Significance
The design of protein receptors responsive to light stimuli has wide-ranging applications in structure–function studies and optogenetics. We present here the design of a light-sensitive NMDA receptor by introducing through genetic-code expansion a photoreactive moiety carried by a single unnatural amino acid. NMDA receptors form glutamate-gated ion channels that are essential mediators of synaptic plasticity associated with learning and memory. By exploring the molecular mechanisms and subunit dependence of the light-induced receptor inactivation, we find that the two major NMDA receptor subtypes, GluN1/GluN2A and GluN1/GluN2B, differ in their ectodomain-subunit interactions. Besides providing important new information about NMDA receptor molecular structure, our work opens the possibility of using engineered light-sensitive NMDA receptors to explore native receptor function.
Keywords: neurotransmitter receptor, protein structure–function
Abstract
Reprogramming receptors to artificially respond to light has strong potential for molecular studies and interrogation of biological functions. Here, we design a light-controlled ionotropic glutamate receptor by genetically encoding a photoreactive unnatural amino acid (UAA). The photo–cross-linker p-azido-l-phenylalanine (AzF) was encoded in NMDA receptors (NMDARs), a class of glutamate-gated ion channels that play key roles in neuronal development and plasticity. AzF incorporation in the obligatory GluN1 subunit at the GluN1/GluN2B N-terminal domain (NTD) upper lobe dimer interface leads to an irreversible allosteric inhibition of channel activity upon UV illumination. In contrast, when pairing the UAA-containing GluN1 subunit with the GluN2A subunit, light-dependent inactivation is completely absent. By combining electrophysiological and biochemical analyses, we identify subunit-specific structural determinants at the GluN1/GluN2 NTD dimer interfaces that critically dictate UV-controlled inactivation. Our work reveals that the two major NMDAR subtypes differ in their ectodomain-subunit interactions, in particular their electrostatic contacts, resulting in GluN1 NTD coupling more tightly to the GluN2B NTD than to the GluN2A NTD. It also paves the way for engineering light-sensitive ligand-gated ion channels with subtype specificity through the genetic code expansion.
Introducing light-sensitive moieties into proteins provides a powerful approach to investigate molecular mechanisms as well as biological functions with high temporal and spatial resolution (1, 2). An attractive strategy to engineer light responsiveness relies on the use of photoreactive unnatural amino acids (UAAs), allowing site-specific incorporation in a protein target. The methodology relies on the read-through of an unassigned codon (commonly the amber stop codon) in an mRNA by a suppressor tRNA aminoacylated with a desired UAA. Using this approach, UAAs with unique chemical functionalities including light-sensitivity have been successfully incorporated into ion channels and neurotransmitter receptors, significantly contributing to our understanding of receptor function (3, 4). However, the challenging synthesis of the chemically acylated tRNA has limited the general applicability of the approach. The recent development of genetically engineered suppressor tRNA/aminoacyl-tRNA synthetase pairs with altered amino acid specificity allowed for aminoacylation in the expression system in situ. This method provided a major step forward by advancing the UAA technology to the all-genetic–based level, also known as “the genetic-code expansion” (5–7).
Here, we present the design of a light-sensitive ionotropic glutamate receptor (iGluR) through the genetic incorporation of a photoreactive UAA. Our approach takes advantage of the recent development of the genetic-code expansion in Xenopus oocytes (8), which is a classical vehicle for heterologous expression and functional characterization of ligand-gated ion channels (LGICs). We focused on NMDA receptors (NMDARs), which play pivotal roles in brain physiology and pathology (9). NMDARs are obligatory heterotetramers commonly composed of two glycine-binding GluN1 subunits and two glutamate-binding GluN2 subunits. Although GluN1 is encoded by a single gene, there are four types of GluN2 subunits (GluN2A to -D) encoded by four different genes, which endow NMDARs with different properties including channel open probabilities (Po) and sensitivities to allosteric modulators (9). The extracellular region of both GluN1 and GluN2 subunits consists of a tandem of large clamshell-like domains comprising an N-terminal domain (NTD) and an agonist-binding domain (ABD) (Fig. 1A). Besides having essential functions in receptor assembly (10, 11), recent studies of the NTDs have also revealed that the individual GluN2 (12–14) and GluN1 (15) NTDs fine-tune NMDAR gating and pharmacological properties by undergoing large-range conformational changes. The recent X-ray crystal structure of a GluN1/GluN2B NTD complex reveals a unique arrangement of the two NTD protomers with intersubunit interactions distinct from those observed in AMPA and kainate receptors (16, 17). However, the importance of these dimer interfaces in the subunit-specific receptor regulation is poorly understood. We show that encoding the photoreactive UAA p-azido-l-phenylalanine (AzF) at the NTD upper lobe dimer interface in GluN1/GluN2B receptors serves as a photoswitch, triggering irreversible decrease of channel activity upon UV exposure. We further investigated the photo-induced conformational changes at the NTD dimer interfaces, as well as the subunit-dependent regulation, identifying structural determinants that differ between GluN2A- and GluN2B-containing NMDARs. Finally, we applied our approach to mammalian cells, including cultured hippocampal neurons, providing evidence for the transferability of light-sensitive NMDARs to more native cellular environments. Our results not only prove the feasibility of designing light-controlled NMDARs by introducing a genetically encoded photoreactive UAA at a conformational sensitive site, but also reveal aspects of the NMDAR assembly as highly subtype specific.
Fig. 1.
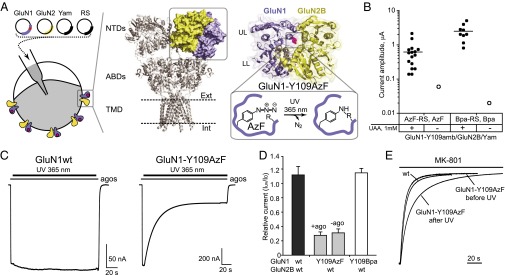
Light inactivation of GluN1/GluN2B NMDARs incorporating a genetically encoded photoactive UAA. (A, Left) Four plasmids encoding the GluN1 subunit with an amber stop codon at position Y109 (red dot), the wt GluN2 subunit, the suppressor tRNA (Yam), and the engineered tRNA synthetase (RS) were coinjected into Xenopus oocytes. (Center) Crystal structure of the GluA2 AMPA receptor (40). The three major domains—N-terminal domain (NTD), agonist-binding domain (ABD) and transmembrane domain (TMD)—are arranged in layers. One NTD dimer is highlighted. (Right) Crystal structure of the NMDAR GluN1/GluN2B NTD heterodimer (16); the ifenprodil molecule is omitted for clarity. LL, lower lobe; UL, upper lobe. The GluN1-Y109 site is highlighted. On UV irradiation, the azide moiety generates a biradical, which subsequently can react with a nearby residue to form a covalent adduct. (B) Current amplitudes from oocytes injected with plasmids as indicated, in the absence or presence of UAAs. For each condition, 20 oocytes were tested. Only currents >10 nA were plotted. (C) Representative current traces showing UV-induced current inhibition of GluN1-Y109AzF/GluN2B receptors but not wt GluN1/GluN2B receptors. (D) UV-induced current modifications at wt GluN1/GluN2B (1.11 ± 0.13; n = 8), GluN1-Y109AzF/GluN2Bwt with (0.28 ± 0.05; n = 16) or without (0.31 ± 0.05; n = 5) agonist, and GluN1-Y109Bpa/GluN2Bwt (1.15 ± 0.06; n = 5) receptors. Error bars, SD. (E) MK-801 inhibition kinetics of wt GluN1/GluN2B and GluN1-Y109AzF/GluN2B receptors before and after UV treatment.
Results
Encoding Photoreactive UAAs in the GluN1 NTD.
To genetically encode photoreactive UAAs, we chose the GluN1-Y109 site, which is situated at the NTD upper lobe–upper lobe (UL/UL) dimer interface according to the X-ray crystal structure of the GluN1/GluN2B NTD dimer (16) (Fig. 1A). Using conventional mutagenesis to introduce point mutations at this site, we found that, depending on the side chain property, receptor gating can be bidirectionally manipulated, as assessed by the inhibition kinetics of MK-801, a selective NMDAR open-channel blocker classically used to index receptor channel Po (15, 18, 19) (SI Appendix, Fig. S1 A and B). In agreement with previous studies (20), we also observed that mutations at this position affect sensitivities to various allosteric modulators, including proton, zinc, and spermine (SI Appendix, Fig. S1 C–E). We therefore hypothesized that encoding photoreactive UAAs at this site may affect receptor activity by perturbing the NTD conformation under light stimuli.
Based on our recent establishment of the genetic-code expansion in Xenopus laevis oocytes (8), we generated GluN1-Y109amb containing an amber stop codon at position Y109. Robust NMDAR currents were observed after coinjection with cDNAs encoding GluN1-Y109amb, wild-type GluN2B, orthogonal suppressor tRNA (Yam), and mutant aminoacyl tRNA synthetase (RS) together with the corresponding UAA [AzF or the photo–cross-linker p-benzoyl-l-phenylalanine (Bpa)] added in the external medium (Fig. 1B). In contrast, oocytes incubated in the absence of UAAs generated no or only tiny responses (<100 nA) (Fig. 1B), confirming successful and efficient incorporation of AzF and Bpa at the GluN1-Y109 site through genetic-code expansion. Continuous UV (365 nm) illumination of maximally activated GluN1-Y109AzF/GluN2B receptors produced strong receptor inhibition, characterized by a progressive and irreversible current reduction (Iuv/I0 = 0.28 ± 0.05, n = 16; current decrease time constant τ = 25.6 ± 4.6 s, n = 8) (Fig. 1 C and D). In contrast, wild-type receptors were almost unaffected (Iuv/I0 = 1.11 ± 0.12, n = 8) (Fig. 1 C and D), confirming that the UV treatment per se doesn’t cause significant functional changes or photo damaging. To test the influence of agonists on the UV-induced inactivation, we applied UV treatment in the absence of agonists. The Iuv/I0 ratio did not differ with or without agonists (Fig. 1D), indicating that the UV-induced channel inhibition is independent of receptor activation (i.e., agonist binding). Surprisingly, no UV-dependent channel inhibition was observed in the case of GluN1-Y109Bpa (Fig. 1D), demonstrating that, at this site, the photoswitching is specific to the azido moiety of AzF.
Photoswitching Locks the Receptor in a Low Po Mode.
The UV-induced current reduction may either render receptors completely inactive or decrease their activity. To distinguish between these two possibilities, we assessed the channel activity of photoswitched receptors. After UV treatment, MK-801 inhibition kinetics were significantly slowed, indicating a marked decrease of Po (Fig. 1E and SI Appendix, Fig. S2A). This result indicates that UV illumination does not completely silence the receptors but rather switches them into a low-activity mode. MK-801 inhibition kinetics for the GluN1-Y109AzF mutant before UV treatment were similar to those of wild-type receptors, suggesting that the introduction of AzF at this position causes minimal change in receptor function. In contrast, channel activity of GluN1-Y109Bpa/GluN2B receptors both before and after UV exposure was significantly lower compared with wild-type receptors (SI Appendix, Fig. S2A).
To evaluate further the impact of photo-illumination on GluN1-Y109AzF/GluN2B receptor properties, we tested receptor sensitivity to allosteric modulators. We first tested sensitivity to ifenprodil, the prototypical GluN2B-selective inhibitor, which binds the dimer interface between GluN1 and GluN2B NTDs (16). Ifenprodil sensitivity of photo-treated AzF-encoded receptors was markedly reduced compared with the situation before UV treatment (∼10-fold increase in IC50) (SI Appendix, Fig. S2B). In contrast, the sensitivity of UV-treated receptors to Zn2+, an allosteric inhibitor binding to the GluN2B NTD interlobe cleft and presumably promoting its closure (12, 21), was not affected, suggesting that UV did not lock GluN2B NTD in a closed state (SI Appendix, Fig. S2C). We also observed that the glutamate sensitivity was moderately increased after UV treatment whereas glycine sensitivity was unaffected (SI Appendix, Fig. S2 D and E). These results are in line with previous findings (15) that an alteration of GluN1 (or GluN2) NTD conformation primarily affects binding of glutamate, but not of glycine. In all measurements, the AzF receptors before UV treatment showed similar values as wild-type receptors, confirming that introduction of AzF creates no significant perturbation.
Photoswitching Is Receptor Subtype-Specific.
We next paired GluN1-Y109AzF mutant subunit with the wild-type GluN2A subunit—the most abundant GluN2 subunit in the adult brain (9). Because the GluN1 subunit is an obligatory subunit shared by all NMDAR subtypes, similar photoinactivation between GluN2A and GluN2B receptors could be anticipated. Strikingly, however, GluN1-Y109AzF/GluN2A receptors were almost completely insensitive to UV illumination (Iuv/I0 = 1.10 ± 0.22, n = 6) (Fig. 2). The incorporation of AzF in GluN2A receptors was confirmed by comparing oocytes incubated in the presence or absence of AzF (SI Appendix, Fig. S3A). Moreover, the channel Po remained similar before and after UV treatment (SI Appendix, Fig. S3B), providing additional evidence that the photoswitching does not occur in GluN2A receptors. To further assess the role of the GluN2B NTD in photoswitching, we generated chimera constructs. First, two previously generated GluN2A-GluN2B chimeras with swapped NTD (12) were applied, namely 2A-2BNTD and 2B-2ANTD. Second, two GluN2A chimeric receptors were developed, 2A-2BUL-loop and 2A-2BUL, which contain progressively smaller portions of the GluN2B NTD (Materials and Methods). The UV-induced channel inhibition was significantly recovered in the 2A-2BNTD-containing receptors (Iuv/I0= 0.43 ± 0.08, n = 6) (Fig. 2) whereas it was completely abolished in 2B-2ANTD (SI Appendix, Fig. S4A). Between 2A-2BUL-loop and 2A-2BUL chimeras (Fig. 2 and SI Appendix, Fig. S4A), only 2A-2BUL-loop restored the UV effect to a detectable level. There are thus structural determinants absent in GluN2A receptors and specific to GluN2B NTD that confer light sensitivity to AzF-incorporated receptors.
Fig. 2.
UV-induced functional changes at the GluN1-Y109 position are GluN2 NTD-specific. (Left) Representative current traces of UV-induced effects on receptors incorporating GluN1-Y109AzF and wt GluN2B, GluN2A-2BNTD, GluN2A-2BUL-loop (the loop represents GluN2B-208MSLDDGD), or wt GluN2A. (Right) Relative currents (Iuv/I0) from receptors containing the GluN1-Y109AzF subunits and 2B (0.28 ± 0.05; n = 16), 2A-2BNTD (0.43 ± 0.08; n = 6), 2A-2BUL-loop (0.78 ± 0.04; n = 5), or 2A (1.10 ± 0.22; n = 5) subunit. Error bars, SD.
To further explore for potential differences in the NTD arrangement between GluN2A and GluN2B receptors, we performed experiments using sulfhydryl-modifying MTS compounds, reagents that have been extensively used to investigate the accessibility of a protein site and associated conformational changes (12, 19, 22). We focused on the GluN1-Y109 site, which, modified to a cysteine, retains wild-type functionality (SI Appendix, Fig. S1). Among the four types of MTS compounds tested, only the smallest MTSEA produced a massive current inhibition (SI Appendix, Fig. S5A). Comparing GluN2A, GluN2B, and the different chimeras, the relative inhibitions induced by MTSEA share a strikingly similar trend as the UV-induced inhibitions (SI Appendix, Figs. S4B and S5B), with GluN2B showing the strongest effect and GluN2A the smallest. This parallel trend strengthens the idea that there is a fundamental difference in the NTD dimer assembly between GluN1/GluN2A and GluN1/GluN2B receptors.
Differential GluN1/GluN2 NTD Dimer Assembly.
The crystal structure of the GluN1/GluN2B NTD dimer has revealed two major interfaces of contacts between the two neighboring domains (16). The first one involves helical contacts between GluN1 NTD UL and GluN2B NTD UL, an interface shared by other iGluRs (17). The second one is unique to NMDARs and involves GluN1 NTD UL and GluN2B NTD LL. This interface participates in stabilizing a closed-cleft conformation of the GluN2B NTD clamshell (16). To further elucidate the subunit-specific arrangement in these regions and associated functional effects, we systematically compared these two interfaces between GluN2A and GluN2B receptors by combining biochemical and functional analyses.
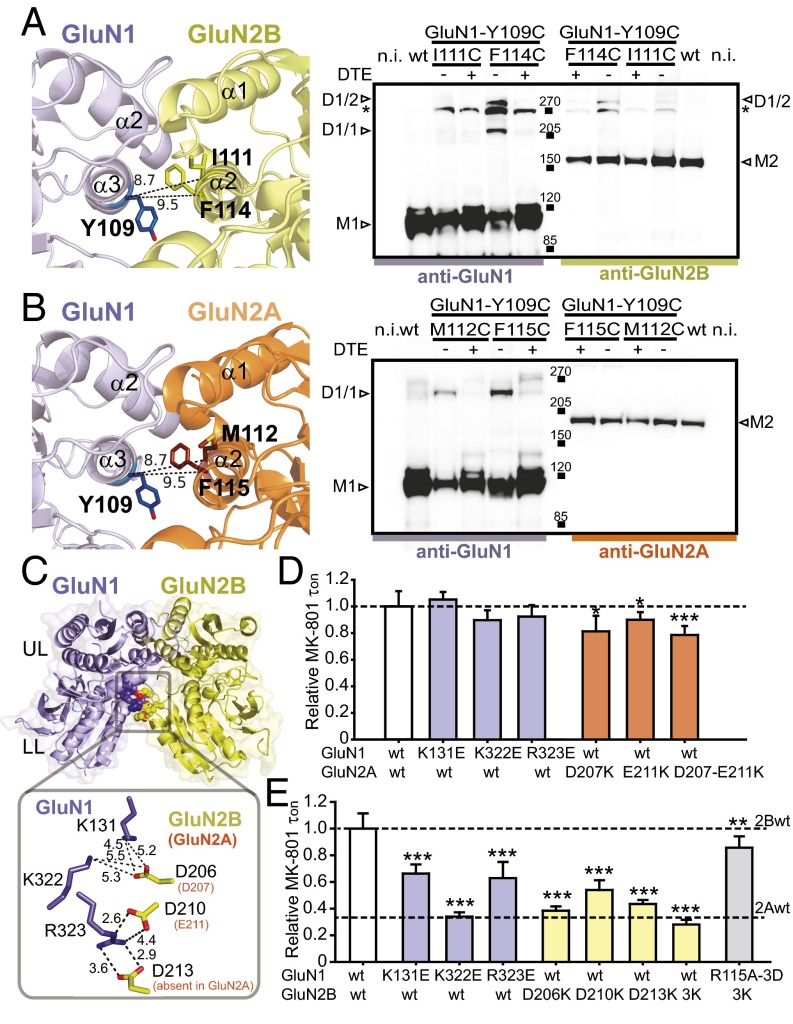
We first probed the UL/UL interface using disulfide cross-linking. The UL/UL interface mainly contains hydrophobic interactions mediated by α2- and α3-helices in GluN1, and α1- and α2-helices in GluN2 (16, 23) (Fig. 3 A and B). In GluN1, we used the GluN1-Y109C mutant because the Y109 site is situated at the center of the α3-helix and has proven a critical site in the interface. In GluN2B, we generated GluN2B-I111C and GluN2B-F114C, both residues pointing toward GluN1-Y109 with Cα-Cα distances of 9.5 and 8.7 Å, respectively (Fig. 3A). In GluN2A, we generated the homologous mutations GluN2A-M112C and GluN2A-F115C (Fig. 3B). Using nonreducing Western blots from oocytes expressing full-length receptors, we observed clear heterodimer formation in the case of the GluN1-Y109C/GluN2B-F114C double mutant (Fig. 3A and SI Appendix, Fig. S6 A and B). The GluN2B-I111C mutant showed weaker heterodimer bands that can be detected occasionally (SI Appendix, Fig. S6B). In contrast, no detectable heterodimer band was observed for either of the two GluN1/GluN2A double cysteine mutants (Fig. 3B and SI Appendix, Fig. S6 C and D). We also observed GluN1 homodimer bands likely originating from intracellular intermediates of receptor biogenesis (11) (Fig. 3 A and B and SI Appendix, Fig. S6). Overall, these results suggest that GluN1/GluN2B receptors form a tighter coupling at the UL/UL interface than GluN1/GluN2A receptors.
Fig. 3.
Structural basis for the differential GluN1/GluN2 NTD dimer interfaces. (A and B, Left) View of the UL/UL interface in the GluN1/GluN2B NTD dimer crystal structure (A) (16) or GluN1/GluN2A NTD dimer model (B). Residues mutated into cysteines are represented in sticks with Cα-Cα distances indicated in Å. (Right) Immunoblots from Xenopus oocytes expressing either wt or mutant subunits. M1 indicates the GluN1 monomer (∼110 kDa); M2 the GluN2A or GluN2B monomer (∼180 kDa); D1/1 the GluN1 homodimer (∼220 kDa); and D1/2 the GluN1/GluN2 heterodimer (∼290 kDa). Treatment with (+) or without (−) DTE is also shown. *, a nonspecific band; n.i., noninjected oocytes. (C) Electrostatic interactions at the GluN1-UL/GluN2-LL interface as observed in the GluN1/GluN2B NTD dimer crystal structure (distances indicated in Å). (D and E) Relative MK-801 inhibition kinetics of receptors incorporating wt or mutant GluN1 and GluN2 subunits. Values are (from left to right) in D, 1.00 ± 0.11, 1.05 ± 0.06, 0.90 ± 0.07, 0.92 ± 0.09, 0.81 ± 0.12, 0.90 ± 0.06, and 0.79 ± 0.07; in E, 1.00 ± 0.13, 0.66 ± 0.07, 0.34 ± 0.03, 0.63 ± 0.12; 0.38 ± 0.03, 0.54 ± 0.07, 0.43 ± 0.03, 0.28 ± 0.04, and 0.85 ± 0.08. n = 4–35. *P < 0.05, **P < 0.01, ***P < 0.001, Student t test. Error bars, SD.
We then probed the UL/LL interface, which can be “locked” closed in GluN1/GluN2B receptors by introducing double cysteine mutations (GluN1-L320C/GluN2B-D210C) (16). We generated GluN2A-E211C at the homologous site and observed spontaneous heterodimer formation in GluN1-L320C/GluN2A-E211C receptors (SI Appendix, Fig. S7 A–C). In addition, we found the initial Po for double cysteine mutants in both GluN1/GluN2A and GluN1/GluN2B receptors to be significantly decreased compared with wild-type and single cysteine mutant receptors, an effect that was reversed by DTE treatment (SI Appendix, Fig. S7 D and E). These results reveal that the UL/LL interface captured in the GluN1/GluN2B NTD dimer crystal structure can also form in GluN1/GluN2A receptors and impact receptor activity. Next, we designed mutations aiming to “force open” this interface. Several charged residues participating in intersubunit electrostatic interactions are present at the UL/LL interface (residues K131, K322, and R323 in GluN1; D206, D210, and D213 in GluN2B; and D207 and E211 in GluN2A based on sequence alignment) (Fig. 3C and SI Appendix, Fig. S7B). We introduced opposite charge-point mutations either on GluN1 NTD UL or GluN2 NTD LL to switch from electrostatic attraction to repulsion. MK-801 assays on GluN1/GluN2A mutants revealed either no change or only slight increases in Po (Fig. 3D). In striking contrast, all mutations significantly increased Po in GluN2B receptors. Among these mutants, GluN1-K322E and GluN2B-D206K displayed the strongest effects, with level of channel activity close to that of “high” Po GluN2A-receptors (Fig. 3E). Finally, when sufficient charges were reversed on both sides of the interface, receptor Po was restored to values close to wild-type GluN2B-receptors (Fig. 3E and SI Appendix, Fig. S8), as expected for direct through-space ionic interactions. These observations indicate that the UL/LL interactions are stronger in GluN1/GluN2B receptors than in GluN1/GluN2A receptors, in agreement with a model in which GluN2B NTD spends most of the time in a closed conformation whereas GluN2A NTD is mostly open (12).
Photo–Cross-Linking Likely Induces NTD Dimer Interface Closure.
The lack of heterodimer formation in Western blots from UV-exposed GluN1-Y109AzF/GluN2B receptors suggests intrasubunit UV-induced cross-linking (SI Appendix, Fig. S9). Upon UV excitation, AzF usually generates a nitrene radical and forms a covalent linkage with a nearby atom at a distance of 3–4 Å (24). Based on the GluN1/GluN2B NTD dimer crystal structure (16), we identified GluN1-I133, which situates in a four-residue loop (K131-H134), satisfying the distance criteria (Fig. 4A). This loop adopts different configurations in various GluN1 NTD crystal structures (11, 16) and displays high flexibility in molecular dynamic simulations (25). To explore the possible role of I133 in photo–cross-linking, we generated a series of double mutant receptors, mutating I133 to G, A, V, or L on the GluN1-Y109amb background. Among all AzF mutants, the smallest mutant, I133G, showed the strongest effect, with only minimal UV-induced receptor inhibition (Iuv/I0 = 0.82 ± 0.08, n = 4) (Fig. 4B). Plotting UV-induced inhibition versus amino acid volume demonstrated that the two parameters were remarkably correlated, with decreasing side chain size at I133 systematically reducing the UV-mediated effect (Fig. 4C). Due to the strong distance dependence in AzF-mediated photo–cross-linking, this correlation hints to GluN1-I133 being the potential cross-linking partner. We suggest that the photo–cross-linking between AzF and GluN1-I133 “locks” the GluN1 K131-H134 loop and restricts its mobility, an effect that subsequently may alter the NTD dimer interfaces.
Fig. 4.

Exploration of the photo–cross-linking site. (A) Local environment around the GluN1-Y109 site in the GluN1/GluN2B NTD dimer (16). The potential candidates on the GluN1-KSIH-loop for UV cross-linking are highlighted, and distances are indicated in Å. The interface residues GluN1-K322 and GluN2B-D206 are also highlighted. (B) Changes in current amplitude after UV illumination on GluN1-Y109AzF/GluN2B wt receptors or receptors incorporating an additional substitution (L, V, A, or G) at position GluN1-I133. Values are 0.28 ± 0.05, 0.28 ± 0.02, 0.37 ± 0.05, 0.48 ± 0.09, and 0.82 ± 0.08; n = 3–16. (Inset) Normalized current traces for GluN1-I133 and its substitutions. (C) The GluN1-133 residue volume and the UV-induced current reductions are strongly correlated (linear regression, R2 = 0.89). (D) Disruption of the UV effect by destabilizing the GluN1-UL/GluN2B-LL electrostatic contacts. Values are (from left to right): 0.28 ± 0.05, 0.32 ± 0.05, 0.40 ± 0.03, 0.28 ± 0.03, 0.85 ± 0.09, 0.43 ± 0.08, and 0.40 ± 0.04. n = 4–16. *P < 0.05, ***P < 0.001, Student t test. Error bars, SD.
Because the GluN1 K131-H134 loop directly participates to the NTD UL/LL dimer interface (Fig. 4A), we hypothesized that the UV-induced conformational switch may involve a strengthening of the UL/LL interface, an effect that would translate into decreased receptor activity. To evaluate this possibility, we tested UV-induced effects on mutants combining the GluN1-Y109AzF and “force open” GluN1 or GluN2B mutations (Fig. 3E). Out of six double mutants, four showed a clear reduction in UV-induced inactivation (Fig. 4D), as expected if the mutations introduced in the UL/LL interface hamper the UV-induced interface tightening. Although modest in GluN1-K322E, GluN2B-D210K, and GluN2B-D213K double mutants, the effect was considerably stronger in the GluN2B-D206K double mutant (Iuv/I0 = 0.85 ± 0.09, n = 9) (Fig. 4D). Overall, these results support a structural mechanism whereby receptor inactivation following UV-induced AzF photo–cross-linking proceeds through closure of the GluN1/GluN2B UL/LL dimer interface. They also reveal the key role of the ionic lock between GluN1-K322E and GluN2B-D206K in controlling receptor Po and transducing the photo–cross-linking–induced conformational switch in the GluN1 subunit to the downstream gating machinery.
Expressing Light-Sensitive NMDARs in HEK Cells and Neurons.
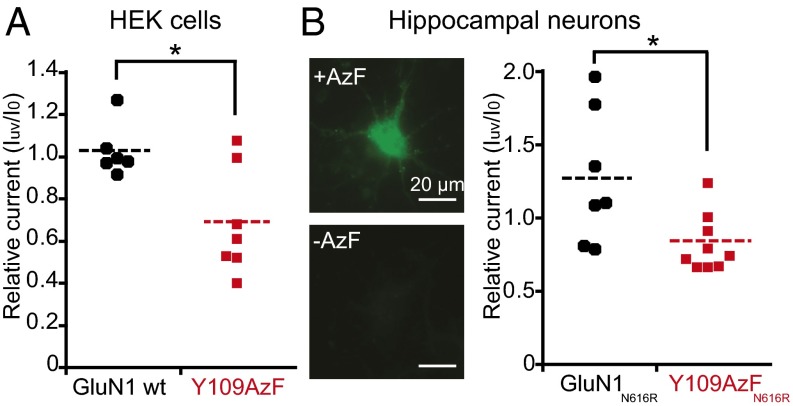
Finally, we sought to test GluN1-Y109AzF NMDARs in mammalian cells, including neurons. To reduce the number of plasmids for transient transfection, we engineered a single bidirectional plasmid coding for both the suppressor tRNA and the synthetase (SI Appendix, Fig. S10 A and B). Moreover, we fused the GluN2B subunit with an N-terminal GFP containing an amber stop codon at position Tyr182 (26) (SI Appendix, Fig. S10A), to use GFP fluorescence as a simple proxy for the successful UAA incorporation. After cotransfection, green positive HEK cells were readily detected when incubated in the presence of AzF, and patch-clamp recordings revealed UV-inactivated NMDAR currents (Fig. 5A). Similar procedures were then applied to primary hippocampal cultured neurons. To distinguish the GluN1-Y109AzF NMDARs from the endogenous NMDARs, we combined the conventional N616R mutation to the GluN1-Y109amb subunit (SI Appendix, Fig. S10 A and C). This mutation confers resistance to Mg2+ block (27), allowing us to inhibit endogenous NMDAR currents with Mg2+. In the presence, but not absence, of AzF, a few bright green positive neurons were observed and displayed NMDAR responses inhibited by UV exposure (Fig. 5B) although the effects were not as pronounced as with Xenopus oocytes or HEK cells, presumably due to the presence of triheteromeric receptor populations (9) with various light sensitivity (GluN1/GluN1-Y109AzF/GluN2B and/or GluN1-Y109AzF/GluN2A/GluN2B receptors). Overall, these results show that expressing light-sensitive NMDARs by genetically encoding a photoreactive UAA is achievable in mammalian cells, including cultured neurons.
Fig. 5.
Expression of UV-sensitive GluN1-Y109AzF NMDARs in mammalian cells. (A) UV-induced current modifications at GluN1 wt/GFP-Y182AzF-GluN2B (1.03 ± 0.12, n = 6) or GluN1-Y109AzF/GFP-Y182AzF-GluN2B (0.69 ± 0.25, n = 7) receptors expressed in HEK cells. (B, Right) UV-induced current modifications at GluN1-N616R/GFP-Y182AzF-GluN2B (1.27 ± 0.45, n = 7) or GluN1-Y109AzF-N616R/GFP-Y182AzF-GluN2B (0.84 ± 0.19, n = 9) receptors expressed in culture hippocampal neurons. (Left) Fluorescence images of neurons transfected with three plasmids encoding GluN1-Y109amb-N616R, GFP-Y182amb-GluN2B, and the suppressor tRNA/AzFRS pair (SI Appendix, Fig. S10A) with (Upper) or without (Lower) AzF in the medium. *P < 0.05, Student t test.
Discussion
We provide here a novel step in the development of light-controlled glutamate receptors. Our approach exploits the genetic code expansion technology to directly introduce into the receptor a light-sensitive moiety carried by a single UAA. Together with a recent study on Bpa-containing AMPA receptors (28), our work opens new avenues for using genetically encoded UAAs to study glutamate receptor structure and mechanisms, an approach likely to extend to other neurotransmitter receptors. The genetic encoding technique is also well-suited for biological applications as it allows for both receptor-subtype selectivity and cell-type specificity. It thus complements the existing posttranslational labeling approach, which relies on photoswitchable ligands tethered to specific cysteines introduced in the receptor (29, 30). Although this latter approach is limited to extracellular sites, it has the essential advantage of being reversible (the receptor can be turned on and off in a reversible manner) and has proven powerful in controlling neuronal firing and synaptic circuits in behaving animals (2, 31). Implementation of the UAA-based approach in vivo remains challenging, but feasibility is within reach as recently demonstrated using a light-activatable potassium channel incorporating a genetically encoded photocaged UAA (26).
We genetically encoded a photo–cross-linking UAA to engineer light-sensitive NMDARs. The signal of the light-controlled channel activity was detected and followed in real time using cellular electrophysiology. We have observed UV-promoted, subunit-specific receptor inactivation mediated by the photo–cross-linker AzF inserted at the GluN1-Y109 site when paired with the GluN2B subunit. By assessing the UV sensitivity of receptor carrying structure-guided mutations, we propose that UV illumination triggers an intrasubunit photo–cross-linking event that leads to tightening of the heterodimer interface between the GluN1 NTD UL and the GluN2B NTD LL. Although we identified GluN1-I133 as a potential cross-linking partner, more direct structural evidence is required for confirmation. We found photoinactivation to be highly receptor subtype-specific, with no effect observed for GluN2A-receptors. We attribute the structural determinants of this effect to be differential NTD dimer interactions, with GluN1 and GluN2B subunits being more strongly constrained than GluN1 and GluN2A subunits. This difference in subunit association is manifested by stronger NTD interactions at both the UL/UL and UL/LL interfaces, the latter interface involving several GluN2B-specific electrostatic contacts. By showing that the GluN1 NTD couples less tightly to the GluN2A NTD than the GluN2B NTD, our results provide insight about the structural determinants controlling subtype-specific properties in NMDARs. In addition, it suggests that the subunit-selective binding of NTD allosteric modulators (e.g., ifenprodil binding to GluN2B receptors) (9, 16) may stem from fundamental differences in the NTD dimer assembly rather than differences in binding pockets per se. This subunit-selective and NTD-driven control of receptor activity appears unique to NMDARs. In AMPA and kainate receptors, the NTDs usually form much more rigid and tightly packed dimer assemblies symmetrically arranged by both UL/UL and LL/LL interactions (17).
Both AzF and Bpa as photo–cross-linking UAAs have been previously applied in mapping protein–protein (6, 28, 32) and protein–ligand (33) interactions. The availability of multiple UAAs with different properties extends the flexibility of the genetic code expansion technique. Although Bpa displays higher stability and more specific photochemistry compared with AzF (24, 33, 34), we have observed no detectable UV-induced functional changes when using Bpa, likely due to its bulky side chain introducing structural perturbations in the receptor before the UV treatment. We also note that AzF is a versatile UAA with multiple applications, including serving as an infrared probe (35) or chemical handle for bioorthogonal conjugations (36).
Although incorporating photoreactive UAAs has been already applied in structure–function studies of LGICs, previous approaches predominantly relied on the delivery of chemically modified tRNAs in oocytes (4). The “all-genetic” strategy, which uses engineered orthogonal aaRS/suppressor tRNA pairs, has significantly enhanced biological and technical convenience (8, 28). We envision further development in the genetic encoding of various photoreactive UAAs into LGICs, such as photo-caged amino acids (26, 37–39), or UAAs containing azo-benzene moieties (7) that adopt two conformations with different wavelengths. Incorporation of the latter would provide the possibility of generating bidirectionally controlled LGICs, which could be switched between on and off states in response to light, as afforded by photoswitchable ligands (1, 2). Furthermore, such protein tools should prove particularly valuable in tracking receptor conformational changes during gating steps or allosteric transitions. Following our demonstration of proof-of-concept in cultured neurons, we also foresee the development of light-sensitive NMDARs in more intact preparations, such as brain slices (26), to achieve optogenetic control of neuronal excitability through remote control of specific NMDAR populations.
Materials and Methods
Incorporation of UAAs in NMDARs expressed in Xenopus oocytes was performed as previously described (8). Expression of UAA-containing NMDARs in HEK cells and rat hippocampal cultured neurons is described in SI Appendix, SI Materials and Methods. Plasmid construction, cell culture and transfection, UAA incubation, two-electrode voltage-clamp of Xenopus oocytes, patch-clamp recording of HEK cells and neurons, UV exposure, and immunoblotting are detailed in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Eszter Kozma, Qian Wang, and Damien Baigl for suggestions on the manuscript. We thank Prof. Thomas Sakmar (The Rockefeller University) for providing suppressor tRNA and engineered synthetase plasmids. This work was supported by Agence Nationale de la Recherche-Jeunes Chercheuses Jeunes Chercheurs (S.Y.), Equipe Fondation pour la Recherche Médicale Grant DEQ2000326520 (to P.P.) and fellowship (to S.Z.), Fondation Pierre-Gilles-de-Gennes (S.Y. and P.P.), and Direction Générale de l'Armement (M.R.). This work also received support from the program “Investissements d’Avenir” (ANR-10-LABX-54 MEMOLIFE; ANR-11-IDEX-0001-02 PSL* Research University).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318808111/-/DCSupplemental.
References
- 1.Fehrentz T, Schönberger M, Trauner D. Optochemical genetics. Angew Chem Int Ed Engl. 2011;50(51):12156–12182. doi: 10.1002/anie.201103236. [DOI] [PubMed] [Google Scholar]
- 2.Szobota S, Isacoff EY. Optical control of neuronal activity. Annu Rev Biophys. 2010;39:329–348. doi: 10.1146/annurev.biophys.093008.131400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pless SA, Ahern CA. Unnatural amino acids as probes of ligand-receptor interactions and their conformational consequences. Annu Rev Pharmacol Toxicol. 2013;53:211–229. doi: 10.1146/annurev-pharmtox-011112-140343. [DOI] [PubMed] [Google Scholar]
- 4.Beene DL, Dougherty DA, Lester HA. Unnatural amino acid mutagenesis in mapping ion channel function. Curr Opin Neurobiol. 2003;13(3):264–270. doi: 10.1016/s0959-4388(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292(5516):498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 6.Davis L, Chin JW. Designer proteins: Applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13(3):168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 7.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 8.Ye S, Riou M, Carvalho S, Paoletti P. Expanding the genetic code in Xenopus laevis oocytes. ChemBioChem. 2013;14(2):230–235. doi: 10.1002/cbic.201200515. [DOI] [PubMed] [Google Scholar]
- 9.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: Impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14(6):383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 10.Hansen KB, Furukawa H, Traynelis SF. Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol. 2010;78(4):535–549. doi: 10.1124/mol.110.067157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farina AN, et al. Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. J Neurosci. 2011;31(10):3565–3579. doi: 10.1523/JNEUROSCI.6041-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459(7247):703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29(39):12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mony L, Zhu S, Carvalho S, Paoletti P. Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. EMBO J. 2011;30(15):3134–3146. doi: 10.1038/emboj.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat Struct Mol Biol. 2013;20(4):477–485. doi: 10.1038/nsmb.2522. [DOI] [PubMed] [Google Scholar]
- 16.Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475(7355):249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer ML. Emerging models of glutamate receptor ion channel structure and function. Structure. 2011;19(10):1370–1380. doi: 10.1016/j.str.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen N, Luo T, Raymond LA. Subtype-dependence of NMDA receptor channel open probability. J Neurosci. 1999;19(16):6844–6854. doi: 10.1523/JNEUROSCI.19-16-06844.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talukder I, Borker P, Wollmuth LP. Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J Neurosci. 2010;30(35):11792–11804. doi: 10.1523/JNEUROSCI.5382-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuko T, et al. A regulatory domain (R1-R2) in the amino terminus of the N-methyl-D-aspartate receptor: Effects of spermine, protons, and ifenprodil, and structural similarity to bacterial leucine/isoleucine/valine binding protein. Mol Pharmacol. 1999;55(6):957–969. doi: 10.1124/mol.55.6.957. [DOI] [PubMed] [Google Scholar]
- 21.Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28(24):3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlin A, Akabas MH. Substituted-cysteine accessibility method. Methods Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- 23.Lee CH, Gouaux E. Amino terminal domains of the NMDA receptor are organized as local heterodimers. PLoS ONE. 2011;6(4):e19180. doi: 10.1371/journal.pone.0019180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddington SC, et al. Different photochemical events of a genetically encoded phenyl azide define and modulate GFP fluorescence. Angew Chem Int Ed Engl. 2013;52(23):5974–5977. doi: 10.1002/anie.201301490. [DOI] [PubMed] [Google Scholar]
- 25.Burger PB, et al. Mapping the binding of GluN2B-selective NMDA receptor negative allosteric modulators. Mol Pharmacol. 2012;82(2):344–59. doi: 10.1124/mol.112.078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang JY, et al. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron. 2013;80(2):358–370. doi: 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wollmuth LP, Kuner T, Sakmann B. Adjacent asparagines in the NR2-subunit of the NMDA receptor channel control the voltage-dependent block by extracellular Mg2+ J Physiol. 1998;506(Pt 1):13–32. doi: 10.1111/j.1469-7793.1998.013bx.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klippenstein V, Ghisi V, Wietstruk M, Plested AJ. Photoinactivation of glutamate receptors by genetically encoded unnatural amino acids. J Neurosci. 2014;34(3):980–991. doi: 10.1523/JNEUROSCI.3725-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volgraf M, et al. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol. 2006;2(1):47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janovjak H, Szobota S, Wyart C, Trauner D, Isacoff EY. A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nat Neurosci. 2010;13(8):1027–1032. doi: 10.1038/nn.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyart C, et al. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature. 2009;461(7262):407–410. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato S, et al. Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid. Biochemistry. 2011;50(2):250–257. doi: 10.1021/bi1016183. [DOI] [PubMed] [Google Scholar]
- 33.Grunbeck A, Huber T, Sakmar TP. Mapping a ligand binding site using genetically encoded photoactivatable crosslinkers. Methods Enzymol. 2013;520:307–322. doi: 10.1016/B978-0-12-391861-1.00014-9. [DOI] [PubMed] [Google Scholar]
- 34.Knowles JR. Photogenerated reagents for biological receptor-site labeling. Acc Chem Res. 1972;5(4):155–160. [Google Scholar]
- 35.Ye S, et al. Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature. 2010;464(7293):1386–1389. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]
- 36.Huber T, Naganathan S, Tian H, Ye S, Sakmar TP. Unnatural amino acid mutagenesis of GPCRs using amber codon suppression and bioorthogonal labeling. Methods Enzymol. 2013;520:281–305. doi: 10.1016/B978-0-12-391861-1.00013-7. [DOI] [PubMed] [Google Scholar]
- 37.Miller JC, Silverman SK, England PM, Dougherty DA, Lester HA. Flash decaging of tyrosine sidechains in an ion channel. Neuron. 1998;20(4):619–624. doi: 10.1016/s0896-6273(00)81001-6. [DOI] [PubMed] [Google Scholar]
- 38.England PM, Lester HA, Davidson N, Dougherty DA. Site-specific, photochemical proteolysis applied to ion channels in vivo. Proc Natl Acad Sci USA. 1997;94(20):11025–11030. doi: 10.1073/pnas.94.20.11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gautier A, Deiters A, Chin JW. Light-activated kinases enable temporal dissection of signaling networks in living cells. J Am Chem Soc. 2011;133(7):2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462(7274):745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.