Significance
Expanded polyglutamine (polyQ) tracts in proteins are associated with many neurodegenerative diseases, including Huntington disease. The structure and dynamics of polyQ peptides are difficult to study due to the homogenous nature of the sequence and their high propensity to aggregate into amyloid fibrils. In this manuscript, we study mixtures of isotope-labeled peptides with 2D IR spectroscopy to resolve the structure of individual monomers within the polyQ. These results, in combination with spectra calculated from molecular-dynamics simulations, determine the dominant structure of polyQ fibrils consists of stacked β-hairpins.
Keywords: Huntington disease, isotope dilution, antiparallel β-sheets, two-dimensional infrared spectroscopy
Abstract
Polyglutamine (polyQ) sequences are found in a variety of proteins, and mutational expansion of the polyQ tract is associated with many neurodegenerative diseases. We study the amyloid fibril structure and aggregation kinetics of K2Q24K2W, a model polyQ sequence. Two structures have been proposed for amyloid fibrils formed by polyQ peptides. By forming fibrils composed of both 12C and 13C monomers, made possible by protein expression in Escherichia coli, we can restrict vibrational delocalization to measure 2D IR spectra of individual monomers within the fibrils. The spectra are consistent with a β-turn structure in which each monomer forms an antiparallel hairpin and donates two strands to a single β-sheet. Calculated spectra from atomistic molecular-dynamics simulations of the two proposed structures confirm the assignment. No spectroscopically distinct intermediates are observed in rapid-scan 2D IR kinetics measurements, suggesting that aggregation is highly cooperative. Although 2D IR spectroscopy has advantages over linear techniques, the isotope-mixing strategy will also be useful with standard Fourier transform IR spectroscopy.
Numerous neurodegenerative diseases, including Huntington disease, are associated with the mutational expansion of CAG repeats in specific genes. This expansion causes an increase in the length of normally benign polyglutamine (polyQ) tracts in expressed proteins (1, 2). In vivo, polyQ tracts occur embedded within larger proteins, such as the huntingtin protein. Full length proteins are impractical for detailed structural studies, and so isolated polyQ sequences are often used instead in experiments and simulations (3–5). Although isolated polyQ sequences may not aggregate in precisely the same manner as they would in full-length proteins (3, 6, 7), it has been established that such models yield data that are relevant to some, if not all, polyQ pathologies (8). Additionally, proteolytic polyQ-containing fragments, rather than full-length proteins, may be the primary toxic species in Huntington disease (9) and some types of spinocerebellar ataxia (10). As a consequence, simple polyQ peptides remain an attractive alternative to more complicated proteins for understanding polyQ-mediated aggregation.
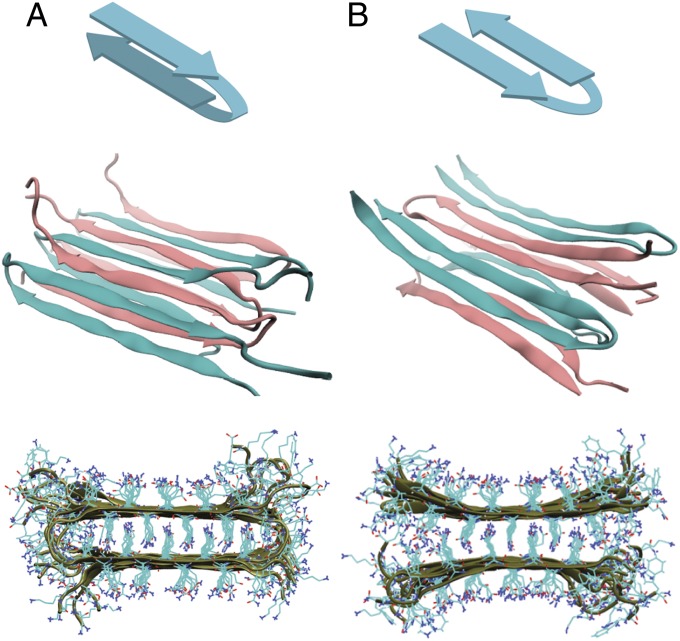
A plethora of structures have been suggested for polyQ aggregates, ranging from α-helical coiled coils (11) or β-helices (12) to amyloid fibrils comprising stacked, linear β-sheets (13). Current consensus favors fibrillar structures, but the precise arrangement of the peptides within the fibril remains unclear. Most amyloid proteins form parallel β-sheets within fibrils, although antiparallel sheets do occur (14). Parallel sheets have been observed for glutamine-rich hexapeptides (15), but NMR studies (16), as well as the results presented here, suggest that sequences approaching the pathological length of 40 glutamines adopt an antiparallel structure. Fig. 1 shows two models that have been proposed for how 20- to 30-residue polypeptides might form fibrils with such antiparallel β-sheets (17, 18). In the β-arc model (Fig. 1A), each monomer forms two β-strands that contribute to the formation of separate β-sheets. The backbone structure of the β-arc model is reminiscent of fibrils of amyloid-β (Aβ) (14) and human islet amyloid polypeptide (hIAPP) (19), but with antiparallel rather than parallel alignment of the strands within each β-sheet. In the β-turn model (Fig. 1B), each monomer adopts a β-hairpin conformation and contributes two β-strands to the same β-sheet. It is thought that these sheets stack on top of one another by zippering and hydrogen bonding of the glutamine side chains, although the number of stacked sheets is not known. Although the structures of the monomers in each model are very different, to a first approximation the two models predict the same backbone structure of the β-sheets; they differ only in whether the loop connects strands in the same β-sheet or in opposite β-sheets.
Fig. 1.
Visualizations of fibril models used in MD simulations. Schematic of monomer conformation (Top), one-half of the fibril unit cell (Middle), and fibril cross-section (Bottom) for the (A) β-arc and (B) β-turn fibril models.
Although infrared (IR) spectroscopy is a good tool for measuring secondary structure, in this case it cannot distinguish between the two structural models. The amide I mode, which is generated primarily by backbone carbonyl stretching motions, is sensitive to protein secondary structure because the carbonyl groups couple to one another. Coupling causes their vibrational motions to delocalize along the backbone so that multiple carbonyl groups vibrate in unison (20). This delocalization changes the IR intensities and frequencies in characteristic ways for different protein structures. Uncoupled or disordered structures such as random coils have a broad absorption near 1,645 cm−1 that is characteristic of little delocalization. In contrast, amyloid β-sheets structures absorb around 1,620 cm−1 because the vibrational modes delocalize across multiple residues.
In the case of polyQ, the β-sheet structures of the two fibril models are so similar that their IR spectra would be indistinguishable, with only negligible differences expected due to variations in the loop structure. If it were possible to measure the spectrum of an individual monomer within the fibril, rather than the spectrum of the fibril itself, then this problem would be circumvented because the spectra of β-arc and β-turn monomers should be very different from one another. The IR spectrum of a β-arc monomer would be dictated primarily by the positive couplings along the β-strands (20), which would produce a shift to higher frequencies relative to the random coil frequency. In contrast, the spectrum of a β-turn monomer would be dominated by the negative couplings associated with hydrogen-bonded β-sheets, which are significantly larger in magnitude than the positive couplings along the β-strands. These negative couplings would cause a shift to lower frequencies. Thus, by identifying a positive versus a negative frequency shift, one could identify the fibril structure.
In this paper, we use mixtures of isotopically labeled peptides to obtain two-dimensional infrared (2D IR) spectra of the individual folded monomers within the fibrils. We apply this isotope dilution approach to examine the fibrillar structure of the polyglutamine sequence K2Q24K2W, which is a standard model peptide for studies of polyQ fibrils (16, 17). The experimental spectra exhibit a negative frequency shift, which is consistent with the β-turn model as the dominant fibrillar structure under our experimental conditions. Quantitative simulations of the 2D IR spectra from molecular-dynamics (MD) models of the two fibril structures confirm this assignment. The glutamine side chains form hydrogen-bonded columns that couple strongly to the backbone vibrations. In contrast, K2Q24K2W is largely unstructured before aggregation. Importantly, we observe no spectrally distinct intermediate species during aggregation, suggesting that the aggregation of this peptide is highly cooperative.
Results
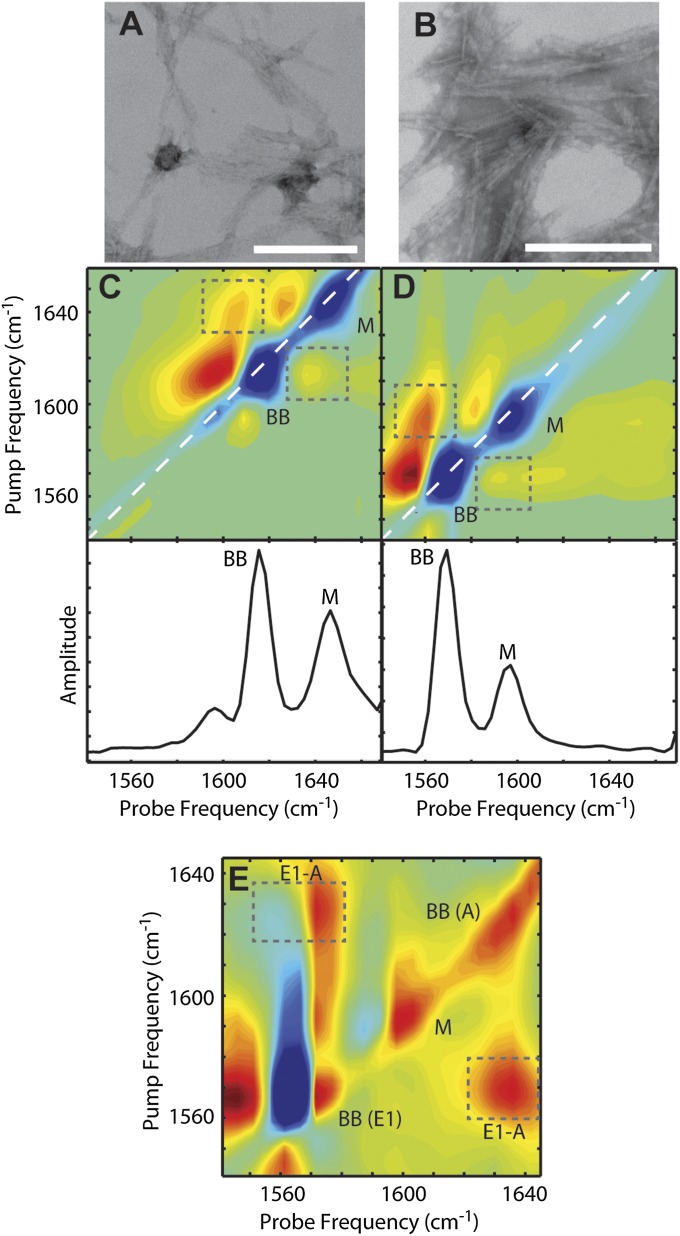
The sequence K2Q24K2W (henceforth referred to as Q24) was expressed in Escherichia coli grown in either natural abundance or 13C-enriched media. The peptides were fully disaggregated using the protocol established by Chen and Wetzel (21), then lyophilized and reconstituted in deuterated phosphate buffer at pD ∼7.4 to initiate aggregation. Electron micrographs of the resulting Q24 fibrils (Fig. 2 A and B) show broad, nontwisted ribbons, similar to those observed for other simple polyglutamine peptides (17).
Fig. 2.
Transmission electron microscopy and experimental 2D IR spectra of Q24 fibrils. Electron micrographs of (A) unlabeled and (B) 13C-labeled Q24 fibrils. In each image, the white scale bar represents 200 nm. Two-dimensional IR spectra and diagonal intensity slices of (C) unlabeled and (D) 13C-labeled Q24 fibrils. (E) Polarization difference spectrum of 13C-labeled Q24 fibrils. Diagonal peaks are labeled BB for backbone modes and M for mixed backbone–side-chain modes. Symmetry labels are added for backbone modes in the difference spectrum. Cross-peaks between diagonal modes are highlighted with boxes. Frequencies are summarized in Table S1.
Q24 Forms Antiparallel β-Sheets.
Two-dimensional IR spectra of the fibrils in the amide I region are shown in Fig. 2 C and D for unlabeled and uniformly 13C-labeled peptides. In the spectrum of unlabeled Q24 aggregates (Fig. 2C), the most intense features are a pair of peaks (BB) appearing at a pump frequency of 1,616 cm−1. These peaks are a characteristic feature of amyloid β-sheets and result from vibrational coupling between the backbone amide groups (22). A second intense pair of peaks (M), which are unique to polyglutamine fibrils, appear at 1,644 cm−1. This feature has been attributed previously to the amide I stretching mode of the glutamine side chains (17, 23, 24). However, this assignment is ambiguous due to the possibility of strong coupling between side chain and backbone carbonyls. Interdigitation of side chains between stacked β-sheets, which is a key feature of both the β-arc and β-turn fibril models, leads to the close spatial proximity of backbone and side-chain amide groups and could cause the amide vibrations to delocalize across both the side chains and backbone. The intense cross-peaks between the BB and M diagonal peaks (Fig. 2C, boxes) indicate strong coupling between the modes, and simulations suggest that the M peaks may in fact result from mixed modes (Fig. S1). Finally, a weak pair of peaks are observed near 1,590 cm−1, which probably arise from side-chain bending modes (24).
Fig. 2D shows the 2D IR spectrum of uniformly 13C-labeled Q24 aggregates. This spectrum is nearly identical to that of the unlabeled peptide, except that the amide features are downshifted uniformly by 44 cm−1. Such a frequency shift is consistent with the change in effective mass upon 13C substitution of the amide carbon and has been observed in other fully labeled peptide samples (25). The similarity between the spectra indicates that uniform 13C labeling does not disrupt the structure or coupling of the peptides. As a consequence, we can treat the 2D IR spectra of 12C and 13C peptides interchangeably by taking into account the frequency shift.
It is difficult to tell whether Q24 is forming parallel or antiparallel β-sheets from the spectral features discussed above. Both types of β-sheets display a sharp low-frequency peak around 1,620 cm−1, which is caused by a backbone normal mode, E1, with oscillating dipoles perpendicular to the β-strands. In principle, the two structures can be differentiated by the presence of a second high-frequency peak around 1,680 cm−1, which is only present for antiparallel sheets and is caused by a backbone normal mode, A, with an oscillating dipole parallel to the strands (26, 27). In our spectra, the A band is not readily apparent, and so we use cross-peaks to identify if an A band is present by subtracting 2D IR spectra collected with parallel versus perpendicular polarizations. A study by Hahn et al. (28) showed that subtraction produces strong cross-peaks for antiparallel sheets, but weak or no cross-peaks for parallel sheets. In Fig. 2E, we show the polarization difference spectrum for 13C-labeled Q24 fibrils. Very intense cross peaks (E1-A, boxes) appear at 1,572 and 1,630–1,635 cm−1, which correspond to the characteristic E1 and A modes of an antiparallel β-sheet when taking into account the 44 cm−1 isotope shift. Thus, our Q24 peptides do indeed form antiparallel β-sheets, in agreement with previous studies on similar peptides (16, 29).
Atomistic Structural Models.
We used atomistic MD simulations to construct models of the β-arc and β-turn structures. The model systems consisted of a 12-peptide assembly with water and counterions represented explicitly. The model fibril was aligned longitudinally with its periodic image to generate a pseudoinfinite amyloid fibril. The initial configuration for the β-arc model was adapted from the structure proposed by Schneider et al. (16). Each Q24 peptide contributes one strand to each of two stacked, antiparallel β-sheets, and the sheets are held together by chain reversals connecting the strands (Fig. 1A). The sheets are further stabilized by tight interdigitation of the side chains within the fibril interior. As for the β-turn model, no detailed structural model exists in the literature; however, the structure of the core β-sheet region is essentially identical to that of the β-arc. The difference lies in the arrangement of the monomers; rather than donating a single strand to both of the fibril β-sheets, each monomer donates two strands to a single β-sheet. With this in mind, the initial configuration for the β-turn model was adapted from the fully equilibrated β-arc configuration by leaving the β-strands intact and reattaching the residues linking β-strands on opposite sheets so as to link pairs of adjacent β-strands within a sheet (Fig. 1B). Upon equilibration, the ends of the β-turn model curve apart because they are no longer held in place by the loop, but the fibril remains intact for the duration of the simulations (100 ns; Fig. S2). Full details of the MD simulations can be found in SI Text.
Comparison of Simulations and Experiments Differentiates the Two Fibril Models.
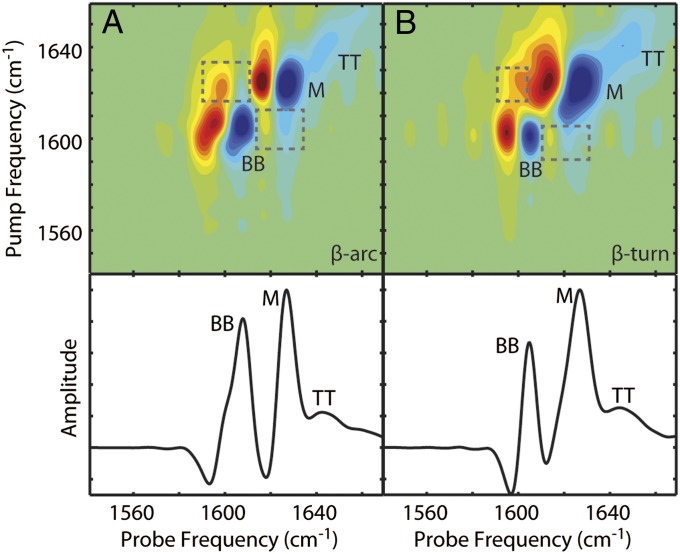
The calculation of amide I 2D IR spectra within the mixed quantum-classical approximation has been described in previous publications and applied to peptides with a variety of secondary structures (30–32). In Fig. 3, we show the simulated 2D IR spectra and diagonal slices for both the β-arc and β-turn models of unlabeled Q24 fibrils. Comparing to experiment (Fig. 2C), the simulations of both models reproduce the spectral features of the experiments. Both models produce a set of sharp peaks for the backbone β-sheet mode (BB) and another set for the mixed backbone–side-chain modes (M). Strong cross-peaks are also present between the backbone and side-chain features (boxes), as observed experimentally, indicating that the coupling between the backbone and side chains is approximately correct. The frequencies of both bands are 10–15 cm−1 lower than observed experimentally and the intensity ratio is reversed. These differences may indicate, respectively, that the semiempirical frequency maps are not perfectly calibrated to the electrostatics of these amyloid fibrils, and that the transition dipole coupling model describes the couplings between the side-chain and backbone residues somewhat inaccurately. Another possibility is that the simulation results, which are based on the simulation of a single fibril, do not properly account for multiple fibrils stacking into a bundle; in recent work, such stacking effects were found to increase the amide I frequency (33). Even so, the agreement is remarkable, considering that these calculations include no adjustable parameters. The similarity between the simulations for the β-arc and β-turn models underscores the fact that IR spectroscopy unaided by isotope labeling cannot distinguish between the two fibril morphologies.
Fig. 3.
Simulated 2D IR spectra of Q24 fibril models. Simulated 2D IR spectra and diagonal intensity slices for the (A) β-arc and (B) β-turn models of unlabeled Q24 fibrils. Diagonal peaks are labeled BB for backbone modes, M for mixed backbone–side-chain modes, and TT for modes from the disordered turns and termini. Cross-peaks between the BB and M modes are highlighted with boxes. Frequencies are summarized in Table S1.
The major difference between the two fibril models is the number of β-strands each polypeptide contributes to a single β-sheet: Each monomer contributes only one strand per sheet in the β-arc structure, whereas each monomer contributes two strands in the β-turn structure. Using this fact, we can test which of these models best fits the experimental data by using isotope dilution. As discussed in the Introduction, the amide I vibrational modes for an amyloid β-sheet are delocalized across many strands. Couplings between residues on the same β-strand are typically weak and positive, whereas couplings between residues on adjacent strands are strong and negative. Thus, the negative couplings between strands dominate and the number of strands over which the mode is delocalized is inversely correlated with the frequency of the mode. As a result, delocalization across a single strand produces a very different spectral signature than across two strands. In a mixture of 10% 13C-labeled peptides with 90% unlabeled peptides, it is statistically unlikely for two 13C-labeled peptides to be stacked consecutively within a fibril. Because delocalization is largely limited to consecutive strands of the same isotope type (20, 34), the 13C-labeled peptides will behave like monomers in the spectrum, and the frequency of the 13C amide I bands will directly reflect the number of β-strands that each monomer contributes to a β-sheet. We have previously used a similar approach to estimate the number of strands that each γd-crystallin monomer contributes to fibril β-sheets (25).
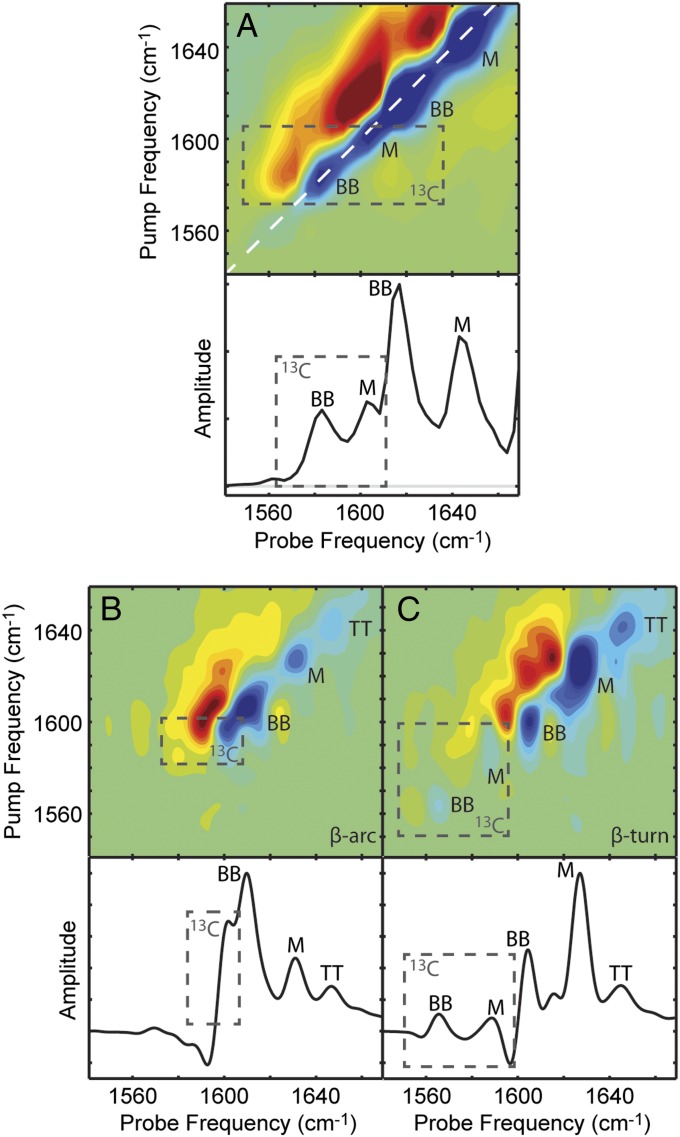
For the experiment, we prepared a disaggregated solution of 10% uniformly 13C-labeled Q24 with 90% unlabeled Q24 and allowed the peptides to aggregate together. The 2D IR spectrum of the mixture (Fig. 4A) exhibits four peaks. The two most intense peak pairs appear at the same frequencies as those of pure unlabeled Q24 fibrils (Fig. 2C). It is not surprising that these modes would be unaffected because the addition of only 10% labeled peptides does not significantly disrupt the delocalization of the unlabeled vibrational excitons (25). The two weaker peak pairs (box) are 13C-Q24 modes and appear at 1,584 and 1,604 cm−1, shifted 12 and 4 cm−1 higher, respectively, than the corresponding modes in pure 13C-labeled fibrils (Fig. 2D). The positive frequency shift upon isotope dilution of the backbone mode confirms that the dominant couplings between monomers are negative, as is typical for amyloid fibrils. The positive frequency shift of the mixed backbone–side-chain peak indicates that its coupling constants are also predominantly negative, which suggests that the side-chain amides form hydrogen-bonded chains similar to the backbone amides.
Fig. 4.
Experimental and simulated 2D IR spectra of isotope diluted fibrils. (A) Experimental 2D IR spectrum and diagonal intensity slice of 10% 13C-labeled Q24 fibrils. Simulated 2D IR spectra and diagonal intensity slices for (B) β-arc and (C) β-turn models of ∼8% labeled Q24 fibrils. Diagonal peaks are labeled with BB for backbone modes, M for mixed backbone–side-chain modes, and TT for disordered modes from the turns and termini. Peaks arising from 13C-labeled peptides are highlighted with boxes. Peak frequencies are summarized in Table S2.
To simulate the isotope dilution experiments, 1 of the 12 monomers in each fibril model was treated as 13C labeled by downshifting the diagonal frequency of all of the amino acids in that monomer by 44 cm−1. As shown in Fig. 4B, the β-arc model generates a spectrum that differs dramatically from the experimental results. The backbone (BB) and mixed (M) peaks of the unlabeled peptides are altered substantially (compare Fig. 4B to Fig. 3A), so much so that the BB peak is now much more intense than the M peak. This dramatic change may arise because the isotope labeling of a single monomer of the β-arc influences the excitonic coupling of both β-sheets, effectively localizing the vibrational modes of the entire fibril. Moreover, the β-arc model predicts only a single 13C feature (Fig. 4B, box), and that single feature is at such a high frequency that it overlaps the 12C peak of the β-sheet. There are no distinct features below 1,600 cm−1, despite the spectrum calculations generally underestimating amide I frequencies. When we systematically examine couplings within the vibrational Hamiltonian, we find that the intramonomer couplings for the β-arc model are largely positive and thus tend to shift the normal mode frequencies above the local-mode frequencies (Fig. S3). As isotope dilution effectively eliminates all couplings except for those between chromophores in the same monomer, this likely accounts for the unusually high frequency of the 13C feature in Fig. 4B. Simply put, the β-arc model fails to reproduce even the qualitative features of the experimental isotope dilution spectrum.
In contrast, the relative intensities of the backbone and mixed modes in the β-turn model (Fig. 4C) are largely unaffected by isotope labeling, in agreement with experiment, because the excitonic coupling is only disrupted along one β-sheet. Furthermore, the β-turn model properly produces the two 13C features (Fig. 4C, box) expected from the experimental 13C signal. As with isotopically pure fibrils, we attribute the lowest 13C feature (peak BB) to backbone motions and the upper 13C feature (peak M) to mixed backbone–side-chain modes. Again, all of the features appear at lower frequency than in experiment. The 13C backbone mode shifts less upon dilution in theory than in experiment, indicating that theory underestimates the delocalization of the backbone modes. Nevertheless, the qualitative success of the β-turn model and the qualitative failure of the β-arc model provide convincing evidence that polyQ forms fibrils with a β-turn topology.
No Intermediate Structures Are Observed During Aggregation.
In the work above, we only obtained 2D IR spectra of mature Q24 fibrils. Comparing experimental and simulated 2D IR spectra allowed us to determine that mature Q24 fibrils exhibit a conformation consistent with the β-turn model and inconsistent with the β-arc model. With better understanding of the fibril structure, we can now monitor structural changes during aggregation to determine whether any spectrally distinct intermediate species exist.
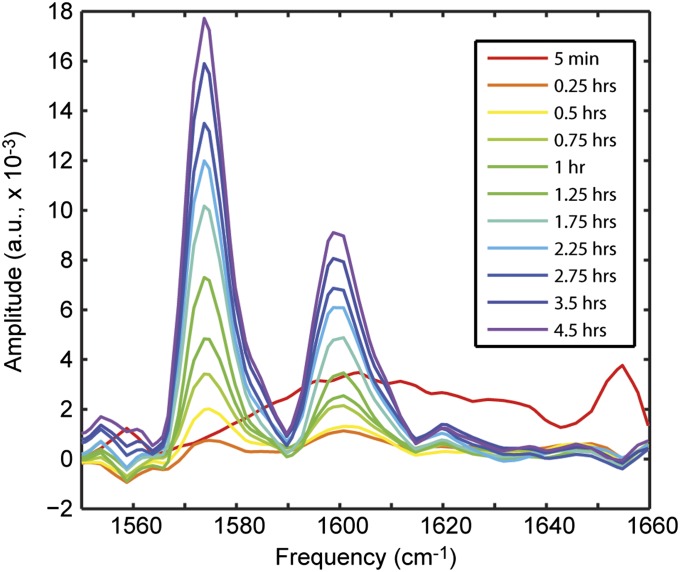
Fig. 5 shows a series of diagonal slices obtained from 2D IR spectra of uniformly 13C-labeled Q24 (Fig. S4) over 4.5 h of aggregation. The initial slice (red) was taken within 5 min of initiating aggregation and shows a weak, broad feature centered around 1,604 cm−1. Adding 44 cm−1 to mimic an unlabeled peptide would place this mode at 1,648 cm−1, which is the characteristic frequency of a disordered peptide (35). Thus, we can confirm that the sample has been fully disaggregated. To highlight the spectral changes that occur during the aggregation process, the subsequent diagonal slices are plotted as difference slices, calculated by subtracting the initial red slice from the slice at that time point. Within 15 min of initiating aggregation, two peaks appear at 1,574 and 1,600 cm−1. These peaks match those observed in samples of 13C-Q24 fibrils that had equilibrated for 12 h (Fig. 2D). As time progresses, these features gain intensity and no other features are observed. In particular, there is no evidence of individual β-hairpins. From the isotope dilution experiments (Fig. 4A), we know the spectral signatures of these β-hairpins to be peaks at 1,584 and 1,604 cm−1, which are clearly absent in the kinetics data. This suggests that β-hairpins in solution do not constitute a significant fraction of the conformational ensemble and certainly do not exist as a kinetically stable intermediate species.
Fig. 5.
Kinetics of Q24 aggregation. Difference slices obtained from 2D IR spectra of 13C-Q24. The initial slice (red) was subtracted from subsequent slices to highlight changes.
We also plot the intensity of these peaks as a function of time (Fig. S5). When the intensity traces are normalized, it is clear that the two features grow in at the same rate, suggesting that the backbone and side chains come into alignment at the same time. In all, the kinetics provide evidence that Q24 aggregation is highly cooperative. As suggested by Kar et al. (17), metastable individual β-hairpins likely form only during the initial nucleation phase, whereas subsequent fibril growth involves the cooperative addition and folding of monomers onto the fibril ends without the formation of a significant population of intermediate species.
Discussion and Conclusions
In this paper, we use a combination of experimental and theoretical spectroscopy to determine the structure of amyloid fibrils formed from simple polyglutamine peptides. Two-dimensional IR spectra of isotopically pure samples show that Q24 forms fibrils with antiparallel β-sheets, which agrees with the general findings of other studies (16, 17, 36). Two possible fibril structures (Fig. 1) have been proposed to accommodate antiparallel sheets (18). Constraints obtained from solid-state NMR were interpreted to support the β-arc structure, in which each monomer contributes β-strands to more than one β-sheet (16), whereas others have argued in support of the β-turn structure, which comprises stacks of β-hairpins (17, 36). We find that the two models produce 2D IR spectra that are basically indistinguishable for isotopically pure samples (Fig. 3). Because there is essentially no difference in the fundamental β-sheet structure, the local-mode frequencies and coupling strengths in the vibrational Hamiltonians are nearly identical for both models. Thus, IR spectroscopy alone is insufficient to differentiate between the two possible structures.
However, isotope dilution experiments reveal a dramatic difference between the two models. The β-turn model reproduces the qualitative features of the experimental spectrum quite well, whereas the β-arc model fails to produce even the correct number of spectral features (Fig. 3). This difference can be explained based on the number of strands each monomer contributes to a single β-sheet. Isotope dilution essentially isolates the dilute labeled monomers, decoupling them from the bulk unlabeled peptides. For the 13C-labeled peptides, only intramonomer couplings remain intact. Typically, the strongest couplings in a β-sheet are between amide groups that are stacked along the fibril axis (28), whereas coupling between modes on the same β-strand is much weaker. Thus, if a monomer contributes only a single β-strand per sheet, as in the β-arc model, the negative shift of the amide I mode is eliminated. We see this in the calculated spectrum for the β-arc (Fig. 3A). In contrast, if a monomer contributes multiple, adjacent strands to a β-sheet, some of the strong couplings between strands are retained. In the calculated spectrum for the β-turn model, we see that the couplings between the aligned backbone residues of a single monomer suffice to qualitatively reproduce the double-peaked feature observed in experiment. Thus, we find that Q24 forms fibrils with a β-turn structure. We arrived at this conclusion mostly by examining the diagonal peaks in the 2D IR spectra. Thus, although there are many advantages to using 2D IR spectroscopy to make these measurements [such as the nonlinearity of the signals with the transition dipole strength (37)], absorption or Fourier transform IR spectroscopy could also be used in conjunction with mixed isotopes.
Next, we examine the suggestion made by other studies that simple polyQ peptides aggregate into fibrils without populating conformationally distinct intermediates (38, 39). Two-dimensional IR kinetic studies further support this hypothesis. Isotope dilution allowed us to identify the spectral features of an individual β-hairpin peptide, the fundamental building block of the β-turn fibril model. Kinetic studies revealed that disordered, monomeric Q24 aggregates directly into fibrils with no evidence of monomeric β-hairpins in solution. This suggests that the aggregation of Q24 is highly cooperative.
Why is the β-turn structure favored? One explanation may be related to curvature of the turn region. According to our MD structures, the turn region is tight enough in the β-turn model that backbone hydrogen bonds can form between turn residues, but the turn region is wider in the β-arc model and so less hydrogen bonding appears to occur. That being said, amyloid polymorphs are a common occurrence and so it would not be surprising if other structures are possible. It will be interesting to apply this isotope-labeling strategy in the future to longer glutamine sequences, including those above the 40-repeat threshold typically required for the development of disease symptoms. Longer polyQ monomers may simply form longer β-hairpins, or the monomers might adopt a formation with additional turns (16). These additional turns may simply add strands to the same β-sheet, in an extension of the β-turn model; alternatively, one could envision a more complex fold in which a single monomer forms stacked β-hairpins that are each in separate sheets. Without the use of isotope-labeled samples, IR spectroscopy would probably not be able to discriminate between these possibilities, but we have shown here that isotope dilution provides a mean to test such structures. Adding additional strands to the same β-sheet would increase the negative couplings within a monomer, resulting in a red shift of the isotope diluted features, whereas the formation of stacked β-hairpins in separate β-sheets would generate the positive intramonomer couplings observed in the β-arc model. Isotopic dilution provides critical insights into the structure of monomers within amyloid fibrils, and could shed light on the structures and assemblies of other amyloid-forming proteins.
Methods
A full description of methods is given in SI Text. Briefly, K2Q24K2W was expressed in Escherichia coli grown in either natural abundance or 13C-enriched media. The peptides were fully disaggregated in a 1:1 mixture of deuterated hexafluoroisopropanol and trifluoroacetic acid (21), then lyophilized and reconstituted in phosphate buffer at pD ∼7.4 to initiate aggregation. The final total peptide concentration was 0.5 mM for isotopically pure samples or 1 mM for 10% isotope-diluted samples.
MD simulations were performed using the GROMACS 4.5 package (40, 41). The model systems consisted of a 12-peptide assembly, represented atomistically except for methylene hydrogens, with water and counterions represented explicitly. The simulated fibril was aligned with its periodic image along the fibril axis to generate a pseudoinfinite amyloid fibril. The coordinates of the β-arc model were kindly provided by Prof. Marc Baldus (Utrecht University, Utrecht, The Netherlands) (17). The initial configuration for the β-turn model was adapted from the fully equilibrated β-arc configuration by relocating residues linking β-strands on opposite sheets to link pairs of adjacent β-strands within a sheet.
Amide I 2D IR spectra were calculated within the mixed quantum-classical approximation (30), using frequency maps for backbone and side-chain chromophores developed by Wang et al. (30). For 13C-labeled chromophores, the frequencies were shifted down by 44 cm−1. The frequency shifts and couplings between nearest-neighbor backbone amides were calculated from the relevant Ramachandran angles (42), whereas transition dipoles and couplings between all other chromophore pairs were calculated using the transition dipole coupling scheme of Torii and Tasumi (43).
Supplementary Material
Acknowledgments
We thank Prof. Marc Baldus at Utrecht University for kindly providing the coordinates of the β-arc structure proposed by Schneider et al. (16). We also thank Randall Massey at the University of Wisconsin Medical School Electron Microscope Facility for his help with transmission electron microscopy. Support for this research was provided by the National Science Foundation through Graduate Research Fellowship Program Grant DGE-0718123 (to L.E.B. and A.M.F.), Grant CHE-0840494 (to J.K.C. and J.L.S.), and Grant CBET-1264021 (to J.J.d.P.), and by the National Institutes of Health through Grant DK79895 (to A.J.H., S.D.M., and M.T.Z.) and Grant DK088184 (to J.K.C. and J.L.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401587111/-/DCSupplemental.
References
- 1.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Martin JB. Molecular basis of the neurodegenerative disorders. N Engl J Med. 1999;340(25):1970–1980. doi: 10.1056/NEJM199906243402507. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya AM, Thakur AK, Wetzel R. Polyglutamine aggregation nucleation: Thermodynamics of a highly unfavorable protein folding reaction. Proc Natl Acad Sci USA. 2005;102(43):15400–15405. doi: 10.1073/pnas.0501651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walters RH, Murphy RM. Examining polyglutamine peptide length: A connection between collapsed conformations and increased aggregation. J Mol Biol. 2009;393(4):978–992. doi: 10.1016/j.jmb.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitalis A, Lyle N, Pappu RV. Thermodynamics of β-sheet formation in polyglutamine. Biophys J. 2009;97(1):303–311. doi: 10.1016/j.bpj.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thakur AK, et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol. 2009;16(4):380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellisdon AM, Thomas B, Bottomley SP. The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J Biol Chem. 2006;281(25):16888–16896. doi: 10.1074/jbc.M601470200. [DOI] [PubMed] [Google Scholar]
- 8.Kar K, Jayaraman M, Sahoo B, Kodali R, Wetzel R. Critical nucleus size for disease-related polyglutamine aggregation is repeat-length dependent. Nat Struct Mol Biol. 2011;18(3):328–336. doi: 10.1038/nsmb.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125(6):1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Haacke A, et al. Proteolytic cleavage of polyglutamine-expanded ataxin-3 is critical for aggregation and sequestration of non-expanded ataxin-3. Hum Mol Genet. 2006;15(4):555–568. doi: 10.1093/hmg/ddi472. [DOI] [PubMed] [Google Scholar]
- 11.Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143(7):1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc Natl Acad Sci USA. 2002;99(8):5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma D, Shinchuk LM, Inouye H, Wetzel R, Kirschner DA. Polyglutamine homopolymers having 8-45 residues form slablike β-crystallite assemblies. Proteins. 2005;61(2):398–411. doi: 10.1002/prot.20602. [DOI] [PubMed] [Google Scholar]
- 14.Tycko R. Molecular structure of amyloid fibrils: Insights from solid-state NMR. Q Rev Biophys. 2006;39(1):1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 15.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435(7043):773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider R, et al. Structural characterization of polyglutamine fibrils by solid-state NMR spectroscopy. J Mol Biol. 2011;412(1):121–136. doi: 10.1016/j.jmb.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 17.Kar K, et al. β-hairpin-mediated nucleation of polyglutamine amyloid formation. J Mol Biol. 2013;425(7):1183–1197. doi: 10.1016/j.jmb.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajava AV, Baxa U, Steven AC. β arcades: Recurring motifs in naturally occurring and disease-related amyloid fibrils. FASEB J. 2010;24(5):1311–1319. doi: 10.1096/fj.09-145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luca S, Yau W-M, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46(47):13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strasfeld DB, Ling YL, Gupta R, Raleigh DP, Zanni MT. Strategies for extracting structural information from 2D IR spectroscopy of amyloid: Application to islet amyloid polypeptide. J Phys Chem B. 2009;113(47):15679–15691. doi: 10.1021/jp9072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, Wetzel R. Solubilization and disaggregation of polyglutamine peptides. Protein Sci. 2001;10(4):887–891. doi: 10.1110/ps.42301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buchanan LE, Dunkelberger EB, Zanni MT (2011) Examining Amyloid Structure and Kinetics with 1D and 2D Infrared Spectroscopy and Isotope Labeling. Protein Folding and Misfolding: Shining Light by Infrared Spectroscopy, eds Fabian H and Naumann D (Springer, Heidelberg), pp 217–237.
- 23.Smith MH, et al. Polyglutamine fibrils are formed using a simple designed β-hairpin model. Proteins. 2010;78(8):1971–1979. doi: 10.1002/prot.22713. [DOI] [PubMed] [Google Scholar]
- 24.Jayaraman M, et al. Slow amyloid nucleation via α-helix-rich oligomeric intermediates in short polyglutamine-containing huntingtin fragments. J Mol Biol. 2012;415(5):881–899. doi: 10.1016/j.jmb.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran SD, et al. Two-dimensional IR spectroscopy and segmental 13C labeling reveals the domain structure of human γD-crystallin amyloid fibrils. Proc Natl Acad Sci USA. 2012;109(9):3329–3334. doi: 10.1073/pnas.1117704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee C, Cho M. Local amide I mode frequencies and coupling constants in multiple-stranded antiparallel β-sheet polypeptides. J Chem Phys B. 2004;108(52):20397–20407. [Google Scholar]
- 27.Mukherjee S, Chowdhury P, Gai F. Effect of dehydration on the aggregation kinetics of two amyloid peptides. J Phys Chem B. 2009;113(2):531–535. doi: 10.1021/jp809817s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn S, Kim S-S, Lee C, Cho M. Characteristic two-dimensional IR spectroscopic features of antiparallel and parallel beta-sheet polypeptides: Simulation studies. J Chem Phys. 2005;123(8):084905. doi: 10.1063/1.1997151. [DOI] [PubMed] [Google Scholar]
- 29.Thakur AK, Wetzel R. Mutational analysis of the structural organization of polyglutamine aggregates. Proc Natl Acad Sci USA. 2002;99(26):17014–17019. doi: 10.1073/pnas.252523899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Middleton CT, Zanni MT, Skinner JL. Development and validation of transferable amide I vibrational frequency maps for peptides. J Phys Chem B. 2011;115(13):3713–3724. doi: 10.1021/jp200745r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Skinner JL. Thermally induced protein unfolding probed by isotope-edited IR spectroscopy. J Phys Chem B. 2012;116(32):9627–9634. doi: 10.1021/jp304613b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woys AM, et al. Parallel β-sheet vibrational couplings revealed by 2D IR spectroscopy of an isotopically labeled macrocycle: Quantitative benchmark for the interpretation of amyloid and protein infrared spectra. J Am Chem Soc. 2012;134(46):19118–19128. doi: 10.1021/ja3074962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welch WRW, Kubelka J, Keiderling TA. Infrared, vibrational circular dichroism, and Raman spectral simulations for β-sheet structures with various isotopic labels, interstrand, and stacking arrangements using density functional theory. J Phys Chem B. 2013;117(36):10343–10358. doi: 10.1021/jp4056126. [DOI] [PubMed] [Google Scholar]
- 34.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Two-dimensional infrared spectra of isotopically diluted amyloid fibrils from Abeta40. Proc Natl Acad Sci USA. 2008;105(22):7720–7725. doi: 10.1073/pnas.0802993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shim S-H, et al. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc Natl Acad Sci USA. 2009;106(16):6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong K, Punihaole D, Asher SA. UV resonance Raman spectroscopy monitors polyglutamine backbone and side chain hydrogen bonding and fibrillization. Biochemistry. 2012;51(29):5822–5830. doi: 10.1021/bi300551b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grechko M, Zanni MT. Quantification of transition dipole strengths using 1D and 2D spectroscopy for the identification of molecular structures via exciton delocalization: Application to α-helices. J Chem Phys. 2012;137(18):184202. doi: 10.1063/1.4764861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S, Ferrone FA, Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc Natl Acad Sci USA. 2002;99(18):11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein FAC, et al. Pathogenic and non-pathogenic polyglutamine tracts have similar structural properties: Towards a length-dependent toxicity gradient. J Mol Biol. 2007;371(1):235–244. doi: 10.1016/j.jmb.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 40.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 41.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 42.la Cour Jansen T, Dijkstra AG, Watson TM, Hirst JD, Knoester J. Modeling the amide I bands of small peptides. J Chem Phys. 2006;125(4):44312. doi: 10.1063/1.2218516. [DOI] [PubMed] [Google Scholar]
- 43.Torii H, Tasumi M. Ab initio molecular orbital study of the amide I vibrational interactions between the peptide groups in di- and tripeptides and considerations on the conformation of the extended helix. J Raman Spectrosc. 1998;29(1):81–86. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.