Significance
Many forms of muscular dystrophy produce muscle weakness through injury to skeletal muscle myofibers and specifically disruption of the muscle plasma membrane. Using a mouse model of muscular dystrophy in a genetically diverse background, a genome-wide scan for genetic modifiers was undertaken. A modifier locus that altered plasma membrane leak was interrogated, and a splice site variant in Anxa6, encoding annexin A6, was identified. The Anxa6 splice site produces a truncated annexin A6 protein. The truncated annexin A6 protein was found to inhibit membrane repair by disrupting the formation of the normal annexin A6-rich cap and repair zone. These data demonstrate annexin A6's role in muscle membrane leak and repair in muscular dystrophy.
Keywords: dystrophin, muscle, plasma membrane
Abstract
Many monogenic disorders, including the muscular dystrophies, display phenotypic variability despite the same disease-causing mutation. To identify genetic modifiers of muscular dystrophy and its associated cardiomyopathy, we used quantitative trait locus mapping and whole genome sequencing in a mouse model. This approach uncovered a modifier locus on chromosome 11 associated with sarcolemmal membrane damage and heart mass. Whole genome and RNA sequencing identified Anxa6, encoding annexin A6, as a modifier gene. A synonymous variant in exon 11 creates a cryptic splice donor, resulting in a truncated annexin A6 protein called ANXA6N32. Live cell imaging showed that annexin A6 orchestrates a repair zone and cap at the site of membrane disruption. In contrast, ANXA6N32 dramatically disrupted the annexin A6-rich cap and the associated repair zone, permitting membrane leak. Anxa6 is a modifier of muscular dystrophy and membrane repair after injury.
Dystrophin and its associated proteins stabilize the plasma membrane of muscle fibers (1). Mutations that disrupt dystrophin produce a fragile sarcolemma susceptible to contraction-induced damage and muscle fiber disruption (2). The dystrophin complex, which includes dystroglycan and the sarcoglycans, provides a mechanically strong link between the cytoskeleton and the extracellular matrix (3). Genetic loss of the sarcoglycan complex similarly weakens the sarcolemma, rendering it leaky and susceptible to damage (4). Dystroglycan adheres directly to the extracellular matrix in a manner dependent on its glycosylation status, underscoring the necessity of the membrane-matrix link for normal sarcolemmal function (5).
Genetic modifiers can alter the outcome in muscular dystrophy (MD). The primary gene mutation in Duchenne and Becker muscular dystrophy (DMD/BMD) is the strongest determinant of outcome, but other genes also influence clinical manifestations. Mutations in γ-sarcoglycan (SGCG) cause Limb-Girdle mucular dystrophy 2C (LGMD2C), which is characterized by MD and cardiomyopathy similar to DMD. The SGCG mutation Δ521-T is associated with a variable age of ambulatory loss and severity of cardiopulmonary involvement (6). The γ-sarcoglycan (Sgcg) mouse model closely mimics the phenotype seen in humans with LGMD2C (7). The severity of disease is highly dependent on genetic background pointing to a role for genetic modifiers (8).
The major cause of death in MD is cardiopulmonary failure. The major respiratory muscle, the diaphragm, is severely diseased in DMD and the LGMDs, a process well replicated in the mdx and Sgcg mouse models (7, 9). In DMD patients, accessory muscles become recruited to maintain breathing, and this includes the intercostal and abdominal muscles (10). In MD, cardiopulmonary involvement is highly variable. In brothers with dystrophin mutations, approximately half the sibships had significantly different courses of respiratory involvement (11).
We used quantitative trait locus (QTL) mapping in the Sgcg mouse model of MD to identify modifiers. A region on chromosome 11 was found to significantly modify membrane damage in the abdominal muscles and also modify right ventricle mass, genetically linking traits relevant to cardiopulmonary function. Whole genome sequencing combined with RNA sequencing identified variation in Anxa6 that correlated with these traits. The severe DBA/2J allele creates a cryptic splice donor site that truncates the annexin A6 protein generating a carboxy-terminally truncated annexin A6, called ANXA6N32. The annexins are calcium-dependent membrane binding proteins involved in membrane repair (12). Live cell imaging demonstrated that annexin A6 was recruited rapidly to sites of sarcolemma disruption after laser induced damage, forming a tight cap over a vesicle-rich repair zone. ANXA6N32 disrupts the cap and repair zone, causing visible leak. Anxa6 modifies MD because annexin A6 orchestrates sarcolemmal repair.
Results
Chromosome 11 Modifies Abdominal Muscle Membrane Damage and Right Ventricle Mass.
QTL mapping was performed using the Sgcg-null mouse model of LGMD2C (7). The Sgcg-null mutation in the DBA/2J (D2) mouse background results in severe disease with significant muscle membrane damage and fibrosis (8). In contrast, the Sgcg mutation on the 129T2/SvEmsJ (129) background is milder. A phenotypically and genetically diverse F3 QTL mapping cohort was created by interbreeding severe D2 (SgcgD2) and mild 129 (Sgcg129) Sgcg-null mice (Fig. S1). One hundred sixty-three SgcgD2/129 F3 animals were phenotyped at 8 wk of age by measuring mass of muscle groups and Evans blue dye content as a marker of membrane leak (13).
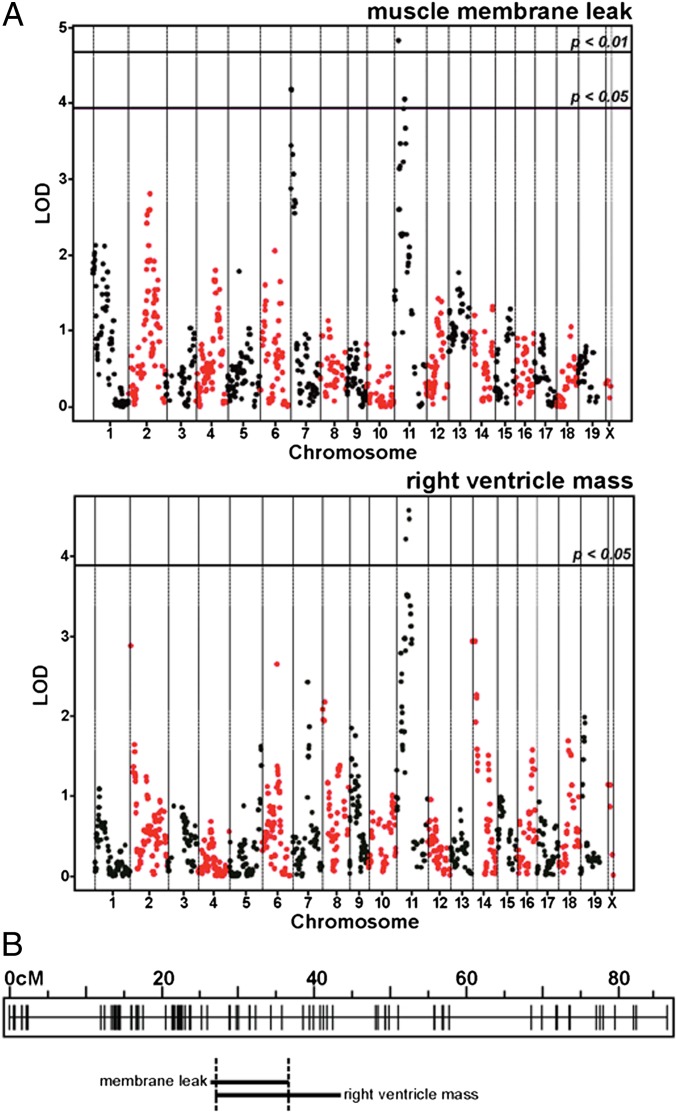
The F3 cohort was genotyped with nearly 2,000 SNP markers. QTLRel was used to identify modifier loci (14, 15). A chromosome 11 locus associated with abdominal muscle dye uptake, a measure of membrane damage. An overlapping chromosome 11 QTL was found to associate with right ventricle mass (Fig. 1). The confidence interval for abdominal muscle membrane damage extended from 25.9 to 36.4 cM. The confidence interval for the right ventricle mass QTL spanned from 27.3 to 43.2 cM. The QTLs overlapped in a region that extends from 27.3 to 36.4 cM. To identify genomic variation, we compared genome sequences of the DBA/2J and 129T2/SvEmsJ parental strains. The DBA/2J sequence derived from the Wellcome Trust database [24.7× coverage, ∼4.46 million SNPs, and 868,000 indels vs. the C57BL/6J reference (16)]. The 129T2/SvEmsJ substrain genome sequence was determined and aligned to the C57BL/6J (B6) mouse reference genome, version mm9; 52.5× coverage was achieved with ∼5.2 million SNPs and 1.9 million indel vs. the referent genomes. To identify polymorphisms between the 129 and D2 genomes, the DNA variants from each genome were compared to the referent B6 genome (Fig. S2).
Fig. 1.
QTL mapping identified a muscular dystrophy modifier locus on chromosome 11. (A) A mouse model of muscular dystrophy was used to map genetic modifiers using QTL mapping. Shown are the QTL plots for Evans blue dye uptake in the abdominal muscles (Upper) and right ventricle mass (Lower). Evans blue dye uptake is a measure of membrane leak. Normal muscle excludes dye. In muscular dystrophy, myofibers take up dye because of sarcolemmal instability. Increased right ventricle mass is a reflection of pathology including hypertrophy and edema. A region on chromosome 11 significantly associated with abdominal muscle membrane leak. The chromosome 11 region also modified right ventricle mass. (B) The locations of the chromosome 11 QTLs are shown with the genetic map of markers used for analysis. The membrane leak QTL interval overlaps with the right ventricular mass QTL.
Genic Variation in the Chromosome 11 QTL Identified Anxa6 as a Modifier.
QTL mapping targeted a smaller region of the chromosome 11 interval, which derived from an analysis using 2,313 markers and was completely embedded within the larger chromosome 11 interval. The interval spans 7.8 Mbp and contains 116 protein-coding genes, 80 of which are named genes (Table S1). Olfactory receptor genes (44 genes) were excluded from this count and analysis because of the low likelihood of their involvement in muscle and heart disease. Of the 80 genes, 20 genes contained genic variation between the parental strains (Fig. S2). RNA sequence data from WT and Sgcg hearts was compared using Tophat and Cufflinks (17, 18). The most highly expressed gene with protein coding variation was Anxa6.
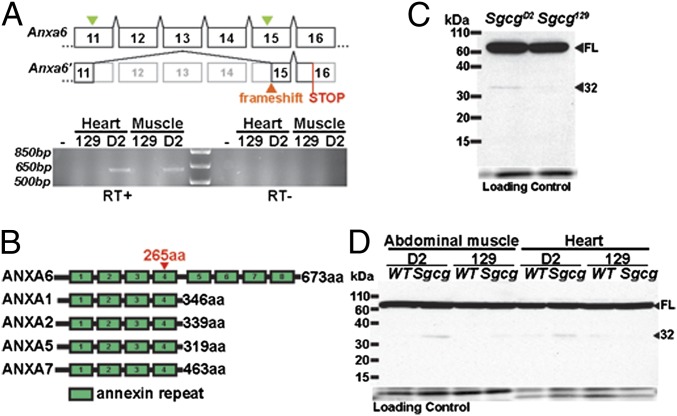
Anxa6 variants between the parental strains are indicated in Fig. S3. To examine how these variants altered Anxa6 transcripts, RT-PCR was performed using RNA from heart and abdominal muscle of WT and mutant animals from each strain (WTD2, WT129, SgcgD2, and Sgcg129). The entire coding mRNA of Anxa6 (exons 2–26) was interrogated, and an additional RT-PCR product, Anxa6′, was identified from heart and abdominal muscle from the DBA/2J strain. Anxa6′ derived from a splice junction between exons 11 and 15, disrupting the normal Anxa6 reading frame (Fig. 2A). Primers specific to the novel exon 11–15 junction confirmed this alternative product in mRNA from DBA/2J muscle and heart (Fig. 2A). The alternative splice junction occurs in the middle of each exon and is consistent with the use of cryptic splice donor and acceptor sites. Anxa6′ was expressed at much lower levels than the conventional transcript.
Fig. 2.
A splice variant of Anax6 is differentially expressed between the severe D2 strain and the mild 129 strain in heart and muscle. (A) Anxa6 contains a number of variants between the D2 and 129 genomes. Two synonymous variants occur in exons 11 and 15 (green arrows). Sanger sequencing of the additional RT-PCR product (Anxa6’) identified a splice junction between the middle of exon 11 and the middle of exon 15. The exon 11–15 junction deletes 124 amino acids and disrupts the reading frame of the transcript resulting in a premature STOP codon in exon 16. RT-PCR specific for the alternate splice form shows the alterative splice form in muscle and heart cDNA only in the D2 background. (B) The annexins are calcium-dependent membrane binding proteins. Annexin A6 is highly expressed in heart and muscle and uniquely contains eight annexin repeats (21). The alternate splice form of ANXA6 observed in D2 mice is predicted to truncate annexin A6 at amino acid 265 (red arrowhead) removing four carboxyl-terminal annexin repeats. (C) Immunoblotting with an anti-annexin A6 antibody recognizing an amino-terminal epitope detects high level expression of full-length annexin A6 in Sgcg hearts [68 kDa, full length (FL)]. An additional protein, ∼32 kDa (32), referred to as ANXA6N32, was present at higher levels in Sgcg hearts from the severe D2 background compared with those from the mild 129 background. (D) ANXA6N32 was expressed in abdominal muscle at higher levels in Sgcg from the D2 background. On longer exposure, ANXA6N32 could be detected in Sgcg129 tissues at lower levels. Sgcg tissues expressed higher levels of ANXA6N32 than WT; 129 WT tissue expressed no or very little ANXA6N32.
Anxa6′ is associated with rs26961431 in Anxa6 exon 11, and this G/A variant predicts an alternate exonic splice donor in exon 11 (Fig. S3). A second variant, 18 bp upstream of the Anxa6 alternate splice junction in exon 15, may also contribute because these are found together. The 162 Sgcg animals used for mapping were genotyped for rs26961431 to determine correlation with muscle membrane damage and right ventricle mass. Both traits were significantly different based on genotype (one-way ANOVA, P = 0.0013 and P = 0.0168, respectively; Fig. S4). The DBA/2J G allele associated with more severe phenotype for both traits, increased muscle membrane damage, and greater right ventricle mass. Increased mass in the right ventricle, or hypertrophy, is a known pathological trait (19). For right ventricle mass, the G variant had a dominant effect. The mode of inheritance of rs26961431 on muscle membrane damage is less clear. The chromosome 11 region containing Anxa6 was found to associate with right ventricular hypertrophy when analyzing an independent cohort of Sgcg mice in a mixed D2 and MRL background (Fig. S4) (20).
Smaller ANXA6 Protein Product Is Detected in DBA/2J.
The annexins are a family of calcium-dependent phospholipid binding proteins (21). Annexins typically contain four annexin repeats with the exception of ANXA6, which contains eight annexin repeats (Fig. 2B). Anxa6′ encodes a protein truncated from 673 to 265 amino acids, removing the four carboxyl-terminal annexin repeats. An amino-terminal anti-annexin A6 antibody detected robust expression of annexin A6 in Sgcg hearts from both the D2 and 129 backgrounds. An additional 32-kDa product, referred to as ANXA6N32, was detected in SgcgD2 hearts (Fig. 2C). ANXA6N32 protein was expressed at lower levels than full-length annexin A6 but was consistently detected in SgcgD2 hearts and was not seen with a carboxyl-terminal antibody. ANXA6N32 was also detected in SgcgD2 abdominal muscle (Fig. 2D). On longer exposure, the ANXA6N32 protein product could be detected in WTD2 heart and in Sgcg129 abdominal and heart muscle. Expression of the alternate transcript and protein in the 129 strain is not surprising given the nature of the sequences surrounding the Anxa6 splice junctions. The level of ANXA6N32 was always higher in tissues from the D2 strain. The synonymous variant present in D2 creates a cryptic splice donor, albeit an inefficient one, and it is possible that the A allele is also used as a splice donor but at even lower frequency.
DBA/2J and C57BL/6J Muscle Is More Susceptible to Membrane Damage.
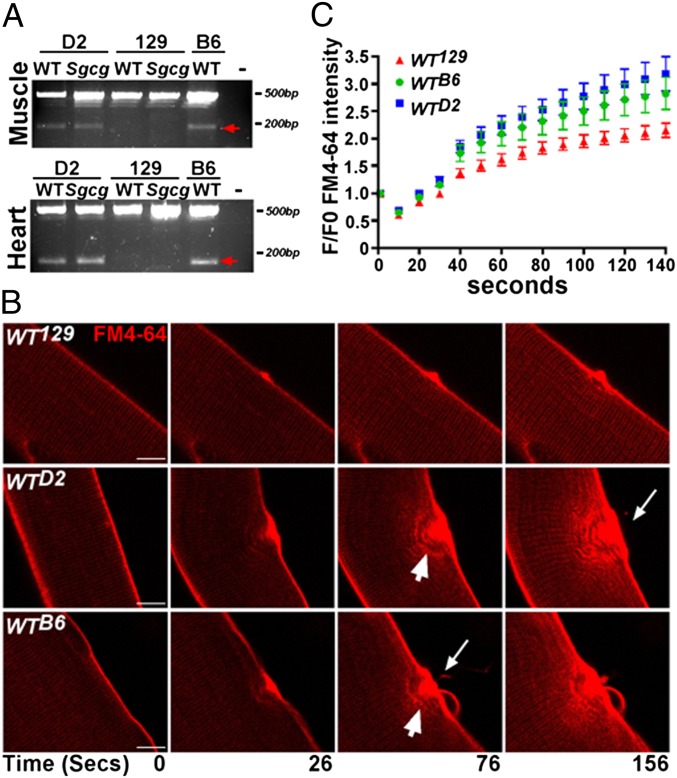
The C57BL/6J (B6) and DBA/2J (D2) inbred strains share a haplotype in the Anxa6 region of chromosome 11 so that B6 has the same Anxa6 variants as D2. Expression of the Anxa6 exon 11–15 splice form was seen in B6 heart and muscle (Fig. 3A), confirming a relationship between the genetic interval and the alternative transcript. Strains expressing the alternate Anxa6 transcript and protein are expected to have increased sarcolemmal damage. To assess sarcolemmal damage and repair, the plasma membrane of isolated muscle fibers was disrupted with a laser in the presence of FM4-64 dye (22, 23). FM4-64 is a lipophilic dye that exhibits low fluorescence in water and does not cross intact membranes (24). Myofiber fluorescence before injury is low, and FM4-64 increases fluorescence intensity when exposed to lipid membranes during injury. After laser damage, FM4-64 dye uptake was minimal in WT129 myofibers (Fig. 3 B, Top, and 3C). WTD2 and WTB6 myofibers, both of which express the ANXA6N32 protein product, demonstrated significantly elevated FM4-64 dye fluorescence compared with WT129 myofibers (Fig. 3 B, Middle and Bottom, white arrowheads, and C) (n > 10 fibers per genotype, n = 3 isolations). Representative time lapse movies of FM4-64 uptake after laser damage are included for WT129, WTD2, and WTB6 myofibers as Movies S1–S3, respectively. Fibers from B6/129 intercrossed mice displayed slower repair compared with 129 fibers, indicating that low levels of Anxa6′ are sufficient to reduce membrane repair (Fig. S5). Additionally, WTD2 and WTB6 myofibers had excessive FM4-64 leak (white arrows shown in Fig. 3B), suggestive of defective membrane patching. Thus, there was slowed muscle membrane repair in the WTD2 and WTB6 strains expressing ANXA6N32.
Fig. 3.
The presence of ANXA6N32 correlates with increased laser-induced membrane damage and slower membrane repair. (A) The D2 and B6 inbred strains of mice share a haplotype distinct from 129 in the chromosome 11 region of Anxa6. B6 mice also carry the SNPs that produce the Anxa6 alternative splice form. RT-PCR was performed on abdominal muscle and heart samples. In abdominal muscle, the alternate product is detected in only D2 and B6 samples (red arrows). The expected PCR product from full-length Anxa6 at ∼500 bp is detected in all samples. The same results are seen in heart tissue (Lower). (B) To assess sarcolemmal damage and repair, laser damage was induced in the presence of a lipophilic dye FM4-64. FM4-64 fluorescence increases until membrane resealing is complete. Damaged WT129 myofibers had minimal FM4-64 influx at 156 s after laser induced membrane disruption. WTD2 and WTB6 myofibers, both of which express endogenous Anxa6′ splice form, had dramatic FM4-64 dye influx during the same time after injury (156 s, arrowheads). FM4-64 extravasation was also detected in WTD2 and WTB6 fibers (white arrows) consistent with ineffective and inefficient membrane repair. (Scale bar, 5 µm.) (C) The amount of FM4-64 entry into myofibers was quantified after laser damage. Fibers from the 129 strain resealed more efficiently than those from the D2 or B6 strains.
ANXA6N32 Disrupts Membrane Repair.
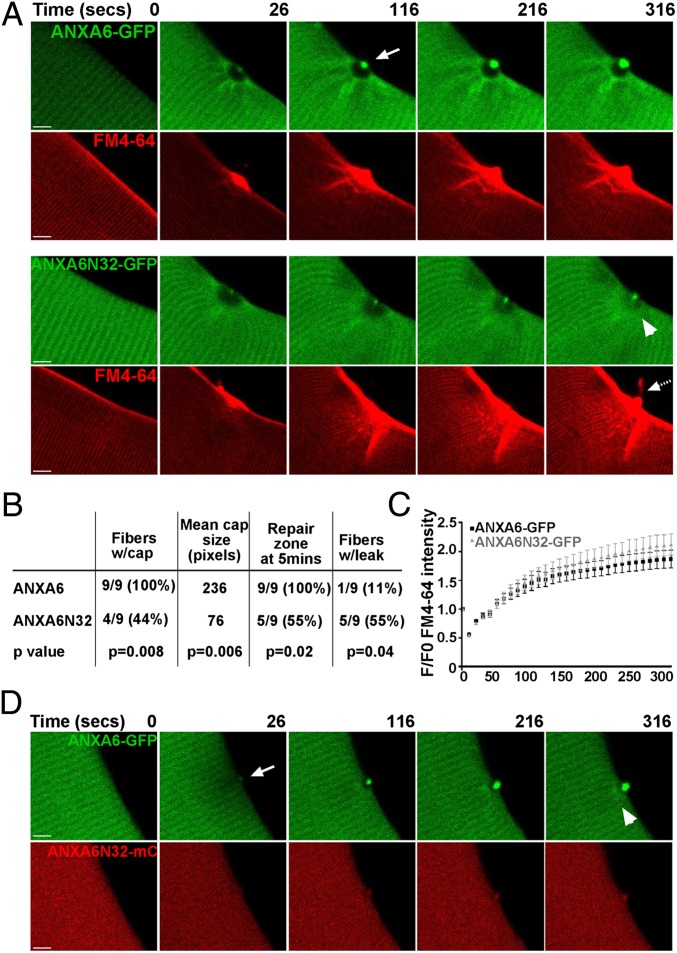
ANXA6-GFP and ANXA6N32-GFP plasmids were electroporated into myofibers to examine protein translocation in live cells after laser wounding. WT129 fibers were used to avoid endogenous ANXA6N32 protein, and FM4-64 was used to mark membrane damage. Before laser wounding, both ANXA6-GFP and ANXA6N32-GFP localized in a distinct, striated pattern at the Z-band, consistent with the known localization of endogenous annexin A6 (21) (Fig. 4A). Within 30 s of laser disruption, ANXA6-GFP was recruited to a cap at the site of sarcolemma damage (Fig. 4A, top row) in all fibers damaged (n = 9/9, 100%). This area correlated with the FM4-64 accumulation, representing vesicles recruited to the site of injury. In comparison, ANXA6N32-GFP formed a repair cap poorly or not at all (in four of nine FM4-64–positive fibers, 44%; P = 0.008; Fig. 4B). A representative ANXA6N32-GFP fiber forming a repair cap is shown (Fig. 4A, third row). In the 44% of ANXA6N32-GFP fibers that formed a repair cap, the cap was significantly smaller than the cap formed by full-length ANXA6-GFP (Fig. 4B; P < 0.006).
Fig. 4.
ANXA6N32 disrupts membrane resealing after membrane injury. WT129 myofibers were electroporated with ANXA6-GFP or ANXA6N32-GFP for live cell imaging after laser damage. (A) ANXA6-GFP (green) readily formed a distinct cap at the site of membrane disruption (arrow). This cap was visible by 26 s and persisted at 316 s after laser disruption. Concurrent imaging for FM4-64 showed that the cap forms over a vesicle-rich area devoid of annexin A6, referred to as the repair zone. In contrast, truncated annexin 6, ANXA6N32-GFP, formed a much smaller cap. The FM4-64 rich repair zone with ANXA6N32 was disorganized and hazy (compare GFP images at 316 s, arrowhead) and was frequently associated with leak of FM4-64 (dashed arrow). (Scale bar, 5 µm.) (B) Quantitation of laser experiments indicating that ANXA6N32 was associated with statistically smaller repair caps and leak. Full-length ANXA6-GFP formed a repair cap in 100% of damaged fibers. ANXA6N32-GFP formed a repair cap in only 44% of damaged fibers with a smaller mean size of the repair cap in those fibers that formed a cap (P < 0.006, n = 9). The repair zone of ANXA6-GFP persisted through 5 min in 100% of fibers, whereas the repair zone in ANXA6N32-GFP fibers persisted in only 55% of fibers. FM4-64 leak from the fiber occurred in 55% of ANXA6N32-GFP fibers at the site of damage, whereas only one ANXA6-GFP fiber (11%) showed FM4-64 efflux. (C) FM4-64 entry was quantified showing delayed repair with ANXA6N32 compared with ANXA6. (D) Expression of ANXA6-GFP along with ANXA6N32-mcherry (mC) demonstrated that ANXA6N32 disrupted the repair zone and cap formed by ANXA6, consistent with a dominant negative effect.
At the site of membrane disruption immediately under the annexin cap, there was a GFP-negative area enriched for FM4-64 dye, which we termed the repair zone. The repair zone was always distinctly seen with ANXA6-GFP and was buttressed by a clear striated pattern underlying the zone. In contrast, truncated ANXA6N32-GFP formed a hazy repair zone, characterized by a disrupted striated pattern and premature closure (arrowhead) in 55% (n = 5/9 fibers). This premature closure of ANXA6N32-GFP repair zones differed from ANXA6-GFP repair zones where 100% (n = 9/9) repair zones persisted past 5 min of imaging (P = 0.02; Fig. 4B and Movies S4–S7). Strikingly, five of nine (55%) ANXA6N32-GFP fibers leaked FM4-64 dye, whereas only one of nine (11%) of ANXA6-GFP fibers leaked dye (P = 0.04), consistent with ineffective repair of the disrupted membrane. ANXA6N32 induced delayed repair, seen as increased internalized FM4-64 (Fig. 4C). Fibers were scored for low and high level ANXA6N32-GFP expression, and membrane repair was equally slow in both, indicating that very low levels of ANXA6N32 are sufficient to disrupt repair (Fig. S5). Expression of both full-length ANXA6-GFP and ANXA6N32-mCherry demonstrates that the presence of ANXA6N32 interfered with normal membrane repair (Fig. 4D).
Altered ANXA6 Expression in MD.
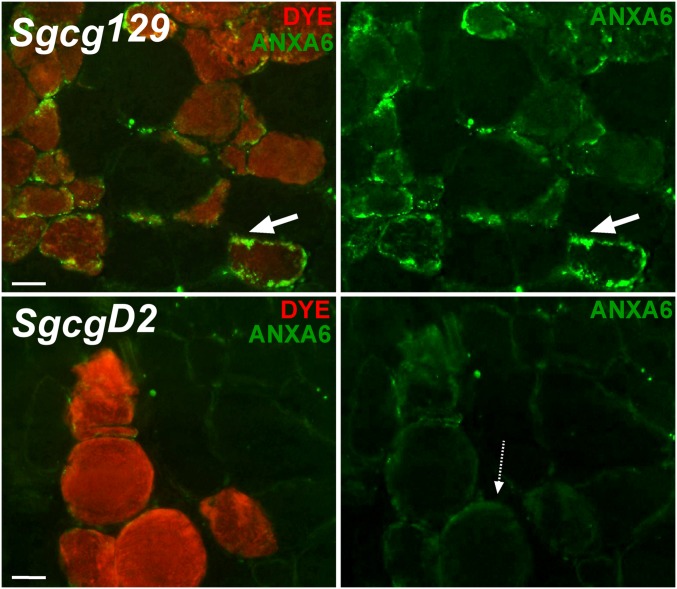
Sgcg muscle from the severe DBA/2J (SgcgD2) and mild 129T2/SvEmsJ (Sgcg129) strains was studied using an antibody to the amino terminus of annexin A6 (Fig. 5). Fibers with Evans blue dye uptake (red opacification indicative of a disrupted sarcolemma) showed membrane-associated annexin A6 staining in Sgcg129 muscle. Membrane-associated annexin A6 was reduced from the sarcolemma in the SgcgD2 muscle compared with the Sgcg129 muscle (Fig. 5). A similar pattern was seen in Sgcd muscle lacking δ-sarcoglycan (25). Sgcd129 muscle showed more membrane associated annexin A6 immunoreactivity, whereas SgcdB6, with the same Anxa6 haplotype as the D2 strain, showed less membrane-associated annexin A6 (Fig. S6). Dysferlin, a protein implicated in membrane repair, colocalized with annexin A6 (Fig. S6). Together these data are consistent with ANXA6N32 interfering with the membrane recruitment of full-length annexin A6.
Fig. 5.
Altered annexin A6 intracellular localization in myofibers with disrupted membranes. Sgcg129 and SgcgD2 mice were injected with Evans blue dye (Dye, red) to mark damaged muscle fibers. Shown are representative images of Sgcg129 and SgcgD2 muscle immunostained with an antibody to the amino terminus of annexin A6 (green). Annexin A6 strongly localized in discrete patches on the periphery of dye-positive Sgcg129 fibers (top row, white arrow). Annexin A6 was only weakly present on the periphery of dye-positive SgcgD2 fibers (dotted arrow).
Discussion
Annexin A6 as a Modifier of Injury and Sarcolemmal Repair.
The annexins are calcium-dependent membrane binding proteins found at the Z band or transverse tubules in heart and skeletal muscle (26). Annexin A6 is an atypical family member with eight instead of four annexin domains. Annexins bind both calcium and phospholipids, and with sarcolemmal disruption there is both an influx of calcium and exposure to distinct phospholipids (12). Using a zebrafish model, it was shown recently that dysferlin, a membrane repair protein, and annexins A1, A2, and A6 assemble at the site of sarcolemmal damage (27). Dysferlin mutations lead to MD, and loss of dysferlin is associated with slower membrane resealing after laser-induced disruption (22). Dysferlin was also previously shown to interact with annexins A1 and A2 to help regulate membrane resealing (28). These data support a role for annexin A6 broadly in membrane injury beyond muscle disease.
The dystrophin complex regulates stability of the sarcolemma, as the absence of dystrophin alters mechanical compliance rendering muscle more susceptible to damage (1). Dysferlin deficient muscle is defective in fiber repair rather than its promotion of muscle injury. Dysferlin deficiency exacerbates the phenotype in the dystrophin-deficient mdx mouse (29). In zebrafish, the combination of dysferlin and annexin 6 depletion generated a more severe myopathic process (27) suggesting that Anxa6 is modifier of multiple forms of MD.
Annexin A6 in MD.
Because genetic modifiers act in a non-Mendelian manner to alter the phenotype in question, the comparatively low levels of Anxa6′ and ANXA6N32 are highly consistent with its role as a modifier of MD and sarcolemmal injury. The annexin repeats that make up the core domain of the annexin proteins contain calcium-binding sites and are thought to be the site of interactions between the protein and membrane structures (30). Expression of even low levels of ANXA6N32 was associated with impaired membrane resealing. The level of expression of ANXA6N32 was ∼2% of the total annexin A6 protein, and the modifier effect explains ∼7% of the variance. Therefore, even small amounts of ANXA6N32 at the site of membrane damage decrease efficiency of membrane repair.
Although annexin A6 was identified as a modifier affecting abdominal muscles and ventricular mass, it would be expected to act in most if not all muscle groups. The D2 strain was previously noted to disrupt myoblast fusion in the mdx mouse model, and there are other QTLs that modify muscular dystrophy in the D2 strain (31, 32). Membrane repair and membrane fusion share some of the same molecular machinery (33). Because membrane fusion is necessary for myoblast fusion and muscle growth, it is possible that truncated annexin A6 imparts some of its effect by reducing muscle regeneration. Inefficient regeneration would exacerbate muscle damage in the context of increased demand for membrane resealing.
Modifiers of Cardiopulmonary Function in Muscle Disease.
With disruption of the dystrophin complex, the heart and respiratory musculature is susceptible to the same injury program as limb skeletal muscles. Membrane damage, marked by dye uptake, is thought to reflect an early stage in pathogenesis because it signifies sarcolemmal interruption. Right ventricular hypertrophy is pathological in most settings (19). It is possible that the increase in mass from Anxa6′ arises because of respiratory muscle impairment, deriving from failed diaphragm and abdominal muscles, as accessory muscles of respiration. Severe muscular dystrophy is sufficient to induce cardiomyopathy (34) and gene therapy to rescue the diaphragm muscle improves cardiac function, underscoring the interrelationship in cardiopulmonary disease (35).
Anxa6 is highly expressed in cardiomyocytes and so the effect of Anxa6 on ventricular mass may be a direct effect. Loss of Anxa6 in cardiomyocytes is associated with an increase in the degree of shortening and rates of contraction and relaxation (36). Thus, altered annexin A6 may actually promote damage directly in the ventricular myocytes, especially in the context of a weakened sarcolemma. In this setting, increase in ventricular mass may also reflect enhanced edema, as a consequence of increased sarcolemmal rupture. These data highlight the role of membrane repair in the process of muscular dystrophy and cardiomyopathy and underscore the importance of using sensitized strains to identify modifiers. Although these studies were designed to map modifiers of muscular dystrophy, given the known roles and patterns of expression of annexin A6, it is likely that Anxa6 is a general modifier and mediator of muscle and heart injury. SNPs within human ANXA6 have been associated with inflammatory phenotypes like psoriasis, but this genetic effect is thought to be due to the neighboring TNIP1 gene (37). Whether these SNPs, or others within ANXA6, will correlate with muscle or heart phenotypes is yet to be determined.
Materials and Methods
Animals.
Sgcg-null animals were described previously (8). F3-null SgcgD2/129 animals were generated from 22 unique breeding pairs comprised of null F2 SgcgD2/129 animals. All mice were housed in uniform conditions in a single pathogen-free barrier facility. Animals were housed and treated in accordance with the standards set by the University of Chicago Animal Care and Use Committee. F3 SgcgD2/129 animals used for modifier mapping were killed during their eighth week of life. Evans blue dye content was measured in muscle groups as previously described (8).
Genome-Wide Genotyping.
Low-density genotyping was performed on 189 F3 SgcgD2/129 animals using the custom-designed Illumina Golden Gate Mutation Mapping and Developmental Analysis Project (MMDAP) panel (Brigham and Women’s Hospital, Harvard Medical School; Harvard Partners Center for Genetics and Genomics; The Broad Institute). Additional medium density genome-wide genotyping used the Illumina Infinium Mouse Universal Genotyping Array (MUGA) panel (GeneSeek). Quality control thresholds were used based on marker performance data provided by GeneSeek: call frequency ≥ 0.75, minor allele frequency ≥ 0.05, minor genotype frequency ≥ 0.05. Marker segregation distortion was calculated using the geno.table function in R/qtl (38). Markers with P ≥ 1 × 10−7 were removed. The parental origin of each marker was determined using DBA/2J and 129S1 strain genotype data provided by GeneSeek (39). Potentially switched genotypes were identified using the est.rf function in R/qtl and corrected. F3 animals meeting the following criteria were used for mapping: genotype call frequency ≥ 0.75 and total crossover events ≤ 80 (calculated using the countXO function in R/qtl).
QTL Mapping.
The marker map file was created using marker position data provided by GeneSeek. Marker positions in National Center for Biotechnology Information Build 37 were converted to genetic distances in sex-averaged centimorgans using the Jackson Laboratory’s Mouse Map Converter tool (http://cgd.jax.org/mousemapconverter/) (40, 41). The genetic map of the markers used for analysis was plotted using the plot.map function in R/qtl. QTL mapping was performed using QTLRel (14, 15). Log-transformed values were used for phenotypes with nonnormal distributions. Identity coefficients were calculated using cic; the argument df = 0 was used. Default was used for all other arguments. Genetic matrices were derived using genMatrix and default arguments. The variance component was estimated with estVC. For the variance component, AA, DD, and EE were considered. Defaults were used for all other arguments. Missing genotypic data were imputed using default arguments for genoImpute. Genotype probabilities were calculated using genoProb and the Haldane method, gr = 2, and all other arguments with default settings. Genome scans were conducted using scanOne with default argument values and minorGenoFreq = 0.05. Sex was used as a noninteractive covariate for all phenotypes. Significance thresholds were determined by 1,000 permutation tests. QTL support intervals were estimated using lodci with cv = 2.5, lod = 1.5, and drop = 2.
Whole Genome Sequencing.
Genomic DNA was isolated from the liver of four WT 129T2/SvEmsJ animals. Sequencing libraries were constructed per Illumina protocol, except the final PCR amplification was for four cycles. The libraries were quantified using the Library Quant kit from Kapa Biosystems. Paired-end 2 × 100-bp sequencing was performed on the HiSeq2000 (Illumina). Alignment to the mouse reference genome mm9, NCBI Build 37 used the Burrows-Wheeler Alignment (BWA) tool (42), with the following arguments: −l 32 −t 4. Alignment files were converted to the SAM file format using BWA sampe. SAM files were converted to the BAM file format using SAMtools (43) view −bS. Alignments were sorted by position using Picard SortSam (http://picard.sourceforge.net). Duplicate reads were removed using SAMtools rmdup with argument –S. Variants between 129 and the mouse reference were called using SAMtools mpilup with argument −uf and bcftools with arguments view −bNvcg and filtered with samtools varFilter −D80. Variants between D2 and the mouse reference were obtained from the Wellcome Trust Sanger Institute’s Mouse Genomes Project (16). Alignment and variant statistics were calculated using bamtools and vcf− stats. Variants distinguishing the D2 and 129 strains were determined by comparing variants called against the reference genome. Variants were annotated using Ensembl’s Variant Effect Predictor (44) and SnpEff (45).
RNA Sequencing.
RNA sequencing libraries, starting with 500 ng total RNA, were constructed with the TruSeq RNA Sample Prep Kits v2 from Illumina on hearts from Sgcg and strain-matched controls. Five RNA samples were indexed with different adapters and pooled for paired-end 2 × 100-bp sequencing in Illumina HiSeq2000. RNA-seq reads were aligned with TopHat v2.0.2 (17) to the mouse genome, version mm9. The Tophat alignment rate was 89%, resulting in an average of 27 million reads per sample. Transcripts were assessed and quantities were determined by Cufflinks (18), using a GTF file based on Ensembl mouse NCBI37. Comparison expression levels were made using FPKM values using Cuffdiff from the Cufflinks package.
Laser Damage and Electroporation.
The flexor digitorum brevis (FDB) muscle bundle was dissected and placed in DMEM with BSA plus collagenase solution. Dissociated fibers were plated on Matek confocal microscopy dishes (P35G-1.5-14-C; Matek). FM4-64 dye (T-13320; Molecular Probes) was added at 2.5 μM before imaging. Fibers were irradiated using Bleach point in the fluorescence recovery after photobleaching (FRAP) wizard protocol in LAS AF Leica Imaging Software using a 405-nm laser set at 80% power for 3 s on a Leica SP5 2 photon microscope. Single images were acquired before damage, on laser damage, every 2 s after damage, and then one image every 10 s for a total of 176 or 326 s. AVI files were compiled in Image J from individual images. Basal FM4-64 fluorescence was lower in B6 fibers than in D2 and 129 strains. Therefore, for quantitative analysis, FM4-64 fluorescence was measured at the site of injury in individual frames using Image J and adjusted to baseline fluorescence at time 0 (F/F0), allowing comparison of all strains. Flexor digitorum brevis (FDB) fibers were transfected by in vivo electroporation methods described in detail in ref. 46. Muscle fibers were isolated as above and studied 7 d after electroporation to allow for recovery and protein expression in the electroporated muscles.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants U54AR052626, T32HL007381, T32HD007009, R01HL061322, R01NS047726, and P30 AR057230 and Parent Project Muscular Dystrophy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324242111/-/DCSupplemental.
References
- 1.Rahimov F, Kunkel LM. The cell biology of disease: Cellular and molecular mechanisms underlying muscular dystrophy. J Cell Biol. 2013;201(4):499–510. doi: 10.1083/jcb.201212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA. 1993;90(8):3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rybakova IN, Patel JR, Ervasti JM. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol. 2000;150(5):1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lapidos KA, Kakkar R, McNally EM. The dystrophin glycoprotein complex: Signaling strength and integrity for the sarcolemma. Circ Res. 2004;94(8):1023–1031. doi: 10.1161/01.RES.0000126574.61061.25. [DOI] [PubMed] [Google Scholar]
- 5.Goddeeris MM, et al. LARGE glycans on dystroglycan function as a tunable matrix scaffold to prevent dystrophy. Nature. 2013;503(7474):136–140. doi: 10.1038/nature12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNally EM, et al. Mild and severe muscular dystrophy caused by a single gamma-sarcoglycan mutation. Am J Hum Genet. 1996;59(5):1040–1047. [PMC free article] [PubMed] [Google Scholar]
- 7.Hack AA, et al. Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J Cell Biol. 1998;142(5):1279–1287. doi: 10.1083/jcb.142.5.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heydemann A, Huber JM, Demonbreun A, Hadhazy M, McNally EM. Genetic background influences muscular dystrophy. Neuromuscul Disord. 2005;15(9-10):601–609. doi: 10.1016/j.nmd.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Stedman HH, et al. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature. 1991;352(6335):536–539. doi: 10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- 10.Romei M, et al. Low abdominal contribution to breathing as daytime predictor of nocturnal desaturation in adolescents and young adults with Duchenne Muscular Dystrophy. Respir Med. 2012;106(2):276–283. doi: 10.1016/j.rmed.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Birnkrant DJ, et al. Cardiac and pulmonary function variability in Duchenne/Becker muscular dystrophy: An initial report. J Child Neurol. 2010;25(9):1110–1115. doi: 10.1177/0883073810371003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerke V, Creutz CE, Moss SE. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6(6):449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 13.Straub V, Rafael JA, Chamberlain JS, Campbell KP. Animal models for muscular dystrophy show different patterns of sarcolemmal disruption. J Cell Biol. 1997;139(2):375–385. doi: 10.1083/jcb.139.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng R, Abney M, Palmer AA, Skol AD. QTLRel: An R package for genome-wide association studies in which relatedness is a concern. BMC Genet. 2011;12:66. doi: 10.1186/1471-2156-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng R, et al. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics. 2010;185(3):1033–1044. doi: 10.1534/genetics.110.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haddad F, Hunt SA, Rosenthal DN, Murphy DJ. Right ventricular function in cardiovascular disease, part I: Anatomy, physiology, aging, and functional assessment of the right ventricle. Circulation. 2008;117(11):1436–1448. doi: 10.1161/CIRCULATIONAHA.107.653576. [DOI] [PubMed] [Google Scholar]
- 20.Heydemann A, et al. The superhealing MRL background improves muscular dystrophy. Skelet Muscle. 2012;2(1):26. doi: 10.1186/2044-5040-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camors E, Monceau V, Charlemagne D. Annexins and Ca2+ handling in the heart. Cardiovasc Res. 2005;65(4):793–802. doi: 10.1016/j.cardiores.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Bansal D, et al. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423(6936):168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 23.Cai C, et al. MG53 nucleates assembly of cell membrane repair machinery. Nat Cell Biol. 2009;11(1):56–64. doi: 10.1038/ncb1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128(5):779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hack AA, et al. Differential requirement for individual sarcoglycans and dystrophin in the assembly and function of the dystrophin-glycoprotein complex. J Cell Sci. 2000;113(Pt 14):2535–2544. doi: 10.1242/jcs.113.14.2535. [DOI] [PubMed] [Google Scholar]
- 26.Camors E, et al. Association of annexin A5 with Na+/Ca2+ exchanger and caveolin-3 in non-failing and failing human heart. J Mol Cell Cardiol. 2006;40(1):47–55. doi: 10.1016/j.yjmcc.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Roostalu U, Strähle U. In vivo imaging of molecular interactions at damaged sarcolemma. Dev Cell. 2012;22(3):515–529. doi: 10.1016/j.devcel.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Lennon NJ, et al. Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J Biol Chem. 2003;278(50):50466–50473. doi: 10.1074/jbc.M307247200. [DOI] [PubMed] [Google Scholar]
- 29.Han R, Rader EP, Levy JR, Bansal D, Campbell KP. Dystrophin deficiency exacerbates skeletal muscle pathology in dysferlin-null mice. Skelet Muscle. 2011;1(1):35. doi: 10.1186/2044-5040-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerke V, Moss SE. Annexins: From structure to function. Physiol Rev. 2002;82(2):331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 31.Fukada S, et al. Genetic background affects properties of satellite cells and mdx phenotypes. Am J Pathol. 2010;176(5):2414–2424. doi: 10.2353/ajpath.2010.090887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swaggart KA, Heydemann A, Palmer AA, McNally EM. Distinct genetic regions modify specific muscle groups in muscular dystrophy. Physiol Genomics. 2011;43(1):24–31. doi: 10.1152/physiolgenomics.00172.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han R, Campbell KP. Dysferlin and muscle membrane repair. Curr Opin Cell Biol. 2007;19(4):409–416. doi: 10.1016/j.ceb.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Megeney LA, et al. Severe cardiomyopathy in mice lacking dystrophin and MyoD. Proc Natl Acad Sci USA. 1999;96(1):220–225. doi: 10.1073/pnas.96.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crisp A, et al. Diaphragm rescue alone prevents heart dysfunction in dystrophic mice. Hum Mol Genet. 2011;20(3):413–421. doi: 10.1093/hmg/ddq477. [DOI] [PubMed] [Google Scholar]
- 36.Song G, et al. Altered mechanical properties and intracellular calcium signaling in cardiomyocytes from annexin 6 null-mutant mice. FASEB J. 2002;16(6):622–624. doi: 10.1096/fj.01-0892fje. [DOI] [PubMed] [Google Scholar]
- 37.Sun LD, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42(11):1005–1009. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 39.Consortium CC. Collaborative Cross Consortium The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190(2):389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox A, et al. A new standard genetic map for the laboratory mouse. Genetics. 2009;182(4):1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shifman S, et al. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4(12):e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiFranco M, Quinonez M, Capote J, Vergara J. DNA transfection of mammalian skeletal muscles using in vivo electroporation. J Vis Exp. 2009 doi: 10.3791/1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.