Understanding biological function in the profusion of proteins containing significant levels of intrinsic disorder depends on how accurately we can describe their conformational behavior (1). Recently, Wang et al. used molecular dynamics (MD) techniques to study the molecular recognition element of the C-terminal domain of the measles virus nucleoprotein (MeV-NTAIL), an example of this enigmatic family of intrinsically disordered proteins (IDPs) (2). In justifying their approach, the authors state that “in general, it is not feasible to characterize IDPs by an ensemble averaged method due to the underlying structural heterogeneity.” Although the potential advantages of restraint-free MD in terms of dynamic time scales are evident, numerous developments in the field also exploit ensemble-averaged NMR data to derive molecular descriptions of IDPs.
NMR ensemble and time averaging are determined by the chemical shift time scale, whereby conformational states interconverting on rates faster than tens of microseconds contribute to average resonance peaks. The conformational dependence of experimental measurements [e.g., chemical shifts (CSs) and residual dipolar couplings (RDCs)] can be predicted for independent states, and the average interpreted in terms of statistical ensembles. Ensemble-based analyses of experimental NMR data from IDPs (3) have thus been used to highlight propensities for local folding or tertiary contacts that may be linked to function. One such approach—ASTEROIDS—uses a statistical-coil model to sample accessible states, from which actual contours of conformational space are delineated using experimental data (4).
The degrees of freedom available to IDPs provides clear potential for overfitting, making calibration procedures essential, if only to determine the potential accuracy with which combinations of complementary experimental data can describe conformational sampling. The ability of ensemble descriptions to predict independent experimental measurements provides a simple test of consistency. It is therefore instructive to compare the predictive ability of NMR-based ensembles with MD simulation performed on the same system.
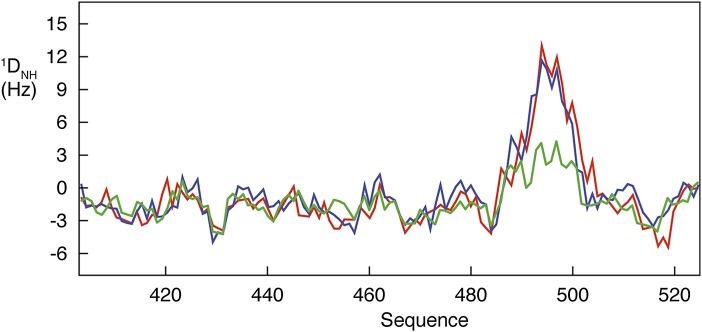
ASTEROIDS was applied to the MeV-NTAIL system studied by Wang et al. (2), using a combination of 13Cα, 13Cβ, 13C′, 15N, and 1H CSs that has been shown to map distinct regions of Ramachandran space (4). RDCs are also dependent on backbone dihedral sampling, reporting in addition on medium- and long-range order and therefore representing a more demanding test of the validity of conformational descriptions. RDCs were therefore predicted on the basis of ensembles descriptions derived from experimental CSs (5) and from the MD-based ensemble (Fig. 1) to compare the predictive nature of the two approaches. Although the authors claim remarkable agreement with experimental data, the representative ensemble described by Wang et al. from MD predicts RDCs significantly worse than the NMR-based ensembles, possibly because of the tendency to form two separate helices that orient in open turn conformations, inducing orientational order to which RDCs are particularly sensitive. Although agreement with RDCs does not prove validity of the ASTEROIDS ensemble, it does demonstrate consistency with independent data. More generally, this comparison supports the premise that NMR is indeed well suited to the characterization of IDPs—precisely because it is so diversely sensitive to conformational heterogeneity.
Fig. 1.
Comparison of the ability of the MD-based (green) and NMR-based ensemble (blue) to reproduce experimental 1DNH RDCs (red). The x axis indicates the primary sequence of MeV-NTAIL (molecular recognition element comprises residues 485–504). The molecular dynamics-based representative ensemble of the protein with associated populations for each substate was kindly provided by Jin Wang (Jilin University, China; State University of New York, Stony Brook, NY). The NMR-based free-energy description was developed using the ensemble selection algorithm ASTEROIDS on the basis of experimental chemical shifts. RDCs from the MD-based ensemble were predicted by constructing statistical coil chains from the termini of the region defined in the MD simulation of the molecular recognition element. This accounts for the long-range effects of the unfolded N- and C-terminal chains on the measured RDCs. One thousand structures were constructed for each of 70 substates constituting the representative MD ensemble, and RDCs were averaged for each substate and then were coadded using the associated population weighting from Wang et al. Prediction of RDCs from the 50,000-strong ASTEROIDS ensemble was achieved using previously published approaches (4). Finally, calculated RDCs were uniformly scaled to reproduce experimental data in both cases. Experimental uncertainty is on the order of 1 Hz throughout. Although the NMR CS-based ensemble selection reproduces the data within the experimental uncertainty in the helical segment, the χ2 for the MD-based ensemble is ∼30 times higher.
Footnotes
The authors declare no conflict of interest.
References
- 1.Tompa P. Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21(3):419–425. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, et al. Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein. Proc Natl Acad Sci USA. 2013;110(40):E3743–E3752. doi: 10.1073/pnas.1308381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17(1):3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Ozenne V, et al. Mapping the potential energy landscape of intrinsically disordered proteins at amino acid resolution. J Am Chem Soc. 2012;134(36):15138–15148. doi: 10.1021/ja306905s. [DOI] [PubMed] [Google Scholar]
- 5.Jensen MR, et al. Intrinsic disorder in measles virus nucleocapsids. Proc Natl Acad Sci USA. 2011;108(24):9839–9844. doi: 10.1073/pnas.1103270108. [DOI] [PMC free article] [PubMed] [Google Scholar]