Significance
The C-terminal domain (CTD) of the largest subunit of RNA polymerase II is responsible for coordinating a wide range of cotranscriptional functions. Although tandem repeats of a 7-aa motif comprise the CTD in model organisms, the domain is highly unordered in many other species. To our knowledge, this study represents the most comprehensive investigation of CTD diversity and evolution to date, and finds that the CTD’s tandem structure likely existed in the last eukaryotic common ancestor; that unordered CTDs have resulted from extensive, lineage-specific sequence modifications; and that tandem heptads have been lost and reinvented many times. We also highlight interesting parallels in CTD evolution that appear to be associated with the requirements of developmental complexity and adaptations to parasitism.
Keywords: development, parasitism, splicing, transcription
Abstract
In model eukaryotes, the C-terminal domain (CTD) of the largest subunit of DNA-dependent RNA polymerase II (RNAP II) is composed of tandemly repeated heptads with the consensus sequence YSPTSPS. The core motif and tandem structure generally are conserved across model taxa, including animals, yeasts and higher plants. Broader investigations revealed that CTDs of many organisms deviate substantially from this canonical structure; however, limited sampling made it difficult to determine whether disordered sequences reflect the CTD’s ancestral state or degeneration from ancestral repetitive structures. Therefore, we undertook, to our knowledge, the broadest investigation to date of the evolution of the RNAP II CTD across eukaryotic diversity. Our results indicate that a tandemly repeated CTD existed in the ancestors of each major taxon, and that degeneration and reinvention of this ordered structure are common features of CTD evolution. Lineage-specific CTD modifications appear to be associated with greater developmental complexity in multicellular organisms, a pattern taken to an extreme in fungi and red algae, in which the CTD has undergone dramatic to complete alteration during the transition from unicellular to developmentally complex forms. Overall, loss and reinvention of repeats have punctuated CTD evolution, occurring independently and sometimes repeatedly in various groups.
The RNA polymerase II (RNAP II) largest subunit (RPB1) has a unique C-terminal domain (CTD) that, in its canonical form, is composed of tandemly repeated heptapeptides with the consensus YSPTSPS. It has been more than a quarter century since the CTD was first described in yeast (1), in which global functions and constraints on its evolution are most thoroughly understood (2, 3). In yeast and animals, the CTD mainly functions as a docking platform to recruit transcription and processing factors at appropriate stages of the transcription cycle (4, 5). Research to date has revealed that CTD-associated factors have a variety of functions, such as mRNA 5′ capping and 3′ processing, pre-mRNA splicing, histone modification, and snRNA processing (6–8). Moreover, the CTD uses different codes to recruit different protein factors (9–11). Reversible phosphorylation of Ser2 and Ser5 residues are the primary CTD codes, and are crucial for regulating transcription and binding mRNA processing factors (12); the major kinases responsible for these phosphorylations are conserved from yeast to metazoans (13). The CTD adopts additional modifications to enrich its functions, including Tyr1 (14), Ser7 (15), and Thr4 phosphorylations (16, 17), as well as cis/trans isomerization of Pro3 and Pro6 (18).
Despite its essential nature and the conservation of multiple core functions, when and in what form the CTD originated remains unclear, as do reasons for the remarkable diversity in CTD sequences and structures across eukaryotic organisms. The last major explicitly phylogenetic treatment of broad-scale CTD evolution was published more than 10 y ago and suggested the presence of a “CTD clade,” all descended from a common ancestor, in which canonical CTD heptads and functions are invariably conserved (19). This, in turn, suggested that a “critical mass” of CTD–protein interactions could have coalesced in the common ancestor of this group, after which the canonical CTD became indispensable to cellular function. With the acceleration of DNA sequencing during the past decade, the diversity of CTD sequences available has grown substantially. It is now clear that evolutionary processes leading to CTD conservation and degeneration are far more complicated than suggested by earlier studies (7, 20, 21). Moreover, recent combined experimental and comparative analyses of the yeast CTD revealed that many fungi have experienced changes across the domain that are incompatible with functional requirements established in Saccharomyces cerevisiae (3). Given the CTD’s centrality to the entire RNAP II transcription cycle, this degree of degeneration is surprising. Therefore, we undertook a comprehensive investigation of CTD evolution within and among major eukaryotic phyla.
Results
The CTD Originated with Tandemly Repeated Heptads.
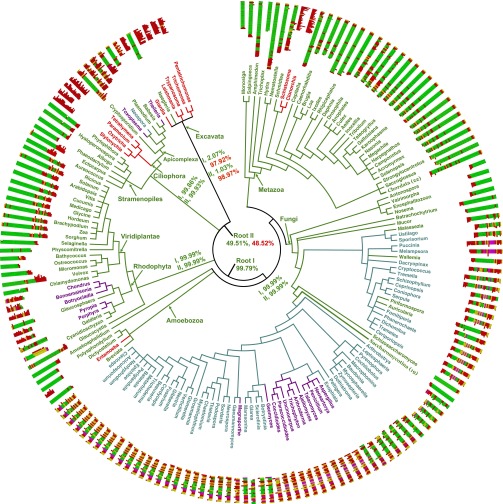
A global phylogenetic tree reflecting current best estimates of eukaryotic relationships was constructed based on the Tree of Life Web Project and National Center for Biotechnology Information (NCBI) Taxonomy. The tree included all genera for which CTD sequences were available, and overall CTD structures were mapped onto the tree (Fig. 1). Interestingly, in all major lineages except the Ciliophora and Excavata, ancestral taxa have the least modified CTD structures; that is, the most deeply branching species contain CTDs mostly of uniform tandem repeats. In contrast, indels, substitutions, or even wholesale degeneration of this structure tend to occur in later diverging taxa, particularly in more complex, multicellular forms. Thus, it is reasonable that a tandemly repeated CTD structure was present in the ancestors of all major taxa currently recognized, and that degeneration of this initial tandem structure is a common feature of CTD evolution.
Fig. 1.
CTD diversity in eukaryotes. The tree shows consensus relationships of the 205 eukaryotes with CTD sequences mapped to each taxon. Sequences are oriented with N-termini at the outer edge and C-termini toward the center. Most CTD sequences are shown from the first obvious heptad to the C-terminal end; those with few or without heptads are shown from a supposed first heptad position, based on typical linker lengths, to the C-terminal end (the same convention is used in other figures). The 22 chordates are collapsed into one branch as their CTD sequences are nearly identical; the same was done for the 19 saccharomycete species. The annotated CTD structure for each genus is shown around the tree. Genus names and their branches are shown in four different colors based on their CTD states (Materials and Methods); state 3, green; state 2, teal; state 1, purple; and state 0, red. Roots I and II reflect alternative rootings of the eukaryotic tree for character state analyses. The probability that the ancestor of descending clades in state 0 (completely disorganized CTD) or state 3 (mostly tandem repeats) are shown separately in red and green.
We addressed this hypothesis more rigorously through maximum-likelihood (ML) character evolution analysis, using four assigned states based on the overall structure of each CTD sequence (Materials and Methods). Analyses were performed by using two commonly suggested roots of the eukaryotic tree, the Excavata and between the Unikonta and Bikonta (22). With the former rooting, ML analysis indicated a 49.51% probability that the eukaryotic common ancestor had a CTD with tandemly repeated heptads, vs. a 48.52% probability of a random sequence; however, the inferred ancestors of all major taxa except the Excavata had 99.96% or greater probabilities of containing a tandemly repeated CTD. The latter rooting resulted in a 99.79% likelihood of a tandem CTD structure in the eukaryotic ancestor. Therefore, contrary to early conclusions based on more limited sampling (20, 23), it appears that the CTD originated as tandemly repeated heptads before the divergence of all (or at least most) extant eukaryotic taxa, and that those taxa without recognizable CTD repeats have undergone degeneration.
The CTD Has Expanded and Diversified with Developmental Complexity in Animals and Plants.
Animals and land plants have achieved the greatest developmental diversity and complexity in the eukaryotic world, and, interestingly, they have parallel patterns of CTD evolution. Animal CTDs are conserved to different degrees in different taxa. In chordates, all 22 genera examined have almost identical CTD sequences with 52 tandem repeats, although serine codon use (TCx or AGC/T) is slightly different in proximal heptads among more distantly related organisms. Likewise, three nematodes (Caenorhabditis to Loa; Fig. 1) have similar CTD structures and codon use, with two from the same family highly conserved. Interestingly, the two available choanoflagellates (Monosiga, Salpingoeca), the closest relatives of metazoans (24), have similar tandemly repeated CTD structures with only subtle codon differences. In contrast, arthropods (Ixodes to Solenopsis) display somewhat more variable levels of CTD conservation across orders and families.
In general, CTD length in animals appears positively correlated with greater developmental complexity, but this is not absolute, as the more deeply branching and morphologically simple animal, Hydra, has the longest set of heptad repeats among all known CTDs (n ∼ 60). Given the generally dynamic nature of the CTD, however, it is likely that Hydra amplified extra repeats recently, and has not yet lost them to a random mutation that would reset the CTD back to a more typical length. In fact, the extremely degenerated distal region of the inferred Hydra CTD appears to reflect this very process. We also found that the tendency toward canonical repeats in proximal regions with substitutions and/or indels in distal regions, first noted in mammalian CTDs, is consistent across metazoan diversity, albeit most prominent in more developmentally complex animals like arthropods and chordates.
Previous broad scale sampling suggested that, in groups like metazoans with highly intricate and well-programmed gene expression, a multiplicity of CTD–protein interactions prevent loss of an overall tandem CTD structure (25); however, CTD sequences from two flatworms, Clonorchis and Schistosoma, show that this is not the case. Neither displays almost any vestige of a canonical CTD, so far a unique condition within the Metazoa. Interestingly, their nearest available relative, Schmidtea, has a more typical metazoan CTD. Clonorchis and Schistosoma are parasitic trematodes, whereas Schmidtea is a free-living turbellarian; this highlights another interesting but not absolute association, that of parasitic lifestyles with extreme CTD modifications (as detailed later).
Generally, CTD evolution in green plants has been analogous to that of animals. Five unicellular green algae available (Chlamydomonas to Bathycoccus; Fig. 1) show similar tandemly repeated heptads, but with largely different serine codon use. Likewise, the CTDs of two early-diverging land plant genera, Physcomitrella and Selaginella, have few or no modifications of distal repeats. More derived and developmentally complex angiosperms (Sorghum to Ricinus), however, contain longer heptad regions with more frequent distal substitutions or indels. There is general conservation of CTD structure and codon use in monocots (Sorghum to Hordeum) and dicots (Glycine to Ricinus), with subtle differences between them. Interestingly, CTD modifications associated with greater developmental complexity even seem to be present in green algae; sequences from unicellular genera (e.g., Chlamydomonas) consist mostly of tandem heptads, whereas the more developmentally complex, colonial genus Volvox contains a longer, more modified CTD.
Parallel CTD Evolution in Fungi and Red Algae.
The CTDs of available Chytridiomycetes (Batrachochytrium) and Zygomycetes (Mucor), representatives of the ancestors of true fungi, contain nearly uniform tandemly repeated heptads (Fig. S1). The same is true for all microsporidian parasites (Antonospora to Nosema; Fig. S1), although their classification as ancient fungi remains controversial (26). In the more derived Ascomycota (Schizosaccharomyces to Claviceps), unicellular yeasts in the Saccharomycotina have simple tandemly repeated CTDs. In the Pezizomycotina (Arthrobotrys to Claviceps), however, there are numerous alterations resulting in regions that would be dysfunctional in yeast based on mutational analyses (3). This is especially striking in the Eurotiomycetes (Exophiala to Coccidioides), in which few typical heptads and no CTD functional units (as characterized in yeast) occur. Based on the tandemly structured CTDs in more ancestral fungi, developmentally complex ascomycetes have lost repetitive heptads through substitutions and indels during their evolutionary diversification. This could parallel lineage-specific adaptive modifications in the distal CTD regions of complex animals and plants, only without a comparable retention of more canonical proximal repeats. Similar but less extreme patterns of heptad modifications are found in other pezizomycete classes. Interestingly, the overall structural patterns within these CTDs, even in serine codon use, are largely conserved at the taxonomic level of classes, and even more so within orders (Fig. S1). This suggests that coadapted molecular processes that underlie conserved developmental patterns reflected in systematic classification, also are reflected in conservation of CTD–protein interactions that regulate RNAP II driven gene expression.
The Basidiomycota (Malassezia to Ceriporiopsis; Fig. 1) is comparable to the Ascomycota in diversity, but far fewer CTDs are known. Nevertheless, available basidiomycete sequences show varied degrees of modifications to ancestral heptads and, given the limited sampling, structural patterns and serine codon use also seem to be conserved within orders. For example, members of the Polyporales (Trametes, Ceriporiopsis, Dichomitus) have highly similar CTD structures (Fig. S1) and codon use. Thus, despite sparser data, it is reasonable to expect that CTD evolution in basidiomycetes has proceeded comparably to what is observed in better-sampled ascomycetes.
With respect to broad-scale CTD evolution in fungi, it is intriguing that the basidiomycetes and pezizomycetes are predominantly multicellular fungi, often with complex developmental programs. In contrast, microsporidians, chytrids, zygomycetes, and saccharomycetes are unicellular or simpler multicellular forms. Thus, our results indicate that there are two distinct evolutionary trajectories for the CTD in fungi. Simple forms tend to retain canonical heptad repeats, albeit with varied differences in serine codon use suggesting that heptads were lost and regained regularly. In contrast, morphologically complex fungi generally underwent extreme modifications in their CTDs, which then were largely conserved at higher (i.e., order) classification levels. This perhaps reflects the evolution of strongly conserved lineage-specific CTD/protein interactions. Unlike in plants and animals, however, there appears to be no strong selection in multicellular fungi to maintain long stretches of uniform tandem repeats.
It appears that CTD evolution in red algae followed a remarkably similar pattern to what occurred in fungi, based on available sequences from eight genera. Unicellular forms (Glaucosphaera, Cyanidioschyzon, Galdieria) all have simple tandem CTD structures, although Cyanidioschyzon has a surprising series of nonapeptide repeats (YSPSSPNVA), unique in all CTD sequences known. In contrast, CTDs from five multicellular rhodophytes have almost no canonical heptads. Although taxon sampling is much weaker, this suggests that, as in fungi, large-scale modifications of ancestral heptads, along with reduced purifying selection on a tandem structure, are correlated with the evolution of developmental complexity in red algae. It also is interesting that Pyropia yezoensis has a very similar CTD to several Porphyra species, although the two genera have proven to be genetically distant (27). This indicates another interesting parallel with the Fungi. Although highly modified, CTD structures are relatively conserved at the order level (Bangiales), in this case correlating with conserved life history and development that traditionally placed Pyropia and Porphyra in the same genus (Porphyra, sensu lato).
CTD Diversity Across Protist Groups.
Stramenopiles (Aureococcus to Phytophthora; Fig. 1) comprise a diverse group of eukaryotes with a broad range of morphologies and ecological habits. The group includes photosynthetic algae ranging from unicellular diatoms to giant kelp, as well as heterotrophic oomycetes and protists (28). At present, complete and well-annotated RPB1 sequences are available from only six genera; these are the diatom Phaeodactylum and pelagophyte Aureococcus, the multicellular brown alga Ectocarpus, and the filamentous oomycetes Hyaloperonospora, Albugo, and Phytophthora. All six have CTDs of long tandemly repeated heptads (YSPTSPA) with few modifications.
Four ciliate CTDs are known, and none displays a discernible tandem structure or even recognizable individual heptads. In contrast, of the four CTD sequences available from amoebozoans, only the parasite Entamoeba lacks tandem repeats. The Excavata is a eukaryotic supergroup composed of diverse unicellular forms. At present, CTD sequences are available from five excavate genera adapted to parasitism, none of which has discernible heptads except for a single YSPASPL present in the trichomonad Pentatrichomonas. In contrast, the predominantly free-living genus Naegleria contains 23 typical heptad repeats.
CTD Evolution in the Apicomplexa.
As in most eukaryotic lineages, the CTD of the deepest branching apicomplexan, Cryptosporidium, has a long array of tandemly repeated heptads. Beyond that, CTD evolution has been unusually fluid in this group. CTDs from Neospora, Theileria, and Toxoplasma all are highly degenerate with few recognizable heptads, whereas Babesia contains numerous tandem repeats in its middle region, but with a different consensus sequence from those in Cryptosporidium. CTD evolution within the genus Plasmodium has been particularly dynamic (Fig. S2). Although the proximal and distal CTD regions are highly conserved across the genus, at least two independent acquisitions of tandem repeats (YSPTSPK) have occurred in primate-infecting species (29), one in the lineage containing Plasmodium fragile, P. knowlesi, and P. vivax, the other in the common ancestor of P. falciparum and P. reichenowi. Even more interesting, the reamplified heptads vary in number not only between species, but also among different strains of P. falciparum and P. vivax. Thus, it appears that tandem heptad degeneration and reinvention have occurred repeatedly in the Apicomplexa, reflecting, in microcosm, the global pattern of CTD evolution across the whole of eukaryotic diversity.
Discussion
Our comprehensive analyses show that the phylogenetic distribution of tandemly repeated sequences does not support earlier hypotheses of a CTD clade, in which some critical mass of CTD–protein interactions coalesced to place strong purifying selection on a canonical, repetitive structure (19). In fact, tandemly structured CTDs are scattered across the eukaryotic tree of life, and appear to have been lost and reamplified from different heptads on numerous occasions. Although it is possible that some CTD variation reflects horizontal gene transfer among unrelated taxa, horizontal gene transfer generally is not favored in genes encoding core informational proteins with multiple complex interactions (30), and we find no empirical evidence of it in the sequences we analyzed. Broader sampling has demonstrated that the CTD can degenerate completely in members of groups, for example, multicellular animals, previously suggested to be incapable of surviving without a well-ordered CTD. Our findings show that tandemly repeated CTDs have been subject to a dynamic process of birth, modification or degeneration, and rebirth throughout eukaryotic evolution.
The Origin of the CTD.
Based on a more limited sample of CTD sequences and differences in serine codon use, Chapman et al. proposed that CTD heptads were built up initially from smaller motifs (YSPx or SPxY, with “x” representing any amino acid), and then amplified independently in various different eukaryotic lineages (20). Our comprehensive investigation of CTD evolution indicates that the extended CTD, present in all RPB1 sequences known to date, originated as tandemly repeated heptads before the divergence of extant eukaryotic groups. It is interesting to note that ML analyses inferred the ancestral presence of a repetitive CTD even in groups in which no well-ordered sequence currently is known. For example, although all ciliates examined to date have fully degenerate CTDs, ancestral tandem repeats are inferred at more than 99% probability, regardless of how the tree is rooted (Fig. 1). Therefore, differences in consensus heptads and codon use reflect the extremely dynamic evolution of tandem repeats rather than their independent origins.
A very early origin of the CTD through rapid amplification of one or more initial heptads raises a provocative question: what was the initial functional advantage of this new domain? The fact that an extended RPB1 C-terminus was never lost from any eukaryotic lineage suggests the CTD was, from its origin, connected to an essential function that also evolved in the common ancestor of extant eukaryotes. Thus, the most likely candidates are CTD-associated processes that are widely distributed across eukaryotic diversity. It also seems most reasonable that initial selection was on a single function rather than complexes of proteins involved in more complicated pathways, and that it favored longer C-terminal extensions rather than a single binding domain. Given these caveats, we argue that the most likely ancestral function for CTD tandem repeats was as a platform for cotranscriptional pre-mRNA splicing. It is believed that the last common ancestor of all extant eukaryotes contained an extremely high density of introns in its protein-coding genes (31), apparently the result of a rapid invasion by group II parasitic self-splicing introns at the dawn of the eukaryotic domain. The spliceosome likely evolved as a mechanism to efficiently remove group II introns that lost the ability to self-splice (32). It is reasonable, that the extended CTD evolved to permit spliceosomes to function cotranscriptionally, thereby increasing splicing efficiency and the overall rate of RNAP II transcription. Experimental results linking the CTD to exon recognition and the earliest stages of spliceosome assembly (33) suggest the two could have coevolved in this manner. Effectively, the CTD could have originated as part of a genomic immune response to a massive invasion of genetic parasites.
Another possibility for the ancestral CTD function is as a platform for 5′ capping, which also appears to be conserved across eukaryotes. Lethal CTD substitutions in fission yeast can be complemented by fusing capping enzyme to the CTD, suggesting that 5′ capping could be the only essential CTD function in that system (34). Because only a single docking site is required; however, capping provides a less compelling explanation for why an extended array of repeats would have been favored from the outset. In any case, once the domain was in place, it quickly proved to be an attractive binding platform for a wide variety of protein partners.
We propose the following scenario for the CTD’s origin and early evolution. First, as suggested by Chapman et al. (20), submotifs such as YSP and SP evolved at the end of the ancestral RPB1 H domain through random mutations, finally in combination resulting in formation of one or more initial YSPxSPx motifs. These heptads then were tandemly duplicated to create the first major RPB1 C-terminal extension. Such an origin is consistent with numerous more recent CTD expansions through tandem duplications, for example those well-documented in Plasmodium parasites (29), as well as nearly identical codon use in many tandem CTD motifs across the breadth of eukaryotic diversity (the most prominent examples are proximal repeat regions in more evolutionarily derived animals and plants). As the CTD grew longer to extend more prominently from the core of RNAP II, the heptads in the linker region degenerated. The former presence of typical CTD heptads is reflected in the modern distribution of SP submotifs, which, on average, are nearly 30 times more abundant in linker regions than in RPB1 domains A through H (Fig. S3).
The Evolution of the CTD Across Eukaryotic Diversity.
The remarkable sequence diversity and variable serine codon use in CTD sequences across eukaryotes highlight the domain’s extraordinarily dynamic evolution. Although more deeply branching genera in nearly all major eukaryotic taxa contain clear tandem CTD heptads, it is unlikely that these specific repeats were conserved from an ancient common ancestor. Selection appears to have worked on the overall tandem structure of the CTD in ancestral eukaryotes, but not necessarily on their underlying primary sequences. In other words, as long as a structurally unordered and reversibly modifiable docking platform was maintained, slightly different heptapeptides were functionally interchangeable, an observation that has been supported experimentally through evolutionary complementation for CTD function in yeast (35). Once present, tandemly repeated sequences are easy to lose and amplify during DNA replication (7). The process most likely involves expansion of the CTD by repeated tandem duplications, balanced by degeneration of distal sequences after random mutations introduced new 3′ stop codons.
Two independent heptad expansions in plasmodium parasites demonstrate how a tandemly structured CTD can be reinvented, even from a largely unordered sequence, when required by the addition of new functions. The specific advantage conferred by these new repeats is unclear, but could involve the coincident acquisition of chromatin remodeling pathways not present in other apicomplexans (36). In any case, they show that the CTD is extremely plastic in response to selection. Given the diversity of CTD–protein interactions across eukaryotes (7), it seems unlikely that modifications specific to any given lineage will prove to be generally applicable. Rather, analogous selective pressures likely have yielded parallel patterns of CTD evolution.
CTD evolution in multicellular eukaryotes is most tantalizing. The domain grew longer in developmentally complex animals and plants, with tandemly repeated proximal regions and somewhat modified distal heptads in both groups. Presumably, this was accomplished not by adding distal nonrepetitive motifs, but by adaptive evolution of ancestral heptads toward specific functions, combined with simultaneous or later additions of new repeats upstream to permit more diverse and overlapping protein binding. In contrast, although CTD heptads underwent various levels of modification in multicellular fungi and red algae, generally more severe than those in land plants and animals, neither group retained or reamplified proximal tandem repeats. Thus, it appears that the evolution of multicellular complexity is associated with specific alterations of the CTD resulting in deviations from the ancestral tandem structure. In organisms that exhibit the greatest levels of cell and tissue differentiation, such as animals and land plants, transcription and processing functions associated with RNAP II are likely too varied and complex to be accommodated without an enlarged CTD, including a repetitive region that permits flexible, redundant function. An association of modified CTD regions with greater transcriptional efficiency required for multicellular development is supported by the observations that only nonconsensus repeats 1–3 and 52 are essential for proliferation of mammalian cell cultures (37), whereas removal of other modified heptads causes retarded growth and increased neonatal lethality during development (38). Presumably multicellular fungi and red algae also evolved lineage-specific functions that modified ancestral heptads; however, perhaps based on a lesser overall need for complexity in gene expression, they did not re- or coevolve tandemly repeated regions for more generalized CTD–protein interactions.
Unfortunately, there are no empirical data that tie specific functions to modified CTD regions in most organisms. Nevertheless, studies of specific CTD alterations in animals provide evidence that some modifications could be related to conserved, lineage-specific functions. For example, an investigation of the role of R1810 (an Arg7) in the human CTD indicates it is involved specifically in regulating expression of snRNA and small nucleolar RNA (39). This could represent a more broadly applicable lineage-specific function because the Arg7 modification is conserved at a comparable position across chordates. Intriguingly, a distal Arg7 also is found in some invertebrate genera, but a specifically conserved position is not apparent outside the chordate lineage.
It is unknown why developmentally complex fungi and red algae lost the need for tandemly repeated heptads as their CTDs underwent extensive modifications associated with the evolution of multicellularity. It may not be coincidental, however, that both groups have relatively simpler developmental programs involving the elaboration of filaments. Unlike metazoans and land plants, they do not exhibit coordinated cellular development required to elaborate highly differentiated tissues and organs. It also is interesting that, thus far, the patterns we highlight are compatible with CTD evolution in stramenopiles, another group that has evolved complex multicellular forms. All unicellular stramenopiles (e.g., Aureococcus, Phaeodactylum) examined to date have relatively uniform tandemly repeated CTDs, as do mycelial oomycete species and the simple, filamentous alga Ectocarpus. The group as a whole, however, has evolved more complex cellular differentiation, including rudimentary vascularization (40). We predict that CTD evolution in stramenopiles will prove to be more similar to animals and green plants than to fungi and red algae; that is, more developmentally complex brown algae like kelp will have longer CTDs with greater numbers of modifications in distal repeats.
Extensive modifications and relaxed purifying selection on the CTD can be associated with the transition to a parasitic lifestyle (21). Remarkably this even extends to several parasitic animals, in which a well-ordered CTD is otherwise invariably conserved. It also is clear that a parasitic lifestyle is not synonymous with CTD decay. Microsporidians, arguably the most derived of all eukaryotic parasites, retain tandemly repeated CTDs. Furthermore, the relationship between parasitism and CTD structure is more complicated in apicomplexan parasites, in which tandem repeats have been lost and reinvented multiple times.
In conclusion, the CTD most likely originated as a tandemly repeated structure, which has been maintained, modified, and/or lost during broad-scale evolution of eukaryotes. The result is a remarkable diversity of sequences, which undoubtedly reflect a comparable diversity of underlying CTD–protein interactions. Some CTD-associated proteins could have undergone related changes to allow continued interactions with changing CTD structures. For example, although both bind to the CTD, mammalian and yeast capping enzymes read CTD codes differently (41). Even so, it is likely that only a handful of CTD functions, if any, are conserved across all eukaryotes. Nevertheless, given that parallel patterns of CTD evolution can be found among unrelated taxa, investigations like those in apicomplexan parasites can help to elucidate more broadly applicable mechanisms of CTD evolution.
Materials and Methods
Data Collection.
RPB1 protein sequences from 205 genera were collected from NCBI and individual genome project databases. We excluded sequences with apparent annotation errors, keeping only reliably interpreted sequences in our analyses. Evolutionary relationships used to interpret patterns of CTD evolution are based on the Tree of Life Web Project and NCBI Taxonomy Database.
CTD Annotation.
Previous analyses in budding and fission yeasts indicated that essential functions of the CTD are conferred by repeated domains, and that minimum essential units of function are contained within heptad pairs (3, 42). To better highlight patterns of CTD conservation and degeneration, we developed graphics for each CTD based on these results with the following color annotations. Green regions contain essential CTD functional units identified in budding yeast (3); that is, paired heptads are present within conserved essential sequence elements (YSPxSPxYSP or SPxYSPxSPxY). Yellow designates individual canonical CTD heptads (YSPxSPx) that are not part of such a CTD functional unit. Red regions have no conserved heptad structure or contain substitutions that are incompatible with CTD function as defined in yeast. Purple heptads have the sequence FSPxSPx that is lethal (if present universally) in budding yeast but turns out to be very common in many fungal genera.
Character Evolution Analysis.
Each CTD was assigned a character state ranging from 0 to 3. CTDs with at least eight consecutive tandem heptads were assigned state 3; examples are the CTDs of yeasts, most animals, and plants. CTD sequences that contain functional units, but fewer than eight uninterrupted heptads (the minimum for viability in yeast), were assigned state 2; examples include CTDs of most sordariomycetes (e.g., Sordaria). Sequences with few to no functional regions, but still with recognizable heptads, were assigned state 1 (e.g., eurotiomycetes). CTDs with no discernible heptads were assigned state 0 (e.g., ciliates). The program Mesquite (version 2.75; http://mesquiteproject.org) was used to carry out ML character state analysis, using the Mk 1 Model, to estimate likelihoods of each state at key nodes and at the root of the eukaryotic tree.
Supplementary Material
Acknowledgments
This study was supported by National Science Foundation Grant 0849586.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323616111/-/DCSupplemental.
References
- 1.Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 2.West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140(4):1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu P, Kenney JM, Stiller JW, Greenleaf AL. Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Mol Biol Evol. 2010;27(11):2628–2641. doi: 10.1093/molbev/msq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartkowiak B, Mackellar AL, Greenleaf AL. Updating the CTD story: From tail to epic. Genet Res Int. 2011;2011:623718. doi: 10.4061/2011/623718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26(19):2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corden JL. RNA polymerase II C-terminal domain: Tethering transcription to transcript and template. Chem Rev. 2013;113(11):8423–8455. doi: 10.1021/cr400158h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113(11):8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 9.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24(6):280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Jasnovidova O, Stefl R. The CTD code of RNA polymerase II: A structural view. Wiley Interdiscip Rev RNA. 2013;4(1):1–16. doi: 10.1002/wrna.1138. [DOI] [PubMed] [Google Scholar]
- 11.Zhang DW, Rodríguez-Molina JB, Tietjen JR, Nemec CM, Ansari AZ. Emerging views on the CTD code. Genet Res Int. 2012;2012:347214. doi: 10.1155/2012/347214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829(1):55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Bartkowiak B, Greenleaf AL. Phosphorylation of RNAPII: To P-TEFb or not to P-TEFb? Transcription. 2011;2(3):115–119. doi: 10.4161/trns.2.3.15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer A, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336(6089):1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 15.Chapman RD, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318(5857):1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 16.Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334(6056):683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hintermair C, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31(12):2784–2797. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werner-Allen JW, et al. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem. 2011;286(7):5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiller JW, Hall BD. Evolution of the RNA polymerase II C-terminal domain. Proc Natl Acad Sci USA. 2002;99(9):6091–6096. doi: 10.1073/pnas.082646199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24(6):289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Stump AD, Ostrozhynska K. Selective constraint and the evolution of the RNA polymerase II C-Terminal Domain. Transcription. 2013;4(2):77–86. doi: 10.4161/trns.23305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr Biol. 2003;13(17):R665–R666. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- 23.Stiller JW, Hall BD. Sequences of the largest subunit of RNA polymerase II from two red algae and their implications for rhodophyte evolution. J Phycol. 1998;34(5):857–864. [Google Scholar]
- 24.Lang BF, O’Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr Biol. 2002;12(20):1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 25.Guo Z, Stiller JW. Comparative genomics and evolution of proteins associated with RNA polymerase II C-terminal domain. Mol Biol Evol. 2005;22(11):2166–2178. doi: 10.1093/molbev/msi215. [DOI] [PubMed] [Google Scholar]
- 26.James TY, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443(7113):818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 27.Sutherland JE, et al. A new look at an ancient order: Generic revision of the Bangiales (Rhodophyta) J Phycol. 2011;47(5):1131–1151. doi: 10.1111/j.1529-8817.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 28.Riisberg I, et al. Seven gene phylogeny of heterokonts. Protist. 2009;160(2):191–204. doi: 10.1016/j.protis.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Kishore SP, Perkins SL, Templeton TJ, Deitsch KW. An unusual recent expansion of the C-terminal domain of RNA polymerase II in primate malaria parasites features a motif otherwise found only in mammalian polymerases. J Mol Evol. 2009;68(6):706–714. doi: 10.1007/s00239-009-9245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA. 1999;96(7):3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koonin EV. Intron-dominated genomes of early ancestors of eukaryotes. J Hered. 2009;100(5):618–623. doi: 10.1093/jhered/esp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogozin IB, Carmel L, Csuros M, Koonin EV. Origin and evolution of spliceosomal introns. Biol Direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13(10):1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell. 2011;43(2):311–318. doi: 10.1016/j.molcel.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stiller JW, McConaughy BL, Hall BD. Evolutionary complementation for polymerase II CTD function. Yeast. 2000;16(1):57–64. doi: 10.1002/(SICI)1097-0061(20000115)16:1<57::AID-YEA509>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 36.Kishore SP, Stiller JW, Deitsch KW. Horizontal gene transfer of epigenetic machinery and evolution of parasitism in the malaria parasite Plasmodium falciparum and other apicomplexans. BMC Evol Biol. 2013;13:37. doi: 10.1186/1471-2148-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapman RD, Conrad M, Eick D. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol Cell Biol. 2005;25(17):7665–7674. doi: 10.1128/MCB.25.17.7665-7674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Litingtung Y, et al. Growth retardation and neonatal lethality in mice with a homozygous deletion in the C-terminal domain of RNA polymerase II. Mol Gen Genet. 1999;261(1):100–105. doi: 10.1007/s004380050946. [DOI] [PubMed] [Google Scholar]
- 39.Sims RJ, 3rd, et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332(6025):99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charrier B, et al. Development and physiology of the brown alga Ectocarpus siliculosus: Two centuries of research. New Phytol. 2008;177(2):319–332. doi: 10.1111/j.1469-8137.2007.02304.x. [DOI] [PubMed] [Google Scholar]
- 41.Ghosh A, Shuman S, Lima CD. Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell. 2011;43(2):299–310. doi: 10.1016/j.molcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwer B, Sanchez AM, Shuman S. Punctuation and syntax of the RNA polymerase II CTD code in fission yeast. Proc Natl Acad Sci USA. 2012;109(44):18024–18029. doi: 10.1073/pnas.1208995109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.