Significance
Staphylococcal pathogenicity islands (SaPIs) are highly mobile and carry and disseminate superantigen and other virulence genes. Here we report a remarkable example of molecular parasitism in which the SaPIs hijack the packaging machinery of the phages they victimize, using two unrelated and complementary mechanisms. Phage packaging starts with the recognition in the phage DNA of a specific sequence, termed “pac” or “cos” depending on the phage type. The SaPI strategies involve carriage of the helper phage pac- or cos-like sequences in the SaPI genome, which ensures SaPI packaging in full-sized phage particles, depending on the helper phage machinery. These strategies interfere with phage reproduction, which ultimately is a critical advantage for the bacterial population by reducing the number of phage particles.
Keywords: interference, horizontal gene transfer, PICIs, molecular parasitism
Abstract
Staphylococcal pathogenicity islands (SaPIs) are the prototypical members of a widespread family of chromosomally located mobile genetic elements that contribute substantially to intra- and interspecies gene transfer, host adaptation, and virulence. The key feature of their mobility is the induction of SaPI excision and replication by certain helper phages and their efficient encapsidation into phage-like infectious particles. Most SaPIs use the headful packaging mechanism and encode small terminase subunit (TerS) homologs that recognize the SaPI-specific pac site and determine SaPI packaging specificity. Several of the known SaPIs do not encode a recognizable TerS homolog but are nevertheless packaged efficiently by helper phages and transferred at high frequencies. In this report, we have characterized one of the non–terS-coding SaPIs, SaPIbov5, and found that it uses two different, undescribed packaging strategies. SaPIbov5 is packaged in full-sized phage-like particles either by typical pac-type helper phages, or by cos-type phages—i.e., it has both pac and cos sites—a configuration that has not hitherto been described for any mobile element, phages included—and uses the two different phage-coded TerSs. To our knowledge, this is the first example of SaPI packaging by a cos phage, and in this, it resembles the P4 plasmid of Escherichia coli. Cos-site packaging in Staphylococcus aureus is additionally unique in that it requires the HNH nuclease, carried only by cos phages, in addition to the large terminase subunit, for cos-site cleavage and melting.
Staphylococcal pathogenicity islands (SaPIs) are highly mobile and parasitize temperate helper phages to enable their reproduction and their highly efficient encapsidation in infectious phage-like particles. Previously characterized SaPIs and their helper phages use the pac mechanism for DNA encapsidation and encode small terminase subunit (TerS) homologs which complex with the phage-coded large terminase subunit and thus determine SaPI packaging specificity. This process is aided by a SaPI-coded protein, Ppi, which complexes with the phage TerS, blocking its function (1). Many of the SaPIs also encode a capsid morphogenesis module consisting of two proteins, CpmA and -B, which cause the formation of small procapsids that match the smaller size of the SaPI genome (2).
Among the ∼20 SaPIs originally identified were several that lacked a TerS homolog and an identifiable morphogenetic module. At the time, these were thought to be defective and were not studied further. More recently, several newly identified SaPIs of bovine mastitis origin were also found to lack these genes; nevertheless, they were efficiently packaged and transferred by helper phages, suggesting that they are not defective after all (3). The prototype of these recently identified islands is SaPIbov5 (3). In this report we decipher a strategy of molecular piracy by which the non–terS-coding SaPIs can be efficiently packaged and transferred.
Results
Phage-Specific SaPI Packaging and Transfer.
SaPIbov1, a prototypical SaPI (4), and SaPIbov5 encode identical SaPI Stl repressors, and therefore, it was predicted that both islands would be induced by the same helper phages (5, 6). A comparison of phage-specific mobilizations of SaPIbov1 and SaPIbov5 revealed that two standard SaPI helper phages, ϕ11 and 80α, induced and mobilized both SaPIs, whereas two additional closely related cos phages, ϕ12 and ϕSLT, not previously known to be SaPI helper phages, induced and mobilized SaPIbov5 but not SaPIbov1 (Fig. 1 and Table 1). SaPIbov1 induction by phages ϕ11 and 80α, which use a headful packaging mechanism, generated the characteristic SaPI-specific band, which represents the SaPI monomers packaged into the small-capsid SaPI particles generated by the SaPI-encoded packaging module (2). No SaPI-specific band was seen with SaPIbov5. This was expected because SaPIbov5 lacks the SaPI packaging module and is packaged in full-sized phage particles (see below). As previously demonstrated (7, 8), deletion of ϕ11 terS eliminated the packaging of phage DNA but had no effect on SaPIbov1 transfer (Table 1). This mutation, however, eliminated SaPIbov5 transfer (Table 1), suggesting that SaPIbov5 contains a pac site that is identical or very similar to that of ϕ11 and explaining the ability of SaPIbov5 to be mobilized by a helper phage despite its apparent lack of any terS gene. Induction and mobilization of SaPIbov5 by ϕ12 and ϕSLT, however, is not so easily explained. ϕ12 and ϕSLT have a putative cos site that is present and shown to be functional in several other related phages (9) (SI Appendix, Fig. S1). Examination of the SaPIbov5 sequence revealed a putative cos site identical to the putative phage cos sites. Several other SaPIs missing the specific packaging module also carry the putative cos site (SI Appendix, Fig. S1). To test these phage cos sites for function, we cloned the ϕ12 and ϕSLT cos sites to a plasmid, pCU1 (10), which was not transferrable by ϕ12, and found that the cloned cos sites, but not the cloned flanking sequences, enabled transfer of the plasmids by ϕ12 (SI Appendix, Table S1) and did not affect their frequency of transfer by ϕ11, which would be by conventional generalized transduction. This result confirmed the identity of these sequences as cos sites. We next deleted the cos site of SaPIbov5 and found that this eliminated transfer of the island by ϕ12 and ϕSLT but not by ϕ11 (Table 1). These results establish that SaPIbov5, unlike any other SaPI thus far characterized, has both pac and cos sites and therefore can be mobilized and packaged by either a pac or a cos helper phage.
Fig. 1.
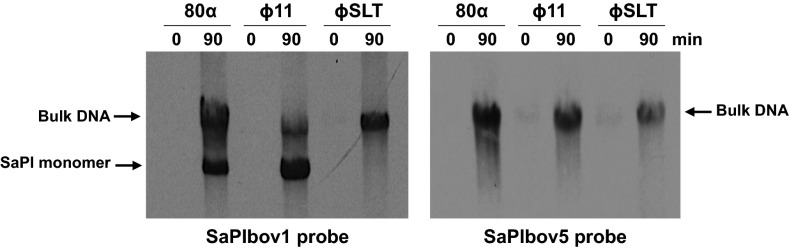
Replication and encapsidation analysis of SaPIbov5. A Southern blot of the different phage lysates carrying SaPIbov1 (Left) or SaPIbov5 (Right), obtained with samples taken 0 or 90 min after MC induction, separated on agarose, and blotted with SaPIbov1- or SaPIbov5-specific probe, is shown. The upper band is “bulk” DNA, including chromosomal, phage, and replicating SaPI DNA. The lower band is SaPI linear monomers released from phage heads.
Table 1.
Effect of phage mutations on phage and SaPI titers
| Donor strain | Titer | |||
| Phage | SaPI | pCN51* | Phage† | SaPI‡ |
| ϕ11 | — | — | 6.6 × 109 | — |
| ϕ11 ΔterS | — | — | <10 | — |
| ϕ11 | SaPIbov1 | — | 2 × 107 | 1 × 107 |
| ϕ11 | SaPIbov5 | — | 2 × 109 | 1.3 × 107 |
| ϕ11 | SaPIbov5 Δcos | — | 1.6 × 109 | 1.1 × 107 |
| ϕ11 ΔterS | SaPIbov1 | — | <10 | 1.9 × 107 |
| ϕ11 ΔterS | SaPIbov5 | — | <10 | <10 |
| ϕ11 ΔterS | SaPIbov5 | pCN51-terSϕ11 | 1.8 × 105 | 2.3 × 105 |
| 80α | — | — | 8 × 1010 | — |
| 80α | SaPIbov1 | — | 3.4 × 109 | 1 × 107 |
| 80α | SaPIbov5 | — | 1.5 × 1011 | 1.2 × 107 |
| ϕ12 | — | — | 8 × 107 | — |
| ϕ12 Δhnh | — | — | <10 | — |
| ϕ12 Δhnh | — | pCN51-hnhϕ12 | 2.2 × 104 | — |
| ϕ12 | SaPIbov1 | — | 1.1 × 109 | <10 |
| ϕ12 | SaPIbov5 | — | 1.1 × 106 | 1.1 × 106 |
| ϕ12 | SaPIbov5 Δcos | — | 1.4 × 107 | <10 |
| ϕ12 Δhnh | SaPIbov5 | — | <10 | <10 |
| ϕ12 Δhnh | SaPIbov5 | pCN51-hnhϕ12 | 1.4 × 104 | 8.1 × 104 |
| ϕSLT | — | — | 6.2 × 106 | — |
| ϕSLT Δhnh | — | — | <10 | — |
| ϕSLT Δhnh | — | pCN51-hnhϕSLT | 3.1 × 104 | — |
| ϕSLT | SaPIbov1 | — | 2.7 × 106 | <10 |
| ϕSLT | SaPIbov5 | — | 8.2 × 105 | 8.7 × 103 |
| ϕSLT | SaPIbov5 Δcos | — | 3.3 × 106 | <10 |
| ϕSLT Δhnh | SaPIbov5 | — | <10 | <10 |
| ϕSLT Δhnh | SaPIbov5 | pCN51-hnhϕSLT | 5.7 × 103 | 1.3 × 103 |
The means of results from three independent experiments are shown. Variation was within ±5% in all cases. —, the strain has not SaPIs or plasmids, and consequently, the transfer of the SaPIs can not be analyzed.
Complemented both the donor and the recipient strains.
PFU/mL induced culture, using RN4220 as the recipient strain.
Number of transductants per milliliter induced culture, using RN4220 as the recipient strain.
Phage-Coded HNH Endonuclease Is Required for Packaging of ϕ12, ϕSLT, and SaPIbov5.
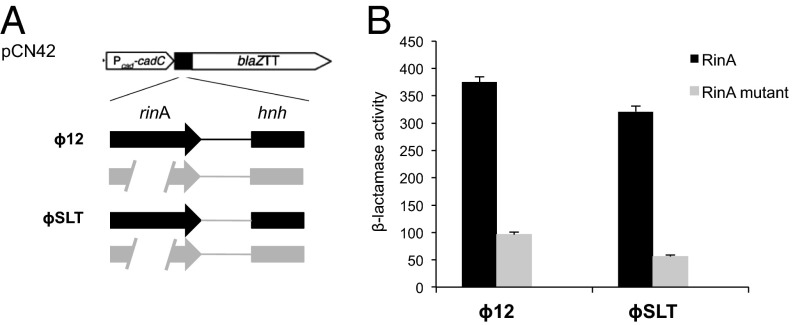
Examination of the packaging modules of phages ϕ12 and ϕSLT revealed an uncharacterized ORF between the late gene regulator rinA (11, 12) and the terminase genes terS and terL (SI Appendix, Fig. S2). The products of these ORFs, ϕ12p28 and ϕSLTp37 (98% identity), contain a region of homology to proteins of the widespread HNH family of endonucleases, and we considered the possibility that these proteins are involved in packaging. If they were involved in packaging, they would presumably be coregulated with the terminase genes—i.e., activated by the late gene regulator RinA (11, 12). Accordingly, we constructed reporter gene fusions with and without rinA and tested them for reporter gene expression. As shown in Fig. 2, the HNH ORF is under RinA control. As its proximity to the terminase genes suggested that it might have a role in phage DNA packaging, we deleted it from both ϕ12 and ϕSLT prophages and tested the deletion derivatives for the production of plaque-forming particles following mitomycin C (MC) induction. As shown in Table 1, neither of these deletion derivatives was capable of producing infectious phage, nor were the ϕ12 and ϕSLT mutants able to mobilize SaPIbov5. Complementation of the hnh mutants re-sorted both the phage and SaPI titers (Table 1). These results suggest that ϕ12p28 and ϕSLTp37 are essential for ϕ12 and ϕSLT reproduction, respectively. Because proteins of the HNH family have endonuclease activity, usually nonspecific, but in some cases directed by a recognition motif to bind and cleave at a specific site, we considered the possibility that these proteins have nuclease activity and that this activity is required for phage encapsidation.
Fig. 2.
RinA proteins control hnh expression. (A) Schematic representation of the different blaZ transcriptional fusions. (B) Derivatives of strain RN4220 containing each of the indicated plasmids were assayed at the midexponential phase for β-lactamase activity under standard conditions. Samples were normalized for total cell mass. Error bars show SEM.
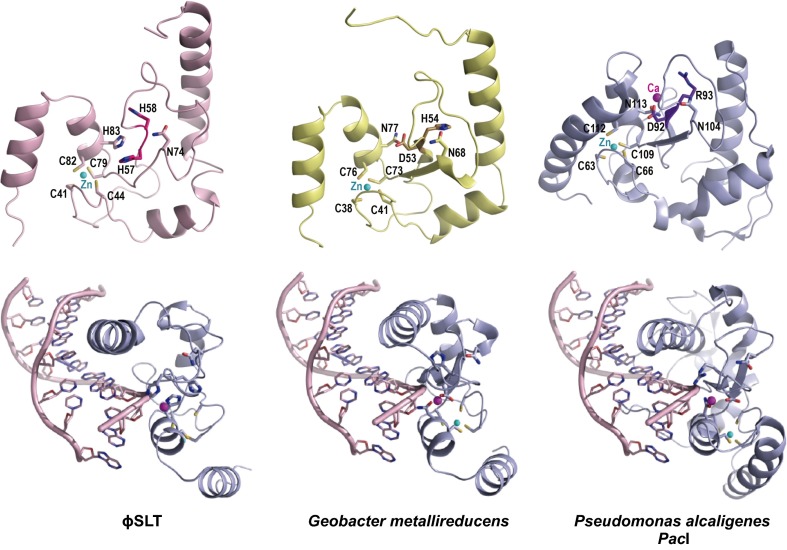
We first analyzed ϕSLTp37 to determine whether it has the structural features of the HNH nucleases by constructing a 3D structural model of the protein using iterative threading assembly refinement (I-TASSER) server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) (13) (Fig. 3). According to this model, the protein folds with similar topology to structures of known HNH nucleases such as that of Geobacter metallireducens GS-15 [Protein Data Bank (PDB) ID code 4H9D] (14) and Pseudomonas alcaligenes PacI endonuclease (PDB ID code 3LDY) (15), despite the low sequence similarity of ϕSLTp37 with these proteins. In the HNH model, the conserved HNH ββα–metal-binding motif comprises residues from 38 to 90 and the catalytic HNH residues are placed in the proper position for catalysis as is observed in structural alignment with other HNH nucleases (SI Appendix, Fig. S3). ϕSLTp37 His58 occupies the place of the histidine general base that activates the water nucleophile attacking the sugar phosphate backbone. ϕSLTp37 His83 and His57 correspond to the metal-binding residues that chelate the catalytic divalent cation (Mg2+ or Mn2+) and Asn74 to the residue that correctly orientates the histidine general base for catalysis. In addition, four cysteine residues, Cys41, Cys44, Cys79, and Cys82 are placed as expected for the tetrahedral coordination of a Zn2+ cation for folding as is observed in previously determined HNH structures. Altogether, the model predicts that ϕSLTp37 and ϕ12p28 are functional HNH nucleases. To confirm this, we expressed the ϕSLTp37 gene in Escherichia coli and tested it for its ability to degrade DNA in vitro. As shown in SI Appendix, Fig. S4 and as reported for other phage-encoded HNH nucleases (16), ϕSLTp37 degrades DNA. Because this degradation is nonspecific, the protein evidently lacks a sequence-recognition domain. We also tested derivatives of ϕSLTp37 with alanine replacements of the two putative catalytic histidines H57 and H58 and found that these also lacked nuclease activity (SI Appendix, Fig. S4). Accordingly, the two proteins are henceforth designated HNHϕSLT and HNHϕ12 and the genes hnhϕSLT and hnhϕ12, respectively. These results suggest that it is the nuclease activity of these proteins that is required for DNA packaging, and that cos-site specificity must be provided by other protein(s).
Fig. 3.
ϕSLT HNH shows the characteristic HNH nuclease fold. ϕSLT HNH structural model (pink) generated with I-TASSER is compared with the experimental structures of G. metallireducens GS-15 (yellow; PDB ID code 4H9D) and P. alcaligenes PacI (blue; PDB ID code 3LDY) HNH nucleases. (Upper) The ribbon representations of the three structures are shown in the same orientation with catalytic and structural relevant residues represented as sticks and colored by atoms (nitrogen, oxygen, and sulfur in blue, red, and yellow, respectively) with carbon in the same color of the corresponding structure. Residues mutated to Ala in ϕSLT HNH (H57 and H58) and the equivalent residues in GS-15 and PacI are highlighted in darker hues. The structural Zn2+ ion is shown as a cyan sphere. (Lower) The DNA–HNH complex is modeled for HNHϕSLT and GS-15 by superimposing these protein structures with PacI HNH in the PacI–DNA experimental complex (PDB ID code 3LDY).
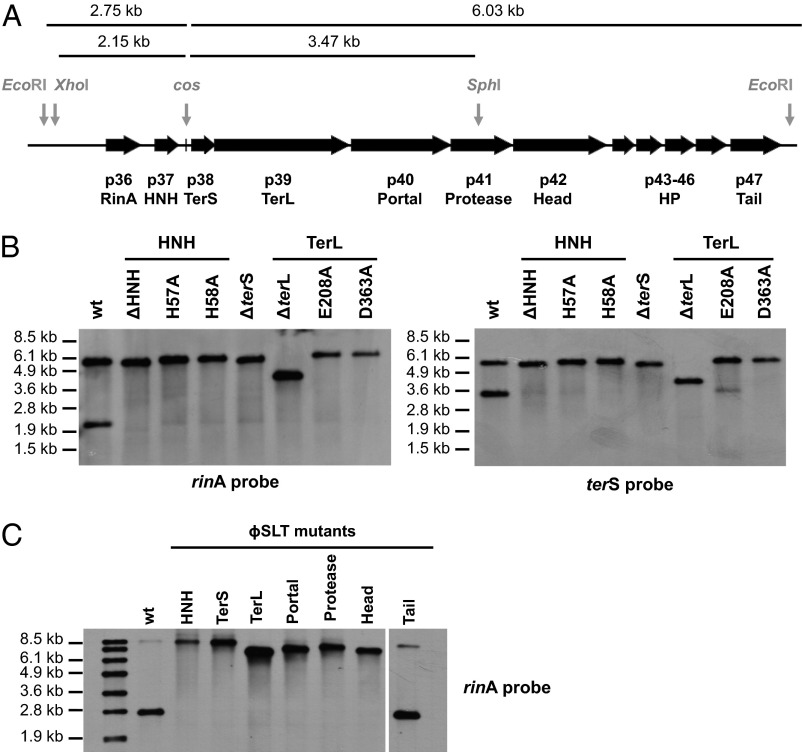
We next analyzed the cos-site cleavage reaction by means of ϕSLT derivatives with mutations in terS, terL, or hnh, using a detoxified derivative of the phage containing a tetracycline-resistance marker (tetM) in the Panton–Valentine leukocidin locus. This provided a sensitive test for packaging, as a single packaged molecule could be detected in the recipient strain as a prophage. For TerL, we included alanine replacements of E208 (motor) and D363 (nuclease) on the basis of previous studies (17) and comparative TerL sequences (SI Appendix, Figs. S5 and S6); and for HNH we included alanine replacements of H57 and H58 (Fig. 3). For these experiments, the WT and mutant prophages were induced with MC and DNA prepared from a 90-min postinduction culture sample and analyzed for cos-site cleavage by means of codigestion with XhoI and SphI and Southern blotting with rinA- and terS-specific probes. In the absence of cos-site cleavage, a 5.62-Kb fragment was expected; following cos-site cleavage, the rinA probe would hybridize with a 2.15-Kb fragment and the terS probe with a 3.47-Kb fragment (see the scheme in Fig. 4A). As shown in Fig. 4B, the smaller fragments produced by cleavage at the cos site appeared with the WT phage but not with any of the hnh, terS, or terL mutants. Cleavage was partial with the WT, and a very faint band, corresponding to the processed species, was seen with most of the mutant phages, suggesting that very weak processing could occur in the absence of any one of the three proteins. Note that cos-site duplexes cannot account for the appearance of partial cleavage or for the weakness of the bands in the mutants because the samples were prepared under conditions that would ensure cos-site melting. This poor processing, however, was not sufficient to enable detectable packaging, which was partially re-sorted after complementation of the different phage mutants (SI Appendix, Table S2). Of interest is the fact that the complementation rate of the single point mutants carrying the plasmids expressing the WT proteins was reduced compared with that observed for the complementation of the deletion mutants (SI Appendix, Table S2). This suggests that the mutant proteins are expressed, forming nonfunctional complexes with the other phage-encoded proteins and interfering with the WT proteins to generate functional phage particles. Taken together, these results suggest that the packaging of this family of phages (and their parasite SaPIs) requires the nuclease activity of both TerL and HNH proteins. We propose here that these enzymes plus TerS form a cooperative nuclease complex to bring about cos-site cleavage, with TerS presumably determining the cleavage site.
Fig. 4.
Role of the different ϕSLT mutants in phage packaging. (A) ϕSLT map. The relevant genes and the cos site are shown. (B and C) Lysogenic strains carrying the different phage mutations were exposed to MC, then incubated in broth at 32 °C. Samples were removed after 90 min and used to isolate DNA, which was digested with XhoI/SphI (B) or with EcoRI (C). DNAs were separated by agarose gel electrophoresis and transferred. Southern blot hybridization patterns of these samples were hybridized overnight with a rinA or terS (to the left and right of the cos site, respectively in A) phage-specific probes. In C, all samples were run on the same gel, but four lanes analysing mutants that are not included in the manuscript have been removed. Noncontiguous lanes are divided by a white line.
The HNH–TerS–TerL Complex Is Not Sufficient to Activate Cos-Site Processing.
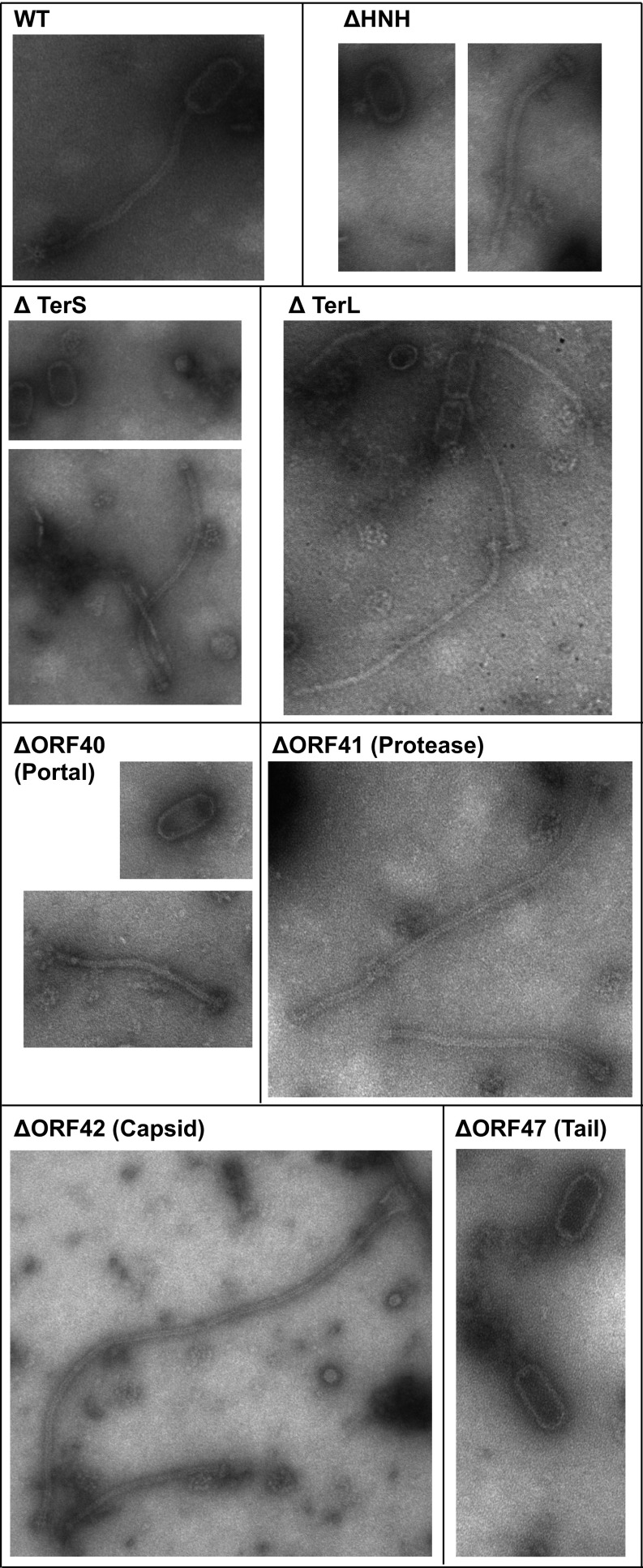
To prevent premature cos-site processing, the λ TerS–TerL complex performs its nuclease function only in the presence of a preformed procapsid (18). We tested this with the HNH–TerS–TerL complex in vivo by mutating various proteins involved in ϕSLT capsid formation and assaying for phage packaging and cos-site cleavage as above (Fig. 4A and SI Appendix, Table S2). This included terS, p40 (portal), p41 (protease), p42 (major capsid protein), and p47 (major tail protein). Electron microscopic analysis of the particles present in lysates prepared with the different deletion-carrying phages confirmed the predicted roles for the deleted genes. Thus, although deletions of the hnh, terS, terL, and portal protein genes resulted in empty capsids and tails, deletions of the protease and capsid genes resulted in only tail structures, whereas the deletion of p47 resulted in capsids only (Fig. 5). Surprisingly, some of the tails observed in the protease and major capsid protein mutants were extremely long, suggesting that the capsid structure also controls the length of the tail. Complementation of the different mutants re-sorted the production of functional phage particles (SI Appendix, Table S2). As shown in Fig. 4C, each of the deletions affecting capsid formation or DNA packaging eliminated cos-site cleavage, whereas deletion of the major tail protein gene did not. This result confirms the prediction that cos-site cleavage by the HNH–TerS–TerL complex is activated by the preformed capsid—which is required for cos but not for pac phages (19).
Fig. 5.
Electron micrographs of ϕSLT mutant lysates.
The Packaging System Involving HNH Proteins Is Widespread in Nature.
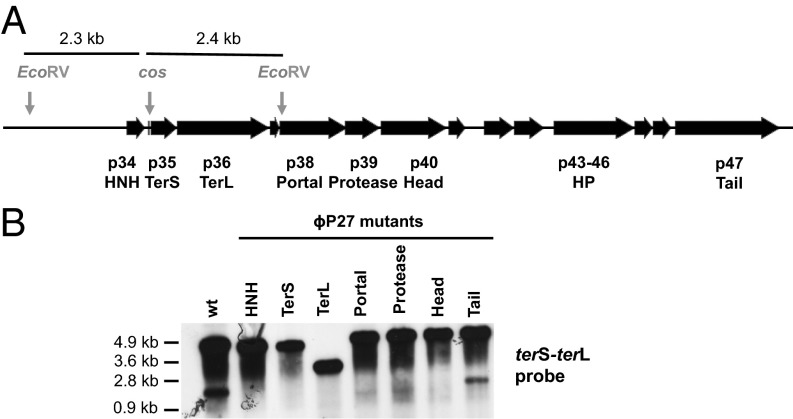
A recent study has reported that many phages, infecting both Gram-positive and -negative bacteria, encode an HNH protein adjacent to the genes encoding the TerS, TerL, and portal proteins (20). Interestingly, all of the characterized phages carrying an hnh gene are cos phages. Our results have demonstrated that not only these proteins but also those encoding the major capsid, the portal, and the protease proteins are required for cos-site cleavage and therefore for DNA packaging. To demonstrate the diversity of this mechanism, we analyzed E. coli phage ϕP27, which is clinically relevant as it encodes the Shiga toxin (Stx) (21). We used a detoxified derivative of phage ϕP27 containing a tetracycline-resistance marker (tetA) inserted into the stx locus. We deleted from the ϕP27 prophage the genes encoding the HNH, TerS, TerL, portal, protease, and major capsid proteins (SI Appendix, Fig. S2). Deletion of these genes did not affect phage DNA replication but completely eliminated phage packaging and infectivity (SI Appendix, Table S3). These defects were partially re-sorted by complementation (SI Appendix, Table S3). In addition, as previously demonstrated for the staphylococcal phages, cos-site cleavage in vivo was observed only after induction of the WT prophage, but not with any of the deletion mutants (Fig. 6), confirming that the supramolecular complex containing HNH–TerS–TerL plus an intact capsid is required for cos-site cleavage.
Fig. 6.
Role of the different ϕP27 mutants in phage packaging. (A) ϕP27 map. The relevant genes and the cos site are shown. (B) Lysogenic strains carrying the different phage mutations were exposed to MC, then incubated in broth at 32 °C. Samples were removed after 90 min and used to isolate DNA, which was digested with EcoRV. DNAs were separated by agarose gel electrophoresis and transferred. Southern blot hybridization patterns of these samples were hybridized overnight with a terS–terL phage-specific probe.
Discussion
Characterization of several of the phage-inducible SaPIs and their helper phages has established that the pac (or headful) mechanism is used for encapsidation, which is exactly what would be expected for a transductional mode of transfer. In keeping with this concept, the SaPIs thus far characterized encode a homolog of TerS, which complexes with the phage-coded large terminase subunit TerL to enable packaging of the SaPI DNA in infectious particles composed of phage proteins (2, 7). These also contain a morphogenesis (cpm) module that causes the formation of small capsids commensurate with the small SaPI genomes (2). Among the SaPI sequences first characterized several years ago, there were several that did not include either a TerS homolog or a cpm homolog, and the same is true of several recently identified SaPIs from bovine sources (3) and for many phage-inducible chromosomal islands from other species. It was assumed, for these several islands, either that they were defective derivatives of elements that originally possessed these genes, or that terS and cpm genes were present but not recognized by homology. We have performed an extensive study of one of these islands, SaPIbov5, and found that the element is fully functional—able to be induced, efficiently packaged, and transferred at high frequency by helper phages, and therefore is not defective, and by implication, nor are the others defective. Rather than encoding unrecognizable TerS and Cpm proteins, SaPIbov5 contains neither. Rather, it is encapsidated by two different mechanisms, both different from the classical TerS/Cpm mechanism used by the previously characterized SaPIs. One of these mechanisms involves induction by pac type helper phages (ϕ11 and 80α), followed by packaging in full-sized phage particles, initiated by the phage terminase. This means that SaPIbov5 must have a pac site that is recognized by at least these two helper phages (which encode virtually identical TerS). The other mechanism involves induction and packaging by different helper phages, ϕ12 and ϕSLT, that we have shown to be cos phages. SaPIbov5 and phages ϕ12 and ϕSLT have similar cos sites and flanking sequences, which accounts for SaPIbov5 DNA encapsidation by ϕ12 and ϕSLT. Thus, SaPIbov5 has two unrelated encapsidation sequences to enable independent packaging by pac and cos phages.
Cos sites were also identified in several other SaPIs that lack terS and cpm, suggesting that cos packaging in full-sized phage particles is a widespread alternative strategy for the SaPIs, which is reminiscent of plasmid P4 encapsidation by coliphage P2 (22). However, in contrast to the P4–P2 interaction (22), SaPIbov5 does not induce its helper phage ϕ12. Interestingly, an evolutionary tree including SaPIs that use the cos and pac packaging strategies revealed that the cos SaPIs clustered together (SI Appendix, Fig. S7). However, some pac SaPIs (like SaPI2 or SaPI4) were more related to the cos SaPIs than to the other pac SaPIs. There are a number of possibilities to explain this. One is that a recombinatorial exchange of packaging modules between pac and cos SaPIs generated the different subbranches observed (SI Appendix, Fig. S7). Another is that some pac SaPIs have acquired a cos site, presumably from a cos phage or a cos-type SaPI and have lost the characteristic operon I present in the pac islands.
We have observed that an important feature of ϕSLT/SaPIbov5 packaging is the requirement for an HNH nuclease, which is encoded next to the ϕSLT terminase module. Proteins carrying HNH domains are widespread in nature, being present in organisms of all kingdoms. The HNH motif is a degenerate small nucleic acid-binding and cleavage module of about 30–40 aa residues and is bound by a single divalent metal ion (23). The HNH motif has been found in a variety of enzymes playing important roles in many different cellular processes, including bacterial killing; DNA repair, replication, and recombination; and processes related to RNA (23). HNH endonucleases are present in a number of cos-site bacteriophages of Gram-positive and -negative bacteria (16, 20), always adjacent to the genes encoding the terminases and other morphogenetic proteins. However, their biological role in the phage life cycle had not been determined.
We have demonstrated that the HNH nucleases encoded by ϕ12 and the closely related ϕSLT have nonspecific nuclease activity and are required for the packaging of these phages and of SaPIbov5. We have shown that HNH and TerL are jointly required for cos-site cleavage; as a different member of the HNH family has been shown to have single-stranded nicking activity (20), it is possible that cos-site cleavage in these phages involves the nicking of one strand by HNH and the other strand by TerL; because spontaneous melting of the 100% G + C cos-site duplex generated by staggered nicks would be inefficient, the HNH–TerS–TerL complex may also catalyze this melting, as occurs with the λ phage gpNu1-gpA complex (24). Alternatively, melting may be facilitated by a different protein, corresponding to the E. coli integration host factor (24).
We have also observed that only cos phages of Gram-negative as well as of Gram-positive bacteria encode HNH nucleases, consistent with a special requirement for cos-site cleavage as opposed to pac-site cleavage, which generates flush-ended products. The demonstration that HNH nuclease activity is required for some but not other cos phages suggests that there is a difference between the TerL proteins of the two types of phages—one able to cut both strands and the other needing a second protein to enable the generation of a double-stranded cut. The biological significance of these alternative packaging mechanisms remains to be determined. In addition, previous studies with phage SPP1 have demonstrated that the DNA end not used for encapsidation is degraded in vivo whereas the TerSL–DNA complex protects the DNA end that will be packaged. This protection mechanism, which is operative even in absence of subsequent genome packaging (25), is not yet understood. Because our experiments demonstrated that both ϕSLT DNA ends are resistant to host nucleases, it is tempting to speculate that the HNH–TerS–TerL complex can also be involved in this process.
It has been classically assumed that transfer of mobile genetic elements (MGEs) involves pac phages but not cos phages. Our results demonstrate that not only pac but also cos phages can have a relevant role in spreading MGEs with their virulence and resistance genes. Our study also establishes a strategy used by the SaPIs to hijack and interfere with the phage biology. HNH-mediated cos-site packaging thus represents a remarkable and novel mechanism of MGE transfer which may have important implications for phage-mediated horizontal gene transfer in general.
Materials and Methods
Bacterial Strains and Growth Conditions.
The bacterial strains used in these studies are listed in SI Appendix, Table S4. The procedures for preparation and analysis of phage lysates, in addition to transduction and transformation of Staphylococcus aureus, were performed essentially as previously described (5, 6, 26).
DNA Methods.
General DNA manipulations were performed using standard procedures. DNA samples were heated at 75 °C for 10 min before the electrophoresis to ensure cos-site melting. The plasmids and oligonucleotides used in this study are listed in SI Appendix, Tables S5 and S6, respectively. The labeling of the probes and DNA hybridization were performed according to the protocol supplied with the PCR-DIG DNA labeling and Chemiluminescent Detection Kit (Roche). To produce the phage and SaPI mutations, we used plasmid pMAD, as previously described (5).
Complementation of the Mutants.
The different phage genes under study were PCR amplified using the oligonucleotides listed in SI Appendix, Table S6. PCR products were cloned into pCN51 (S. aureus) (27) or pBAD18 (E. coli) (28) and the resulting plasmids (SI Appendix, Table S5) were introduced into the appropriate recipient strains (SI Appendix, Table S4).
Enzyme Assays.
β-Lactamase assays, using nitrocefin as substrate, were performed as described (5, 6), using a Thermomax (Molecular Devices) microtiter plate reader. Cells were obtained in exponential phase. β-Lactamase units are defined as (Vmax)/OD650.
Electron Microscopy.
Electron microscopy was performed as previsouly described (1) and performed at Centro de Investigación Príncipe Felipe (Valencia, Spain).
Structural Modeling of HNHϕSTL and TerLϕSTL Nucleases.
Models of the 3D structure of HNHϕSTL and TerLϕSTL nucleases were generated using the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) (13). Models with higher a C score (−0.55 and −1.21 for HNHϕSTL and TerLϕSTL, respectively) were selected for further structural analysis using Collaborative Computational Project No. 4 suite (29) and Crystallographic Object-Oriented Toolkit software (30).
Supplementary Material
Acknowledgments
This work was supported by Grants Consolider-Ingenio CSD2009-00006, BIO2011-30503-C02-01, and Eranet-Pathogenomics PIM2010EPA-00606 (to J.R.P.); Ministerio de Ciencia e Innovación Grant BIO2010-15424 (to A.M.); and National Institutes of Health Grant R01AI022159 (to R.P.N. and J.R.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320538111/-/DCSupplemental.
References
- 1.Ram G, et al. Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism. Proc Natl Acad Sci USA. 2012;109(40):16300–16305. doi: 10.1073/pnas.1204615109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ubeda C, et al. SaPI operon I is required for SaPI packaging and is controlled by LexA. Mol Microbiol. 2007;65(1):41–50. doi: 10.1111/j.1365-2958.2007.05758.x. [DOI] [PubMed] [Google Scholar]
- 3.Viana D, et al. Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol Microbiol. 2010;77(6):1583–1594. doi: 10.1111/j.1365-2958.2010.07312.x. [DOI] [PubMed] [Google Scholar]
- 4.Ubeda C, et al. SaPI mutations affecting replication and transfer and enabling autonomous replication in the absence of helper phage. Mol Microbiol. 2008;67(3):493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 5.Tormo-Más MÁ, et al. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature. 2010;465(7299):779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tormo-Más MÁ, et al. Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. Mol Cell. 2013;49(5):947–958. doi: 10.1016/j.molcel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 7.Tormo MA, et al. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J Bacteriol. 2008;190(7):2434–2440. doi: 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quiles-Puchalt N, Martínez-Rubio R, Ram G, Lasa I, Penadés JR. Unravelling bacteriophage ϕ11 requirements for packaging and transfer of mobile genetic elements in Staphylococcus aureus. Mol Microbiol. 2014;91(3):423–437. doi: 10.1111/mmi.12445. [DOI] [PubMed] [Google Scholar]
- 9.García P, et al. Functional genomic analysis of two Staphylococcus aureus phages isolated from the dairy environment. Appl Environ Microbiol. 2009;75(24):7663–7673. doi: 10.1128/AEM.01864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Augustin J, et al. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992;204(3):1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- 11.Ferrer MD, et al. RinA controls phage-mediated packaging and transfer of virulence genes in Gram-positive bacteria. Nucleic Acids Res. 2011;39(14):5866–5878. doi: 10.1093/nar/gkr158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quiles-Puchalt N, et al. A super-family of transcriptional activators regulates bacteriophage packaging and lysis in Gram-positive bacteria. Nucleic Acids Res. 2013;41(15):7260–7275. doi: 10.1093/nar/gkt508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu S-Y, et al. Structure determination and biochemical characterization of a putative HNH endonuclease from Geobacter metallireducens GS-15. PLoS ONE. 2013;8(9):e72114. doi: 10.1371/journal.pone.0072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen BW, et al. Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI. Structure. 2010;18(6):734–743. doi: 10.1016/j.str.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moodley S, Maxwell KL, Kanelis V. The protein gp74 from the bacteriophage HK97 functions as a HNH endonuclease. Protein Sci. 2012;21(6):809–818. doi: 10.1002/pro.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smits C, et al. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 2009;10(6):592–598. doi: 10.1038/embor.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sippy J, Feiss M. Initial cos cleavage of bacteriophage lambda concatemers requires proheads and gpFI in vivo. Mol Microbiol. 2004;52(2):501–513. doi: 10.1111/j.1365-2958.2004.03990.x. [DOI] [PubMed] [Google Scholar]
- 19.Chai S, et al. Molecular analysis of the Bacillus subtilis bacteriophage SPP1 region encompassing genes 1 to 6. The products of gene 1 and gene 2 are required for pac cleavage. J Mol Biol. 1992;224(1):87–102. doi: 10.1016/0022-2836(92)90578-8. [DOI] [PubMed] [Google Scholar]
- 20.Xu S-Y, Gupta YK. Natural zinc ribbon HNH endonucleases and engineered zinc finger nicking endonuclease. Nucleic Acids Res. 2013;41(1):378–390. doi: 10.1093/nar/gks1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muniesa M, Recktenwald J, Bielaszewska M, Karch H, Schmidt H. Characterization of a shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect Immun. 2000;68(9):4850–4855. doi: 10.1128/iai.68.9.4850-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindqvist BH, Dehò G, Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol Rev. 1993;57(3):683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H, Yuan HS. The conserved asparagine in the HNH motif serves an important structural role in metal finger endonucleases. J Mol Biol. 2007;368(3):812–821. doi: 10.1016/j.jmb.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 24.Yang Q, Catalano CE. Kinetic characterization of the strand separation (“helicase”) activity of the DNA packaging enzyme from bacteriophage lambda. Biochemistry. 1997;36(35):10638–10645. doi: 10.1021/bi970689t. [DOI] [PubMed] [Google Scholar]
- 25.Cornilleau C, et al. The nuclease domain of the SPP1 packaging motor coordinates DNA cleavage and encapsidation. Nucleic Acids Res. 2013;41(1):340–354. doi: 10.1093/nar/gks974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29(2):527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 27.Charpentier E, et al. Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl Environ Microbiol. 2004;70(10):6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.