Significance
Long-term morphological stasis is a major feature of the paleontological record, but the explanation for this pattern has been controversial. Here, we use the species-rich plant clade Malpighiaceae to determine whether long-term floral stasis is maintained by selection or developmental and genetic constraint. Our results, which use an explicit phylogenetic framework and comparative methods, strongly support selection. We hypothesize that this floral morphology has been maintained over tens of millions of years via their specialized pollinator interaction with oil-collecting bees. To our knowledge, this study in which stasis has been connected to such a plant-pollinator mutualism, is unique, and opens the door to future research on how this association may have enhanced diversification in this plant lineage.
Keywords: phylogeny, flower morphology, comparative methods, Centridini, diversification rate
Abstract
Many major branches in the Tree of Life are marked by stereotyped body plans that have been maintained over long periods of time. One possible explanation for this stasis is that there are genetic or developmental constraints that restrict the origin of novel body plans. An alternative is that basic body plans are potentially quite labile, but are actively maintained by natural selection. We present evidence that the conserved floral morphology of a species-rich flowering plant clade, Malpighiaceae, has been actively maintained for tens of millions of years via stabilizing selection imposed by their specialist New World oil-bee pollinators. Nine clades that have lost their primary oil-bee pollinators show major evolutionary shifts in specific floral traits associated with oil-bee pollination, demonstrating that developmental constraint is not the primary cause of morphological stasis in Malpighiaceae. Interestingly, Malpighiaceae show a burst in species diversification coinciding with the origin of this plant–pollinator mutualism. One hypothesis to account for radiation despite morphological stasis is that although selection on pollinator efficiency explains the origin of this unique and conserved floral morphology, tight pollinator specificity subsequently permitted greatly enhanced diversification in this system.
Long-term morphological stasis is a hallmark of the fossil record, but we still lack general explanations for this pattern. The presence of morphological stasis is especially perplexing when clades have diversified in terms of their ecology (1–6). There have been two main competing hypotheses to explain clades that accumulate phenotypic variation at a slow rate. The first hypothesis invokes genetic and developmental constraints that restrict the production of phenotypic variation (1, 7, 8). The second major explanation is that stasis is a result of stabilizing selection (3, 9, 10). Here, we investigate the species-rich clade (∼1,300 species) Malpighiaceae to explore the relative importance of intrinsic (e.g., developmental and genetic) vs. extrinsic (e.g., stabilizing selection due to ecological interactions) factors in reducing a clade’s rate of phenotypic change. Members of this clade are widespread and locally common elements in the forests and savannas of the Old and especially New World tropics and subtropics (11). The members exhibit remarkable variety in habit (i.e., herbs, shrubs, trees, and vines) and fruit morphology (i.e., berries, drupes, nutlets, and samaras). The floral morphology of these members, however, is highly conserved throughout the New World range (Fig. 1). It has been hypothesized by Anderson (12), although never explicitly tested, that this floral stasis has been actively maintained by their principle pollinators. These plants' characteristic, bilaterally symmetrical flowers have specialized glands borne on the calyx, which secrete oil that is the primary reward offered to their specialist pollinators: females of ∼300 oil-bee species in the tribes Centridini, Tapinotaspidini, and Tetrapediini (13–15, Fig. 1). These solitary bees are restricted to the Neotropics and use floral oils, often mixed with pollen, for larval provisioning and nest cell lining (16). Species in six other plant families are known to have at least some Neotropical representatives that produce floral oils and are visited by these three oil-collecting bee tribes. None of them, however, are as diverse as Malpighiaceae, which are also the oldest of the seven oil-bee–pollinated clades (17).
Fig. 1.
Stereotyped floral morphology of the ∼1,300 species of New World Malpighiaceae showing the prominent dorsal banner petal that is highly differentiated from the other four petals (A). All petals are conspicuously clawed at the base (A), which allows their primary oil-bee pollinators access to the large paired, multicellular abaxial oil glands, borne on four or all five of the sepals (B) (15, 25). A female oil bee orients toward the banner petal (C) and lands at the center of the flower and grasps the thickened claw of that petal with her mandibles (D). She then reaches between the claws of the lateral petals and scrapes the oil from the glands with her modified front and mid legs. She then transfers the oil and pollen to her hairy hind legs and takes the mixture to her nest, where it serves in nest construction and as food for her larvae. A and B are of Mezia angelica W. R. Anderson, courtesy of C. Gracie (New York Botanical Garden). C and D are of a Centridini oil bee visiting Malpighia emarginata DC, courtesy of G. Gerlach (Munich Botanical Garden, Munich).
Malpighiaceae provide a natural test of the role of intrinsic versus extrinsic factors in generating morphological stasis; this is particularly relevant in the context of Anderson’s earlier hypothesis outlined above, but also more generally in the context of developmental constraints that have been hypothesized to limit floral form (18). The evolution of floral symmetry within the large asterid clade, which includes snapdragons and mints, is highly homoplastic, but a limited subset of the possible floral forms are found to occur, most of which have five corolla lobes, two dorsal (adaxial) petals, and a medially positioned ventral (abaxial) petal (18). Although developmental constraint has been invoked to explain these findings, this hypothesis has not been critically tested. An alternative hypothesis to explain uniform floral symmetry patterns is selection on pollinator efficiency (19), which may subsequently lead to increased diversification rates via species selection on pollinator specificity (20). Here, we analyze morphological diversification patterns in Malpighiaceae to test the hypothesis that floral morphological stasis is driven and maintained by their specialist oil-bee pollinators. This finding may in turn shed light on the significant increase in net species diversification rate that coincided with the origin of the Malpighiaceae and their pollinator mutualism (21).
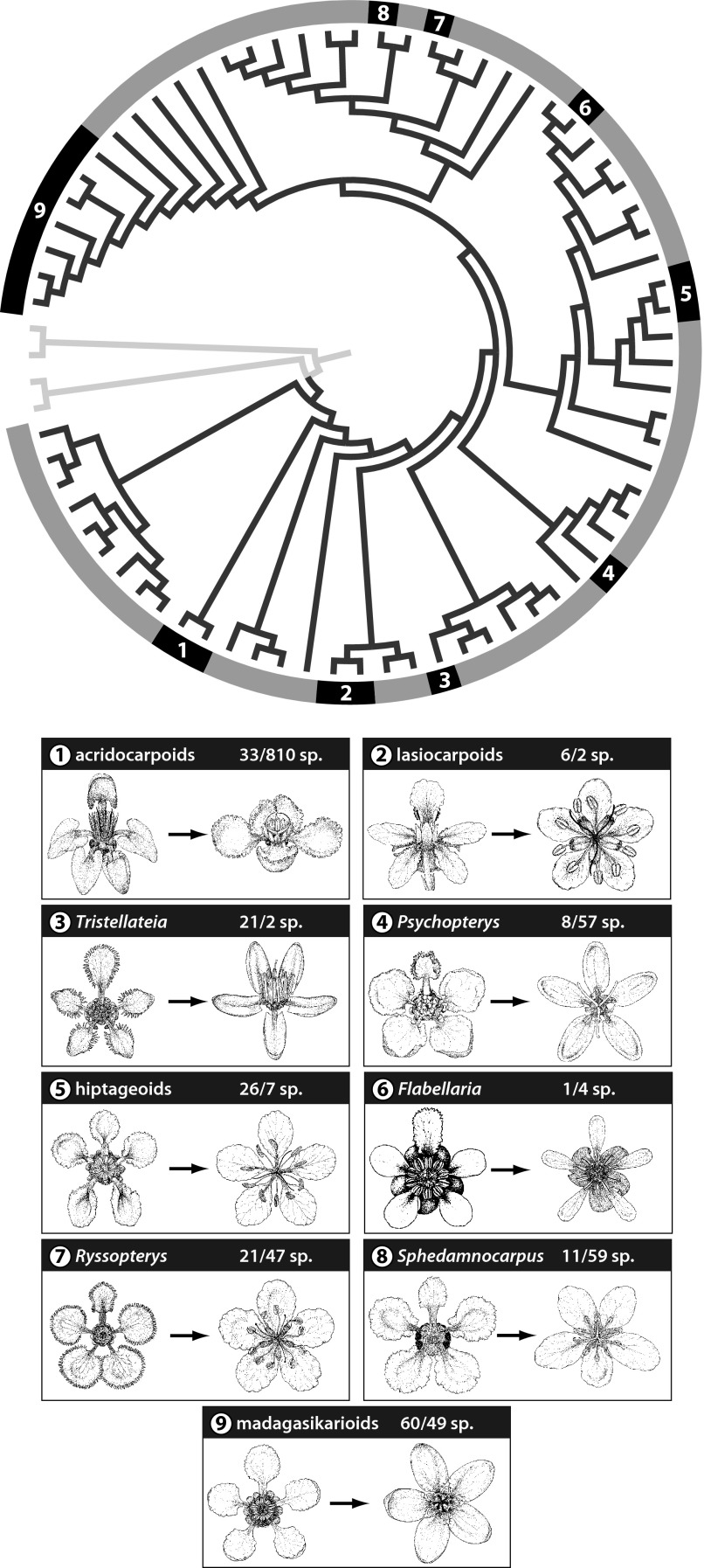
In Malpighiaceae the Old World members of the family are derived from seven independent migrations from the New World (22; Fig. 2, nos. 1, 3, 5–9). In each case specialist oil-bee pollination was lost because oil-collecting bees, which visit Malpighiaceae, do not occur in the Old World (12, 16, 23). In addition, two New World clades, Psychopterys (Fig. 2, no. 4) and the lasiocarpoids (i.e., the genera Lasiocarpus and Ptilochaeta) (Fig. 2, no. 2), have also lost the oil-bee association, although the evolutionary and ecological circumstances of their shift away from oil-bee pollination are unclear. These nine losses of specialized pollinators provide a rare opportunity to examine the cause and consequences of morphological stasis (24). The stereotypical floral morphology of New World Malpighiaceae could be due to genetic or developmental constraints that have restricted the evolution of other floral forms, or the result of ongoing stabilizing selection in response to their specialized oil-bee pollinators (25). If genetic or developmental constraints were primarily responsible for the stereotypical floral morphology of New World Malpighiaceae, we would expect only minor changes in floral architecture in clades that have lost the oil-bee association. Alternatively, if selection by oil bees were actively maintaining the New World floral morphology, we would expect floral morphology to be significantly more labile following release from these specialized pollinators, particularly for traits specifically associated with this mutualism.
Fig. 2.
Phylogeny and floral evolution in Malpighiaceae. The nine panels below the phylogeny illustrate the stereotypical floral morphology of the New World oil-bee associated flowers (Left) contrasted with a representative of its sister clade that has lost the oil-bee association (Right). These nine clades are numbered 1–9 and in black around the periphery of the phylogeny; the remaining oil-bee–associated Malpighiaceae are highlighted in gray; the branches of the non-Malpighiaceae outgroup species are white. The numbers to the left of the backslash represent the species diversities of the nine clades that have independently lost the oil-bee association, and numbers to the right of the backslash represent species diversity of their oil-bee associated sister clades. See Fig. S2 for full chronogram.
Results and Discussion
A robust, reliably dated phylogeny from four genes (11) (Materials and Methods) provides the context for assessing whether the nine clades that lost their association with oil-bee pollinators experienced accelerated rates of floral evolution (Fig. 2). To properly root and date our phylogeny, we first clarified the closest relatives of Malpighiaceae. Previous phylogenetic studies (26–28) have identified the radially flowered, species poor clades Centroplacaceae (∼5 species) and Elatinaceae (35 species) as successive sisters to the species rich Malpighiaceae (∼1,300 species) (species diversity counts are from refs. 29 and 30). Centroplacaceae was not placed with confidence, however. To resolve this uncertainty, we collected nucleotide sequence data from the plastid inverted repeat region (IR; i.e., 19 plastid genes plus intervening spacers; ∼25,000 base pairs), which has been shown (31) to have great utility for resolving ancient rapid radiations like Malpighiales (32). Results from our analyses (Fig. S1) were identical to recent studies but were better supported in key parts of the phylogeny (21, 28). Here, Elatinaceae + Malpighiaceae is established as a well-supported (86 maximum-likelihood bootstrap percentage, 1.0 Bayesian posterior probability) sister clade to Centroplacaceae.
Sixty-five morphological characters were scored for Malpighiaceae (15) in three groups (vegetative, n = 8; flowers/inflorescence, n = 52; fruits, n = 5) (Table S1) and examined in a likelihood framework using GEIGER (33) (Materials and Methods). A likelihood ratio test indicated a dramatic increase in the rates of floral evolution in non–oil-bee clades (P value < 0.0001; mean increase in floral trait variation is ∼10-times higher in non–oil-bee–associated clades). In contrast, the data supported a single-rate model for vegetative and fruit evolution (all P values > 0.05). Although this finding suggests an absence of accelerated morphological evolution for these trait classes, we cannot exclude that this was because of the smaller number of vegetative and fruit characters scored (χ2 test for potential sample size bias between vegetative, floral, and fruit characters, P value = 0.3). However, it is clear that eight floral differences showed significantly elevated rates of change in clades that have lost oil-bee pollinators. Specifically, we found elevated rates of change in the following traits: oil glands (Table S1, trait 34), presence or absence of petal claws (Table S1, trait 38), floral symmetry (Table S1, trait 40), breeding system (Table S1, trait 26), inflorescence structure (Table S1, trait 15), and style and stigma morphology (Table S1, traits 65, 66, and 68). Importantly, the first three of these traits are thought to be crucial to successful pollination by oil bees (15, 25). Floral glands have been lost entirely or converted to producing sugar rather than oil (34) in all of the Old and New World clades that have lost oil-bee pollination. Likewise, clawed petals have been greatly reduced or lost entirely in many of the oil-bee free clades (Fig. 2). Although more work is needed on the pollination of non–oil-bee–associated taxa, it is likely that in some cases pollen is the principal pollinator reward (35–37).
A particularly interesting trait in this regard is floral symmetry. In five of the Old World clades [i.e., the hiptageoids (i.e., Flabellariopsis plus Hiptage) (Fig. 2, no. 5), Flabellaria (Fig. 2, no. 6), Ryssopterys (Fig. 2, no. 7), and the madagasikarioids sensu (11) (Fig. 2, no. 9)] and mainland African sphedamnocarpoids (i.e., Philgamia plus Sphedamnocarpus, not illustrated), and the two New World clades that have lost oil-bee pollination [i.e., the lasiocarpoids (Fig. 2, no. 2) and Psychopterys (Fig. 2, no. 4)], the corolla has become radially symmetrical. The remaining three Old World clades [i.e., the acridocarpoids sensu (11) (Fig. 2, no. 1), Tristellateia (Fig. 2, no. 3), and Malagasy sphedamnocarpoids (Fig. 2, no. 8)] have maintained bilaterally symmetrical corollas, but the flowers have been reoriented such that they display two dorsal (adaxial) petals and a ventrally positioned medial petal that may serve as a landing platform, rather than a single dorsal banner petal and no clear landing platform. Although such reorientations appear to be rare across angiosperms (18), they may be more readily accomplished in Malpighiaceae, where two dorsal petal primordia are the norm early in development and the dorsal medial position of the mature banner petal appears to be achieved by a twisting of the flower stalk later in development (23). Indeed, recent developmental genetic studies (38–40) have shown that the expression of CYCLOIDEA2-like (CYC2-like) genes, which are responsible for establishing floral symmetry in a wide range of angiosperms, has been altered in Malpighiaceae. This finding provides a potential mechanistic basis for changes in floral symmetry coinciding with the loss of oil-bee pollinators (39), and shows that development per se does not significantly constrain changes in this trait.
One additional floral trait, anther vesture (Table S1, trait 46), shows a lower rate of evolution in clades that have lost the oil-bee association. This lower rate may be because anther hair type is not under significant stabilizing selection in oil-bee taxa, but under strong stabilizing selection in relation to non–oil-bee pollinators. In any case, it is clear that the general pattern is conservatism in floral form among taxa that are oil-bee pollinated, with “release” in taxa that have lost this association.
Together, these results firmly support the hypothesis by Anderson (12) that selection by oil-collecting bees has actively maintained the conservative floral form of most New World Malpighiaceae throughout their evolutionary history. Direct fossil evidence dates this mutualism to at least 35 Myr (41), but molecular dating estimates for oil bees and their host plants suggest that this association likely arose much earlier, at least 75 Myr (17, 22, 42, 43). These results argue against a primary role for intrinsic factors, such as genetic or developmental constraints, in shaping floral evolution in Malpighiaceae. If these morphologies were intrinsically constrained we would not expect such dramatic floral evolution in clades that have lost their oil-bee pollinators. Moreover, the major architectural rearrangements we document following the loss of oil-bee pollinators are widely distributed across the timeline of Malpighiaceae diversification, occurring as recently as the Miocene, which indicates that such changes can occur within relatively short periods of time (Fig. S2). It is possible that our findings may also apply to the asterids, where developmental constraint has previously been suggested (18). Perhaps, instead, the limited number of floral forms in this large clade reflects active maintenance by more specialized pollinators, particularly bees, which appear to have radiated with eudicots (43).
Our analysis demonstrates that floral stasis in Malpighiaceae is better explained by extrinsic (e.g., stabilizing selection due to ecological interactions) rather than intrinsic (e.g., developmental and genetic) factors. Studies of adaptive radiation have commonly highlighted cases in which phenotypic change and net diversification rate (i.e., speciation–extinction) are positively correlated (44). These examples mainly involve competitive ecological interactions and sexual selection, with classic examples including the Caribbean Anolis lizards and African rift lake cichlids. Examples of putative adaptive radiations also extend to plant–pollinator interactions, where several studies have identified strong associations between floral traits and species diversification, including shifts from biotic to abiotic pollination (45), nectar spur evolution (46, 47), transitions to bilateral flower symmetry (19), and floral pigment evolution (48).
In contrast, some recent analyses have suggested that the evolution of phenotypic differences and the accumulation of species (diversification) might be inversely correlated during a radiation (49). Malpighiaceae may represent one such example. It has recently been shown that a significant increase in net species diversification rate coincided with the origin of Malpighiaceae and their pollinator mutualism (21). One possibility is that speciation in New World Malpighiaceae is explained by the small geographic range of most solitary bee pollinators and territoriality in male oil bees (50–53), combined with the wide geographic ranges of numerous Malpighiaceae clades (30), which together could facilitate allopatric speciation within the group. Diversification rates in plant clades with zygomorphic flowers like Malpighiaceace have been shown to be significantly increased (19, 54); this has been cited as an example of species selection (20). This finding could be explained by increased oil-bee pollinator specificity in Malpighiaceace clades coupled with a higher probability of speciation in isolated plant populations. Even though future analyses of other traits, like floral oil chemistry, in Malpighiaceae could reveal hitherto overlooked trait differences, our study raises the intriguing possibility that a highly specialized mutualism may explain how apparent morphological stasis can be coupled with species diversification. This possibility should also be investigated in other large plant clades that engage in tight mutualisms, including figs-fig wasps, yuccas-yucca moths, and Phyllantheae-Epicephala moths.
Materials and Methods
Phylogenetic Analyses to Identify the Closest Relatives of Malpighiaceae.
We used the ASAP (amplification, sequencing, and annotation of plastomes) method (55) to sequence the IR from 29 species of Malpighiales. These species were carefully selected to span the basal nodes of all major clades identified from a prior 13-gene analysis of the order (28). We also obtained publicly available IR sequences for Malpighiales [Manihot esculenta Crantz (GenBank accession no. EU117376), Ochna mossambicensis Klotzsch (HQ664566), Phyllanthus calycinus Labill. (HQ664567), and Populus alba L. (AP008956)], and for close relatives of Malpighiales, including Celastrales [Euonymus americanus L. (HQ664608)] and Oxalidales [Oxalis latifolia Kunth (HQ664602)]. Available IR data from Arabidopsis thaliana (L.) Heynh. (AP000423), Beta vulgaris L. (EF534108), Calycanthus fertilis L. (AJ428413), Cucumis sativus L. (AJ970307), Lotus japonicus L. (AP002983), and Vitis vinifera L. (DQ424856) were used as more distant outgroups. Calycanthus was used for rooting purposes based on broader angiosperm relationships (29). Total cellular DNA extractions, PCR amplification, and sequencing protocols, including chromatogram assembly, followed Wurdack and Davis (28).
We analyzed the IR data set using maximum likelihood in RAxML v7.2.6 (56) under the optimal model of sequence evolution [i.e., the General Time Reversible model, with Γ distributed-rate heterogeneity and an estimated proportion of invariable sites (GTR+I+Γ)] as determined by the Akaike Information Criterion criterion using MODELTEST v3.06 (57, 58). Maximum-likelihood bootstrap percentages were estimated from 1,000 bootstrap replicates as a single partition (i.e., a single parameterized model for the entire dataset).
Bayesian analyses were implemented in a parallelized version of BayesPhylogenies v1.1 (59) using a reversible-jump implementation of the mixture model described by Venditti et al. (60). This approach is advantageous because it allows the fitting of multiple models of sequence evolution to the data without a priori partitioning. For each partition, the GTR model plus a Γ distribution with four rate categories was enforced. Two independent Markov-chain Monte Carlo (MCMC) analyses were performed to determine consistency of stationary-phase likelihood values and estimated parameter values between runs. Each MCMC analysis consisted of 10 million generations, with sampling of trees and parameters every 1,000 generations. Convergence was assessed using Tracer v1.5 (61). Bayesian posterior probabilities were determined by building a 50% majority rule consensus tree after discarding the burn-in generations (i.e., the first 2,000 sampled trees).
Phylogenetic and Divergence Time Estimation for Malpighiaceae Using BEAST.
Our analyses of morphological diversification in Malpighiaceae required a time-calibrated phylogeny. We used the Bayesian method implemented in BEAST v1.6.1 (62) to simultaneously estimate the phylogeny and divergence times of Malpighiaceae using a recently published four-gene dataset for the family that included plastid matK, ndhF, and rbcL, and nuclear PHYC (11) (Fig. S2). A likelihood-ratio test rejected a strict clock for the entire dataset (P value < 0.001), and we therefore chose the uncorrelated-rates relaxed-clock model, which allows for clade-specific rate heterogeneity.
One hundred forty-four taxa representing all major clades of Malpighiaceae (i.e., all genera) plus 18 outgroup species were selected for the divergence time analysis (Fig. S2). Centroplacaceae (Bhesa and Centroplacus) and Elatinaceae (Bergia and Elatine) were included as outgroups on the basis of the broader Malpighiales analyses described in the section above. In addition, we included several more distant outgroups to help stabilize the ingroup topology, including other Malpighiales [Chrysobalanaceae (Atuna), Euphorbiaceae (Acalypha and Endospermum), Goupiaceae (Goupia), Ochnaceae (Ochna), Phyllanthaceae (Phyllanthus), Picrodendraceae (Androstachys), Putranjivaceae (Putranjiva), and Violaceae (Hymenanthera)], Celastrales (Denhamia), and Saxifragales (Peridiscus). Peridiscus was used for rooting our phylogeny (29). Our ingroup sampling of Malpighiaceae represents the broadest taxonomic, morphological, and biogeographic diversity within the clade, plus their most closely related non–oil-bee–associated outgroups. These four gene regions were analyzed simultaneously as a single partition using the GTR+I+Γ model as determined using the model selection method described above.
Three fossil calibration points served as minimum age constraints for Malpighiaceae and were fit to a log-normal distribution in our BEAST analysis (Fig. S2). The phylogenetic placement of these calibration points are described in more detail elsewhere (22, 63, 64). A fossil species of Tetrapterys from the early Oligocene (33 Myr) (65) of Hungary and Slovenia provides a reliable age estimate for the stem node of the two Tetrapterys clades (66, 67). Eoglandulosa warmanensis Taylor and Crepet from the Eocene Upper Claiborne formation of northwestern Tennessee (43 Myr) (41, 68) provides a reliable stem node age for Byrsonima. Finally, Perisyncolporites pokornyi Germeraad et al. (69) is found pantropically and provides a reliable stem node age for the stigmaphylloid clade (49 Myr) (70, 71–74). The root node, which we set to a normal distribution of 125 ± 10 Myr, corresponds to the approximate age of the eudicot clade. This finding represents the earliest known occurrence of tricolpate pollen, a synapomorphy that marks the eudicot clade, of which Malpighiales are a member (75).
MCMC chains were run for 10 million generations, sampling every 1,000 generations. Of the 10,001 posterior trees, we excluded the first 2,000 as burn-in. Convergence was assessed using Tracer v1.5 (61). The phylogeny and divergence time estimates simultaneously inferred for Malpighiaceae (Fig. S2) are very similar to those previously published using alternative sampling and methods (11, 22, 63).
Examining Rates of Morphological Trait Evolution of Malpighiaceae.
We applied a model testing approach implemented in GEIGER v1.3.1 (33) using a modified version of the function fitDiscrete to evaluate the rate of morphological trait evolution in clades that have lost their oil-bee pollinator association. These include seven independent clades representing 16 genera that dispersed to the Old World (Acridocarpus + Brachylophon; Aspidopterys + Caucanthus + Digoniopterys + Madagasikaria + Rhynchophora + Triaspis; Flabellaria; Flabellariopsis + Hiptage; Philgamia + Sphedamnocarpus; Ryssopterys; and Tristellateia) and two independent New World clades representing three genera (Lasiocarpus + Ptilochaeta, and Psychopterys) (11) (see also Fig. 2). Those species that were coded as lacking the oil-bee pollination syndrome included the seven Old World clades that occur where the oil bees are not present and the two New World clades that have lost the oil glands and floral orientation crucial to oil-bee pollination (13–15, 25). We tested for elevated rates of morphological evolution in these non–oil-bee–associated clades compared with the rest of the phylogeny. To avoid bias, the species poor sister clades Centroplacaceae and Elatinaceae were excluded from our analysis of morphological trait disparity (i.e., only Malpighiaceae were included).
Morphological data were coded (11) for 144 accessions representing all genera of Malpighiaceae. Seventy-five discrete (binary or multistate) characters, representing the broad diversity of Malpighiaceae morphology, were scored (Table S1). Ten of these characters were invariable and were removed leaving 8 vegetative, 52 floral, and 5 fruit traits. We treated each of the remaining 65 morphological characters as independent, and modeled each as a discrete character with multiple states. We fit a continuous-time Markov model to these data and assumed that transition rates between all possible character states were equal and that each character state was equally likely to be the ancestral state for that clade. If characters were polymorphic for a particular tip, they were treated as having equal probability for each character state at that tip. For each character, we fit two models of evolution: a single-rate model where the rate of character change was constant across all clades, and a two-rate model where clades with and without oil-bee pollinators had distinct rates. For the latter model, we considered the loss of oil-bee pollination to have occurred at the middle of each branch leading to the nine clades that have lost this trait. For each of these two models, we found the maximum-likelihood estimates of the character transition rates. We compared the fit of these two nested models using a likelihood-ratio test.
The single-rate model was rejected in favor of the two-rate model for 9 of the 65 characters (all floral) (Table S1). Of these, all but one trait-described rate increases in lineages that have lost specialized pollinators. There were more significant rate shifts in floral characters than expected given a 5% false-discovery rate under the null hypothesis of no change in rates (Binomial test, P value < 0.01). Furthermore, after applying a very conservative Bonferroni correction, two of these comparisons were still significant, both rate increases. We also compared the number of significant results between the three character types using a χ2 test.
We further compared the fit of both models to assess the overall differences in trait evolution between vegetative, floral, and fruit characters. For each model, we summed the log-likelihoods of all characters in each of these three categories. We then compared the summed likelihoods for each of the two models, again using a hierarchical likelihood-ratio test. Here, the null distribution was a χ2 distribution with n degrees of freedom, where n was the number of characters in a particular set. When we combined these characters, we found significant support for a two-rate model only in floral traits (Δ = 151.4, χ252 P value < 0.0001); we could not reject the single-rate model for the other trait categories (vegetative, Δ = 11.2, χ28 P value = 0.2; fruits, Δ = 5.8, χ25 P value = 0.3).
Supplementary Material
Acknowledgments
We thank W. Anderson, S. Cappellari, E. Kramer, J. Losos, L. Mahler, C. Marshall, C. Webb, W. Zhang, and members of the C.C.D. laboratory for technical assistance and valuable discussions. Funding for this study came from US National Science Foundation Assembling the Tree of Life Grant DEB-0622764, DEB-1120243, and DEB-1355064 (to C.C.D.), and from the Michigan Society of Fellows.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JX977001–JX977020) and TreeBASE, www.treebase.org (accession no. 13486).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403157111/-/DCSupplemental.
References
- 1.Gould SJ. The Structure of Evolutionary Theory. Cambridge, MA: Belknap Press; 2002. p. 1433. [Google Scholar]
- 2.Hunt G. The relative importance of directional change, random walks, and stasis in the evolution of fossil lineages. Proc Natl Acad Sci USA. 2007;104(47):18404–18408. doi: 10.1073/pnas.0704088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estes S, Arnold SJ. Resolving the paradox of stasis: models with stabilizing selection explain evolutionary divergence on all timescales. Am Nat. 2007;169(2):227–244. doi: 10.1086/510633. [DOI] [PubMed] [Google Scholar]
- 4.Futuyma DJ. Evolutionary constraint and ecological consequences. Evolution. 2010;64(7):1865–1884. doi: 10.1111/j.1558-5646.2010.00960.x. [DOI] [PubMed] [Google Scholar]
- 5.Olson ME. The developmental renaissance in adaptationism. Trends Ecol Evol. 2012;27(5):278–287. doi: 10.1016/j.tree.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Stanley SM. Macroevolution: Pattern and Process. San Francisco: W. H. Freeman; 1979. [Google Scholar]
- 7.Smith JM. Macroevolution. Nature. 1981;289(5793):13–14. doi: 10.1038/289013a0. [DOI] [PubMed] [Google Scholar]
- 8.Raff RA. The Shape of Life: Genes, Development, and the Evolution of Animal Form. Chicago: Univ of Chicago Press; 1996. [Google Scholar]
- 9.Charlesworth B, Lande R, Slatkin M. A neo-Darwinian commentary on macroevolution. Evolution. 1982;36(3):474–498. doi: 10.1111/j.1558-5646.1982.tb05068.x. [DOI] [PubMed] [Google Scholar]
- 10.Kirkpatrick M. Quantum evolution and punctuated equilibria in continuous genetic characters. Am Nat. 1982;119(6):833–848. [Google Scholar]
- 11.Davis CC, Anderson WR. A complete generic phylogeny of Malpighiaceae inferred from nucleotide sequence data and morphology. Am J Bot. 2010;97(12):2031–2048. doi: 10.3732/ajb.1000146. [DOI] [PubMed] [Google Scholar]
- 12.Anderson WR. The origin of the Malpighiaceae—The evidence from morphology. Mem N Y Bot Gard. 1990;64:210–224. [Google Scholar]
- 13.Sazima M, Sazima I. Oil-gathering bees visit flowers of eglandular morphs of the oil-producing Malpighiaceae. Bot Acta. 1989;102(1):106–111. [Google Scholar]
- 14.Sigrist MR, Sazima M. Pollination and reproductive biology of twelve species of neotropical Malpighiaceae: Stigma morphology and its implications for the breeding system. Ann Bot (Lond) 2004;94(1):33–41. doi: 10.1093/aob/mch108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel S. Ölblumen und ölsammelnde Bienen. Tropische und Subtropische Pflanzenwelt. 1974;7:283–547. [Google Scholar]
- 16.Michener CD. The Bees of the World. 2nd Ed. Baltimore, MD: The Johns Hopkins Univ Press; 2007. [Google Scholar]
- 17.Renner SS, Schaefer H. The evolution and loss of oil-offering flowers: New insights from dated phylogenies for angiosperms and bees. Phil Trans R Soc. Lond Ser B. 2010;365(1539):423–435. doi: 10.1098/rstb.2009.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donoghue MJ, Ree RH. Homoplasy and developmental constraint: A model and an example from plants. Am Zool. 2000;40(5):759–769. [Google Scholar]
- 19.Sargent RD. Floral symmetry affects speciation rates in angiosperms. Proc Biol Sci. 2004;271(1539):603–608. doi: 10.1098/rspb.2003.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabosky DL, McCune AR. Reinventing species selection with molecular phylogenies. Trends Ecol Evol. 2010;25(2):68–74. doi: 10.1016/j.tree.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Xi Z, et al. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci USA. 2012;109(43):17519–17524. doi: 10.1073/pnas.1205818109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis CC, Bell CD, Mathews S, Donoghue MJ. Laurasian migration explains Gondwanan disjunctions: Evidence from Malpighiaceae. Proc Natl Acad Sci USA. 2002;99(10):6833–6837. doi: 10.1073/pnas.102175899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel S. History of the Malpighiaceae in the light of pollination ecology. Mem N Y Bot Gard. 1990;55:130–142. [Google Scholar]
- 24.Glor RE. Phylogenetic insights on adaptive radiation. Annu Rev Ecol Evol Syst. 2010;41(1):251–270. [Google Scholar]
- 25.Anderson WR. Floral conservatism in neotropical Malpighiaceae. Biotropica. 1979;11(3):219–223. [Google Scholar]
- 26.Davis CC, Chase MW. Elatinaceae are sister to Malpighiaceae; Peridiscaceae belong to Saxifragales. Am J Bot. 2004;91(2):262–273. doi: 10.3732/ajb.91.2.262. [DOI] [PubMed] [Google Scholar]
- 27.Tokuoka T, Tobe H. Phylogenetic analyses of Malpighiales using plastid and nuclear DNA sequences, with particular reference to the embryology of Euphorbiaceae sens. str. J Plant Res. 2006;119(6):599–616. doi: 10.1007/s10265-006-0025-4. [DOI] [PubMed] [Google Scholar]
- 28.Wurdack KJ, Davis CC. Malpighiales phylogenetics: Gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am J Bot. 2009;96(8):1551–1570. doi: 10.3732/ajb.0800207. [DOI] [PubMed] [Google Scholar]
- 29. Stevens PF (2003) Angiosperm Phylogeny Web site. Available at www.mobot.org/mobot/research/apweb/. Accessed January 10, 2012.
- 30. Anderson WR, Anderson C, Davis CC (2006) Malpighiaceae. Available at http://herbarium.lsa.umich.edu/malpigh. Accessed November 5, 2009.
- 31.Jian SG, et al. Resolving an ancient, rapid radiation in Saxifragales. Syst Biol. 2008;57(1):38–57. doi: 10.1080/10635150801888871. [DOI] [PubMed] [Google Scholar]
- 32.Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ. Explosive radiation of Malpighiales supports a mid-cretaceous origin of modern tropical rain forests. Am Nat. 2005;165(3):E36–E65. doi: 10.1086/428296. [DOI] [PubMed] [Google Scholar]
- 33.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: Investigating evolutionary radiations. Bioinformatics. 2008;24(1):129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- 34.Lobreau-Callen D. Les Malpighiaceae et leurs pollinisateurs. Coadaptation ou coévolution [The Malpighiaceae and their pollinators. Coadaptation and coevolution] Bulletin du Muséum National d'Histoire Naturelle. Série 4, section B Adansonia. 1989;11(1, ser. 4):79–94. [Google Scholar]
- 35.Davis CC. Madagasikaria (Malpighiaceae): A new genus from Madagascar with implications for floral evolution in Malpighiaceae. Am J Bot. 2002;89(4):699–706. doi: 10.3732/ajb.89.4.699. [DOI] [PubMed] [Google Scholar]
- 36.Yampolsky C, Yampolsky H. Distribution of sex forms in the phanerogamic flora. Bibliotheca Genetica. 1922;3:1–62. [Google Scholar]
- 37. Anderson WR (2002) Dioecy in the Malpighiaceae. Annual Meeting of the Botanical Society of America 2002, August 2–7, Madison, WI.
- 38.Zhang W, Kramer EM, Davis CC. Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc Natl Acad Sci USA. 2010;107(14):6388–6393. doi: 10.1073/pnas.0910155107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Kramer EM, Davis CC. Similar genetic mechanisms underlie the parallel evolution of floral phenotypes. PLoS ONE. 2012;7(4):e36033. doi: 10.1371/journal.pone.0036033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, Steinmann VW, Nikolov L, Kramer EM, Davis CC. Divergent genetic mechanisms underlie reversals to radial floral symmetry from diverse zygomorphic flowered ancestors. Front Plant Sci. 2013;4:302. doi: 10.3389/fpls.2013.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor DW, Crepet WL. Fossil floral evidence of Malpighiaceae and an early plant-pollinator relationship. Am J Bot. 1987;74(2):274–286. [Google Scholar]
- 42.Cardinal S, Straka J, Danforth BN. Comprehensive phylogeny of apid bees reveals the evolutionary origins and antiquity of cleptoparasitism. Proc Natl Acad Sci USA. 2010;107(37):16207–16211. doi: 10.1073/pnas.1006299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cardinal S, Danforth BN. Bees diversified in the age of eudicots. Proc R Soc B. 2013;280(1755):20122686. doi: 10.1098/rspb.2012.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schluter D. The Ecology of Adaptive Radiation. New York: Oxford Univ Press; 2000. p. 288. [Google Scholar]
- 45.Dodd ME, Silvertown J, Chase MW. Phylogenetic analysis of trait evolution and species diversity variation among angiosperm families. Evolution. 1999;53(3):732–744. doi: 10.1111/j.1558-5646.1999.tb05367.x. [DOI] [PubMed] [Google Scholar]
- 46.Whittall JB, Hodges SA. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature. 2007;447(7145):706–709. doi: 10.1038/nature05857. [DOI] [PubMed] [Google Scholar]
- 47.Ree RH. Detecting the historical signature of key innovations using stochastic models of character evolution and cladogenesis. Evolution. 2005;59(2):257–265. [PubMed] [Google Scholar]
- 48.Smith SD, Miller RE, Otto SP, FitzJohn RG, Rausher MD, editors. The Effects of Flower Color Transitions on Diversification Rates in Morning Glories (Ipomea subg. Quamoclit, Convolvulaceae) Beijing, China: Higher Education Press; 2010. [Google Scholar]
- 49.Adams DC, Berns CM, Kozak KH, Wiens JJ. Are rates of species diversification correlated with rates of morphological evolution? Proc R Soc B. 2009;276(1668):2729–2738. doi: 10.1098/rspb.2009.0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frankie GW, Vinson SB, Newstrom LE, Barthell JF. Nest site habitat preferences of Centris bees in the Costa Rican dry forest. Biotropica. 1988;20(4):301–310. [Google Scholar]
- 51.Zurbuchen A, et al. Maximum foraging ranges in solitary bees: Only few individuals have the capability to cover long foraging distances. Biol Conserv. 2010;143(3):669–676. [Google Scholar]
- 52.Raw A. Territoriality and scent marking by Centris males (Hymenoptera, anthophoridae) in Jamaica. Behaviour. 1975;54(3–4):311–321. doi: 10.1163/156853975x00290. [DOI] [PubMed] [Google Scholar]
- 53.Gathmann A, Tscharntke T. Foraging ranges of solitary bees. J Anim Ecol. 2002;71(5):757–764. [Google Scholar]
- 54.Vamosi JC, Vamosi SM. Factors influencing diversification in angiosperms: At the crossroads of intrinsic and extrinsic traits. Am J Bot. 2011;98(3):460–471. doi: 10.3732/ajb.1000311. [DOI] [PubMed] [Google Scholar]
- 55.Dhingra A, Folta KM. ASAP: Amplification, sequencing & annotation of plastomes. BMC Genomics. 2005;6:176. doi: 10.1186/1471-2164-6-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 57.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 58.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53(5):793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 59.Pagel M, Meade A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Syst Biol. 2004;53(4):571–581. doi: 10.1080/10635150490468675. [DOI] [PubMed] [Google Scholar]
- 60.Venditti C, Meade A, Pagel M. Phylogenetic mixture models can reduce node-density artifacts. Syst Biol. 2008;57(2):286–293. doi: 10.1080/10635150802044045. [DOI] [PubMed] [Google Scholar]
- 61.Rambaut A, Drummond AJ. 2007. Tracer: MCMC Trace Analysis Tool, Version 1.5. Available at http://tree.bio.ed.ac.uk/software/tracer/. Accessed October 1, 2012.
- 62.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5):e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis CC, Fritsch PW, Bell CD, Mathews S. High latitude Tertiary migrations of an exclusively tropical clade: evidence from Malpighiaceae. Int J Plant Sci. 2004;165(4):S107–S121. [Google Scholar]
- 64.Davis CC, Bell CD, Fritsch PW, Mathews S. Phylogeny of Acridocarpus-Brachylophon (Malpighiaceae): Implications for tertiary tropical floras and Afroasian biogeography. Evolution. 2002;56(12):2395–2405. doi: 10.1111/j.0014-3820.2002.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 65.Hably L, Manchester SR. Fruits of Tetrapterys (Malpighiaceae) from the Oligocene of Hungary and Slovenia. Rev Palaeobot Palynol. 2000;111(1–2):93–101. doi: 10.1016/s0034-6667(00)00019-1. [DOI] [PubMed] [Google Scholar]
- 66.Doyle JA, Donoghue MJ. Phylogenies and angiosperm diversification. Paleobiology. 1993;19(2):141–167. [Google Scholar]
- 67.Magallón S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55(9):1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 68.Potter FW, Jr, Dilcher DL. Biostratigraphic analysis of Eocene clay deposits in Henry County, Tennessee. In: Dilcher DL, Taylor TN, editors. Biostratigraphy of Fossil Plants. Stroudsburg, PA: Dowden, Hutchinson & Ross; 1980. pp. 211–225. [Google Scholar]
- 69.Germeraad JH, Hopping CA, Muller J. Palynology of Tertiary sediments from tropical areas. Rev Palaeobot Palynol. 1968;6(3-4):189–348. [Google Scholar]
- 70. Lowrie SR (1982) The palynology of the Malpighiaceae and its contribution to family systematics. PhD dissertation (Univ of Michigan, Ann Arbor, MI) (University Microfilms #82-24999)
- 71.Davis CC, Anderson WR, Donoghue MJ. Phylogeny of Malpighiaceae: Evidence from chloroplast ndhF and trnl-F nucleotide sequences. Am J Bot. 2001;88(10):1830–1846. [PubMed] [Google Scholar]
- 72.Berggren WA, Kent DV, Swisher CC, II, Aubry MP. A revised Cenozoic geochronology and chronostratigraphy. In: Berggren WA, Kent DV, Aubry MP, Hardenbol J, editors. Geochronology, Time Scales and Global Stratigraphic Correlation. Tulsa, OK: SEPM, Society for Sedimentary Geology; 1995. pp. 129–212. [Google Scholar]
- 73.Jaramillo CA. Response of tropical vegetation to Paleogene warming. Paleobiology. 2002;28(2):222–243. [Google Scholar]
- 74.Jaramillo CA, Dilcher DL. Middle Paleogene palynology of Central Colombia, South America: A study of pollen and spores from tropical latitudes. Palaeontographica Abteilung B Paläophytologie. 2001;258:87–213. [Google Scholar]
- 75.Magallón S, Crane PR, Herendeen PS. Phylogenetic pattern, diversity, and diversification of eudicots. Ann Mo Bot Gard. 1999;86(2):297–372. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.