Significance
This study expands our understanding of head motion in brain imaging. Complementing the common knowledge that motion artifacts can distort brain connectivity measures by causing artifacts, this study identifies a neurobiological trait that is linked to differences in motion. This correlate of head motion consists of reduced distant functional connectivity primarily in the default network areas in individuals with high head motion. Importantly, this correlate is stable within individuals across time. These findings may revise the interpretations of imaging findings in many neurodevelopmental, aging, and neuropsychiatric studies. In addition, differentiating the true disease effects from the correlates of motion tendency as reported here is critical for using connectivity markers in the clinical arena because correlates of motion may reduce specificity of biomarkers.
Keywords: resting-state fMRI, motion correction, connectome
Abstract
Individual differences in brain metrics, especially connectivity measured with functional MRI, can correlate with differences in motion during data collection. The assumption has been that motion causes artifactual differences in brain connectivity that must and can be corrected. Here we propose that differences in brain connectivity can also represent a neurobiological trait that predisposes to differences in motion. We support this possibility with an analysis of intra- versus intersubject differences in connectivity comparing high- to low-motion subgroups. Intersubject analysis identified a correlate of head motion consisting of reduced distant functional connectivity primarily in the default network in individuals with high head motion. Similar connectivity differences were not found in analysis of intrasubject data. Instead, this correlate of head motion was a stable property in individuals across time. These findings suggest that motion-associated differences in brain connectivity cannot fully be attributed to motion artifacts but rather also reflect individual variability in functional organization.
Head motion has long been known as a confounding factor in brain imaging including MRI (1, 2), PET (3, 4), single-photon emission computerized tomography (5, 6), and near infrared spectroscopy (7), but has raised particular concerns recently following the growing prominence of resting-state functional connectivity MRI. Studies found that head motion can vary considerably across individuals and often demonstrates systematic group effects when contrasting different populations, especially in neurodevelopmental (8–10), aging (11, 12), and neuropsychiatric studies (13). Some recent work reported that head motion augmented local coupling of the blood oxygenation level-dependent (BOLD) signal but reduced distant coupling (14–16). These correlations between connectivity measures and head motion have raised appropriate concern that previously observed differences in connectivity are due to artifact induced by differences in head motion. For example, developmental changes in functional connectivity might also be predicted by head motion (15). The assumption has been that head motion causes distorted connectivity measurements that must be addressed through improved motion-correction techniques (15). However, this correlation could be driven by causal factors in the other direction. Specifically, individual differences in brain connectivity could determine how well a subject can lie still in the scanner. This is not unreasonable as individual differences in structural connectivity can predict trait anxiety and can be related to attention deficits (17, 18) and individual differences in resting-state functional MRI (fMRI) measures may relate to various behavioral differences, including impulsivity (19–22). In such a scenario, certain intersubject differences in connectivity measures could persist even after the most rigorous motion correction, as has been suggested in several earlier studies (23, 24).
To explore the relation between head motion and brain connectivity, we examined functional connectivity in different subject groups selected on the basis of head motion parameters from a large database of 3,000+ participants, many of whom were scanned multiple times. These cohorts allowed us to compare intersubject and intrasubject differences in connectivity in high- versus low-motion scans. If motion causes connectivity differences, these should be similar both inter- and intrasubject. However, if connectivity differences include a stable trait that predisposes to head motion, then these differences should be present between subjects but not within subjects.
Results
We first investigated whether head motion is a reliable behavioral measure across MRI sessions. In 118 subjects scanned on 2 separate days (mean interval 128.59 ± 151.34 d), head motion across sessions was significantly correlated (r = 0.54, P < 0.001, Fig. S1). This reproducibility suggests that the tendency to move is generally a stable trait across subjects (16).
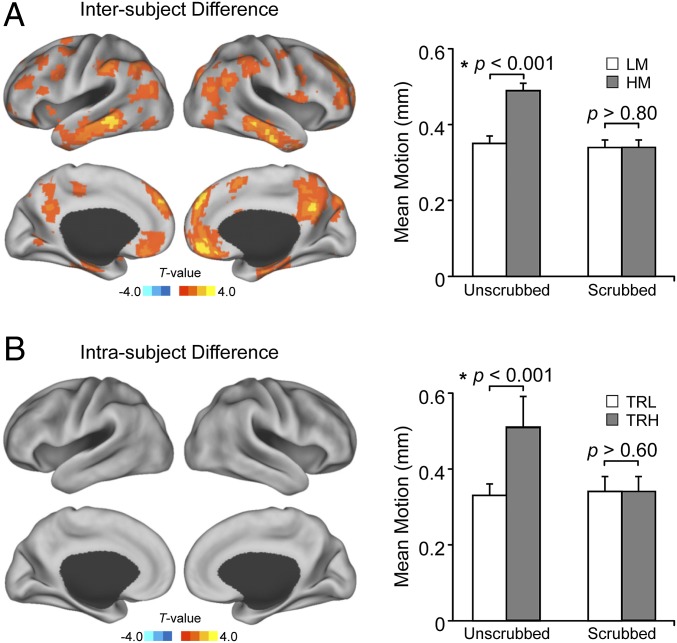
Next we investigated the intersubject difference in connectivity. First, we carefully selected 26 pairs of healthy young adults who were matched in all scanning parameters and demographics but differed in the averaged head motion (0.049 vs. 0.035 mm, P < 0.001, Table S1). To ensure that functional connectivity can be accurately estimated without being confounded by motion-related artifacts, image frames with a motion level (root mean square) exceeding 0.06 mm were scrubbed from the high-motion subjects (15), and then the same time points were also removed from the matched low-motion subjects. Therefore, the frames removed in the low-motion subjects were not necessarily the motion-contaminated frames (Materials and Methods). After this scrubbing operation, no difference in head motion was found between these two groups (0.034 vs. 0.034 mm, P > 0.80, Table S1). The scrubbing had resulted in a loss of 23.5% of the data in both the high-motion and low-motion subjects, but all subjects included in the further analyses had at least 120 time points retained. The degree of distant connectivity was then calculated for each voxel by counting the number of voxels outside the immediate neighborhood (a sphere with the radius of 12 mm) (25) that were significantly correlated with the seed voxel [exceeding the threshold of P < 0.01 after the false discovery rate (FDR) correction]. Distant functional connectivity in a distributed set of regions, primarily in the default network, was found to be significantly greater in the 26 subjects with low head motion than in the 26 matched subjects with high head motion (P < 0.05, FDR-corrected, two-tailed, two-sample t test, Fig. 1A). These regions can be considered a potential marker of head motion.
Fig. 1.
Functional connectivity in a set of marker regions is a trait that predisposes to head motion but remains stable within subjects. (A) Distant connectivity in some cognitive regions (shown in red and yellow), mainly in the default network, was significantly greater in 26 subjects with low head motion than in 26 matched subjects with high motion (P < 0.05, FDR-corrected, two-tailed, two-sample t test). The motion level of the two groups was significantly different before data scrubbing (P < 0.001) but was the same after scrubbing (P > 0.80) (Right). (B) In an independent dataset of 26 subjects who exhibited a different level of sporadic head motion in two scanning sessions, no significant difference in distant connectivity was observed between the low-motion session and the high-motion session (no voxels reached the significance level of P < 0.05, FDR-corrected, two-tailed paired t test). The motion level of the two sessions was also significantly different before data scrubbing (P < 0.001) but was the same after scrubbing (P > 0.80) (Right). The motion level of this test–retest dataset was also matched to that of intersubject dataset (discovery sample). LM/HM, low-/high-motion individuals in the discovery sample; TRL/TRH, low-/high-motion sessions in the test–retest data.
Next we compared these intersubject differences in motion-related connectivity to intrasubject differences. We selected an independent dataset of 26 subjects who were scanned twice on separate days (mean interval 114.50 ± 126.52 d) but exhibited more sporadic head motion in one scanning session than in the other (Table S1). The motion level of this intrasubject dataset was matched to that of the intersubject dataset: there was no significant difference between the two high-motion groups (motion before scrubbing: 0.049 vs. 0.051 mm, P = 0.24; motion after scrubbing: 0.034 vs. 0.034 mm, P = 0.79) or between the two low-motion groups (motion before scrubbing: 0.035 vs. 0.033 mm, P = 0.06; motion after scrubbing: 0.034 vs. 0.034 mm, P = 0.44). Any residual motion artifacts not removed by motion-correction techniques including scrubbing should be similar in the intersubject and intrasubject comparisons. However, potential neurobiological traits should show up only in the intersubject comparison.
Unlike the analysis of the intersubject dataset, no difference in distant connectivity was found between the low-motion sessions and the high-motion sessions of the same group of subjects (no voxel reached the significance level of P < 0.05, FDR-corrected, two-tailed paired t test, Fig. 1B). This result suggests that the intersubject difference shown in Fig. 1A likely includes a neurobiological trait effect beyond a residual motion artifact (see SI Text for discussions on the potential influence of anatomical variability).
To confirm that this neurobiological marker of head motion is not specific to one subject cohort, we replicated our findings in an independent dataset consisting of 30 pairs of demographically matched subjects (r = −0.39, P < 0.005, Fig. S2). We also used this replication dataset to confirm that regions showing the trait effect localized primarily to the default network (see SI Text and Fig. S3).
Although scrubbing is a useful method to minimize the impact of motion-related artifacts and improve distant coupling (15), this operation will cause loss of data and could lead to a biased estimate of distant connectivity. We found that the loss of data could inflate distant coupling, but primarily in the visual cortices and some posterior parietal regions (SI Text and Fig. S4). In the present results, we always scrubbed the data of matched groups in the same way; therefore, the inflation of distant coupling would have minimal impact on our results. However, to further examine whether the results in Fig. 1 were affected by data scrubbing, we repeated all analyses with unscrubbed data. The findings that distant connectivity in the potential biomarker regions correlates with intersubject difference (r = −0.80, P < 0.001), but not intrasubject variation in head motion (r = −0.14, P > 0.30), were largely replicated (see Fig. S5 for the inter- and intrasubject differences). In addition, the findings were replicated in a subset of the subjects matched based on a different scrubbing threshold (SI Text and Fig. S6), suggesting that this connectivity correlate of the trait for head motion is not contingent upon data scrubbing.
Although the image frames immediately contaminated by motion artifacts were removed, potential confounds might still arise from the movement-induced BOLD response, which could have a delay of ∼10 s after the movement. To investigate how the delayed BOLD response may affect the connectivity marker, we performed a stringent scrubbing procedure on the data of 56 pairs of matched subjects (including the discovery sample and the 30 pairs of subjects in the replication samples). Whenever the head motion exceeded a threshold of 0.06 mm, two preceding frames (6 s) and five succeeding frames (15 s) were removed in addition to the noisy frames, ensuring that the delayed BOLD responses and other forms of extended motion effect were largely eliminated from the data. Taking the marker regions (shown in Fig. 1A) as a mask (binarized as shown in Fig. S7A), distant connectivity was computed based on the stringently scrubbed data and then averaged within the mask. Distant connectivity derived from the stringently scrubbed data still strongly predicted head motion (r = −0.67, P < 0.001, Fig. S7B), indicating that the delayed BOLD responses and temporally-extended artifacts had a minimal effect on the connectivity–motion correlation. In a further control analysis, we treated the time course of mean motion of each subject as the representation of a continuous movement task and then convolved this time course with the canonical hemodynamic response function. General Linear Model (GLM) analysis was performed to identify regions activated by this “continuous movement task,” and then the maps were compared between the low-motion subjects and the high-motion subjects. No significant difference was observed between the two matched groups (no voxel reached the significance threshold of P < 0.05, FDR-corrected), indicating that the BOLD response induced by continuous movement may not account for the group difference identified in Fig. 1A.
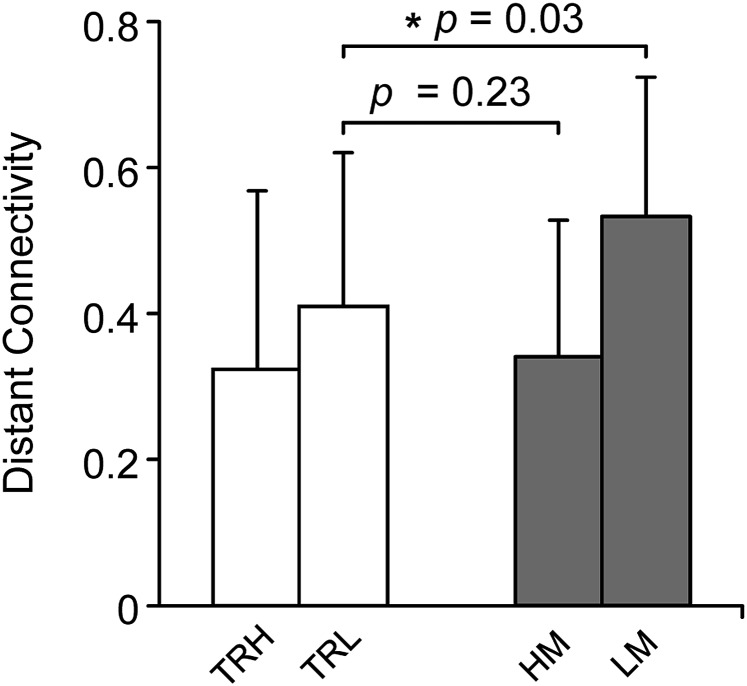
We found that the marker for the tendency of head motion remained stable within individuals, although the intrasubject variability of motion over time could be large in some cases. If these subjects showed little head motion in one session but were not able to refrain from moving in another session, a further question is whether they possess a connectivity signature similar to those who move excessively or similar to those who consistently move less. To address this question, we compared the 26 test–retest subjects with high intrasubject variability of motion to the discovery sample. Distant connectivity was averaged in a mask derived from the independent replication sample by comparing the 30 low-motion with the 30 high-motion subjects (P < 0.05, FDR-corrected, two-sample t test). In the test–retest subjects, distant connectivity derived from the low-motion session was significantly weaker than the low-motion subjects of the discovery sample (P = 0.03, two-sample t test, Fig. 2), but similar to the high-motion subjects (P = 0.23, two-sample t test, Fig. 2). These findings suggested that the subjects who could not persistently remain still had a neural connectivity signature resembling those who tend to have excessive motion, even during the scan when they were still. The findings that the two groups with matched head motion showed a significant difference in distant connectivity, whereas the two groups with significantly different motion showed no difference in connectivity, further supported that the trait effect is dominant over residual motion artifacts after appropriate corrections. Although intrasubject variability of head motion was seen in these few subjects, the majority of the sample showed a consistent level of head motion (as shown in Fig. S1). We examined the distant connectivity in 18 test–retest subjects with consistent levels of head motion (motion difference <0.01 mm between two sessions). In these subjects, distant connectivity averaged in the marker region was significantly correlated with the head motion both in session 1 (r = −0.50, P = 0.03) and in session 2 (r = −0.51, P = 0.04) (Fig. S8), and the correlation was not different between the two sessions (ANCOVA, P > 0.95). These results indicate that the connectivity marker could reliably predict head motion in subjects with a consistent level of head motion, who constitute the majority of the population.
Fig. 2.
Distant connectivity in the test–retest subjects demonstrating higher variability in head motion resembled those subjects with excessive motion. Distant connectivity was averaged in a mask derived from the independent replication sample by comparing the low-motion with the high-motion subjects (P < 0.05, FDR-corrected, two-sample t test). Distant connectivity in the 26 test–retest subjects was significantly different from the 26 subjects in the discover sample with low head motion (P = 0.03), but similar to the 26 subjects with high head motion (P = 0.23).
Discussion
The present study suggests a correlate of head motion during brain imaging that reflects a neurobiological trait rather than simply a technical artifact. The bidirectional relationship between head motion and brain connectivity could have substantial implications for functional connectivity studies.
Distant connectivity in a distributed set of cortical regions, primarily in the default network, was significantly stronger in subjects with low head motion than in subjects with high head motion. Because similar differences were not seen with intrasubject differences in head motion, this connectivity property appears to be a neurobiological trait of head motion tendency. Special attention is needed when interpreting findings of individual differences that overlap with these marker regions. For example, alterations of connectivity in the default network have been suggested in many neuropsychiatric and neurological diseases (26). Determining whether these changes are specific to a disorder or a secondary effect of factors that relates to the tendency of head motion is critical for identifying the treatment target and the development of treatment strategy. If head motion is generally elevated in a patient population, a disease biomarker independent from the marker of motion as identified in the current study might be expected to provide better specificity.
It is intriguing that the marker for the tendency of motion involves the association cortices but not the primary sensorimotor cortex. The default network has been suggested to subserve internal mentation (e.g., ref. 27). Strong distant connectivity within this network may reflect the effective monitoring of internal states. Our data indicated that subjects with greater distant connectivity in the marker region could consistently refrain from moving, whereas subjects with weak distant connectivity in the marker region could not persistently remain still. Head motion therefore may be an indicator of specific cognitive control capacity in the individual brain. Future investigations may explore whether this control of motor behavior or some other indirect correlate of head motion (e.g., respiration) is predictive of other higher cognitive capabilities that are subserved by the same brain regions.
Remarkably, the connectivity marker for the tendency of head motion was stable within individuals. The fact that distant connectivity in the default network explains intersubject difference but not the intrasubject variability in head motion strongly suggests that the marker identified in this study was not due to motion artifacts. This finding has direct implications for the development of motion-correction techniques.
Although the connectivity marker reported in the present study correlates with interindividual difference in head motion and may reflect a trait of motor restlessness, the results should be interpreted with some caveats. First, these results should not be taken as an argument against motion correction. The cause-and-effect relationship between functional connectivity and head motion is two-way. The difference in motion artifacts can lead to the difference in connectivity estimates whereas the neural connectivity difference could also cause difference in motion. One should not simply focus on one side of this relationship and ignore the other. Although interindividual differences in connectivity as shown in Fig. 1A are dominated by the trait effect, it is likely that this map includes a residual motion artifact that persists despite rigorous scrubbing. Our analyses suggest that any remaining artifact effect is small relative to the trait effect (Fig. 1B and Fig. S5B); however, full dissociation of artifact and trait effects may be possible with future advances in motion-correction technology. Some recent development in this direction, such as a more specific model of the motion artifact (28) or separating the artifact component from the neural component based on fMRI data acquired using multiple echo times (TEs) (29), could eventually improve the specificity of biomarkers. Second, our analyses were based on a specific type of connectivity measure, distant connectivity, and therefore the results should not be extrapolated to all types of connectivity measures without careful examination. Finally, the influence of physiological functions such as respiratory and cardiac rates was not monitored in the current study (30). The aliased respiration- and cardiac-related low-frequency variations, as well as the arterial carbon dioxide concentration and blood pressure variations, cannot be removed by the low-pass filtering strategy (31). These physiological processes may also have neurobiological correlates (31). Although the marker regions for the tendency of head motion identified here are largely distinct from the regions demonstrating significant respiratory-induced BOLD signal change (32, 33), the potential physiological confounds on the connectivity marker as reported in this study warrant further investigation.
Materials and Methods
Participants.
Data were acquired as part of the Brain Genomics Superstruct Project. Participants were enrolled by multiple local laboratories, all acquiring similar data on matched 3 Tesla MRI scanners located at Harvard and at the Massachusetts General Hospital. All participants were native English speakers and had normal or corrected-to-normal vision. Participants were excluded with a history of neurological or psychiatric illness. Data were selected using the following criteria: 12-channel head coil, age between 18 and 35 y, and fMRI slice-based temporal signal-to-noise ratio >100. All participants provided written informed consent in accordance with guidelines set by Institutional Review Boards of Harvard University or Partners Healthcare. Portions of these data have been previously reported (16, 34, 35).
Datasets.
The analyses described in this article used three separate resting-state fMRI datasets. Unless specifically noted, these datasets were treated identically in processing and analysis.
Dataset 1 consisted of a total of 56 pairs of subjects from a pool of 3,000+ participants (34); each pair of subjects was matched in scanner, coil, scanning date (difference <105 d), ethnicity, sex (37F/19M), handedness (5L/51R), age (≤3 y, P > 0.80), and education (≤3 y, P > 0.95), but not in head motion (mean motion: 0.036 ± 0.002 vs. 0.053 ± 0.009 mm, P < 0.001). All subjects had two BOLD runs (each run had 124 time points). Twenty-six pairs were regarded as the discovery sample (Table S1), and the remaining 30 pairs were used as the replication sample.
Dataset 2 consisted of a total of 26 test–retest subjects with inconsistent levels of head motion during two scanning sessions (sex: 17 females/9 males; handedness: 2L/24R; age: 20.3 ± 2.38 y; education: 14.04 ± 1.89 y; mean motion difference >0.01 mm between the low-motion sessions and high-motion sessions, P < 0.001), and each session had one or two BOLD runs. The interval between two sessions was 114.50 ± 126.52 (between 2 and 446) days, and only three subjects had a delay exceeding 1 y. The dataset was matched with the discovery sample in demographics as well as the head motion before and after motion scrubbing (Table S1).
Dataset 3 consisted of a total of 18 test–retest subjects with consistent level of head motion (sex: 9F/9M; handedness: 5L/13R; age: 19.33 ± 1.71 y; education: 13.44 ± 1.62 y; mean motion: 0.039 ± 0.013 mm; mean motion difference <0.01 mm between two sessions, P > 0.95), and each session has one or two runs.
Data Acquisition and Preprocessing.
All data were collected on matched 3T Tim Trio scanners (Siemens) using a 12-channel phased-array head coil. Images were acquired using a gradient-echo echo-planar pulse sequence sensitive to BOLD contrast [repetition time (TR) = 3,000 ms, TE = 30 ms, flip angle = 85°, 3-mm isotropic voxels, field of view (FOV) = 216; 47 slices collected with interleaved acquisition with no gap between slices]. Slices were aligned to the anterior commissure–posterior commissure plane using an automated alignment procedure to ensure consistency across subjects (36). Each resting-state (eyes open) fMRI run lasted 6 min and 12 s. Structural data used a high-resolution multiecho T1-weighted magnetization-prepared gradient-echo image (TR = 2,200 ms; inversion time = 1,100 ms; TE = 1.54 ms for image 1 to 7.01 ms for image 4; flip angle = 7°; 1.2-mm isotropic voxels; and FOV = 230). Subjects were instructed to stay awake, keep their eyes open, and minimize head movement; no other task instruction was provided.
Resting-state fMRI data were processed using previously described procedures (16). The first four volumes of each run were discarded to allow for T1-equilibration effects. The following steps were performed: (i) slice timing correction (SPM2, Wellcome Department of Cognitive Neurology, London); (ii) rigid body correction for head motion with the FSL package (Functional MRI of the Brain, Oxford); (iii) atlas registration with an EPI template in the Montreal Neurological Institute atlas space, resampling to 2-mm isotropic voxels and spatially smoothing using a 6-mm full-width half-maximum Gaussian kernel; (iv) normalization for global mean signal intensity across runs; (v) linear detrend and low-pass temporal filtering (<0.8 Hz); and (vi) regression of nuisance variables including the six parameters obtained by rigid-body head-motion correction, global signal, ventricular and white matter signals, and the first temporal derivatives of all of the above.
Motion Scrubbing.
In the current study, we used mean motion as the central metric of head motion (15), which represents the mean absolute displacement of each brain volume compared with the previous volume and was estimated from the translation parameters in the x (left/right), y (anterior/posterior), and z (superior/inferior) directions.
To ensure that the data were minimally affected by motion-related artifacts, motion scrubbing was used to remove frames with high motion (15). Importantly, head motion was matched after scrubbing across all groups including the low-motion and high-motion groups in the discovery sample, replication sample, and test–retest samples (Table S1). In addition, head motion was further matched across the high-motion datasets before scrubbing. The details are described below.
For dataset 1 (discovery sample and replication sample), frames with a motion level exceeding 0.06 mm were scrubbed from the high-motion data, and then the same time points were scrubbed from the matched low-motion data. Therefore, the frames removed in the low-motion subjects were not necessarily the frames of higher motion. Removing the same frames in the high-motion and the low-motion data ensured that the effects caused by data loss were comparable in these two groups. After scrubbing, the low-motion data and high-motion data were matched in head motion (P > 0.80). This operation had resulted in an exclusion of 23.5% of the data, but all subjects included in the analyses had at least 120 time points left.
Similarly, the frames to be removed were based on the high-motion sessions exceeding thresholds for dataset 2 (26 test–retest sample), and the corresponding time points were also scrubbed from the low-motion sessions. After scrubbing, the low-motion sessions and the high-motion sessions were matched in head motion (P > 0.60). This operation resulted in an exclusion of 26.0% of the data, but all sessions included in the analyses had at least 80 time points left.
Frames with a motion level exceeding 0.06 mm were removed for both sessions of dataset 3 (18 test–retest sample). After scrubbing, each subject still showed a consistent level of head motion between two sessions (mean motion difference <0.01 mm, P > 0.90). This operation resulted in an exclusion of 12.4% of the data, but all sessions included in the analyses had at least 100 time points left.
Characterizing Distant Functional Connectivity with Distant Degree Connectivity Map.
Degree of distant functional connectivity was calculated for each subject or session as described in detail elsewhere (25). Degree centrality is a network measure that quantifies the number of edges that are connected to a node in a graph (37). Here brain voxels are the nodes, and positive correlations between voxels with P < 0.01 (FDR-corrected) are the edges. To speed up the computation, data were down-sampled to 4-mm isotropic voxels. Distant connectivity was calculated for each voxel by counting the number of edges to other voxels outside the immediate neighborhood (12-mm radius). The output was a whole-brain distant degree connectivity map for each subject. The maps were then standardized by Z-score transformation so that maps across participant maps could be compared (25, 38). In the present study, all of the difference maps (as shown in Fig. 1 and Figs. S4 and S5) were whole-brain–corrected for multiple comparisons at a significance level of P < 0.05 via FDR.
Estimating Intersubject Difference in Distant Connectivity Related to Head Motion.
Distant connectivity map was computed for each subject of the discovery sample. To estimate intersubject difference in distant connectivity related to head motion, the distant connectivity maps of the 26 low-motion subjects were compared with those of the 26 high-motion subjects of the discovery sample by two-tailed, two-sample t tests (Fig. 1A).
Replication analysis was conducted on the replication sample consisting of 30 pairs of subjects from dataset 1. The distant connectivity map was computed for each subject. Taking the brain regions showing significant intersubject connectivity difference in the discover study as a mask, distant connectivity was averaged within the mask for each subject, and then correlation analysis was performed between the distant connectivity and head motion (Fig. S2A).
Estimating Intrasubject Variation in Distant Connectivity Related to Head Motion.
Dataset 2, consisting of 26 test–retest subjects with an inconsistent level of head motion in two scanning sessions, was used to estimate intrasubject variation in distant connectivity related to head motion. The distant connectivity map was computed for each session, and the maps of the low-motion sessions were compared with those of the high-motion sessions by two-tailed paired t tests (Fig. 1B). Furthermore, taking the brain regions showing significant intersubject connectivity difference in the discover study as a mask, correlation analysis was performed between the distant connectivity averaged within the mask and head motion for these test–retest subjects (Fig. S2B).
Scrubbing Effect Estimation and Replication of the Main Finding in the Unscrubbed Data.
Motion scrubbing will cause the loss of data and may introduce spurious distant connectivity. To understand the impact of scrubbing on distant connectivity, we compared the distant connectivity maps of the scrubbed data to those of the unscrubbed data based on the low-motion subjects in the discovery sample using two-tailed paired t tests (Fig. S4A). Similar analysis was conducted on the data of the low-motion sessions of the test–retest subjects (Fig. S4B). To further test the reliability of the main finding, we used the unscrubbed data to estimate intersubject and intrasubject difference in distant connectivity related to head motion (Fig. S5).
Testing the Impact of Movement-Induced BOLD Responses on Intersubject Difference in Distant Connectivity Related to Head Motion.
To minimize the impact of movement-induced BOLD responses on connectivity measurements, a stringent motion scrubbing was performed. When head motion exceeded the threshold of 0.06 mm, two preceding frames (6 s) and five succeeding frames (15 s) were removed in addition to the immediate frames to ensure the delayed BOLD responses were eliminated from the data. Due to limited sample size, the entire dataset 1, including 56 pairs of matched subjects, was included. After the stringent scrubbing, only 26 pairs of subjects had no less than 80 time points and were used in the following analysis. Among them, 14 pairs of subjects were overlapping with the subjects in the discovery sample. Distant connectivity was computed based on the stringently scrubbed data and averaged within the mask described above. Correlation analysis was then performed between the distant connectivity and head motion. In the next control analysis, the mean motion curve of each subject was treated as the stimulus function and then was convolved with the canonical hemodynamic response function. The GLM analyses were performed on the unscrubbed data of the discovery sample without temporal filtering and regression processing. Brain regions where the BOLD signals corresponded to the response induced by the head movement were identified, and then the maps were compared between the low-motion subjects and high-motion subjects.
Characterizing the Distant Connectivity in the Test–Retest Subjects.
To characterize the distant connectivity in the 26 test–retest subjects with inconsistent level of head motion, we compared the distant connectivity of the test–retest subjects with that of the discover sample. Distant connectivity was averaged in the mask described above and then was compared with that of the 26 subjects with low head motion and that of the 26 subjects with high head motion (Fig. 2).
Distant connectivity was then examined in 18 test–retest subjects with consistent level of head motion in two sessions (dataset 3). Correlation analysis was performed between the distant connectivity averaged in the mask described above and head motion for two scanning sessions, respectively. The connectivity–motion correlation in two sessions was compared using ANCOVA (Fig. S8).
Visualization.
All imaging results were projected onto the inflated PALS cortical surface using CARET for the purpose of visualization (39).
Supplementary Material
Acknowledgments
Data were provided by the Brain Genomics Superstruct Project of Harvard and Massachusetts General Hospital (R.L.B., J. L. Roffman, and J. W. Smoller). This work was supported by National Institute of Neurological Disorders and Stroke Grant K25NS069805, National Alliance for Research on Schizophrenia and Depression Young Investigator Grant, National Basic Research Program of China (2011CB707802), and the Postgraduate Study Abroad Program of China Scholarship Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317424111/-/DCSupplemental.
References
- 1.Friston KJ, Williams S, Howard R, Frackowiak RSJ, Turner R. Movement-related effects in fMRI time-series. Magn Reson Med. 1996;35(3):346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- 2.Rohde GK, Barnett AS, Basser PJ, Marenco S, Pierpaoli C. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn Reson Med. 2004;51(1):103–114. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- 3.Catana C, et al. MRI-assisted PET motion correction for neurologic studies in an integrated MR-PET scanner. J Nucl Med. 2011;52(1):154–161. doi: 10.2967/jnumed.110.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picard Y, Thompson CJ. Motion correction of PET images using multiple acquisition frames. IEEE Trans Med Imaging. 1997;16(2):137–144. doi: 10.1109/42.563659. [DOI] [PubMed] [Google Scholar]
- 5.Fulton RR, Eberl S, Meikle SR, Hutton BF, Braun M. A practical 3D tomographic method for correcting patient head motion in clinical SPECT. IEEE Trans Nucl Sci. 1999;46(3):667–672. [Google Scholar]
- 6.Kyme AZ, Hutton BF, Hatton RL, Skerrett DW, Barnden LR. Practical aspects of a data-driven motion correction approach for brain SPECT. IEEE Trans Med Imaging. 2003;22(6):722–729. doi: 10.1109/TMI.2003.814790. [DOI] [PubMed] [Google Scholar]
- 7.Robertson FC, Douglas TS, Meintjes EM. Motion artifact removal for functional near infrared spectroscopy: A comparison of methods. IEEE Trans Biomed Eng. 2010;57(6):1377–1387. doi: 10.1109/TBME.2009.2038667. [DOI] [PubMed] [Google Scholar]
- 8.Dosenbach NUF, et al. Prediction of individual brain maturity using fMRI. Science. 2010;329(5997):1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fair DA, et al. The maturing architecture of the brain’s default network. Proc Natl Acad Sci USA. 2008;105(10):4028–4032. doi: 10.1073/pnas.0800376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fair DA, et al. Development of distinct control networks through segregation and integration. Proc Natl Acad Sci USA. 2007;104(33):13507–13512. doi: 10.1073/pnas.0705843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrews-Hanna JR, et al. Disruption of large-scale brain systems in advanced aging. Neuron. 2007;56(5):924–935. doi: 10.1016/j.neuron.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damoiseaux JS, et al. Reduced resting-state brain activity in the “default network” in normal aging. Cereb Cortex. 2008;18(8):1856–1864. doi: 10.1093/cercor/bhm207. [DOI] [PubMed] [Google Scholar]
- 13.Greicius M. Resting-state functional connectivity in neuropsychiatric disorders. Curr Opin Neurol. 2008;21(4):424–430. doi: 10.1097/WCO.0b013e328306f2c5. [DOI] [PubMed] [Google Scholar]
- 14.Satterthwaite TD, et al. Impact of in-scanner head motion on multiple measures of functional connectivity: Relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60(1):623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59(3):2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Dijk KRA, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59(1):431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashtari M, et al. Attention-deficit/hyperactivity disorder: A preliminary diffusion tensor imaging study. Biol Psychiatry. 2005;57(5):448–455. doi: 10.1016/j.biopsych.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 18.Kim MJ, Whalen PJ. The structural integrity of an amygdala-prefrontal pathway predicts trait anxiety. J Neurosci. 2009;29(37):11614–11618. doi: 10.1523/JNEUROSCI.2335-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis FC, et al. Impulsivity and the modular organization of resting-state neural networks. Cereb Cortex. 2013;23(6):1444–1452. doi: 10.1093/cercor/bhs126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koyama MS, et al. Resting-state functional connectivity indexes reading competence in children and adults. J Neurosci. 2011;31(23):8617–8624. doi: 10.1523/JNEUROSCI.4865-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jäncke L, Langer N, Hänggi J. Diminished whole-brain but enhanced peri-sylvian connectivity in absolute pitch musicians. J Cogn Neurosci. 2012;24(6):1447–1461. doi: 10.1162/jocn_a_00227. [DOI] [PubMed] [Google Scholar]
- 22.Cole MW, Yarkoni T, Repovs G, Anticevic A, Braver TS. Global connectivity of prefrontal cortex predicts cognitive control and intelligence. J Neurosci. 2012;32(26):8988–8999. doi: 10.1523/JNEUROSCI.0536-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fair DA, et al. Distinct neural signatures detected for ADHD subtypes after controlling for micro-movements in resting state functional connectivity MRI data. Front Syst Neurosci. 2012;6:80. doi: 10.3389/fnsys.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satterthwaite TD, et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sepulcre J, et al. The organization of local and distant functional connectivity in the human brain. PLOS Comput Biol. 2010;6(6):e1000808. doi: 10.1371/journal.pcbi.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox MD, Greicius M. Clinical applications of resting state functional connectivity. Front Syst Neurosci. 2010;4:19. doi: 10.3389/fnsys.2010.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews-Hanna JR. The brain’s default network and its adaptive role in internal mentation. Neuroscientist. 2012;18(3):251–270. doi: 10.1177/1073858411403316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan CG, et al. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kundu P, et al. Integrated strategy for improving functional connectivity mapping using multiecho fMRI. Proc Natl Acad Sci USA. 2013;110(40):16187–16192. doi: 10.1073/pnas.1301725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birn RM, Smith MA, Jones TB, Bandettini PA. The respiration response function: The temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage. 2008;40(2):644–654. doi: 10.1016/j.neuroimage.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy K, Birn RM, Bandettini PA. Resting-state fMRI confounds and cleanup. Neuroimage. 2013;80:349–359. doi: 10.1016/j.neuroimage.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C, Glover GH. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. Neuroimage. 2009;47(4):1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21(4):1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Wang D, Buckner RL, Liu H. Cerebellar asymmetry and its relation to cerebral asymmetry estimated by intrinsic functional connectivity. J Neurophysiol. 2013;109(1):46–57. doi: 10.1152/jn.00598.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo BTT, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Kouwe AJW, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. Neuroimage. 2008;40(2):559–569. doi: 10.1016/j.neuroimage.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage. 2010;52(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Buckner RL, et al. Cortical hubs revealed by intrinsic functional connectivity: Mapping, assessment of stability, and relation to Alzheimer’s disease. J Neurosci. 2009;29(6):1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Essen DC. A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. Neuroimage. 2005;28(3):635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.