidation
Abstract
Commensal and pathogenic bacteria must deal with many different stress conditions to survive in and colonize the human gastrointestinal tract. One major challenge that bacteria encounter in the gut is the high concentration of bile salts, which not only aid in food absorption but also act as effective physiological antimicrobials. The mechanism by which bile salts limit bacterial growth is still largely unknown. Here, we show that bile salts cause widespread protein unfolding and aggregation, affecting many essential proteins. Simultaneously, the bacterial cytosol becomes highly oxidizing, indicative of disulfide stress. Strains defective in reducing oxidative thiol modifications, restoring redox homeostasis, or preventing irreversible protein aggregation under disulfide stress conditions are sensitive to bile salt treatment. Surprisingly, cholate and deoxycholate, two of the most abundant and very closely related physiological bile salts, vary substantially in their destabilizing effects on proteins in vitro and cause protein unfolding of different subsets of proteins in vivo. Our results provide a potential mechanistic explanation for the antimicrobial effects of bile salts, help explain the beneficial effects of bile salt mixtures, and suggest that we have identified a physiological source of protein-unfolding disulfide stress conditions in bacteria.
Bile salts, the cation-compounded form of bile acids, are amphipathic cholesterol metabolites that are produced in the liver and released into the duodenum upon food intake (1). The most prevalent primary and secondary bile acids found in the human intestine are cholic acid [i.e., cholate (CHO)] and its closely related derivative deoxycholic acid [i.e., deoxycholate (DOC)] (Fig. 1A). Both bile acids can reach high millimolar concentrations in the small intestine (2). In addition to their well-characterized role in the solubilization and absorption of lipids, bile salts are known for their highly efficient antimicrobial activity, particularly potent against Gram-positive bacteria (3). Cirrhotic patients, who secrete significantly lower amounts of bile salts than healthy individuals, contain higher levels of bacteria in the intestine (i.e., ileum and cecum) and show an increased incidence of systemic infections (4–6). Studies in cirrhotic rats revealed that oral administration of bile salts reduces this bacterial overgrowth and increases survival (6).
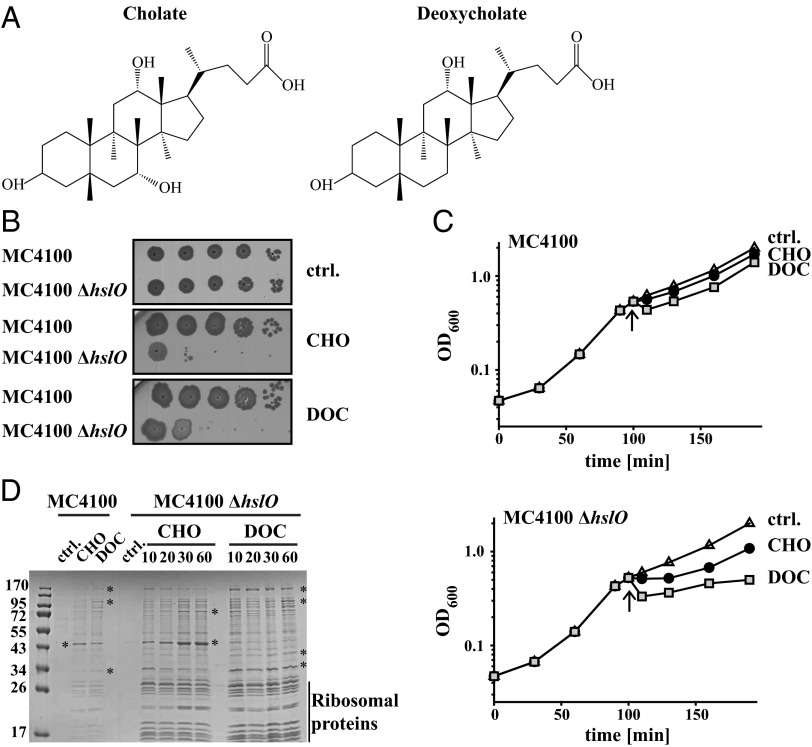
Fig. 1.
Bile salts cause extensive protein aggregation in vivo. (A) Molecular structure of CHO and DOC (3). (B) MC4100 and MC4100 ΔhslO were grown at 37 °C to an OD600 of 0.5, serially diluted, and spot-titered onto LB plates containing either 5 mM CHO or 1.5 mM DOC. Plates were incubated at 37 °C for 18 h. (C) MC4100 or MC4100 ΔhslO were grown in LB at 37 °C. At an OD600 of 0.5 (arrow), cultures were split and either supplemented with buffer (open triangles), 14 mM CHO (closed circles), or 5 mM DOC (squares). Growth at OD600 was monitored. (D) Cultures of MC4100 or MC4100 ΔhslO were grown in LB to an OD600 of 0.5, split, and treated with either nothing, 14 mM CHO, or 5 mM DOC. At the indicated time points (or after 60 min of incubation for MC4100), aliquots were taken, and the aggregated proteins were prepared. Noticeable differences in the protein distribution pattern of cells treated with CHO versus DOC are indicated with a star.
It has long been suggested that the amphipathic character of bile salts allows for their interaction with the lipid bilayer of cells, causing changes in membrane integrity. Indeed, cell lysis upon exposure to bile salts has been reported in erythrocytes (7). However, apart from gene expression studies, which revealed the up-regulation of genes involved in maintaining membrane integrity in bacteria, relatively little is known about the effects that bile salts exert on microbial membranes (8–10). Bile salts are also known to enter the bacterial cytosol via a flip-flop mechanism (11) and, once in the cytosol, cause the up-regulation of a number of genes involved in DNA repair. These results suggest that bile salts potentially cause oxidative DNA damage (12). It is currently unknown, however, to what extent DNA damage contributes to bile salt-mediated cell death.
Here, we demonstrate that bile salts cause widespread protein unfolding and in vivo disulfide stress conditions in Escherichia coli. Our in vitro studies reveal that bile salts partially unfold a number of different proteins, thereby substantially increasing their tendency to aggregate. In vivo, bile salts not only trigger widespread protein aggregation but also cause a significant prooxidizing shift in the ratio between reduced glutathione (GSH) and oxidized GSH (GSSG), indicative of disulfide stress. To defend themselves against these insults, bacteria contain the cytosolic chaperone Hsp33, which is rapidly activated by disulfide bond formation, and effectively mitigates bile salt-mediated protein aggregation in wild-type E. coli. We conclude from these results that bile salts are a physiological source of protein-unfolding disulfide stress conditions in bacteria.
Results
Bile Salts Cause Extensive Protein Aggregation in Vivo.
Mainly because of its 0.15% bile salt content, MacConkey agar has been successfully used for many years as a selective media, supporting growth of Gram-negative enteric bacteria while inhibiting growth of most Gram-positive bacteria. This selective advantage for Gram-negative bacteria is thought to be largely due to components of their outer membrane, which decrease the permeability of bile salts and hence improve survival (13). It was therefore unexpected when we observed that absence of the cytosolic chaperone Hsp33 (gene name: hslO) causes a temperature-sensitive phenotype in Vibrio cholerae when cultivated on MacConkey plates (14). MacConkey medium contains a mixture of bile salts, as well as other components, such as crystal violet, which might also affect cell growth. We therefore decided to test the growth of different bacterial strains (E. coli MC4100, E. coli BL21, V. cholerae O395) and their isogenic hslO-deletion mutants on LB plates or in LB medium, supplemented with physiological concentrations of selected bile salts. We chose CHO and DOC, two of the most common bile salts found in the human gut (3). For each tested strain background, we made the same observation: deletion of the hslO gene increased the bile salt sensitivity even under non–heat-shock conditions (Fig. 1 B and C and Fig. S1 A and B). The bile salt concentrations that showed phenotypes on plates (5 mM CHO, 1.5 mM DOC) or in liquid cultures (14 mM CHO, 5 mM DOC) are very close to the critical micelle concentration of the respective bile salts and comparable to the bile salt concentrations that are typically reached in the small intestine (3). These results suggest that bile salts cause cellular damage that can be effectively prevented by the presence of the cytosolic chaperone Hsp33.
Hsp33 is a highly specialized chaperone, whose chaperone function depends on the redox status of its four cysteines. Inactive when reduced, Hsp33 is quickly activated by stress conditions that cause disulfide bond formation and oxidative protein damage (15). Bacteria experience such stress conditions, for instance, when they are exposed to hypochlorous acid (HOCl), a physiological antimicrobial, which is an effective thiol oxidant and protein-unfolding reagent (16). Activated Hsp33 prevents HOCl-induced protein aggregation and increases cell survival (16). The realization that Hsp33 increases bacterial resistance toward CHO and DOC treatment raised the obvious question of whether Hsp33’s protective action is through its ability to prevent protein aggregation in the cytosol. We therefore grew MC4100 wild-type and hslO-deletion strains to mid-log phase, treated the cultures with either CHO or DOC, and took samples at defined time points before and after the treatment. Protein aggregates were separated from the soluble proteins using an established protocol (17). As shown in Fig. 1D, we found substantially elevated levels of protein aggregates in CHO- or DOC-treated hslO-deletion strains. Protein aggregation was clearly detectable within 10 min of the stress treatment and further increased with prolonged incubation in bile salts. In the presence of Hsp33 (i.e., in wild-type E. coli), many of the same proteins precipitated in response to each bile salt, but the extent of aggregation was significantly reduced (Fig. 1D and Fig. S1C). These results suggest that Hsp33 increases bile salt resistance by protecting proteins against bile salt-mediated protein aggregation. Consistent with this conclusion was the observation that DOC treatment, which causes a more pronounced growth inhibitory effect than CHO treatment in the hslO-deletion strain (Fig. 1C), leads to higher levels of protein aggregates particularly at the early time points of the treatment (Fig. 1D). We concluded from these results that physiological concentrations of bile salts cause extensive in vivo protein aggregation, which is effectively prevented by the cytosolic, redox-regulated chaperone Hsp33.
Bile Salt-Mediated Aggregation Targets Cytosolic Proteins.
Comparison of the proteins that aggregate in MC4100 ΔhslO in response to CHO or DOC treatment revealed some striking differences, with several proteins aggregating to a much higher extent in response to one bile salt than the other (Fig. 1D and Fig. S1C, indicated with stars). This was an unexpected finding, given that the only chemical difference between CHO and DOC is the absence of one hydroxyl group in DOC (Fig. 1A). To determine which proteins are generally sensitive to bile salt-mediated protein aggregation, and to uncover potential bile salt specificities, we performed stable isotope labeling by amino acids in cell culture (SILAC) experiments. Because the same proteins that aggregate in the wild-type strain also aggregate in the hslO-deletion strain but to a much higher extent, we decided to directly compare protein aggregation in MC4100 ΔhslO strain treated with either CHO or DOC. We grew the mutant strain in 3-(N-morpholino)propanesulfonic acid (Mops) minimal media supplemented with either isotopically light [12C6]l-Arg or isotopically heavy [13C6]l-Arg and stressed one culture with CHO and the other one with DOC (Fig. S2A). We took equivalent cell aliquots 30 min after the respective bile salt treatments, prepared the aggregated proteins as before (Fig. S2B), mixed equal volumes, and analyzed the proteins by liquid chromatography–tandem MS (LC-MS/MS). We identified a total of 83 proteins that aggregated under both stress conditions, and an additional six proteins that were only detected in cells treated with DOC (Table 1).
Table 1.
In vivo aggregated proteins upon DOC and CHO treatment
| Gene | Protein | Ratio of aggregates in DOC/CHO |
| Ribosomal and ribosome-associated proteins | ||
| rpsA | 30S ribosomal protein S1 | 2.4 |
| rpsB* | 30S ribosomal protein S2 | 1.1 |
| rpsC | 30S ribosomal protein S3 | 1.0 |
| rpsD | 30S ribosomal protein S4 | 0.9 |
| rpsE* | 30S ribosomal protein S5 | 1.4 |
| rpsF* | 30S ribosomal protein S6 | 0.8 |
| rpsG | 30S ribosomal protein S7 | 1.0 |
| rpsI* | 30S ribosomal protein S9 | 2.3 |
| rpsJ* | 30S ribosomal protein S10 | 0.3 |
| rpsK | 30S ribosomal protein S11 | 3.6 |
| rpsM* | 30S ribosomal protein S13 | 2.7 |
| rpsR | 30S ribosomal protein S18 | 1.3 |
| rplB* | 50S ribosomal protein L2 | 0.6 |
| rplC* | 50S ribosomal protein L3 | 0.4 |
| rplD* | 50S ribosomal protein L4 | 0.9 |
| rplE | 50S ribosomal protein L5 | 0.3 |
| rplF* | 50S ribosomal protein L6 | 0.3 |
| rplI | 50S ribosomal protein L9 | 0.7 |
| rplK | 50S ribosomal protein L11 | 0.5 |
| rplM* | 50S ribosomal protein L13 | 0.5 |
| rplN* | 50S ribosomal protein L14 | 0.5 |
| rplO* | 50S ribosomal protein L15 | 0.8 |
| rplP | 50S ribosomal protein L16 | 0.3 |
| deaD | ATP-dependent RNA helicase | 0.4 |
| rho | Transcription termination factor Rho | 1.0 |
| rapA* | RNA polymerase-associated protein RapA | 1.0 |
| pcnB | Poly(A) polymerase | 1.4 |
| nusG* | Transcription antitermination protein NusG | 1.8 |
| rpoB* | DNA-directed RNA polymerase β | 3.5 |
| rpoC* | DNA-directed RNA polymerase β′ | 4.7 |
| rpoD* | RNA polymerase σ factor | 0.7 |
| tig | Trigger factor | 3.6 |
| tuf2* | Elongation factor Tu-2 | 0.3 |
| fusA* | Elongation factor G | 0.5 |
| infB* | Translation initiation factor IF-2 | 1.0 |
| srmB | ATP-dependent RNA helicase SrmB | 1.0 |
| infC | Translation initiation factor IF-3 | 2.7 |
| ffh | Signal recognition particle protein | 1.6 |
| typA | GTP-binding protein TypA/BipA | 0.8 |
| Peptidyl-tRNA synthetases | ||
| proS* | Prolyl-tRNA synthetase | 0.9 |
| pheT* | Phenylalanyl-tRNA synthetase β chain | 6.0 |
| argS* | Arginyl-tRNA synthetase | DOC only |
| glyS | Glycyl-tRNA synthetase β SU | 2.2 |
| Protein Folding and Proteostasis | ||
| hslU | ATP-dependent protease ATPase SU HslU | 0.1 |
| clpB | Heat shock protein ClpB | 1.1 |
| hslK/A | Modulator of FtsH protease HflK | 1.3 |
| dnaJ | Chaperone protein DnaJ | 1.6 |
| secF | Export membrane protein SecF | 2.3 |
| secY* | Preprotein translocase SU SecY | 9.8 |
| secD* | Protein-export membrane protein SecD | 10.6 |
| lon | ATP-dependent protease La | 0.9 |
| DNA and RNA binding proteins | ||
| mukB | Chromosome partition protein MukB | 1.2 |
| topA* | DNA topoisomerase | 1.1 |
| Crp | Catabolite gene activator | 1.2 |
| nemR | HTH-type transcriptional repressor | 1.6 |
| recA | Protein RecA | 0.2 |
| YfiF | Hypothetical tRNA/rRNA methyltransferase | 3.3 |
| rne | Ribonuclease E | 3.2 |
| gyrA* | DNA gyrase SU A | DOC only |
| Metabolic enzymes | ||
| pykF | Pyruvate kinase | <0.1 |
| thrA | Aspartokinase I | 0.2 |
| prsA | Ribose-phosphate pyrophosphokinase | 0.6 |
| speA | Biosynthetic arginine decarboxylase | 0.9 |
| pflB | Formate acetyltransferase 1 | 1.1 |
| metH | Methionine synthase | 1.2 |
| asnA | Aspartate-ammonia ligase | 1.4 |
| accC* | Acetyl CoA carboxylase, biotin carboxylase SU | 1.4 |
| atpD | ATP synthase SU β | 1.4 |
| ndh | NADH dehydrogenase | 1.5 |
| serA | d-3-phosphoglycerate dehydrogenase | 1.5 |
| sucA | 2-oxoglutarate dehydrogenase E1 component | 2.1 |
| pta | Phosphate acetyltransferase | 2.3 |
| plsB | Glycerol-3-phosphate acyltransferase | 3.0 |
| suhB | Inositol-1-monophosphatase | 3.2 |
| adhE | Aldehyde-alcohol dehydrogenase | 4.3 |
| accD* | Acetyl-CoA carboxylase carboxyl transferase β | 5.0 |
| aceE | Pyruvate dehydrogenase E1 component | 4.7 |
| guaA | GMP synthase | 7.0 |
| ptsG | Glucose phosphotransferase | 13.3 |
| Membrane proteins/lipopolysaccharide biosynthesis | ||
| oxaA | Inner membrane protein OxaA | DOC only |
| ompC | Outer membrane protein OmpC | 4.0 |
| kefA | Potassium efflux system KefA | DOC only |
| yeeF | Predicted amino acid transporter | 0.4 |
| mrcB | Penicillin-binding protein 1B | 0.2 |
| wbbJ | O-acetyl transferase WbbJ | DOC only |
| Other | ||
| nrdA | Ribonucleoside-diphosphate reductase 1 α | 0.4 |
| mreB | Actin homolog | 1.2 |
| yicC | Hypothetical protein YicC | DOC only |
| ybeZ | Putative ATP-binding protein in pho regulon | 1.3 |
Universal essential genes found in the aggregated fraction (21).
We noticed that 23 of our identified aggregated proteins are ribosomal proteins or proteins otherwise involved in protein translation (Fig. 2A and Table 1). One of these proteins is the essential elongation factor EF-Tu, a known Hsp33 client protein, whose sensitivity to HOCl-mediated oxidative damage appears to account for the growth defect observed in bleach-treated Hsp33-deletion mutants (14). Moreover, we found β and β′ subunits of RNA polymerase to precipitate, as well as several other proteins known to play a role in mRNA transcription (e.g., RpoD, RapA, NusG), suggesting that both transcription and translation processes are impaired during bile salt stress. Other proteins that we found to precipitate upon in vivo bile salt treatment are proteins known to chaperone (e.g., DnaJ, Tig, SRP) and/or translocate (e.g., SecD, -F, -Y) newly translated proteins or are otherwise involved in maintaining proteostasis (e.g., ClpB, lon) (Fig. 2A and Table 1). The fact that many of the identified proteins are actively involved in the synthesis, maturation, and translocation of nascent polypeptide chains made us then wonder whether bile salts have the capacity to interact with hydrophobic regions of nascent polypeptide chains, thereby precipitating actively translating polypeptides. We therefore prepared cell lysates of wild-type E. coli and treated them with either CHO or DOC for the indicated times in vitro. As shown in Fig. S2C, we could not detect any significant difference in the aggregation pattern, indicating that active translation plays at most a very minor role in bile salt-mediated protein aggregation. Another possibility is that the high cellular abundance of some of these proteins contributes to their identification in protein aggregates. However, aggregation studies conducted in either heat- or oxidative stress-treated E. coli wild-type, the general chaperone-deficient ΔrpoH strain, or the Hsp33-deficient ΔhslO strains did not reveal any ribosomal proteins apart from S10 to be aggregation-prone or a client protein of Hsp33 (15, 16, 18, 19). These results not only demonstrate that highly abundant proteins are not generally detected in protein aggregates but also suggest that bile salts do not simply destabilize already temperature- or oxidative stress-sensitive proteins but cause the precipitation of a select group of proteins. Because many of the identified proteins that aggregate are essential gene products in E. coli (20) (Table 1, indicated with an asterisk), depletion of any one of them could be responsible for the observed growth defect in bile salt-treated bacteria.
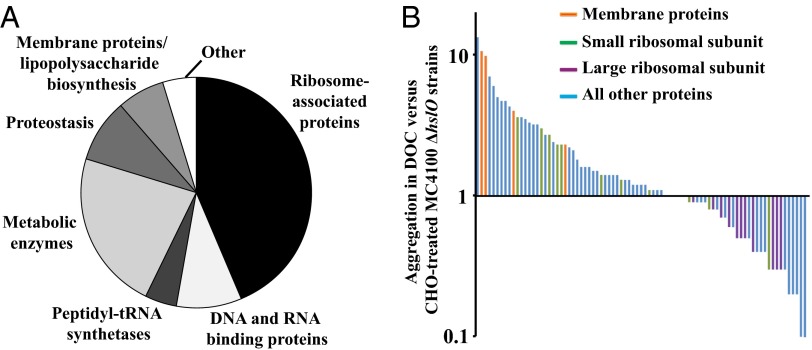
Fig. 2.
Identification of bile salt-sensitive proteins in vivo. (A) Functional distribution of the identified bile salt-sensitive target proteins in MC4100 ΔhslO-deletion strains (Table 1). (B) A SILAC experiment was performed to determine the distribution of aggregated proteins in MC4100 ΔhslO upon treatment with either CHO or DOC. The proteins were identified by MS analysis. After normalization of the spectral counts, the ratio of aggregation in DOC- over CHO-treated cells was calculated for each protein (Table S1).
CHO and DOC Differ in Their Affected Target Proteins.
To assess potential differences in the extent to which proteins aggregate in the presence of DOC or CHO, we normalized the individual spectral counts determined for each identified protein to the total number of spectral counts detected in each sample (Table 1 and Dataset S1), and determined the ratios at which each of the 83 proteins aggregate in DOC- versus CHO-treated cells (Fig. 2B and Table 1). We found that only about 50% of all identified proteins aggregated to approximately the same extent (±factor 2) under both bile salt conditions. Most identified membrane proteins (Fig. 2B, orange bars) and most of the identified small ribosomal subunit proteins (Fig. 2B, green bars), however, aggregated predominantly upon DOC treatment, whereas all of the identified large ribosomal subunits (Fig. 2B, purple bars) aggregated significantly more upon CHO treatment. These results indicate that individual bile salts differ in their extent to which they affect different proteins and protein complexes in vivo. Mixtures of bile salts, such as present in the mammalian intestine, have therefore the potential to target a larger set of proteins and, with that, increase their potency as antimicrobial agents.
CHO and Deoxycholate Act as Protein-Unfolding Agents in Vitro.
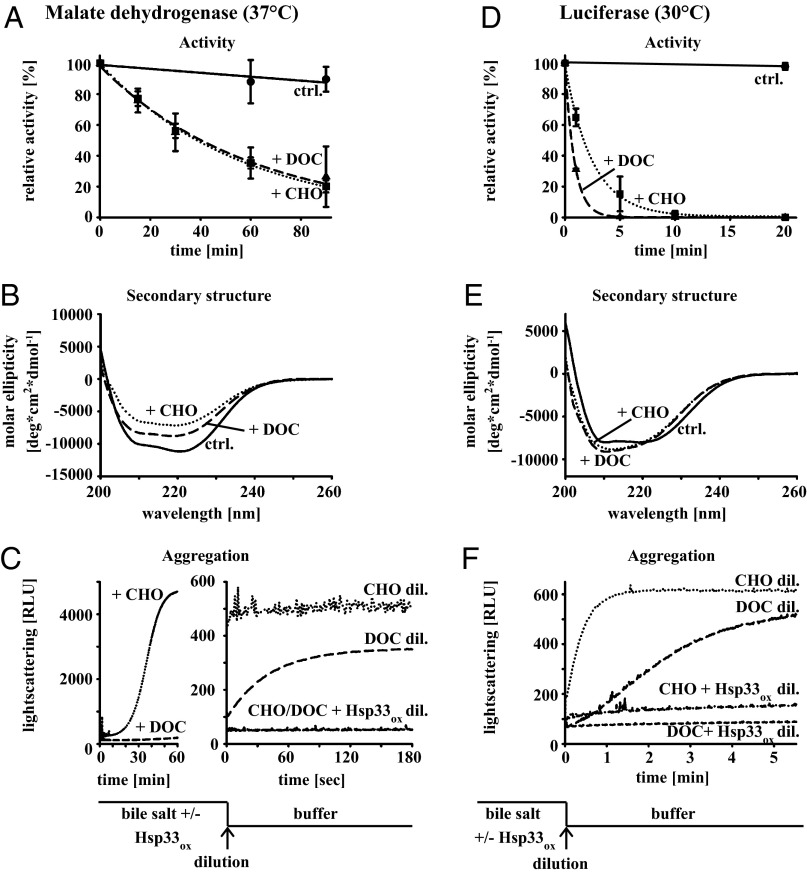
The finding that bile salts cause the aggregation of numerous cytosolic proteins indicated that bile salts might act as protein-destabilizing agents. To directly test this idea, and to identify potential differences in their mechanism of action, we decided to analyze the effects of CHO and DOC on purified proteins in vitro. We started with the dimeric enzyme malate dehydrogenase (MDH), which we incubated with either 14 mM CHO or 5 mM DOC and tested for its enzymatic activity. We observed that the incubation of MDH with either bile salt at 37 °C led to a substantial loss in enzymatic activity (Fig. 3A). Analysis of MDH’s secondary structure by circular dichroism (CD) spectroscopy confirmed these results and revealed substantial changes in the secondary structure of CHO- or DOC-treated enzyme (Fig. 3B). To determine whether these conformational changes are associated with protein aggregation, we conducted light-scattering measurements of MDH in the presence of CHO or DOC. As shown in Fig. 3C, MDH extensively aggregated in the presence of CHO at 37 °C (dotted line) but did not show significant aggregate formation in the presence of DOC (long-dashed line). Once diluted into DOC-free buffer (Fig. 3C, Right), however, DOC-pretreated MDH also rapidly formed insoluble protein aggregates. This might have physiological significance because bacteria are exposed to similarly rapid changes in bile salt concentrations upon transiting from the bile salt-enriched duodenum to the bile salt-deprived colon (21). Presence of in vitro oxidized and activated Hsp33ox during the incubation of MDH with either bile salt fully prevented MDH aggregation, as indicated by the near absence of a light-scattering signal upon dilution of MDH from either CHO or DOC into bile salt-free buffer (Fig. 3C, Right).
Fig. 3.
CHO and DOC are protein-unfolding agents in vitro. (A and D) Influence of bile salts on the enzymatic activity of MDH at 37 °C (A) or luciferase at 30 °C (D) was determined. The enzymes were incubated with either buffer (solid line), 14 mM CHO (dotted line), or 5 mM DOC (dashed line), and enzyme assays were conducted to monitor activity. The average of three independent experiments with SD is shown. (B and E) Influence of bile salts on the secondary structure of MDH at 37 °C (B) or luciferase at 30 °C (E). MDH (0.2 mg/mL) or luciferase (0.3 mg/mL) were incubated in the absence of bile salts (solid line) or in the presence of either 14 mM CHO (dotted line) or 5 mM DOC (dashed line) for 1 h. Then, far-UV–CD spectra were recorded in the presence of the bile salt at the indicate temperature. All spectra are buffer corrected. (C and F) Influence of bile salts on the aggregation propensity of MDH at 37 °C (C) or luciferase at 30 °C (F). MDH (12 µM) or luciferase (12 µM) were incubated with either 14 mM CHO or 5 mM DOC in the absence or presence of a fourfold molar excess (48 µM) of activated Hsp33ox for 1 h. Light scattering was monitored upon a 1:24 dilution of MDH or a 1:160 dilution of luciferase into bile salt-free buffer.
To test whether the protein unfolding effects of CHO and DOC are of a more general nature, we investigated the effects of both bile salts on the structure and function of firefly luciferase, another commonly used chaperone client protein. As with MDH, we found that the presence of either CHO or DOC causes a substantial loss in luciferase activity (Fig. 3D). In contrast to MDH, whose bile salt-mediated structural changes and aggregation sensitivity are significant at 37 °C but negligible at 30 °C (Fig. 3 B and C and Fig. S3 A and B), luciferase activity was already severely impacted by bile salts at 30 °C. Similar to MDH, the changes in luciferase activity upon incubation in bile salts are also paralleled by changes in the protein’s secondary structure, indicative of a loss in α-helical content and an increase in random coil formation (Fig. 3E). Light-scattering measurements revealed that neither bile salt causes significant aggregation of luciferase upon incubation at 30 °C, and yet both treatments trigger immediate aggregation of bile salt-treated luciferase upon its dilution into bile salt-free buffer (Fig. 3F). As observed with MDH, the presence of activated Hsp33ox during the incubation with bile salts prevented the aggregation of luciferase (Fig. 3F). The third protein that we tested was citrate synthase, which we found to be affected in structure and aggregation behavior by CHO at 37 °C but not by DOC (Fig. S3 C and D). These results demonstrate that bile salts cause protein unfolding and aggregation in vitro and generate folding intermediates, which are recognized by the chaperone Hsp33. They furthermore confirm our in vivo results by showing that different bile salts differ in their unfolding efficacies toward individual proteins.
Bile Salts Cause in Vivo Disulfide Stress.
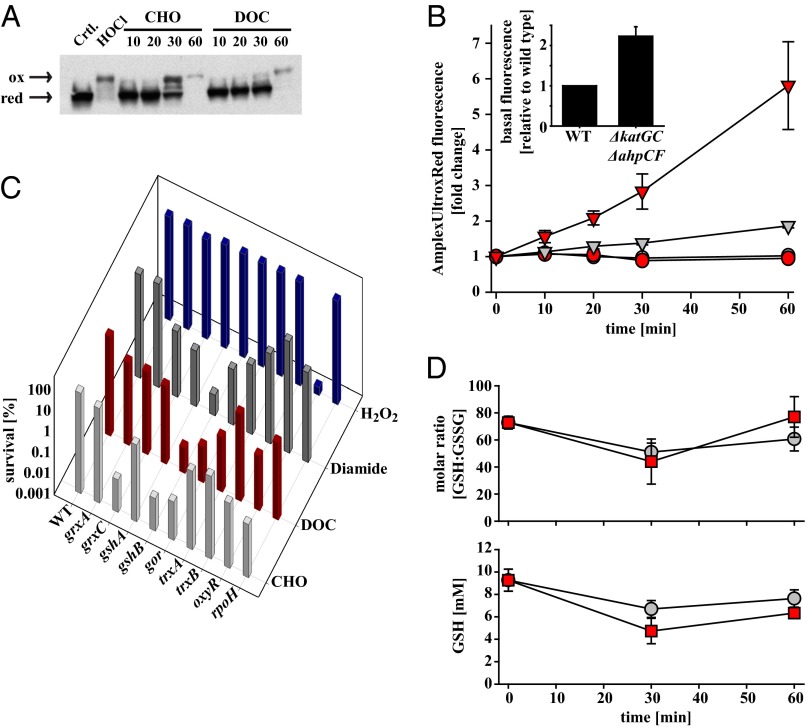
Hsp33 is a redox-regulated chaperone that typically gains its activity upon formation of two disulfide bonds. To test whether bile salts trigger stress conditions that lead to in vivo disulfide bond formation in Hsp33, or work by a different Hsp33-activation mechanism, we conducted differential thiol-trapping experiments of Hsp33 in bile salt-treated bacteria. We labeled all in vivo reduced cysteines with iodoacetamide, and all in vivo oxidized cysteines, upon their ex vivo reduction, with 4-acetamido-4-maleimidylstilbene-2,2′-disulfonic acid (AMS). Each in vivo oxidized cysteine is therefore labeled with a 490-Da AMS moiety, a mass addition that can be easily detected by SDS/PAGE. As shown in Fig. 4A, treatment of E. coli cells with bile salts leads indeed to the accumulation of disulfide-bonded Hsp33. Because bile salts are redox-inactive molecules, which are incapable of directly oxidizing Hsp33 in vitro (Fig. S4A), we concluded from these results that bile salts not only cause protein unfolding but also lead to considerable disulfide bond formation in the bacterial cytosol.
Fig. 4.
Bile salts cause oxidation in vivo. (A) In vivo thiol trapping of Hsp33. Cultures of MC4100 were grown in LB medium to mid-log phase; split into flasks containing 14 mM CHO, 5 mM DOC, or 2 mM HOCl; and incubated at 37 °C for the indicated times. Reduced proteins were alkylated with iodoacetamide, whereas all in vivo oxidized cysteines, upon their reduction, were modified with the 490-Da thiol-alkylating reagent AMS. Hsp33 was visualized by immunoblot using antibodies against Hsp33. (B) Measurement of H2O2 production in E. coli DHB4. Cultures of wild type (circle) or a katGC−ahpCF− triple mutant (triangle) were grown in LB medium to mid-log phase and split into flasks containing buffer, 14 mM CHO (gray), or 5 mM DOC (red). At the indicated time points, cells were spun down, and 50 μL of the supernatant was transferred into a 96-well plate containing nonfluorescent Amplex UltraRed working solution. Fluorescence of Amplex UltraRed was normalized to OD600 and is displayed relative to the untreated sample for each time point. (Inset) Difference in basal H2O2 production of wild type and katGC− ahpCF− mutant. The graph represents the average of at least four independent experiments, and the SEM is shown. (C) DHB4 wild-type and mutant strains were grown in LB medium at 30 °C until mid-log phase; spot-titered onto LB plates containing the indicated concentrations of CHO (light gray), DOC (red), diamide (dark gray), or H2O2 (blue); and subsequently incubated for 24 h at 30 °C. Colony-forming units were counted and normalized to the untreated control. The average (±SEM) of three to five independent experiments is shown. The genes grxA and grxC encode the two glutaredoxins A and C; gshA or gshB encode glutathione synthases A or B; gor encodes glutathione oxidoreductase; trxA encodes thioredoxin A; trxB encodes thioredoxin reductase; oxyR encodes the peroxide-specific transcriptional regulator; and rpoH encodes σ32, the transcriptional regulator of the heat-shock response. (D) Influence of bile salts on the GSH:GSSG ratio and overall GSH content in bile salt-treated E. coli MC4100. Cultures were grown in LB medium to mid-log phase, split into flasks containing either 21 mM CHO or 7.5 mM DOC, and incubated at 37 °C for the indicated times. Samples were taken and reduced, and GSSG were derivatized as described. GSH and GSSG were separated and quantified by HPLC. The GSH:GSSG ratios and overall GSH level for DOC-treated samples are shown in red; CHO treated samples are shown in gray. The average (±SEM) of three independent experiments is shown.
Formation of disulfide bonds in cytosolic proteins occurs in response to physiological oxidants, such as HOCl, as well as nonphysiological oxidants, such as diamide. Both of these oxidants not only directly oxidize protein thiols but also shift the GSH:GSSG ratio toward the oxidized form, thereby increasing protein S-glutathionylation (22, 23). Peroxide, another physiological oxidant that also causes some in vivo protein thiol oxidation, reacts six orders of magnitude more slowly than HOCl with most protein thiols and does not affect the GSH:GSSG ratio in bacteria (24). To begin to understand what type(s) of disulfide bond-forming stress conditions are triggered when E. coli is exposed to bile salts, we first investigated peroxide levels in CHO- or DOC-treated wild-type E. coli. Because peroxide rapidly diffuses through membranes, a commonly used method to measure peroxide production is to incubate cells with Amplex UltraRed and determine the peroxide levels in the media. This method avoids the use of cellular dyes, whose readout is influenced by the extent to which they enter the cells (25). As shown in Fig. 4B (circles), we did not observe any detectable peroxide secretion upon treatment of wild-type E. coli with either 14 mM CHO or 5 mM DOC. These results suggest that peroxide is either not produced at all in response to bile salts or is produced but very effectively detoxified. To distinguish between these two possibilities, we tested an E. coli triple mutant strain, which is unable to detoxify peroxide because of deletions in both catalase genes (katGC) and the genes encoding alkyl hydroperoxide reductase (ahpCF) (25). This strain is highly peroxide-sensitive and contains significantly higher steady-state concentrations of peroxide than wild-type E. coli (25) (Fig. 4B, Inset). As before, we incubated this mutant strain with CHO or DOC and measured peroxide release into the media. Whereas CHO treatment caused only a moderate stimulation of peroxide production (gray triangles), DOC treatment led to a substantial increase of peroxide release over time (red triangles). However, because DOC treatment killed about 90% of these mutant cells over the course of the experiment, it is unclear how much of this peroxide production is directly attributable to DOC treatment or indirectly caused by cell death. We concluded from these results that wild-type E. coli cells produce some peroxide upon bile salt treatment but that peroxide is effectively detoxified by the cellular antioxidant systems.
To independently verify this conclusion, we also measured the extent of protein carbonylation in CHO- or DOC-treated E. coli cells. Oxidation of side chain residues such as lysines and arginines is a hallmark of peroxide or HOCl-mediated oxidative stress (26, 27). As shown in Fig. S4B, we found only a very minor increase in protein carbonylation upon CHO treatment compared with the significant protein carbonylation detected in HOCl-treated cells. DOC treatment caused a more pronounced increase, consistent with our peroxide results. However, most of the protein carbonylation, which is a stable modification, occurred within the first 10 min of bile salt treatment and did not further increase in intensity. In contrast, the extent of disulfide bond formation in Hsp33 increased continuously over time (Fig. 4A), suggesting that other disulfide bond-forming stress conditions might be responsible for Hsp33’s oxidation and activation as a chaperone.
To shed light into the nature of these stress conditions, we decided to test the bile salt sensitivity of a set of isogenic E. coli mutant strains, which lack individual components of bacterial stress response or redox homeostasis systems. We reasoned that analysis of their bile salt phenotypes might reveal the in vivo stress conditions that bacteria experience during CHO and DOC stress. The investigated strains included mutants defective in reducing oxidative thiol modifications, such as disulfide bonds (trxA, trxB, grxA), mutants with diminished capacity to synthesize the small redox buffer component GSH (gshA, gshB), mutants unable to reduce protein S-glutathionylations (grxC) or to maintain the high GSH:GSSG ratio of the bacterial cytosol (gor), and mutants incapable of mounting an effective peroxide stress response (oxyR) or heat shock (i.e., protein unfolding) response (rpoH) (28, 29). We found that the most CHO-sensitive mutants are those lacking grxC, gshB, gor, and rpoH, whereas the absence of the thioredoxin system or OxyR had only very little effect (Fig. 4C, light gray bars). Similar results were obtained when strains were treated with DOC, except that strains lacking trxA or oxyR showed increased sensitivity as well (Fig. 4C, red bars). These findings agreed well with our previous results and indicated that DOC-mediated peroxide production causes some oxidative thiol modifications in addition to protein carbonylation. In general, however, the most highly bile salt-sensitive mutants were those lacking the ability to reduce S-glutathionylations (i.e., grxC) or restore physiological GSH:GSSG levels (i.e., gsh, gor), suggesting that bile salts alter GSH levels and/or the ratio between GSH and GSSG. Indeed, analysis of the GSH:GSSG ratio upon bile salt treatment confirmed these conclusions and revealed that bile salt treatment leads to a rapid, prooxidizing shift in the cellular GSH redox potential by accumulating GSSG over GSH (Fig. 4D, Upper) and decreasing the levels of reduced GSH (Fig. 4D, Lower, and Table S1). These results are reminiscent of the cellular effects caused by diamide, a nonphysiological GSH-oxidizing reagent, commonly used to induce disulfide stress conditions in pro- and eukaryotes (30). In fact, subsequent analysis of the sensitivity of our mutant strains revealed a startling resemblance between diamide sensitivity and bile salt sensitivity (Fig. 4C, dark gray bars), suggesting that, unlike diamide, bile salts are a physiologically relevant source of disulfide stress in bacteria.
Discussion
The fact that bile salts serve as effective physiological antimicrobials that control bacterial colonization in the human intestine has been known for decades. However, the exact mechanism by which bile salts work as antimicrobials has remained largely enigmatic. Based on the known lipid-solubilizing effect of bile salts, and bile salt studies conducted mostly in mammalian cells, it was concluded that bile salts likely cause membrane defects that lead to cell lysis (3). Our results now demonstrate that to survive physiological bile salt concentrations, bacteria rely on at least three different strategies: (i) the ability to prevent protein aggregation under disulfide-stress conditions (e.g., Hsp33); (ii) the ability to effectively reduce oxidized GSSG and oxidative thiol modifications, particularly protein S-glutathionylations (e.g., GrxC); and (iii) the ability to restore redox homeostasis (e.g., Gor). Lack of any one of these components leads to substantially increased bile salt sensitivity in bacteria. Very surprisingly, the same strategies are necessary for bacteria to survive HOCl, the active ingredient of household bleach and a well-known physiological antimicrobial involved in host defense and colonization (31). We conclude from these results that bile salts cause bleach-like physiological effects in bacteria, which explains their highly antimicrobial properties.
CHO and DOC: Two Effective Protein-Unfolding Reagents.
Our studies revealed that CHO and DOC are effective protein-unfolding reagents, which increase the aggregation sensitivity of numerous soluble proteins both in vitro and in vivo. Hsp33, a potent and general chaperone when disulfide bonded and active, prevents the aggregation of numerous essential E. coli proteins and increases bile salt resistance in bacteria. Two important mechanistic questions now remain: how do bile salts unfold proteins and what makes the difference between the unfolding properties of CHO and DOC? It is known that CHO and DOC, which differ by only one hydroxyl group, show very different physicochemical properties. The dihydroxy salt DOC, for instance, traverses membranes much faster (and more efficiently) than the trihydroxy bile salt CHO (11). Moreover, previous studies revealed that some dietary proteins become more protease sensitive in the presence of one bile salt but not the other (32, 33). Our current working model now proposes that the amphipathic character of bile salts interferes with hydrophobic interfaces, causing protein complexes to dissociate and hydrophobic cores to be destabilized. Absence of one hydroxyl group, such as found in DOC, increases the hydrophobicity of the bile salt and with that the ability to disrupt even more hydrophobic interactions. This would explain the higher unfolding efficacy of DOC and potentially the difference in the observed target proteins. In agreement with our in vitro results, we found several groups of proteins that were more extensively affected by DOC than CHO and vice versa. DOC, for instance, predominantly precipitated proteins of the small ribosomal subunits, whereas CHO primarily affected proteins of the large ribosomal subunit. We were unable to identify any specific property that might determine the sensitivity of a protein to DOC and/or CHO and would be common among the identified proteins (e.g., isoelectric point, thermal stability, oxidative stress sensitivity), raising the important question of what features make some proteins very sensitive to CHO or DOC and others not at all. In any case, however, it is tempting to speculate that bile salt mixtures, which differ from individual to individual, serve a beneficial role by affecting a wide and partially nonoverlapping set of proteins.
Bile Salts Are a Physiological Source of Disulfide Stress.
Our investigations were instigated by the unexpected discovery that E. coli and V. cholerae mutants that lack the redox-regulated chaperone Hsp33 are highly sensitive to bile salt treatment. This result was unexpected because activation of Hsp33 requires the oxidation of its four conserved cysteines, which either occurs in response to fast-acting oxidants such as HOCl or in mutant bacteria that lack components of the thioredoxin and glutaredoxin systems (34). These mutant bacteria are characterized by a decreased ratio of reduced to oxidized glutathione (35). Because bile salts are redox-inert and incapable of activating Hsp33 in vitro, we tested whether bacteria experience disulfide stress in response to bile salt treatment. Indeed, we discovered that bile salt-treated bacteria show a prooxidizing shift in their cellular GSH/GSSG redox potential, which is attributable to a combination of increased GSSG oxidation and an overall decrease in cellular GSH levels. Because we found that grxC-deletion mutants are much more sensitive toward bile salt treatment than mutants in the closely related grxA, we concluded that the observed decrease in cellular GSH levels is likely attributable to increased protein S-glutathionylation, a modification that is effectively resolved by GrxC but not by GrxA.
To our knowledge, very few physiologically relevant stressors apart from HOCl have been reported to alter bacterial GSH:GSSG levels as dramatically as bile salts do. In fact, pure disulfide stress, which is defined by decreased ratios of cellular GSH:GSSG ratios and increased amounts of S-glutathionylated proteins, is typically induced by diamide, a thiol-specific oxidant (30). Comparison of the diamide or H2O2 sensitivity of our tested mutant strains revealed that the most bile salt-sensitive strains are also the most diamide-sensitive strains in our collection. These results make it tempting to speculate that we have discovered one important physiological source of disulfide stress conditions in bacteria. One obvious question that remains to be investigated is how bile salts trigger in vivo disulfide stress. Peroxide measurements in bile salt-treated E. coli revealed some initial peroxide generation, but the oxidant appears to be rapidly removed and causes only minor (if any) oxidative damage. This result is also consistent with the fact the peroxide does not cause significant GSH oxidation or protein S-glutathionylation in bacteria (28, 36). One other, more likely, possibility is that one or more proteins involved in maintaining the cellular GSH:GSSG ratio fall victim to the unfolding or inactivating effects of bile salts. Because of the functional redundancies of these systems, significant cytosolic disulfide bond formation, such as found in Hsp33, has only been observed in mutant strains lacking components of both thioredoxin and glutaredoxin systems (34, 35). It is therefore likely that either several individual components of both pathways are affected by bile salts or factor(s), such as NADPH, which are common to both systems. Loss in reductive power will potentially turn oxidoreductases, such as TrxA or GrxA into oxidases, causing a further increase in disulfide bond formation, GSSG levels, and protein S-glutathionylation. It remains now to be tested which factors involved in cellular redox control are affected by bile salts and whether this is a direct or indirect consequence of bile salt treatment.
In summary, our results provide a mechanistic explanation for the antimicrobial effects of bile salts and suggest that we have identified a physiological source of protein-unfolding disulfide stress conditions in bacteria.
Materials and Methods
Growth of E. coli strains and Bile Salt Survival Assays.
If not otherwise noted, wild-type and mutant E. coli strains (for complete strain list, see Table S2) were cultivated in LB medium (Fisher Scientific) at 37 °C until an OD600 of 0.5 was reached. Cells were then treated with either 14 mM Na-CHO (Sigma) or 5 mM Na-DOC (Sigma), and growth curves were monitored at OD600. To determine the effects of bile salt treatment on bacterial growth on plates, exponentially growing (OD600 ∼0.5) wild-type or mutant strains were serially diluted and spotted onto freshly prepared LB agar plates containing various concentrations of CHO, DOC, H2O2 (Fisher Scientific), or diamide (MP Biomedicals). The plates were incubated for 18–48 h at the indicated temperatures, and colony-forming units were determined.
Analysis of in Vivo Protein Aggregation upon Bile Salt Treatment and Identification of Protein Aggregates by SILAC.
Wild-type MC4100 and MC4100 ΔhslO were treated with 14 mM CHO or 5 mM DOC at 37 °C as described above. At defined time points before and after the stress treatment, 5 mL of cells were harvested by centrifugation. Insoluble protein aggregates were prepared (17) and visualized on 14% (wt/vol) SDS/PAGE (Invitrogen). To determine the identity of the aggregated proteins, SILAC experiments were performed. Two cultures of MC4100 ΔhslO cells were grown in Mops minimal media (Teknova) containing all amino acids, supplemented with either 120 µg/mL heavy [13C6]Arg or the isotopically light [12C6]Arg. Cells growing in [13C6]Arg-supplemented Mops media were treated with 42 mM CHO, whereas cells growing in Mops minimal media supplemented with [12C6]Arg were treated with 7.5 mM DOC for 30 min at 37 °C. Under these growth conditions, higher concentrations of CHO and DOC were used to obtain significant growth delays and visible protein aggregation. Aggregated proteins were isolated from each culture as described above, dissolved in DAB buffer [200 mM Tris⋅HCl (pH 7.5), 6 M Urea, 10 mM EDTA, 0.5% SDS] and mixed in a 1:1 ratio. The protein mixtures were then separated on a 12% SDS/PAGE. Protein bands were cut out, trypsin-digested, and analyzed by nano LC-MS/MS (MS Bioworks).
Enzymatic Activity and Aggregation Measurements upon Bile Salt Treatment.
MDH (Roche) or luciferase (Promega) were diluted to a final concentration of 12 µM into 40 mM KH2PO4⋅KOH (pH 7.5) or 40 mM Mops, 50 mM KCl (pH 7.5), respectively, supplemented with either 14 mM CHO or 5 mM DOC and incubated at the indicated temperatures. For activity assays, aliquots were taken at indicated time points and diluted into the respective assay buffers. The enzymatic activity of MDH (125 nM) was determined by measuring the MDH-catalyzed reduction of 1 mM oxaloacetate (Sigma-Aldrich) in the presence of 150 µM NADH (Sigma-Aldrich) at 30 °C. Absorbance at 340 nm was measured over 2 min. Luciferase was diluted to a final concentration of 5 nM into assay buffer [100 mM KH2PO4, 25 mM glycylglycine, 2 mM EDTA (pH 7.5), 70 µM luciferin, 0.5 mg/mL BSA, 2 mM MgATP]. Luminescence was measured in a BMG FLUOstar Omega microplate reader. For aggregation measurements, proteins were incubated in the respective buffers and bile salts for 1 h. Light scattering was either directly monitored during the incubation period or upon further dilution of the proteins into their respective prewarmed, bile salt-free buffers. To determine the effect of Hsp33 on the aggregation behavior of the enzymes, a fourfold molar excess of bleach-activated Hsp33ox (16) to protein was added to the bile salt incubation reaction.
CD Measurements.
To monitor the effects of bile salts on the secondary structure of proteins, 0.2 mg/mL MDH, 0.2 mg/mL citrate synthase, or 0.3 mg/mL luciferase were prepared in 20 mM KH2PO4 (pH 7.5) in the presence or absence of 14 mM CHO or 5 mM DOC and incubated for 1 h at the indicated temperatures. Far-UV–CD spectra were recorded at the indicated temperature in a Jasco-J810 CD spectropolarimeter.
Hsp33 Purification and in Vivo Thiol Trapping of Hsp33.
Wild-type Hsp33 was purified, reduced, and activated (Hsp33ox) according to ref. 16. To visualize the oxidation status of Hsp33 in vivo, 1 mL of MC4100 culture was taken before or after bile salt treatment, and proteins were precipitated with ice-cold trichloroacetic acid (TCA) (final concentration, 10% vol/vol). The protein pellets were resuspended in 50 µL of DAB buffer containing 100 mM iodoacetamide and incubated under shaking (Eppendorf Thermoshaker) for 1 h at 25 °C at 1,300 rpm to label all reduced cysteines. Proteins were TCA-precipitated as before, and the protein pellet was resuspended in 20 µL of 10 mM DTT in DAB buffer (1 h, 25 °C, shaking) to reduce all reversible oxidative thiol modifications. DTT was removed by TCA precipitation, and the protein pellet was finally dissolved in 30 µL of DAB buffer supplemented with 10 mM AMS (Invitrogen) and incubated for 1 h at 25 °C. The samples were supplemented with nonreducing Laemmli buffer and loaded onto a 12% reducing SDS/PAGE gel. Hsp33 was visualized by Western blot using polyclonal antibodies against Hsp33.
Peroxide Detection upon Bile Salt Treatment.
To monitor the effect of bile salts on peroxide production, E. coli MG1655 and the MG1655 katGC−, ahpCF− mutant strain were grown in LB media and treated with CHO and DOC as described above. At the indicated time points, samples were taken, cells were pelleted, and the supernatant was used to measure peroxide levels. Peroxide was detected using the Amplex UltraRed reagent (Molecular Probes). A working solution containing 1× reaction buffer [50 mM NaH2PO4 (pH 7.4)], Amplex UltraRed stock solution, and horseradish peroxidase was prepared according to the manufacturer’s instructions; 50 µL of the working solution was pipetted into a black 96-well microassay plate with transparent bottom (Greiner), to which 50 µL of the supernatant of the bacterial sample was added. After an incubation of 20 min at room temperature (protected from light), the fluorescent product was detected in a BMG FLUOstar Omega Microplate Reader (excitation: 544 nm; emission: 590 nm). Fluorescence was normalized to the measured OD600 and compared with the untreated sample at the respective time point.
Determination of Intracellular GSH Concentrations.
For determination of intracellular reduced and GSSG concentrations, 5 mL of wild-type MC4100 cells were harvested before and after treatment with 21 mM CHO or 7.5 mM DOC for 30 or 60 min at 37 °C. The pellet was resuspended in 75 µL of PBS (Gibco) and mixed with an equal volume of metaphosphoric acid solution (16.8 mg/mL HPO3, 2 mg/mL EDTA, and 9 mg/mL NaCl). The metaphosphoric acid-fixed cells were harvested by centrifugation at 13,000 × g for 10 min at 4 °C to precipitate proteins. The supernatant was transferred, thiols were alkylated with monoiodoacetic acid at a final concentration of 7 mM, and the pH was adjusted to pH 7–8 with saturated K2CO3. After 1 h of incubation [room temperature (RT), in the dark], an equal volume of 2,4-dinitrofluorobenzene solution [1.5% (vol/vol) in absolute ethanol] was added to the mixture and incubated for at least 4 h (RT, in the dark). The N-dinitrophenyl derivatives of GSH and GSSG were separated by HPLC according to ref. 37. To calculate the cellular concentrations of GSH and GSSG, the obtained GSH and GSSG concentrations were adjusted to the number of harvested cells (OD600 of 1 equals 6 × 108 cells per milliliter) and calculated using 1 × 10−15 L as bacterial cell volume.
Supplementary Material
Acknowledgments
We thank James Bardwell and Mike Gray for critically reading the manuscript and helpful discussions. We thank Jordan Rowley for establishing the malate dehydrogenase aggregation assay and Jon Beckwith and James Imlay for providing the mutant strains. Mass spectrometry was performed by MS Bioworks. This work was supported by National Institutes of Health Grants GM065318 (to U.J.) and HL58984 (to R.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401941111/-/DCSupplemental.
References
- 1.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Wells JE, Hylemon PB. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl Environ Microbiol. 2000;66(3):1107–1113. doi: 10.1128/aem.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Ding JW, Andersson R, Soltesz V, Willén R, Bengmark S. The role of bile and bile acids in bacterial translocation in obstructive jaundice in rats. Eur Surg Res. 1993;25(1):11–19. doi: 10.1159/000129252. [DOI] [PubMed] [Google Scholar]
- 5.Ogata Y, et al. Role of bile in intestinal barrier function and its inhibitory effect on bacterial translocation in obstructive jaundice in rats. J Surg Res. 2003;115(1):18–23. doi: 10.1016/s0022-4804(03)00308-1. [DOI] [PubMed] [Google Scholar]
- 6.Lorenzo-Zúñiga V, et al. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 2003;37(3):551–557. doi: 10.1053/jhep.2003.50116. [DOI] [PubMed] [Google Scholar]
- 7.Albalak A, Zeidel ML, Zucker SD, Jackson AA, Donovan JM. Effects of submicellar bile salt concentrations on biological membrane permeability to low molecular weight non-ionic solutes. Biochemistry. 1996;35(24):7936–7945. doi: 10.1021/bi960497i. [DOI] [PubMed] [Google Scholar]
- 8.Prouty AM, Van Velkinburgh JC, Gunn JS. Salmonella enterica serovar typhimurium resistance to bile: Identification and characterization of the tolQRA cluster. J Bacteriol. 2002;184(5):1270–1276. doi: 10.1128/JB.184.5.1270-1276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merritt ME, Donaldson JR. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J Med Microbiol. 2009;58(Pt 12):1533–1541. doi: 10.1099/jmm.0.014092-0. [DOI] [PubMed] [Google Scholar]
- 10.Ray MC, Germon P, Vianney A, Portalier R, Lazzaroni JC. Identification by genetic suppression of Escherichia coli TolB residues important for TolB-Pal interaction. J Bacteriol. 2000;182(3):821–824. doi: 10.1128/jb.182.3.821-824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabral DJ, Small DM, Lilly HS, Hamilton JA. Transbilayer movement of bile acids in model membranes. Biochemistry. 1987;26(7):1801–1804. doi: 10.1021/bi00381a002. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein H, et al. Activation of the promoters of genes associated with DNA damage, oxidative stress, ER stress and protein malfolding by the bile salt, deoxycholate. Toxicol Lett. 1999;108(1):37–46. doi: 10.1016/s0378-4274(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 13.Provenzano D, Schuhmacher DA, Barker JL, Klose KE. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun. 2000;68(3):1491–1497. doi: 10.1128/iai.68.3.1491-1497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wholey W-Y, Jakob U. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol Microbiol. 2012;83(5):981–991. doi: 10.1111/j.1365-2958.2012.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17(3):381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 16.Winter J, Ilbert M, Graf PCF, Özcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135(4):691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cremers CM, Reichmann D, Hausmann J, Ilbert M, Jakob U. Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. J Biol Chem. 2010;285(15):11243–11251. doi: 10.1074/jbc.M109.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40(2):397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- 19.Ilbert M, et al. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14(6):556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerdes SY, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185(19):5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pauli-Magnus C, Stieger B, Meier Y, Kullak-Ublick GA, Meier PJ. Enterohepatic transport of bile salts and genetics of cholestasis. J Hepatol. 2005;43(2):342–357. doi: 10.1016/j.jhep.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Kosower NS, Kosower EM. Diamide: An oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- 23.Pullar JM, Vissers MC, Winterbourn CC. Glutathione oxidation by hypochlorous acid in endothelial cells produces glutathione sulfonamide as a major product but not glutathione disulfide. J Biol Chem. 2001;276(25):22120–22125. doi: 10.1074/jbc.M102088200. [DOI] [PubMed] [Google Scholar]
- 24.Carr AC, Winterbourn CC. Oxidation of neutrophil glutathione and protein thiols by myeloperoxidase-derived hypochlorous acid. Biochem J. 1997;327(Pt 1):275–281. doi: 10.1042/bj3270275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183(24):7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalle-Donne I, et al. Protein carbonylation: 2,4-dinitrophenylhydrazine reacts with both aldehydes/ketones and sulfenic acids. Free Radic Biol Med. 2009;46(10):1411–1419. doi: 10.1016/j.freeradbiomed.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 27.Nyström T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24(7):1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aslund F, Zheng M, Beckwith J, Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Natl Acad Sci USA. 1999;96(11):6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanemori M, Mori H, Yura T. Induction of heat shock proteins by abnormal proteins results from stabilization and not increased synthesis of sigma 32 in Escherichia coli. J Bacteriol. 1994;176(18):5648–5653. doi: 10.1128/jb.176.18.5648-5653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen RE, Roth D, Winther JR. Quantifying the global cellular thiol-disulfide status. Proc Natl Acad Sci USA. 2009;106(2):422–427. doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray MJ, Wholey WY, Jakob U. Bacterial responses to reactive chlorine species. Annu Rev Microbiol. 2013;67:141–160. doi: 10.1146/annurev-micro-102912-142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gass J, Vora H, Hofmann AF, Gray GM, Khosla C. Enhancement of dietary protein digestion by conjugated bile acids. Gastroenterology. 2007;133(1):16–23. doi: 10.1053/j.gastro.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Robic S, Linscott KB, Aseem M, Humphreys EA, McCartha SR. Bile acids as modulators of enzyme activity and stability. Protein J. 2011;30(8):539–545. doi: 10.1007/s10930-011-9360-y. [DOI] [PubMed] [Google Scholar]
- 34.Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96(3):341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 35.Prinz WA, Aslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem. 1997;272(25):15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 36.Carmel-Harel O, Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol. 2000;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 37.Garg SK, Yan ZH, Vitvitsky V, Banerjee R. Analysis of sulfur-containing metabolites involved in redox and methionine metabolism. In: Das D, editor. Methods in Redox Signaling. New York: Mary Ann Liebert; 2010. pp. 7–11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.