Significance
Acetylation and deacetylation of histone H3K56 is important for genomic stability, replication, and the response to DNA damage and replication stress. One of the key regulators of H3K56 acetylation in budding yeast is Hst3, a cell-cycle– and damage-regulated histone deacetylase for the mark. Previously, Hst3 was known to be transcriptionally regulated. In this paper, we show that Hst3 is controlled posttranscriptionally by protein degradation. Specifically, Hst3 protein is degraded by SCFCdc4 recognizing a small phospho-degron. We also find that the degradation of Hst3 increases in response to replication stress through the same phospho-degron.
Abstract
Hst3 is the histone deacetylase that removes histone H3K56 acetylation. H3K56 acetylation is a cell-cycle– and damage-regulated chromatin marker, and proper regulation of H3K56 acetylation is important for replication, genomic stability, chromatin assembly, and the response to and recovery from DNA damage. Understanding the regulation of enzymes that regulate H3K56 acetylation is of great interest, because the loss of H3K56 acetylation leads to genomic instability. HST3 is controlled at both the transcriptional and posttranscriptional level. Here, we show that Hst3 is targeted for turnover by the ubiquitin ligase SCFCdc4 after phosphorylation of a multisite degron. In addition, we find that Hst3 turnover increases in response to replication stress in a Rad53-dependent way. Turnover of Hst3 is promoted by Mck1 activity in both conditions. The Hst3 degron contains two canonical Cdc4 phospho-degrons, and the phosphorylation of each of these is required for efficient turnover both in an unperturbed cell cycle and in response to replication stress.
Histone H3K56 acetylation is a cell-cycle– and damage-regulated histone modification (1). In Saccharomyces cerevisiae, K56 acetylation is important for replication, genomic stability, chromatin assembly, and the response and recovery from DNA damage (1–6). Because of the critical nature of this chromatin mark, the enzymes that control it are themselves very important.
In the budding yeast S. cerevisiae, Hst3 is a sirtuin histone deacetylase that, along with its close homolog Hst4, removes K56 acetylation (5, 7, 8). Overexpression of Hst3 or deletion of Hst3, along with related deacetylase Hst4, sensitizes cells to replication stress, suggesting that cells must control Hst3 levels precisely (5, 7). Hst3 is regulated both transcriptionally and by protein turnover. During an unperturbed cell cycle, HST3 transcript levels cycle, and Hst3 protein turns over very quickly (5, 9). In response to DNA damage, transcription of HST3 is turned down, and Hst3 protein turnover is even faster (5, 8, 10–12).
Many cell-cycle–regulated proteins are targeted for proteasomal degradation after ubiquitination by a member of the SCF E3 ubiquitin ligase family. SCF ligases are multisubunit ubiquitin ligases composed of Skp1, a cullin (Cdc53), a RING-finger protein (Rbx1), and an F-box protein (13). The F-box protein is the substrate recognition module that confers substrate specificity to SCF ligases, with different F-box proteins recognizing different sets of substrates. The essential F-box protein Cdc4 regulates many cell-cycle–regulated proteins after their phosphorylation, including Sic1, Eco1, Cln3, and many others (14–19).
Here, we have characterized the mechanism of Hst3 protein turnover. We find that Hst3 is targeted for degradation by SCFCdc4 through a multisite phospho-degron. We find that Hst3 turnover is increased in response to replication stress in a Rad53-dependent way. Finally, we show that stabilizing Hst3 leads to misregulation of K56 acetylation in response to DNA damage and that this misregulation affects growth and sensitivity to damage.
Results
Hst3 Is a Substrate of SCFCdc4.
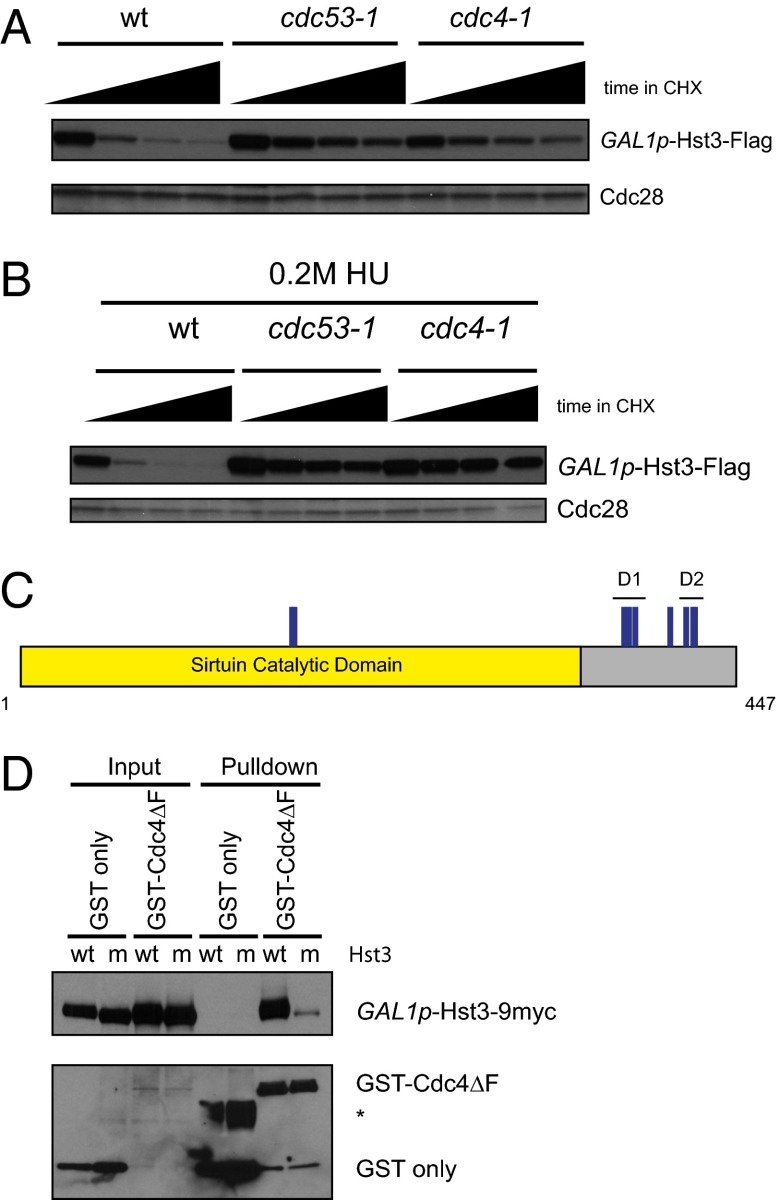
To study Hst3 protein turnover in the absence of its transcriptional regulation, we replaced the promoter of HST3 with the GAL1 promoter. We screened several E3 ubiquitin ligases to identify the one responsible for targeting Hst3 for degradation and found that Hst3 is partially stabilized at the nonpermissive temperature of temperature-sensitive mutants in CDC53 (the cullin scaffold for all SCF ligases) and CDC4 (encoding an essential F-box protein) (Fig. 1A). Inactivation of SCFCdc4 also stabilized Hst3 after treatment with hydroxyurea (HU) to induce replication stress (Fig. 1B).
Fig. 1.
Hst3 is a target of the SCFCdc4 ubiquitin ligase. (A) GAL1p-Hst3-Flag was induced with galactose in wild-type, cdc53-1, and cdc4-1 strains for 3.5 h at 23 °C. Then all strains were shifted to 37 °C for 2.5 h to inactivate the temperature-sensitive alleles before the addition of cycloheximide (CHX). Cycloheximide was added at t = 0, and protein turnover was followed for 45 min, with time points taken every 15 min. Cdc28 is shown as a loading control. (B) The experiment was performed as in A except that after induction with galactose for 7.5 h at 23 °C, cells were prearrested in 0.2 M HU + galactose for 3 h, and then shifted to 37 °C for 2.5 h before addition of CHX. (C) Phosphorylation sites were mapped on Hst3 by mass spectrometry. GAL1p-Hst3-Flag was induced in a cdc4-1 strain grown at the permissive temperature (23 °C) to enrich for phosphorylated forms. Purification was done from untreated cells or cells damaged with 0.05% MMS for 6.5 h. All identified phosphorylation sites are shown in blue. The two degrons identified, degron 1 (D1) and degron 2 (D2), are marked. D1 encompasses S377, T380, and T384. D2 encompasses S416, S420, and S421. The remaining phosphorylation sites shown on the schematic are T172, T173, and S406. (D) Pulldowns of GST or GST-Cdc4∆F from cdc4-1 cells expressing GALp-Hst3wt-9myc (wild-type) or GALp-Hst3d1m/d2m-9myc (m) purified with glutathione Sepharose beads. Hst3d1m/d2m is mutated for all six phosphorylation sites within D1 and D2. (Upper) Probed with an anti-myc antibody. (Lower) Probed with an anti-GST antibody. GST-Cdc4 fusion is expressed at lower levels than GST alone and therefore is not seen in the input with this exposure. The asterisk denotes a background band previously seen with these constructs (17).
SCFCdc4 recognizes its substrates through a phospho-degron (20–22). To understand how Hst3 was targeted for turnover both during an unperturbed cell cycle and after treatment with HU, we mapped replication stress-dependent and -independent phosphorylation sites on Hst3 by mass spectrometry. We found nine phosphorylation sites (Fig. 1C; details are given in Table S1 and spectra in Fig. S1). Seven of the phosphorylation sites that we mapped were located in a small region C-terminal to the conserved sirtuin catalytic domain.
To test whether Hst3 was a direct substrate of Cdc4, we tested whether Hst3 could bind to Cdc4. We used GST-tagged Cdc4 lacking its F-box motif (GST-Cdc4∆F) (17), which cannot interact with the other subunits of the SCFCdc4 complex and therefore allows easier observation of the interaction with its substrates. As shown in Fig. 1D, wild-type Hst3 binds to Cdc4∆F but not to GST alone (lanes 7 and 5). Cdc4 often binds its substrates through a Cdc4 phospho-degron containing closely spaced phosphorylation sites (20–22). Within the Hst3 C terminus are two small clusters of phosphorylation sites that correspond to the di-phosphorylated consensus (20, 21, 23), which in the subsequent analysis we called “degron 1” (D1) and “degron 2” (D2) (shown in Fig. 1C). Mutation of all six residues within D1 and D2 to alanine (Hst3d1m/d2m) blocks the binding of Hst3 to Cdc4∆F (Fig. 1D, lane 8).
The Hst3 C Terminus Functions as a Replication Stress-Responsive Degron.
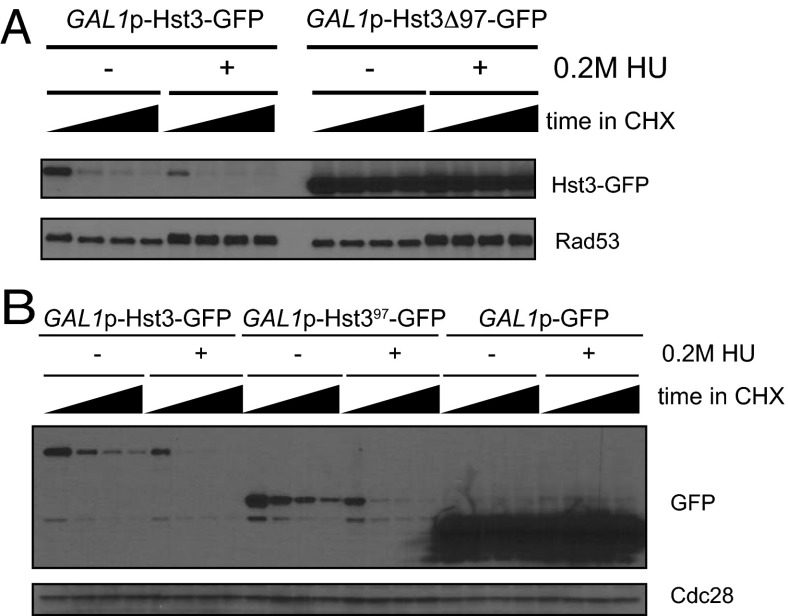
Deletion of the C-terminal 97 amino acids of Hst3 containing seven phosphorylation sites fully stabilized Hst3 (Fig. 2A). In addition, the same 97-amino acid region was sufficient to target the stable protein GFP for turnover, so that its turnover was very similar to that of full-length Hst3-GFP (Fig. 2B). Therefore, the terminal 97 amino acids of Hst3 act as a degron that is both necessary and sufficient for the turnover of Hst3. Moreover, this 97-amino acid degron is responsive to replication stress, suggesting that any changes in Hst3 turnover in response to replication stress are mediated through this degron.
Fig. 2.
The last 97 amino acids of Hst3 comprise a damage-responsive degron. (A) Hst3 full length or lacking the last 97 amino acids was expressed under the control of the GAL1 promoter and tagged with GFP. Hst3 was induced by the addition of galactose, and turnover was followed for 45 min, with time points taken every 15 min. For turnover in 0.2 M HU, cells were grown in galactose for 2.5 h, 0.2 M HU was added for an additional 2.5 h, and then cells were treated with cycloheximide. (B) GFP or GFP fused to either full-length Hst3 or to the last 97 amino acids of Hst3 was expressed under the control of the GAL1 promoter. The experiment was performed as in A.
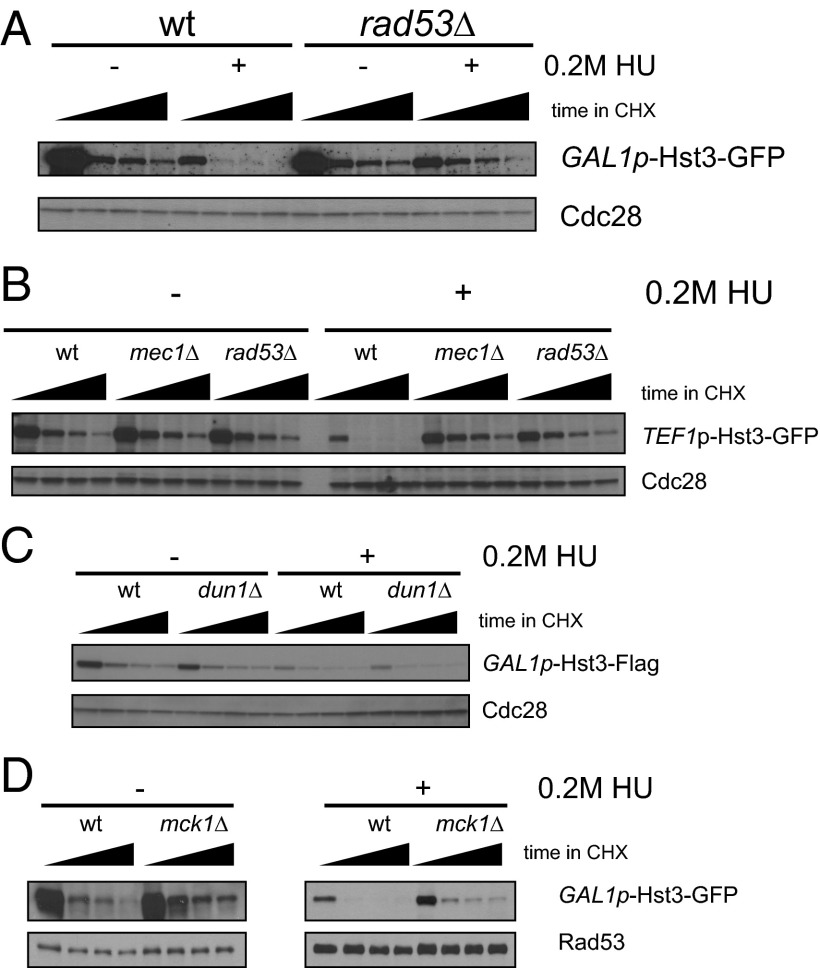
Previously, it was known that the increase in turnover seen in response to DNA damage required Mec1, the homolog of human ATR and an upstream activating kinase of both the DNA damage and replication checkpoint pathways (8). In response to both DNA damage and replication stress, the checkpoint kinase Mec1 phosphorylates and activates the downstream effector kinase Rad53. Rad53 then phosphorylates a number of substrates, including the kinase Dun1 (24, 25). We find that the increase in turnover seen in response to replication stress requires both MEC1 and RAD53 (Fig. 3 A and B and Fig. S2) but not the downstream checkpoint kinase Dun1 (Fig. 3C). To confirm this result independently of the GAL1 promoter, we used Hst3-GFP under the control of the TEF1 promoter (Fig. 3B). We again see a striking increase in turnover in response to HU, although the TEF1 promoter itself is slightly down-regulated in response to checkpoint activation (11). MEC1 and RAD53 are specifically required for the increase in turnover seen in response to replication stress but not for the basal level of Hst3 turnover seen in untreated cells. We see the same change in turnover for GAL1p-Hst3-TAP (Fig. S2), showing that this turnover is not dependent on the GFP tag.
Fig. 3.
Hst3 turnover is increased in response to replication stress. (A) Cycloheximide was added at t = 0, and protein turnover was followed for 45 min, with time points taken every 15 min, in cells expressing Hst3-GFP under the control of the GAL1 promoter (GAL1p-Hst3-GFP). For damaged cells, cells were grown in galactose and then were treated with 0.2 M HU before the addition of cycloheximide. Cdc28 is shown as a loading control. The black triangle indicates the time after the addition of cycloheximide (t = 0, 15, 30, and 45 min). (B) The experiment was performed as in A with cells expressing Hst3-GFP under the control of the TEF1 promoter. For damaged cells, 0.2 M HU was added before the addition of cycloheximide. (C) The experiment was performed as in A, except that Hst3 was tagged with Flag. (D) The experiment was performed as in A with wild-type and mck1∆ strains.
Because we were unable to observe direct phosphorylation of Hst3 by Rad53 in vitro, we investigated whether a downstream kinase might be involved. We chose to investigate the GSK3 homolog Mck1. Mck1 phosphorylates its substrates at a consensus site that contains a priming phosphorylated residue in the +4 position (26). Subsequent phosphorylation by Mck1 generates a di-phosphorylation matching the Cdc4 phospho-degron spacing, and Mck1 has been implicated previously in controlling turnover of several Cdc4 substrates (23, 27–29). Two of the phosphorylation sites we identified by mass spectrometry, T380 and S416, exactly match the Mck1 consensus (with a phosphorylated residue in the +4 position). In addition, an acidic residue replacing a normally phosphorylated residue in the +4 position can also allow GSK3 phosphorylation (30), in which case S420 and S421 also could be phosphorylated by Mck1. Hst3 was stabilized in an mck1∆ mutant both in the absence and presence of replication stress (Fig. 3D). Intriguingly, GSK3 kinases in mammalian cells have increased activity in response to UV and IR (31, 32), and the increase in activity is downstream of ATR (the homolog of yeast Mec1) (31). We propose that Mck1 in budding yeast also may be regulated in a checkpoint-dependent manner in response to DNA damage and replication stress. Consistent with this notion, we find evidence for phosphorylation of many of the sites identified in degrons 1 and 2 even in the absence of RAD53. Notably, Hst3 is not fully stabilized in the mck1∆ mutant, suggesting that additional kinases also may be involved. We note that S420 and S421 fit the casein kinase II consensus sequence (33).
Phosphorylation of the Hst3 Degron Is Required for Turnover.
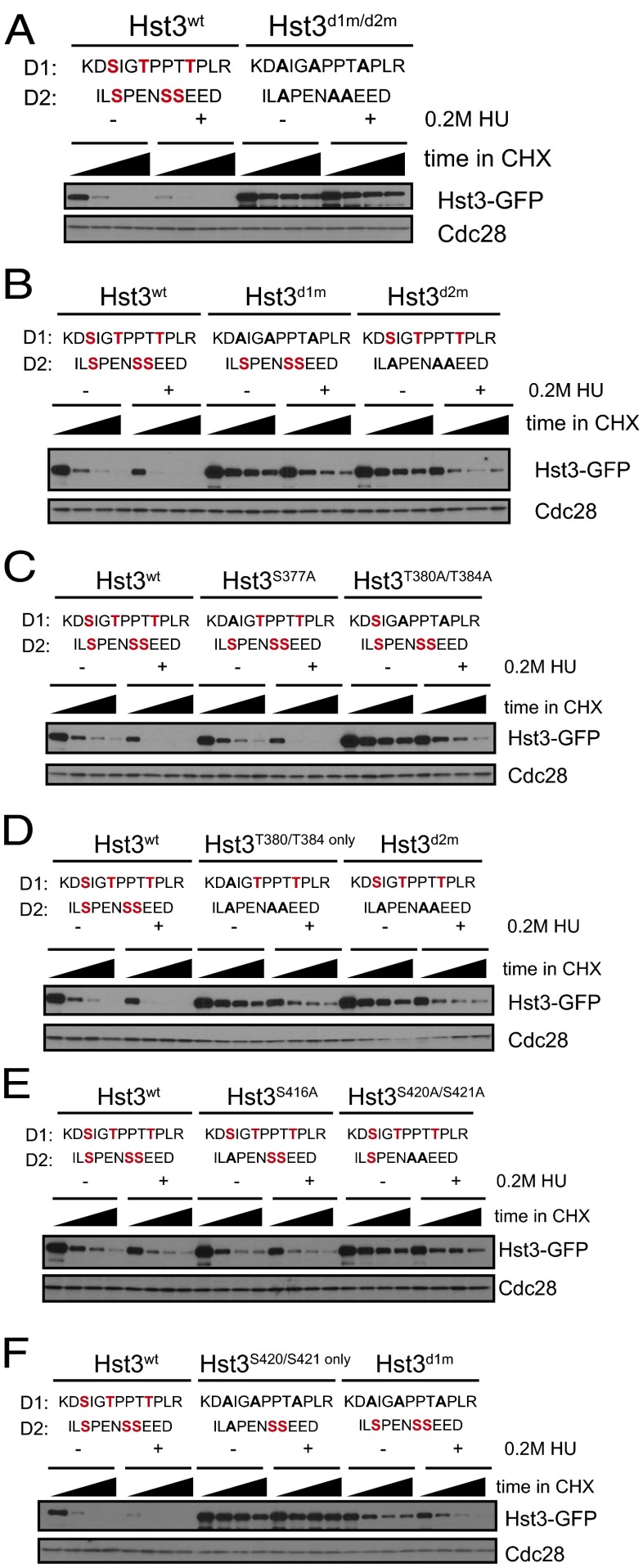
We next analyzed the phosphorylation requirements for Hst3 turnover more specifically. Consistent with our observation that Hst3d1m/d2m failed to interact strongly with Cdc4 (Fig. 1D), mutation of the six phosphorylation sites in D1 and D2 strongly stabilized Hst3 both in the absence and presence of replication stress (Fig. 4A, quantified in Fig. S3). The calculated half-life of wild-type Hst3 is 8 min in the absence of replication stress. In the presence of replication stress its half-life appears to be significantly less, although much more careful analysis will be required to determine this value, given its very low stability under these conditions. The calculated half-life of Hst3d1m/d2m is more than 45 min in both conditions. Mutation of the single remaining phosphorylation site (S406), which is contained within this 97-amino acid region but is not part of D1 or D2, did not affect Hst3 turnover either in untreated or in HU-treated cells (Fig. S4). Mutation of either the first or second degrons alone partially stabilized Hst3 (Fig. 4B, quantified in Fig. S3), although in both mutants we observed faster turnover in cells subjected to replication stress than in untreated cells (the half-life for both mutants was ∼30 min in untreated cells and ∼20 min in the presence of HU). This result suggests that both degrons are involved in targeting Hst3 for turnover and that each cluster alone can respond to replication stress, consistent with there being a potential Mck1 site within each degron.
Fig. 4.
Phosphorylation within two Cdc4 phospho-degrons controls Hst3 turnover. Hst3 was expressed under the control of the GAL1 promoter. Expression was induced by the addition of galactose, and turnover was followed for 45 min after the addition of cycloheximide, with time points taken every 15 min. For turnover in 0.2 M HU, cells were grown in galactose for 2.5 h; then 0.2 M HU was added for an additional 2.5 h (still in the presence of galactose), and cells were treated with cycloheximide. (A–F) Site mutants were used as designated, with wild-type phosphorylated residues in red and mutations in black for each degron (D1 and D2). All gels are quantified in Fig. S3.
Within the first degron, mutation of the first serine (S377A) did not affect Hst3 turnover, but mutation of the two threonines (T380A/T384A) stabilized Hst3 for both its normal and replication stress-induced turnover (Fig. 4C, quantified in Fig. S3). Moreover, the contribution of the first degron is mediated through the phosphorylation of T380 and T384 (Fig. 4D, quantified in Fig. S3), because those two sites are sufficient for turnover similar to the complete first degron. In the context of a wild-type D1, mutation of serine 416 (S416A) in the second degron had no obvious effect, but mutation of the other two serines (S420A/S421A) stabilized Hst3 (Fig. 4E, quantified in Fig. S3). However, in the absence of a functional D1, S420 and S421 are not sufficient for Hst3 targeting by the second degron (Fig. 4F, quantified in Fig. S3).
Hst3 Turnover Is Involved in Proper Regulation of Acetylated H3K56.
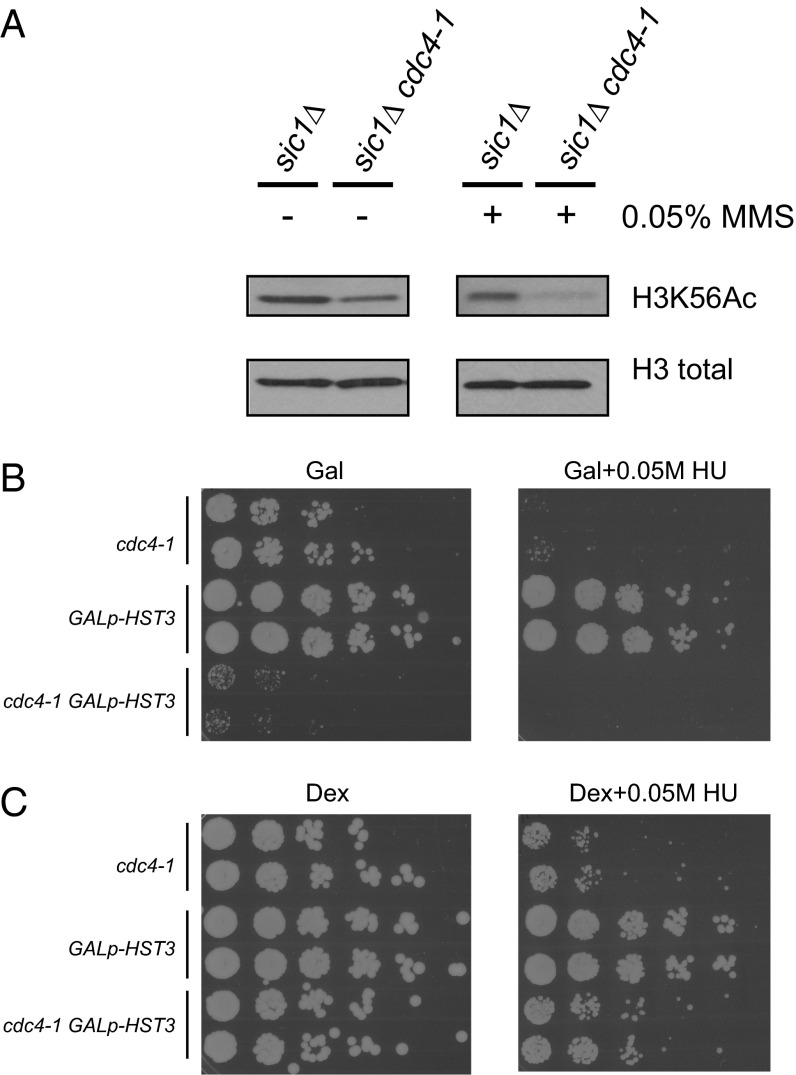
We wanted to test the functional consequence of failing to turn over Hst3 properly. We stabilized Hst3 by mutating CDC4 in the context of sic1∆. Acetylated K56 (K56Ac) levels were lower in cdc4-1 sic1∆ cells than in the sic1∆ controls, both in an asynchronous culture and after exposure to the DNA-damaging agent methyl methane sulfonate (MMS) (Fig. 5A), presumably because of high levels of Hst3 in the cdc4-1 strain. MMS enhances the difference that we observe, presumably because turnover of Hst3 by Cdc4 normally is much faster in MMS, and therefore the difference in Hst3 levels between the wild-type and cdc4-1 strains should be exacerbated. Hst4, the close homolog of Hst3, is also a substrate of Cdc4 (34), can also deacetylate K56Ac (5, 7), and probably also contributes to the low levels of K56Ac seen in cdc4-1 cells. These data suggest that degradation of Hst3 (and Hst4) likely is important for the regulation of K56Ac in an unperturbed cell cycle and for the maintenance of K56Ac in response to replication stress. sic1∆ strains previously have been shown to have DNA damage as observed by Ddc1 focus formation, but these cells do not activate the DNA-damage checkpoint (35).
Fig. 5.
Hst3 degradation is required for proper regulation of H3K56-Ac. (A) sic1∆ and sic1∆ cdc4-1 strains were shifted to 37 °C to inactivate Cdc4 in the absence or presence of 0.05% MMS for 2.5 h. Total histone H3 is shown as a loading control. (B) Shown are fivefold serial dilutions of the indicated strains grown on plates with galactose as the sugar source, which induces high levels of Hst3 under the control of the GAL1 promoter. Plates were grown at 30 °C for 3 d (GAL) or 4 d (GAL+HU) before scanning. (C) The experiment was performed as in B on plates with dextrose as the sugar source, which represses transcription of Hst3 under the control of the GAL1 promoter. Plates were grown at 30 °C for 3 d before scanning.
To test the consequences of misregulating K56Ac on growth and sensitivity to replication stress, we used a strain expressing HST3 under the control of the GAL1 promoter. With this promoter, HST3 is highly overexpressed when grown in the presence of galactose (Fig. 5B), and transcription of HST3 is inhibited when grown in the presence of dextrose (Fig. 5C). As shown in Fig. 5B, cdc4-1 mutants grow slowly and are sensitive to replication stress; however, simultaneous overexpression of HST3 exacerbates the phenotype, leading to even slower growth in the absence of damage and no visible growth in the presence of HU. Although this result is consistent with regulation of Hst3 turnover influencing sensitivity to replication stress, high levels of Hst3 in this context also could interfere with residual Cdc4 function, and the resulting phenotype could be caused, in part, by competition with the ubiquitination of other Cdc4 substrates. Transcriptionally repressing HST3 by growth in the presence of dextrose partially relieves the observed sensitivity of cdc4-1 mutants to HU (Fig. 5C), suggesting that high levels of Hst3 are at least partially responsible for the slow growth and sensitivity. These data are consistent with a role for Hst3 turnover in the proper regulation of K56Ac, because K56Ac misregulation previously has been shown to result in sensitivity to damage and replication stress (5, 7).
Discussion
HST3 expression is controlled by protein turnover. Here, we find that Hst3 turnover is mediated by SCFCdc4 after phosphorylation of a small, replication stress-responsive degron. As noted above, the closely related sirtuin Hst4 that can redundantly remove H3K56-Ac has itself been described as a target of SCFCdc4 (34). The mapped phospho-degron of Hst4 falls in a small region outside the conserved, catalytic sirtuin domain in the N terminus of Hst4. The location of these degrons suggests that the small regions that control degradation of Hst3 and Hst4 evolved later to control the enzymes without altering their catalytic function.
Cdc4 often recognizes its substrates through a phospho-degron containing two phosphorylation sites spaced closely together (20, 21, 23), although data on Sic1 in budding yeast suggests that this spacing may not be strictly required (36). We find that maximal degradation of Hst3 requires two such phospho-degrons, and each phospho-degron contributes to the increased turnover in response to replication stress. The phosphorylation sites we identified are required for turnover both in the presence and in the absence of replication stress, and deletion of the yeast GSK3 homolog MCK1 stabilizes Hst3 in both situations. We propose that Mck1 phosphorylates Hst3 to promote its Cdc4-dependent turnover in both conditions and that, as for mammalian GSK3, Mck1 activity may also be increased in a checkpoint-dependent way in response to replication stress (31, 32). Because an mck1∆ mutant does not fully stabilize Hst3, an additional kinase involved in Hst3 turnover may be differentially regulated by the checkpoint. Whether Hst3 is a direct target of Mck1 remains to be determined.
Recent work on the canonical Cdc4 target Sic1 shows that maximal degradation of Sic1 by SCFCdc4 also requires multiple phospho-degrons (36, 37). We find that in Hst3, two phosphorylation sites—S420 and S421—are required for maximal turnover even though the two sites alone are not sufficient for any turnover. In mammalian cells, cyclin E is recognized for turnover by a dimer of SCFFBW7 through two degrons. Intriguingly, one of the degrons involved is suboptimal and cannot, in the absence of the other degron, promote turnover (38). In Hst3, these two phosphorylation sites, S420 and S421, also have acidic residues in the +4 position, allowing them to function as a suboptimal degron in the absence of S416 phosphorylation. In the presence of S416 phosphorylation, on the other hand, they form a better Cdc4 phospho-degron, and therefore the second degron alone is sufficient for some Hst3 turnover. Thus, dimerization of Cdc4 may be involved in the recognition and ubiquitination of some of its substrates, as in mammalian SCFFBW7 (21, 39, 40).
In mammalian cells NUSAP1 was recently described as a target of an SCF ligase and turns over in an SCF-dependent manner in response to DNA damage (41). Particularly for substrates of SCF ligases, looking for changes in turnover in response to replication stress and DNA damage may be interesting, because phosphorylation is both critical for substrate recognition and a common mode of regulation by the DNA damage and replication checkpoints.
In addition to the regulation of Hst3 protein turnover described here, Hst3 is regulated transcriptionally during the cell cycle and in response to replication stress (5, 9–11). The combination of transcriptional and posttranscriptional control suggests that cells control Hst3 levels tightly during a normal cell cycle and that keeping Hst3 levels low in response to replication stress is critical. Stabilizing Hst3 while simultaneously deregulating its promoter presumably would lead to an even greater deregulation of K56Ac and a stronger cellular phenotype. Many proteins that turn over quickly are also under tight transcriptional regulation. The transcriptional clusters that vary during the cell cycle or in response to different cellular stresses may be a useful starting point for finding more substrates of SCF ligases.
Experimental Procedures
Yeast Methods.
Yeast strains were grown in YM-1 medium (42) with 2% (wt/vol) dextrose at 30 °C unless otherwise noted. Strains were made using standard techniques. Hst3 site mutations were generated using QuikChange Site-Directed Mutagenesis. Strains are listed in Table S2.
Expression from the GAL1 promoter was induced with 2% (wt/vol) galactose. For experiments with temperature-sensitive strains, cells were maintained at 23 °C until the experiment. In the assays shown in Fig. 5A, strains were shifted to 37 °C for 2.5 h to inactivate cdc4-1. For damaged cells in Fig. 5A, 0.05% (vol/vol) MMS was added simultaneously with the 37 °C temperature shift.
For the viability and sensitivity assays shown in Fig. 5 B and C, cells were grown overnight, diluted to the same OD, and plated onto yeast extract/peptone/dextrose (YPD) or yeast extract/peptone/galactose (YPG) plates or to the same plates containing 0.05 M HU. Cells were grown for 3 d (YPD, YPD + 0.05 M HU, and YPG) or 4 d (YPG + 0.05 M HU) and then were scanned.
Western Blots.
From cultures in midlog phase, cell pellets of equivalent optical densities were collected, washed with 1 mL 4 °C H2O, and frozen on dry ice. Pellets were thawed in boiling sample buffer [50 mM Tris (pH 7.5), 5% SDS, 5 mM EDTA, 10% glycerol, 0.5% β-mercaptoethanol, bromophenol blue, 1 µg/mL leupeptin, 1 µg/mL bestatin, 0.1 mM benzamidine, 1 µg/mL pepstatin A, 5 mM NaF, 1 mM Na3VO4, 80 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride]. Cells were boiled for 3 min, bead-beaten with glass beads for 3 min, and clarified by centrifugation. Extracts were analyzed by SDS/PAGE and Western blotting. Western blots were performed with low-salt PBS with Tween-20 (PBS-T) (15 mM NaCl, 1.3 mM NaH2PO4, 5.4 mM Na2HPO4, 0.05% Tween-20). Primary antibody incubations were performed in 5% (wt/vol) milk and low-salt PBS-T. Antibodies were used as follows: α-Cdc28 (yC-20; Santa Cruz Biotechnology) at 1:2,000; α-GFP (clone JL8; Clontech) at 1:1,000; α-Flag (clone M2; Sigma-Aldrich) at 1:2,000; α-Rad53 (DAB001, a gift from Durocher laboratory, University of Toronto, Toronto) at 1:2,000; α-TAP (CAB1001; Thermo Scientific) at 1:1,000; α-H3K56Ac (Upstate Cell Signaling Solutions) at 1:6,000; and α-H3 (ab1791; Abcam) at 1:2,000.
Mass Spectroscopy Analysis.
GAL1p-Hst3-Flag was induced by growth in YM-1 (42) with 2% galactose in a strain with cdc4-1 grown at the permissive temperature (23 °C). For damage-treated samples, cells were treated with 0.05% MMS for 6.5 h (the long time reflects the slow doubling time of cdc4-1 at 23 °C).Then 2 L of cells for each sample were collected. Pellets were lysed in lysis buffer [25 mM Hepes (pH 7.5), 250 mM NaCl, 0.2% Triton-X100, 1 mM EDTA, 10% glycerol, 1 µg/mL leupeptin, 1 µg/mL bestatin, 0.1 mM benzamidine, 1 µg/mL pepstatin A, 5 mM NaF, 1 mM Na3VO4, 80 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride] by bead-beating with glass beads for five to seven 1.5-min intervals in a cold block resting on ice in between intervals. Samples were clarified by centrifugation. Purification was done by immunoprecipitation with α-Flag antibody (clone M2; Sigma-Aldrich, as used for Western blotting) for 3–5 h, rotating at 4 °C. Beads were washed six times with lysis buffer supplemented to 500 mM NaCl. Purified protein was eluted with 150 µg/mL 3-Flag peptide. Samples were concentrated by trichloroacetic acid precipitation and run on SDS/PAGE, and the band corresponding to Hst3 was cut out and sent for mass spectrometry analysis.
Excised gel slices were proteolytically digested by the addition of trypsin as previously described (43, 44). Peptide digests were fractionated online using reversed-phase chromatography followed by analysis on a Thermo Fisher Orbitrap XL mass spectrometer in which MS/MS spectra were collected by data-dependent acquisition as previously described (43, 45). Phosphopeptide identification via database searching, decoy database-based probabilistic filtering, and phosphosite localization were performed using the ProLuCID, DTASelect, and Ascore algorithms, respectively, as implemented in the IP2 proteomic software suite (Integrated Proteomics Applications) (46–49). Phosphosites identified after filtering using a false-positive cutoff of 0.05 are shown in Fig. 1C, and the corresponding MS/MS spectra are shown in Fig. S1.
Cdc4-Binding Experiment.
The Cdc4-binding experiment was performed as described in ref. 17. Cells were grown in C-URA medium with galactose to induce Hst3 (wild-type or d1m/d2m) for 9 h at 23 °C. Cells then were shifted to 37 °C to inactivate cdc4-1 for 1 h. Twenty optical densities of cells were collected and lysed in Hepes lysis buffer [25 mM Hepes (pH 7.6), 400 mM NaCl, 0.2% Triton X-100, 1 mM EDTA, 10% glycerol, 1 µg/mL leupeptin, 1 µg/mL bestatin, 0.1 mM benzamidine, 1 µg/mL pepstatin A, 5 mM NaF, 1 mM Na3VO4, 80 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride] by bead-beating in a cold block for five 1.5-min bursts. Lysate was clarified by centrifugation at 4 °C. Extracts were incubated with 20 µL of 50% glutathione-Sepharose bead slurry in lysis buffer for 2 h at 4 °C. Beads were collected by centrifugation and washed seven times with 1 mL of cold lysis buffer. Proteins then were eluted by boiling in sample buffer and analyzed by Western blotting anti-GST (3818–1; Clontech) and anti-Myc (9E10).
Half-Life Assays.
Cells were grown as for Western blotting to midlog phase. Cycloheximide was added to cultures for a final concentration of 50 µg/mL after collection at the t = 0 time point. Equivalent ODs were collected for each time point and were processed for Western blots as described above. For all experiments with Hst3 under the control of the GAL1 promoter, cells were grown in galactose for several hours, to midlog phase, before the addition of cycloheximide or 0.2M HU. For half-life experiments in the presence of DNA-damaging agents, cells were treated with 0.2 M HU (still in the presence of galactose) for 2.5 h before the addition of cycloheximide. For cdc53-1 and cdc4-1 experiments, cells (including wild-type control) were grown up in galactose for several hours and then were shifted to 37 °C for 2.5–3 h. For the damage turnover in cdc53-1 and cdc4-1, cells were grown up in galactose and then were prearrested in 0.2 M HU for 3 h (while still in galactose) before the temperature shift to 37 °C for 2.5 h.
For quantifications shown in Fig. S3, gels were probed with fluorescent secondary antibodies from LI-COR (IRDye 800CW for both anti-goat and anti-mouse antibodies) and scanned on a LI-COR Odyssey Fc scanner. Quantification was done with Fiji (50). Quantified GFP values were normalized to corresponding Cdc28 loading control values for each blot. All values then were normalized to the wild-type starting level for each. For wild-type Hst3, half-life was calculated on each of the gels from only the 0- and 15-min time points, because levels approach background at the longer time points. The average across all gels is presented in the text. For stabilized mutants of Hst3, half-life was calculated including longer time points.
Supplementary Material
Acknowledgments
We thank D. Durocher for DAB001; C. Gross, H. Madhani, P. O’Farrell, and members of the D.P.T. laboratory for helpful discussions; N. Lyons and B. Cimini for technical assistance; and I. Foe for critical reading of the manuscript. This work was supported by National Science Foundation Grant 1144247 (to E.R.E.), National Institutes of Health Grants GM059691 (to D.P.T.), GM070539 (to D.P.T.), and GM089778 (to J.A.W.), and by funds from the Jonsson Cancer Center at University of California, Los Angeles (J.A.W.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315325111/-/DCSupplemental.
References
- 1.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436(7048):294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 2.Chen CC, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134(2):231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hyland EM, et al. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25(22):10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134(2):244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell. 2006;23(1):109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Ozdemir A, et al. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J Biol Chem. 2005;280(28):25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- 7.Celic I, et al. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16(13):1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Thaminy S, et al. Hst3 is regulated by Mec1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J Biol Chem. 2007;282(52):37805–37814. doi: 10.1074/jbc.M706384200. [DOI] [PubMed] [Google Scholar]
- 9.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9(12):3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edenberg ER, Vashisht A, Benanti JA, Wohlschlegel J, Toczyski DP. Rad53 downregulates mitotic gene transcription by inhibiting the transcriptional activator Ndd1. Mol Cell Biol. 2014;34(4):725–738. doi: 10.1128/MCB.01056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasch AP, et al. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12(10):2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yelamanchi SK, Veis J, Anrather D, Klug H, Ammerer G. Genotoxic stress prevents Ndd1-dependent transcriptional activation of G2/M-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 2014;34(4):711–724. doi: 10.1128/MCB.01090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 14.Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16(19):5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91(2):221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 16.Henchoz S, et al. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11(22):3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landry BD, Doyle JP, Toczyski DP, Benanti JA. F-box protein specificity for g1 cyclins is dictated by subcellular localization. PLoS Genet. 2012;8(7):e1002851. doi: 10.1371/journal.pgen.1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyons NA, Morgan DO. Cdk1-dependent destruction of Eco1 prevents cohesion establishment after S phase. Mol Cell. 2011;42(3):378–389. doi: 10.1016/j.molcel.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91(2):209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 20.Bao MZ, Shock TR, Madhani HD. Multisite phosphorylation of the Saccharomyces cerevisiae filamentous growth regulator Tec1 is required for its recognition by the E3 ubiquitin ligase adaptor Cdc4 and its subsequent destruction in vivo. Eukaryot Cell. 2010;9(1):31–36. doi: 10.1128/EC.00250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26(1):131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414(6863):514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 23.Lyons NA, Fonslow BR, Diedrich JK, Yates JR, 3rd, Morgan DO. Sequential primed kinases create a damage-responsive phosphodegron on Eco1. Nat Struct Mol Biol. 2013;20(2):194–201. doi: 10.1038/nsmb.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elledge SJ. Cell cycle checkpoints: Preventing an identity crisis. Science. 1996;274(5293):1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 25.Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14(2):237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 26.Fiol CJ, Mahrenholz AM, Wang Y, Roeske RW, Roach PJ. Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J Biol Chem. 1987;262(29):14042–14048. [PubMed] [Google Scholar]
- 27.Ikui AE, Rossio V, Schroeder L, Yoshida S. A yeast GSK-3 kinase Mck1 promotes Cdc6 degradation to inhibit DNA re-replication. PLoS Genet. 2012;8(12):e1003099. doi: 10.1371/journal.pgen.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishi T, Ikeda A, Nagao R, Koyama N. The SCFCdc4 ubiquitin ligase regulates calcineurin signaling through degradation of phosphorylated Rcn1, an inhibitor of calcineurin. Proc Natl Acad Sci USA. 2007;104(44):17418–17423. doi: 10.1073/pnas.0704951104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizunuma M, Hirata D, Miyaoka R, Miyakawa T. GSK-3 kinase Mck1 and calcineurin coordinately mediate Hsl1 down-regulation by Ca2+ in budding yeast. EMBO J. 2001;20(5):1074–1085. doi: 10.1093/emboj/20.5.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hergovich A, et al. Priming-dependent phosphorylation and regulation of the tumor suppressor pVHL by glycogen synthase kinase 3. Mol Cell Biol. 2006;26(15):5784–5796. doi: 10.1128/MCB.00232-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JY, Yu SJ, Park YG, Kim J, Sohn J. Glycogen synthase kinase 3beta phosphorylates p21WAF1/CIP1 for proteasomal degradation after UV irradiation. Mol Cell Biol. 2007;27(8):3187–3198. doi: 10.1128/MCB.01461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pontano LL, et al. Genotoxic stress-induced cyclin D1 phosphorylation and proteolysis are required for genomic stability. Mol Cell Biol. 2008;28(23):7245–7258. doi: 10.1128/MCB.01085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meggio F, Marin O, Pinna LA. Substrate specificity of protein kinase CK2. Cell Mol Biol Res. 1994;40(5-6):401–409. [PubMed] [Google Scholar]
- 34.Tang X, et al. Genome-wide surveys for phosphorylation-dependent substrates of SCF ubiquitin ligases. Methods Enzymol. 2005;399:433–458. doi: 10.1016/S0076-6879(05)99030-7. [DOI] [PubMed] [Google Scholar]
- 35.Lengronne A, Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1) Mol Cell. 2002;9(5):1067–1078. doi: 10.1016/s1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- 36.Tang X, et al. Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase. Proc Natl Acad Sci USA. 2012;109(9):3287–3292. doi: 10.1073/pnas.1116455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kõivomägi M, et al. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011;480(7375):128–131. doi: 10.1038/nature10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welcker M, et al. Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 2013;27(23):2531–2536. doi: 10.1101/gad.229195.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang X, et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129(6):1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 40.Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emanuele MJ, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147(2):459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Florens L, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40(4):303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaiser P, Wohlschlegel J. Identification of ubiquitination sites and determination of ubiquitin-chain architectures by mass spectrometry. Methods Enzymol. 2005;399:266–277. doi: 10.1016/S0076-6879(05)99018-6. [DOI] [PubMed] [Google Scholar]
- 45.Wohlschlegel JA. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol. 2009;497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- 46.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 47.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 48.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu T, et al. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5(10):S174. [Google Scholar]
- 50.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.