Significance
Cytosine methylation is an epigenetic mark dynamically regulated during embryogenesis and associated with the establishment of tissue specific enhancers. Despite the close relationship, whether CG hypomethylation can influence enhancer formation and cellular identity remains unclear. Using a DNA methylation mutant mouse model, the Lsh−/− mice with approximately 50% site-specific reduction of CG methylation, we provide evidence for a link between CG hypomethylation and poised enhancers that can acquire functional activity and regulate lineage commitment. Our data suggests a pathway of how alterations in the methylome can influence cellular differentiation.
Abstract
DNA methylation patterns are established in early embryogenesis and are critical for cellular differentiation. To investigate the role of CG methylation in potential enhancer formation, we assessed H3K4me1 modification in murine embryonic fibroblasts (MEFs) derived from the DNA methylation mutant Lsh−/− mice. We report here de novo formation of putative enhancer elements at CG hypomethylated sites that can be dynamically altered. We found a subset of differentially enriched H3K4me1 regions clustered at neuronal lineage genes and overlapping with known cis-regulatory elements present in brain tissue. Reprogramming of Lsh−/− MEFs into induced pluripotent stem (iPS) cells leads to increased neuronal lineage gene expression of premarked genes and enhanced differentiation potential of Lsh−/− iPS cells toward the neuronal lineage pathway compared with WT iPS cells in vitro and in vivo. The state of CG hypomethylation and H3K4me1 enrichment is partially maintained in Lsh−/− iPS cells. The acquisition of H3K27ac and activity of subcloned fragments in an enhancer reporter assay indicate functional activity of several of de novo H3K4me1-marked sequences. Our results suggest a functional link of H3K4me1 enrichment at CG hypomethylated sites, enhancer formation, and cellular plasticity.
Cytosine methylation is an epigenetic mechanism conserved among diverse eukaryotes whereby a family of conserved DNA methyltransferases (DNMTs) is responsible for cytosine methylation at cytosine-phosphate-guanosine (CpG) sites. Deletion of DNMTs in mammals alters DNA methylation patterns and results in early embryonic lethality, indicating a critical role of DNMTs in development (1). Genome-wide methylation changes have been reported after deletion of Uhfr1, a hemimethylation binding protein supporting maintenance of DNA methylation, and after deletion of Lsh (2–4). Lsh and the Arabidopsis thaliana homolog DDM1 belong to the SNF2 family of chromatin remodeling proteins, and DDM1 can cause nucleosomal sliding in vitro (5).
Nucleosome templates provide limited access to DNMTs, and the addition of SNF2 proteins can improve de novo methylation in vitro (6). DDM1 mutants show reduced cytosine methylation at genomic regions rich in the linker histone H1, suggesting that the chromatin remodeling activity of DDM1 promotes access to densely packaged chromatin (7).
Deletion of Lsh in mice is perinatally lethal, indicating that Lsh is not a redundant SNF2 family member and plays a critical role in murine development (8, 9). Lsh promotes de novo methylation of exogenous retroviral sequences and endogenous genomic loci, including stem cell-associated genes (10, 11). Lsh can interact with DNMT3b in vitro and supports the association of DNMT3b to some genomic targets. In addition, deletion of Lsh alters genome-wide cytosine methylation pattern, suggesting a regulatory role for Lsh in the establishment of DNA methylation (3, 4, 12, 13).
DNA methylation patterns are highly dynamic and undergo large transitions during germ cell development, early embryogenesis, and reprogramming into induced pluripotent stem (iPS) cells. Dynamic alterations in DNA methylation and H3K4me1 modifications occur at putative enhancers upon cellular differentiation (14, 15). Despite their close association, the causal relationship of CG methylation changes and the establishment of cis-regulatory regions are only partially understood. Engagement of CCCTC-binding factor or repressor element-1 silencing transcription factor to specific loci causes local CG hypomethylation, indicating that DNA-binding factors, such as transcription factors, can shape the methylome (16). Whether CG hypomethylation in turn can influence the formation of enhancer sites and affect cellular differentiation remains unclear, however.
In the present work, we investigated the relationship between CG hypomethylation and potential enhancer formation using primary murine embryonic fibroblasts (MEFs) derived from Lsh−/− or Lsh+/+ embryos (17). DNA methylation patterns are perturbed in Lsh−/− cells compared with WT cells, and up to 50% cytosine methylation as measured by HPLC analysis is lost in Lsh-deleted tissues (3, 4). CG methylation reduction does not occur evenly across the genome, but rather occurs at specific genomic locations. Thus, deletion of Lsh results in a DNA methylation mutant mouse that allows comparison of site-specific methylation using cells from the same tissue type and developmental stage. We report here perturbation of H3K4me1 modification at CG hypomethylated sites in KO MEFs that are partly enriched at neuronal lineage genes, maintained on reprogramming, and linked to a propensity of KO iPS cells toward the neuronal lineage.
Results
Association of de novo H3K4me1 Peaks with CG Hypomethylation in Lsh KO MEFs.
The histone modification H3K4me1 is commonly used to identify putative regulatory elements and cell type-specific enhancers (18, 19). Using chromatin immunoprecipitation sequencing (ChIP-seq), we assessed H3K4me1 modifications in WT and Lsh−/− (KO) MEFs, selecting only H3K4me1 peaks that did not overlap with transcription start sites and H3K4me3 peaks and thereby omitting promoter regions (20). Using these criteria, we identified approximately 64,870 H3K4me1 peaks in KO MEFs, compared with 57,300 in WT MEFs, indicating a 13% increase in H3K4me1 peak number in KO MEFs over WT MEFs.
We also generated CG methylation maps using whole-genome bisulfite sequencing data in WT MEFs, KO MEFs, and WT ES cells. Whole methylome analysis revealed a reduction of methylated cytosine in KO MEFs that was widespread but locus-specific and not evenly reduced across the genome. De novo H3K4me1 peak formation was evident in genomic regions with profoundly reduced CG methylation in KO MEFs (Fig. 1A). There was no overall increase in H3K4me1 modification in KO MEFs or of known H3K4 histone methyltransferases or demethylases (SI Appendix, Fig. S8D), and the average peak size was slightly smaller in KO MEFs compared with WT MEFs (2.5 kb vs. 3 kb). Approximately 7,760 sites showed significantly increased enrichment in KO MEFs versus WT MEFs [KO-H3K4me1; false discovery rate (FDR) <10−5], whereas approximately 7,830 sites showed significantly increased enrichment in WT MEFs (WT-H3K4me1; FDR <10−5), indicating a perturbation of H3K4me1 enrichment based in part on a shift in the distribution of H3K4me1 modification to novel genomic locations.
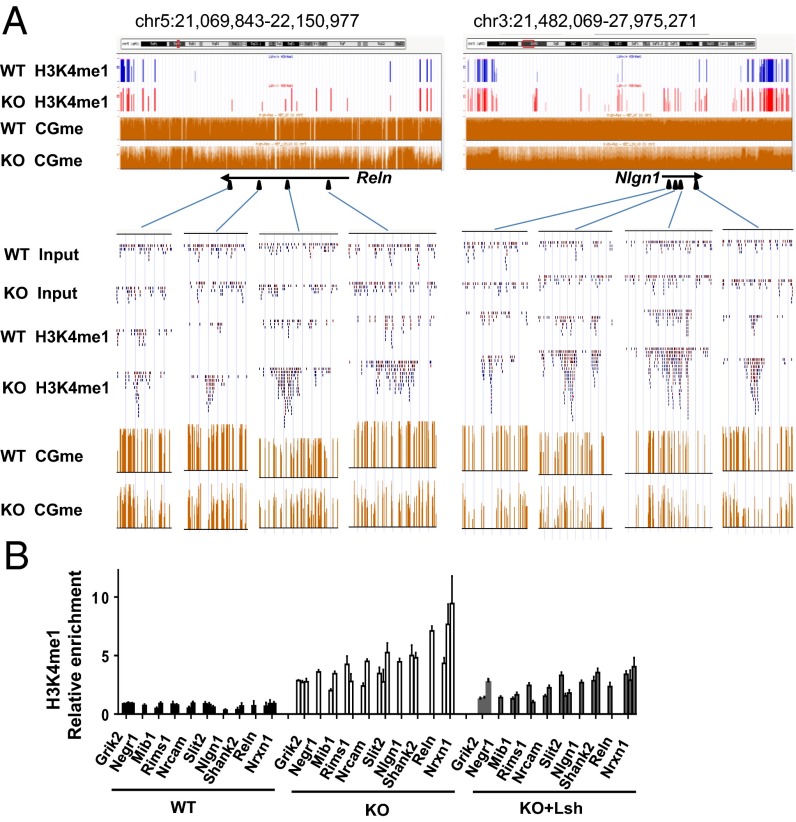
Fig. 1.
De novo H3K4me1 formation at CG hypomethylated regions in Lsh−/− MEFs. (A) UCSC Genome Browser view of specific H3K4me1 peaks in KO MEFs (red) compared with WT MEFs (blue) at selected neuronal genes. CG methylation ratios (CGme) using whole-genome bisulfite sequencing for WT and KO MEFs are shown as orange bars. (Lower) Raw sequence reads of the H3K4me1 signal. (B) qPCR analysis to validate H3K4me1 ChIP-Seq data and assess H3K4me1 marks at KO-H3K4me1 sites after Lsh reexpression in KO MEFs (KO+Lsh) compared with control treated KO and WT MEFs. P < 0.05.
Validation of KO-H3K4me1 ChIP-seq results by conventional ChIP followed by real-time quantitative PCR analysis (qPCR) confirmed a 2- to 10-fold enrichment in KO MEFs compared with WT MEFs (Fig. 1B). Using a conventional bisulfite sequencing technique, we examined CG methylation at sequences located within those KO-H3K4me1 peaks and confirmed substantial differences in CG methylation, ranging between 82% and 94% in WT MEFs and between 28% and 59% in KO MEFs (SI Appendix, Fig. S1 A and B). Despite the difference in DNA methylation, KO MEFs retained CG methylation at some KO-H3K4me1 sites. It is possible that heterogeneity in culture leads to persistence of DNA methylation at sites without H3K4me1 gain, whereas reduced DNA methylation appears at sites with a gain in H3K4me1 modification.
To further explore the relationship between H3K4me1 peak formation and cytosine methylation, we mapped each epigenetic mark around KO-H3K4me1 and WT-H3K4me1 sites (Fig. 2 A–D). H3K4me1 enrichment at KO-H3K4me1 peaks was prominent in KO MEFs but undetectable in WT MEFs (Fig. 2A). CG methylation at flanking regions (−5 kb to +5 kb) of KO-H3K4me1 peaks was significantly higher in WT MEFs than in KO MEFs (0.82 vs. 0.57) (Fig. 2C).
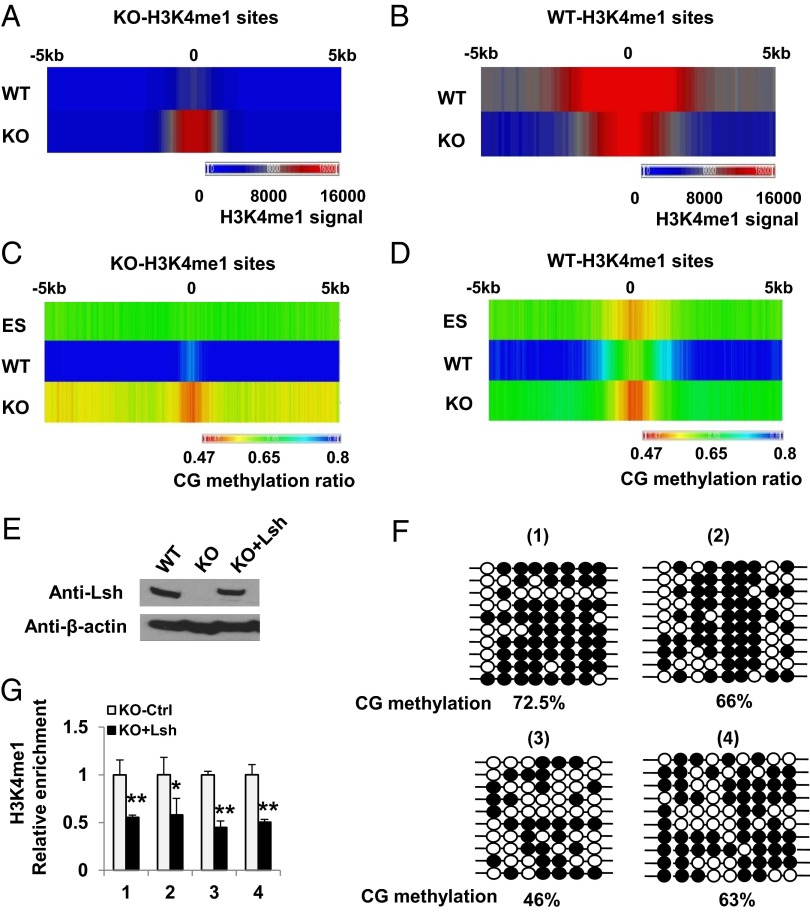
Fig. 2.
Dynamic link between CG hypomethylation and H3K4m1 perturbation. (A–D) Colorimetric representation of H3K4me1 (A and B) and CG methylation (C and D) at KO-H3K4me1 (A and C) and WT-H3K4me1 (B and D) sites comparing WT MEFs (WT), KO MEFs (KO), and WT ES cells (ES). (E) Lsh Western blot analysis after Lsh reexpression (KO+Lsh) in KO MEFs. (F) Bisulfite sequencing representing the CG methylation ratio at four KO-H3K4me1 peaks in Lsh KO that reexpress Lsh (KO+Lsh). The same four sites as in SI Appendix, Fig. S1 are shown for comparison. (G) Relative enrichment of H3K4me1 after Lsh reexpression (KO+Lsh) at four KO-H3K4me1 sites compared to control treated KO MEFs (KO-Ctrl). **P < 0.01; *P < 0.05.
In addition, the center region of the KO-H3K4me1 peaks (−200 bp to +200 bp) showed lower CG methylation in KO MEFs compared with WT MEFs (0.51 vs. 0.76). Moreover, the center of the KO-H3K4me1 peaks displayed significantly lower CG methylation than the global KO methylome (ratio, 0.51 vs. 0.55; P <10–5). Thus, the center region of KO-H3K4me1 peaks is methylated in WT MEFs (CG methyl ratio ≥0.76) and significantly reduced in KO MEFs (CG methyl ratio ≤0.51). This indicates a reciprocal relationship between the increased H3K4me1 modification and the reduced CG methylation at specific sites in KO MEFs compared with WT MEFs. It should be noted that the center region of KO-H3K4me1 peaks was lower in WT MEFs compared with the global WT methylome (0.76 vs. 0.80), indicating that these sites are relatively hypomethylated in WT MEFs. In contrast, WT-H3K4me1 peaks showed, as expected, the strongest CG hypomethylation at the center of WT-H3K4me1 enrichment (Fig. 2 B and D).
Of note, CG methylation ratios differed greatly between KO-H3K4me1 and WT-H3K4me1 sites in WT cells, whereas in KO MEFs, CG methylation ratios and H3K4me1 enrichment were comparable at both locations, with a similar inverse pattern of the two epigenetic marks. Interestingly, the center of KO-H3K4me1 peaks did not display any dip in CG methylation in WT ES cells and showed greater methylation compared with KO MEFs (0.62 vs. 0.51), suggesting the possibility that these sites may have no functional role in undifferentiated ES cells.
To explore whether H3K4me1 peaks and CG methylation are dynamic and can be restored to the WT state, we reintroduced full-length WT Lsh in KO MEFs (Fig. 2E). We observed remethylation at several loci, albeit at lower levels than in WT (Fig. 2F), perhaps because MEFs have reduced DNMT3a and 3b protein levels compared with ES cells (11). Nevertheless, H3K4me1 was decreased concomitantly at these sites (Fig. 2G) and reduced at many other loci that were differentially enriched for H3K4me1 (Fig. 1B), indicating that a partial reversion to the WT state is possible. Furthermore, the data reveal dynamic links among Lsh, CG methylation, and H3K4me1 peak formation.
To test whether CG hypomethylation caused by means other than Lsh deletion could alter H3K4me1 formation, we depleted the maintenance methyltransferase DNMT1 (SI Appendix, Fig. S2). Use of siRNA DNMT1 in WT MEFs effectively reduced protein levels of DNMT1 and CG methylation. Several loci (11 of 16) showed significant H3K4me1 enrichment after DNMT1 depletion compared with control cells. The mean increase in H3K4me1 gain in siDNMT1 samples was 2.1-fold, which is lower than the difference between KO MEFs and WT MEFs for those sites (4.6-fold). It is possible that the overall extent of CG methylation reduction in siDNMT1 MEFs was not sufficient to completely mimic the Lsh KO phenotype, given that the CG methylation level in siDNMT1 samples was between that of WT MEFs and KO MEFs (87% in WT MEFs, 58% in siDNMT1, and 46% in KO MEFs). There are other notable differences as well. For example, DNMT1−/− cells, but not Lsh −/− cells, affect the majority of genomic imprints (1, 21), and DNMT1 depletion reduces CG methylation throughout the genome, whereas Lsh leads to regional CG hypomethylation. In addition, DNMT1−/− ES cells undergo apoptosis on differentiation (22). Nevertheless, the data suggest that at least for a subset of KO-H3K4me1 sites, DNMT1 depletion resulted in H3K4me1 gains, and that these gains occurred at CG hypomethylated sites independent of Lsh modulation.
Association of de novo H3K4me1 Peaks with the Neuronal Lineage Pathway in Lsh KO MEFs.
To explore whether the differentially enriched H3K4me1 peaks were in proximity of genes, we determined genes that were marked by either KO-H3K4me1 or WT-H3K4me1 (located at the gene body and 5-kb flanking sequences) (Dataset S1). Only 3,425 of the 7,760 KO-H3K4me1 peaks were mapped to nearest-neighbor genes. A list of 1,939 genes was compiled with a mean of 1.8 KO-H3K4me1 peaks for each gene, and DAVID functional annotation analysis revealed the highest enrichment for Gene Ontology (GO) terms related to the neuron lineage (e.g., synapse, neuron projection) (Dataset S1) and axon guidance (P < 7.8e-10, KEGG pathway). GO analysis of the biological process (Partek Genomics Suite) detected the greatest enrichment of developmental terms with respect to the neural lineage (SI Appendix, Fig. S3).
We identified approixmately 150 genes (Dataset S2) belonging to the neural lineage pathway, with an average of three peaks per gene. More than 12% of the KO-H3K4me1 peaks were associated with neural lineage genes, indicating that a subset of KO-H3K4me1–marked genes belong to the neural lineage, and that those genes have higher than average enrichment for KO-H3K4me1.
Neuronal lineage-related genes include Reelin (Reln), which is involved in synapsis formation (23) (Fig. 1A); Neuroligin 1 (Nlgn1) (Fig. 1A), which controls synaptic transmission and hippocampus formation (24); Glutamate Receptor; Ionotropic; Kainate 2 (Grik2); and Rims1 (Regulating synaptic membrane exocytosis 1) (Fig.1B and Dataset S2). Only 2% (n = 3) of such neuronal lineage genes were differentially expressed in KO MEFs (Dataset S2), indicating that H3K4me1 marks were not subsequently determined by gene expression differences, and that a gain of H3K4me1 is not sufficient for derepression of these genes. In addition, several H3K4me1-marked sites lacked H3K27ac (see below), suggesting a poised enhancer state in KO MEFs (18).
We next examined whether some of the de novo KO-H3K4me1 peaks at neuronal lineage genes intersected with cis-regulatory sites that are dynamically regulated during development, such as low methylated regions (LMRs) detected during in vitro differentiation of ES cells to neural progenitors (16) or cis-regulatory sites identified based on chromatin states in normal murine brain tissues (20). Although there was no significant overlap of neural lineage gene-associated KO-H3K4me1 peaks with LMRs, we did find substantial overlap with cis-regulatory regions present in normal brain tissue.
The lack of overlap with LMRs may be related to differences in the differentiation protocol or to the use of iPS cells instead of ES cells, leading to variability in neuronal precursor proportions and/or greater variability in enhancer formation. In addition, not all LMRs coincide completely with H3K4me1 modifications, and detection of H3K4me1 modifications as used in this study and in previous studies (20) may be more sensitive in identifying potential regulatory elements. Nevertheless, approximately 40% (168 of 427 peaks) of de novo KO-H3K4me1 peaks that marked neuronal lineage genes overlapped with cis-regulatory elements present in diverse brain tissues (20); for example, several KO-H3K4me1 peaks overlapped with enhancer marks present in cortex, cerebellum, and fetal brain (SI Appendix, Fig. S4). This finding suggests a potential regulatory role for some KO-H3K4me1 sites in the neuronal lineage pathway.
H3K4me1-Enriched Sites in KO iPS Cells and Neuronal Gene Expression.
To test whether some of the KO-H3K4me1–marked sites could become functionally active and serve as enhancers in a neurologic environment, we generated iPS cells that were subsequently challenged to the neuronal lineage pathway. Three sets of iPS clones were derived from independently derived KO MEFs and WT MEFs using a direct reprogramming approach (SI Appendix, Fig. S5). WT and KO iPS clones expressed similar levels of Oct4, Nanog, Sox2, and SSEA-1; were positive for alkaline phosphatase; and showed CG hypomethylation at the Oct4 promoter region. Thus, based on criteria commonly applied for generating iPS cells in vitro, both WT iPS and KO iPS cells manifested characteristics of pluripotency after reprogramming.
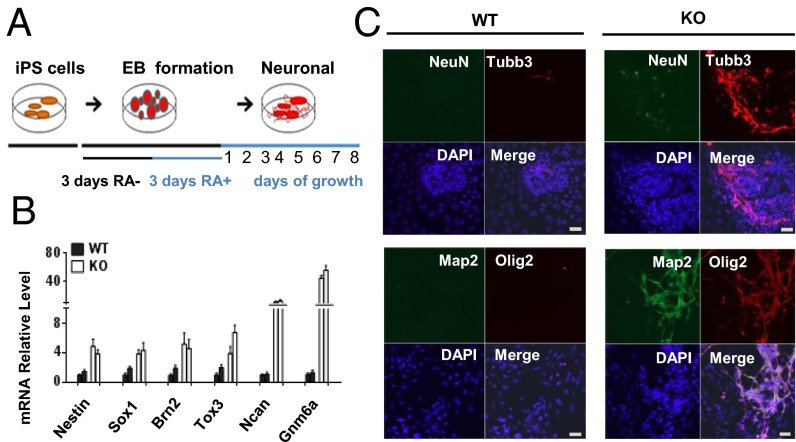
To evaluate the differentiation potential toward the neuron lineage, we aggregated KO and WT iPS cells to form embryoid bodies and treated these bodies with retinoic acid (RA), conditions that favor neural lineage differentiation (Fig. 3A). Using this protocol, we observed an accelerated appearance of cells with neuronal characteristics in KO iPS cultures. After 2 d of RA treatment, KO iPS cells showed increased mRNA expression of several transcription factors that are highly expressed in neuronal progenitor cells (25) (Fig. 3B). After 4 d, KO iPS cells showed enhanced generation of neuronal marker beta-III tubulin (Tubb3/Tuj1), NeuN, Map2, and Olig2 compared with WT iPS cells (Fig. 3C). In addition, we observed augmented neural arborization in three sets of independently derived KO MEFs compared with iPS cells derived from WT MEFs (SI Appendix, Fig. S6 A and B), suggesting that iPS cells derived from CG hypomethylated KO MEFs are prone to enhanced development of neuronal markers during in vitro differentiation.
Fig. 3.
Increased expression of neuronal markers in KO iPS cells. (A) Protocol for neuronal differentiation. (B) mRNA expression analysis by qPCR of neuronal progenitor marker genes after 2 d of RA treatment in two KO iPS and two WT iPS clones. (C) Increased neuronal marker expression and arborization in KO iPS cells compared with WT iPS cells after 4 d of RA treatment. Nuclei were stained with DAPI. (Scale bar: 20 µm.)
To explore whether KO iPS cells also have an enhanced differentiation potential toward the neural lineage in vivo, we injected three sets of independently derived KO iPS and WT iPS cells into immunocompromised mice for teratoma formation. Although teratoma size varied (SI Appendix, Fig. S6C), quantitative mRNA expression of genes specific for neuronal progenitors was significantly higher in samples derived from different KO iPS clones than in samples derived from WT iPS clones (SI Appendix, Fig. S7A). In addition, teratomas derived from KO iPS cells had greater numbers of Tubb3-expressing cells compared with WT iPS cell-derived teratomas of the same size and stage (SI Appendix, Fig. S7 B and C), indicating that KO iPS cells maintained their propensity toward the neural lineage during in vivo differentiation.
We performed an mRNA analysis to investigate whether KO-H3K4me1 marks that are prevalent at specific neuronal lineage genes in KO MEFs are associated with enhanced gene expression in KO iPS cells (Fig. 4A). Uninduced iPS cells showed very little mRNA, and the transcript levels of WT and KO iPS cells were indistinguishable. On differentiation, genes were induced in three independently derived WT and KO iPS clones, but showed greatly increased expression in KO iPS cells, ranging from 2-fold to 10-fold above the levels in WT iPS cells. Thus, neuronal lineage genes that were premarked with H3K4me1 in KO MEFs demonstrated a propensity for increased expression on neuronal differentiation of KO iPS cells.
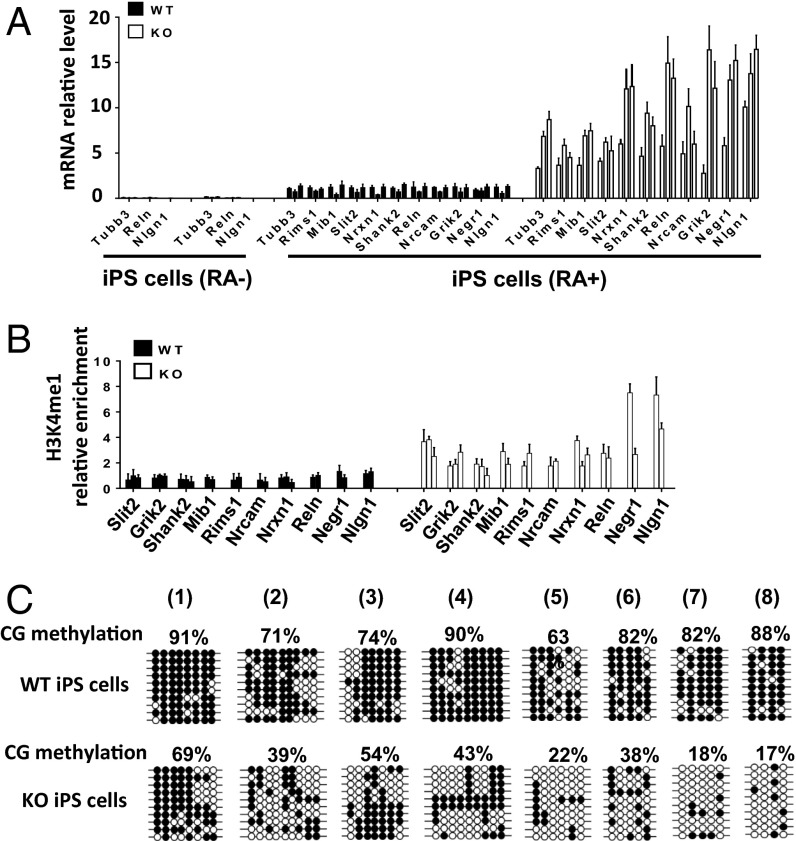
Fig. 4.
Preservation of epigenetic marks in KO iPS cells at neuronal genes. (A) Neuronal lineage gene expression in three sets of KO iPS cells compared with WT iPS cells before and after 4 d of RA treatment. mRNA levels were normalized to one RA-treated WT iPS clone. SD is shown from duplicate experiments for three independent sample sets. (B) H3K4me1 ChIP analysis at 24 different sites of neuronal lineage genes in KO iPS cells compared with WT iPS cells. P < 0.05 (C) Bisulfite sequencing analysis to detect CG methylated sites at KO-H3K4me1 peaks in KO iPS cells compared with WT iPS cells.
Maintenance of Epigenetic Marks in KO iPS Cells.
To further investigate the molecular mechanisms in KO iPS cells that lead to accelerated neuronal gene expression, we examined whether KO-H3K4me1 marks at neuronal lineage genes were preserved after reprogramming. We examined 24 H3K4me1 sites and found that at 23 sites, H3K4me1 was higher in KO iPS cells compared with WT iPS cells, albeit at lower levels than in MEFs (Fig. 4B), suggesting that this histone modification had resisted reprogramming to some degree.
We next asked whether differential CG methylation patterns had been maintained at some of the KO-H3K4me1–marked sites, considering that incomplete demethylation using direct reprogramming with transcription factors has been reported and is associated with the propensity to differentiate into the donor cells (26). Using bisulfite sequencing, we observed differential CG methylation in KO iPS cells compared with WT iPS cells (Fig. 4C), resembling the differences seen in WT and KO MEFs (SI Appendix, Fig. S1A), albeit the CG methylation level were lower in WT and KO samples after reprogramming. Taken together, our data suggest a partial preservation of both epigenetic states throughout reprogramming.
Enhancer Activity of KO-H3K4me1–Enriched Sites.
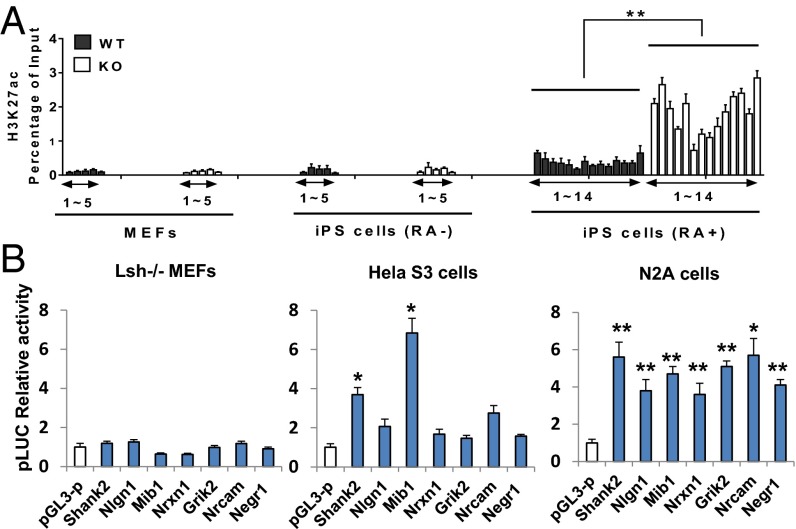
To explore whether the poised KO-H3K4me1 sites seen to partially overlap with enhancers present in brain tissue could indeed switch to an active enhancer state on neuronal differentiation in vitro, we examined the acquisition of H3K27ac, which is considered indicative for functionally active enhancers (20). We examined the sites in those genes that showed increased expression on differentiation in KO iPS cells. MEFs and uninduced iPS cells showed little H3K27ac modification (Fig. 5A, 1–5), consistent with low or undetectable mRNA expression. Of note, after RA treatment, differentiated iPS cells showed increased H3K27ac enrichment at H3K4me1-premarked sites (Fig. 5A, 1–14). Fourteen of the 24 H3K4me1 sites examined showed H3K27ac enrichment, suggesting that at least a fraction (∼60%) of premarked H3K4me1 sites acquired histone acetylation on induction and may function as enhancers in the expression of neuronal lineage genes.
Fig. 5.
Enhancer activity of KO-H3K4me1 sites. (A) H3K27ac ChIPs at KO-H3K4me1 sites of neuronal lineage genes in KO MEFs, WT MEFs, KO iPS cells, and WT iPS cells before and after 4 d of RA treatment. (B) Luciferase activity to assess reporter activity after subcloning of DNA fragments at KO-H3K4me1 sites using Lsh KO MEFs, HeLa, and neuronal cell line N2A. pGL-3P (promoter) served as a negative control.
We then subcloned several fragments containing sequences enriched for H3K4me1 and H3K27ac and tested them in a reporter assay. Although MEFs showed no activity and HeLa cells demonstrated activity of some fragments, all tested fragments showed enhancer activity in the neural cell line Neuro-2A (Fig. 5B). KO-H3K4me1–enriched sites were present in normal development, acquired H3K27ac, and showed enhancer activity in a neural cell line, suggesting that KO-H3K4me1 sites may act as functional enhancers on differentiation into the neuronal lineage.
Transcription Factor Binding at H3K4me1-Enriched Sites.
To investigate the molecular mechanisms in more depth, we performed motif analysis of KO-H3K4me1 sequences at genes with increased expression on differentiation in KO iPS cells. We identified several motifs that could potentially bind transcription factors (TFs) that are present in MEFs but also perform specific roles in neuronal differentiation (Dataset S3). We hypothesized that those TFs are equally expressed in WT and KO MEFs, but may engage differently in the hypomethylated KO genome at KO-H3K4me1 sites. We screened approximately 60 projected binding sites, given the limited capability of motif analysis to predict actual TF binding. At 10 sites, we found specific TF occupancy, along with enrichment of those TFs in KO MEFs (SI Appendix, Fig. S8 A and B). These TFs included Zic1, which is expressed in mesenchymal and neuronal cells during embryogenesis and plays a role in cerebellar development (27); MafB (Kreisler), which is required for hindbrain development (28); Ets1, which has a role in cranial neural crest formation (29); and YY1, which is involved in the regulation of myelination (30). Protein levels of these TFs were comparable in WT and KO MEFs, consistent with our mRNA analysis, suggesting that the differential occupancy of these TFs at KO-H3K4me1 sites was not simply related to differing amounts of TF proteins.
Finally, we examined TF occupancy at some WT-H3K4me1 sites that showed decreased H3K4me1 peaks in KO MEFs but similar enrichment for several of the same TF motifs. We observed a reciprocal binding pattern, with higher TF occupancy in WT MEFs than in KO MEFs (SI Appendix, Fig. S8C), suggesting a redistribution of some TFs from WT-H3K4me1 sites to de novo H3K4me1 peaks in the CG hypomethylated KO MEF genome.
Discussion
Here we report a causal link among Lsh deletion, CG hypomethylation, and perturbed H3K4me1 distribution in KO MEFs, and provide evidence of functional enhancer activity of de novo H3K4me1 peaks in the neuronal lineages. Both epigenetic marks (differential H3K4me1 enrichment and CG hypomethylation) are partially maintained throughout reprogramming, and are associated with increased H3K27ac acquisition and increased mRNA levels. In this way, Lsh-mediated CG methylation can modulate gene activity and influence lineage commitment.
We found a tendency toward neuronal lineage gene expression of KO iPS cells compared with WT iPS cells when challenged to the neuronal pathway. The greatest enrichment for GO analysis revealed conditions for neural development and approximately 12% of gene-associated KO-H3K4me1 peaks clustered at neural lineage genes. However, the list of KO-H3K4me1–marked genes (Dataset S1) also contains many non–neuron lineage-related genes that show low enrichment for GO terms associated with mesoderm-derived lineages (e.g., glomerular basement, gonad, cardiac muscle, smooth muscle, vasculature, ureteric) (SI Appendix, Fig. S3).
To examine whether Lsh deletion leads only to an increased propensity for iPS cells along the neuronal pathway or may efficiently differentiate toward other pathways as well, we challenged Lsh−/− iPS cells into the mesoderm lineage (SI Appendix, Fig. S9 A–C). We found that several markers were equally expressed in KO iPS and WT iPS cells; for example, mRNA expression for mesoderm genes Brachyury, Meox1, and Des was similar in two WT iPS clones and two KO iPS clones, and expression of the mesoderm marker FLK1 was comparable based on FACS analysis. In addition, the development of mesoderm progenies, such as osteoblast-like cells and chondrocyte-like cells, was similar in KO iPS and WT iPS cells, as detected on suitable staining (alizarin red and alcian blue, respectively) (SI Appendix, Fig. S10); thus, we have found no evidence of altered mesoderm differentiation potential. On the other hand, endoderm differentiation revealed increased endoderm marker mRNA expression (Sox17, Gsc, Mixl1) (SI Appendix, Fig. S9 D–F). Our finding of comparable expression of endoderm cell surface marker CXCR4 (as measured by FACS analysis) in KO iPS and WT iPS cells may suggest that despite the increased expression of some early endoderm markers, the definitive endoderm phenotype of KO iPS cells is not significantly affected. It will be exciting to investigate whether the generation of endoderm derivatives in culture (e.g., pancreatic cells, hepatocytes, thyrocytes, lung) is enhanced in KO iPS cells compared with WT iPS cells. In addition, transdifferentiation of MEFs with ectopically expressed TFs may be done to directly assess the full plastic potential of Lsh−/− MEFs. Based on our preliminary studies, CG hypomethylation and increased H3K4me1 modification in Lsh−/− cells may be associated with perturbation of other lineage commitments as well.
We hypothesize that differential occupancy of TFs may represent a molecular mechanism whereby H3K4me1 perturbation may arise at CG hypomethylated sites. In this model, neuronal lineage enhancers may be methylated and “masked” in WT MEFs, but unmasked in the CG hypomethylated genome of KO MEFs, leading to a new equilibrium of TF occupancy. Consistent with this model, we observed differential engagement of a few TFs at KO-H3K4me1 sites in KO MEFs compared with WT MEFs. The TFs were selected because they are expressed in MEFs, and previous studies have demonstrated a role in neural development. Thus, we hypothesize that these TFs may play a role in the expression of neural genes or, alternatively, may be replaced by neuron- specific factors at later stages of neuron differentiation. The differential enrichment of TFs may lead to differential recruitment of H3K4 methyltransferases, such as Setd7 or Mll5, or of H3K4 demethylases, such as Lsd1 (31, 32).
Several mechanisms can be envisioned that may lead to differential TF engagement at CG hypomethylated sites. Some TFs can be directly affected by CG methylation (14, 33–39), whereas others are affected indirectly via association with ternary complexes, in which only one TF may be directly affected by CG methylation (36). In addition, chromatin structure or nucleosomal positioning may be altered at CG hypomethylated sites and thus indirectly influence TF occupancy (40, 41).
Our data indicate that CG hypomethylation influences enhancer formation and thus can affect cellular plasticity. Our results are relevant for regenerative medicine, underscoring the importance of complete reprogramming and resetting of CG methylation patterns to avoid predilection toward a specific tissue. Furthermore, human pathological conditions that involve CG hypomethylation, including cancer, inflammation, and aging, may develop poised enhancers that could switch to an active state on a specific environmental challenge and thus affect cellular identity. Further elucidation of the associations among CG methylation, TF binding, and enhancer formation, and exploration of their role in human diseases, are needed.
Materials and Methods
WT (Lsh+/+) and KO (Lsh−/−) MEFs were maintained in DMEM supplemented with 10% (vol/vol) FBS (Gibco), 2 mM Glutamax (Invitrogen), and 1% penicillin and streptomycin (Invitrogen). Before differentiation, mouse iPS cells were cultured on gelatin-coated plates and maintained in Knockout DMEM supplemented with 15% (vol/vol) Knockout serum replacement (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 0.1 mM MEM nonessential amino acid (Invitrogen), 2 mM Glutamax (Invitrogen), 1% penicillin-streptomycin (Invitrogen), and 1,000 U/ ml of leukemia inhibitory factor (Millipore).
Neuronal differentiation was performed using a modified version of the Mouse Embryonic Stem Cell Neurogenesis Kit (Millipore). In brief, iPS cells were dissociated with accutase, plated onto ultra-low-attachment six-well plates at 5 × 105 cells/well, and cultured in embryonic body (EB) formation medium for 3 d. The culture medium was changed to EB formation medium containing 1.5 μM RA for another 3 d. The EBs were dissociated with accutase, then plated onto 50 μg/mL poly-l-ornithine solution (Sigma-Aldrich) and 5 μg/mL laminin-coated (BD Bioscience) six-well plates or chamber slides (Ibidi). Medium was exchanged with fresh EB formation medium every 2 d for a total of 8 d.
Detailed descriptions of the experiments and computational analyses are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Lino Tessarolo and Dr. Michael Bustin for helpful suggestions on the manuscript. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (Contract HHSN26120080001E). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE56151).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320945111/-/DCSupplemental.
References
- 1.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122(Pt 16):2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107(19):8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myant K, et al. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 2011;21(1):83–94. doi: 10.1101/gr.108498.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao Y, et al. Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc Natl Acad Sci USA. 2011;108(14):5626–5631. doi: 10.1073/pnas.1017000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem. 2003;278(2):823–828. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- 6.Felle M, et al. The USP7/DNMT1 complex stimulates the DNA methylation activity of DNMT1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39(19):8355–8365. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153(1):193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiman TM, et al. Lsh, a SNF2 family member, is required for normal murine development. BBA Gen Subjects. 2001;1526(2):211–220. doi: 10.1016/s0304-4165(01)00129-5. [DOI] [PubMed] [Google Scholar]
- 9.Sun LQ, et al. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18(9):1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xi S, et al. Lsh participates in DNA methylation and silencing of stem cell genes. Stem Cells. 2009;27(11):2691–2702. doi: 10.1002/stem.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H, et al. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25(2):335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi S, et al. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci USA. 2007;104(36):14366–14371. doi: 10.1073/pnas.0703669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28(1):215–226. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153(5):1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gifford CA, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153(5):1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 17.Geiman TM, et al. Lsh, a SNF2 family member, is required for normal murine development. Biochim Biophys Acta. 2001;1526(2):211–220. doi: 10.1016/s0304-4165(01)00129-5. [DOI] [PubMed] [Google Scholar]
- 18.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18(R2):R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan T, Hagan JP, Kozlov SV, Stewart CL, Muegge K. Lsh controls silencing of the imprinted Cdkn1c gene. Development. 2005;132(4):635–644. doi: 10.1242/dev.01612. [DOI] [PubMed] [Google Scholar]
- 22.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122(10):3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 23.Ventruti A, Kazdoba TM, Niu S, D’Arcangelo G. Reelin deficiency causes specific defects in the molecular composition of the synapses in the adult brain. Neuroscience. 2011;189:32–42. doi: 10.1016/j.neuroscience.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 24.Blundell J, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30(6):2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lujan E, Chanda S, Ahlenius H, Südhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci USA. 2012;109(7):2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houtmeyers R, Souopgui J, Tejpar S, Arkell R. The ZIC gene family encodes multi-functional proteins essential for patterning and morphogenesis. Cell Mol Life Sci. 2013;70(20):3791–3811. doi: 10.1007/s00018-013-1285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79(6):1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 29.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y, et al. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat Neurosci. 2010;13(12):1472–1480. doi: 10.1038/nn.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujiki R, et al. GlcNAcylation of a histone methyltransferase in retinoic acid-induced granulopoiesis. Nature. 2009;459(7245):455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 32.Xiao B, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421(6923):652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Kollhoff A, Bergmann A, Stubbs L. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum Mol Genet. 2003;12(3):233–245. doi: 10.1093/hmg/ddg028. [DOI] [PubMed] [Google Scholar]
- 34.Perini G, Diolaiti D, Porro A, Della Valle G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc Natl Acad Sci USA. 2005;102(34):12117–12122. doi: 10.1073/pnas.0409097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rishi V, et al. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci USA. 2010;107(47):20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J. Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol Cell Biol. 2003;23(6):1946–1960. doi: 10.1128/MCB.23.6.1946-1960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polansky JK, et al. Methylation matters: Binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med (Berl) 2010;88(10):1029–1040. doi: 10.1007/s00109-010-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mancini DN, et al. CpG methylation within the 5′ regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998;16(9):1161–1169. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- 39.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: A role for DNA methylation. J Exp Med. 2007;204(7):1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 41.Lazarovici A, et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc Natl Acad Sci USA. 2013;110(16):6376–6381. doi: 10.1073/pnas.1216822110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.