Significance
The gastrointestinal tract directly faces an array of environmental agents, including bacteria and food. Intestine-specific subsets of immune cells maintain gut homeostasis by continuously sampling luminal antigens and maintaining immune tolerance. We here demonstrated that Rbpj, an essential molecule for Notch signaling, is essential for the development of CD11c+CX3CR1+ cells that are crucial for sampling luminal antigens with dendrites projecting through the epithelial cell layer. These findings indicate that Notch signaling is required for maintaining the repertoires of intestinal antigen-presenting cells and suggest new ways to keep immune tolerance by modulating Notch-mediated CD11c+CX3CR1+ cells.
Abstract
The gastrointestinal tract comes into direct contact with environmental agents, including bacteria, viruses, and foods. Intestine-specific subsets of immune cells maintain gut homeostasis by continuously sampling luminal antigens and maintaining immune tolerance. CD11c+CX3CR1+ cells sample luminal antigens in the small intestine and contribute to the trafficking of bacteria to lymph nodes under dysbiotic conditions. The molecular mechanisms crucial for the differentiation of CD11c+CX3CR1+ cells remain unclear. Here we demonstrate that the Notch1– or Notch2–Rbpj axis is essential for the development of CD11c+CX3CR1+ cells. In mice in which Rbpj or Notch1 and Notch2 were deleted from CD11c+ cells, there was a deficit of CD11c+CX3CR1+ cells and an accumulation of CD11clowCX3CR1+ cells. The CD11clowCX3CR1+ cells could not differentiate to CD11c+CX3CR1+ cells, suggesting that CD11clowCX3CR1+ cells represent a lineage distinct from CD11c+CX3CR1+ cells. These data indicate that Notch signaling is essential for lineage fixation of intestinal CD11c+CX3CR1+ cells.
The intestinal mucosa comes into direct contact with a complex environment that includes both foods and microorganisms. Consequently, it has unique subsets of immune cells to protect the host (1–4), and complex immunoregulatory mechanisms determine the immune tolerance to commensal microbes and food antigens (2, 4).
Dendritic cells (DCs) and macrophages in the intestine are required for sensing the presence of invading pathogens. The lamina propria (LP) contains a variety of DCs and macrophage subsets. One such subset expresses integrin αE (CD103) and has the ability to drive IL-17–producing T cells (5, 6). The CX3C chemokine receptor 1 (CX3CR1)+ cells that express integrin αX (CD11c) and a macrophage marker, F4/80 but not CD103 are crucial for the sampling of luminal antigens through their use of projecting dendrites that penetrate the epithelial cell layer (7–9). Furthermore, CD11c+CX3CR1+ cells are involved in the trafficking of commensal bacteria into mesenteric lymph nodes in the absence of Myd88 or under dysbiotic conditions (10) and transfer the luminal soluble antigen to integrin αM (CD11b)+CD103+ DCs via the gap junction to induce oral tolerance (11). The exact molecular mechanism that controls the development or differentiation of CD11c+CX3CR1+ cells remains unclear.
Rbpj is a transcriptional regulator required for Notch signaling (12). Accumulating evidence indicates that Notch signaling is crucial for the development of immune cells and for the functional differentiation of T cells (12–19). Recent studies have revealed that Notch signaling also regulates the development of DCs in the spleen and intestine (20) (21, 22). These data indicate that Rbpj regulates the survival of CD8− DCs in the spleen and that Notch2 signaling controls CD11b+CD103+ DCs in the intestinal LP. A recent study has revealed that Notch2-regulated intestinal DCs produce IL-23 required for controlling Citrobacter rodentium (22). In addition to the development or differentiation of DCs, Notch plays an important role in the activation of DCs and macrophages (23, 24).
Here we demonstrate that Rbpj is required for the development of CD11c+CX3CR1+ cells and that both Notch1 and Notch2 are involved in the development of CD11c+CX3CR1+ cells. The lack of CD11c+CX3CR1+ cells in mice in which Rbpj or Notch1 and Notch2 was deleted from CD11c+ cells was accompanied by an accumulation of CD11clowCX3CR1+ cells that could not differentiate toward CD11c+CX3CR1+ cells. These findings indicate that Notch–Rbpj axis is required for the lineage fixation of CD11c+CX3CR1+ cells and suggest that Notch signaling has a complex role in maintaining repertoires of intestinal antigen-presenting cells.
Results
Deficiency of Rbpj in CD11c+ Cells Reduces CD11c+CX3CR1+ Cells.
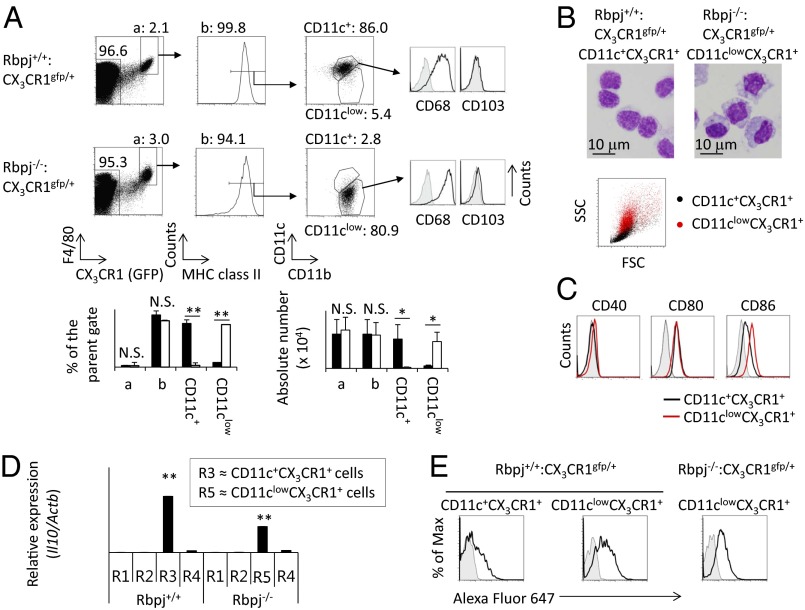
To reveal the involvement of Notch signaling in the development of CD11c+CX3CR1+ cells, Rbpjflox/flox mice were crossed with CD11c promoter-driven Cre transgenic (Rbpj−/−) mice, and this strain was crossed further with Cx3cr1 knockin mice in which Cx3cr1 was replaced by GFP (7). (Hereafter, this strain is called “Rbpj−/−:CX3CR1gfp/+ mice.”) We first investigated CX3CR1+ cells in the small intestine of Rbpj−/−:CX3CR1gfp/+ mice. The expression of CD11c and CD11b was evaluated by gating on F4/80, CX3CR1, and MHC class II+ LP cells from Rbpj−/−:CX3CR1gfp/+ mice and control Rbpj+/+:CX3CR1gfp/+ mice (Fig. 1A). We found a marked reduction of CD11c+CX3CR1+ cells in Rbpj−/−:CX3CR1gfp/+ mice (Fig. 1A). In contrast, in Rbpj−/−:CX3CR1gfp/+ mice, there was an accumulation of CD11clowCX3CR1+ cells that were rare in control Rbpj+/+:CX3CR1gfp/+ mice (Fig. 1A). Both CD11c+CX3CR1+ cells in Rbpj+/+:CX3CR1gfp/+ mice and CD11clowCX3CR1+ cells in Rbpj−/−:CX3CR1gfp/+ mice were negative for CD103 and positive for the macrophage marker CD68 (Fig. 1A). These data indicate that CD11c+CX3CR1+ cells and CD11clowCX3CR1+ cells are not DCs. The sideways scatter (SSC) and forward scatter (FSC) were relatively higher in CD11clowCX3CR1+ cells than in CD11c+CX3CR1+ cells (Fig. 1B). Giemsa staining of cells demonstrated that CD11clowCX3CR1+ cells had larger cytosols with irregular shapes and contained more granules than did CD11c+CX3CR1+ cells (Fig. 1B).
Fig. 1.
Rbpj deficiency in CD11c+ cells disturbs the differentiation of CD11c+CX3CR1+ cells. (A) Small intestinal LP cells from Rbpj−/−:CX3CR1gfp/+ and Rbpj+/+:CX3CR1gfp/+ mice were stained with anti-F4/80, MHC class II, CD11b, CD11c, CD103, and CD68 antibodies. (Upper) Seven-amino actinomycin D–negative (7AAD−) cells were analyzed by flow cytometry. The gray histogram shows the negative control. The numbers shown in each figure indicate the percentage of cells in parental gates. (Lower) The size of each population in Rbpj−/−:CX3CR1gfp/+ mice (open bars) and Rbpj+/+:CX3CR1gfp/+ mice (filled bars) was calculated by counting the total number of small intestinal cells and the percentage of each population. The data are shown as means ± SE. *P < 0.05; **P < 0.01. N.S., not significant. (B) (Upper) Giemsa staining of sorted F4/80+MHC class II+CD11b+CD11clowCX3CR1+ (CD11clowCX3CR1+) cells from Rbpj−/−:CX3CR1gfp/+ mice and F4/80+MHC class II+CD11b+CD11c+CX3CR1+ (CD11c+CX3CR1+) cells from Rbpj+/+:CX3CR1gfp/+ mice. (Original magnification: 1,000×.) (Scale bar: 10 μm.) (Lower) The sizes of CD11clowCX3CR1+ cells from Rbpj−/−:CX3CR1gfp/+ mice (red dots) and CD11c+CX3CR1+ cells from Rbpj+/+:CX3CR1gfp/+ mice (black dots) were assessed by FSC and SSC. (C) CD11clowCX3CR1+ cells from Rbpj−/−:CX3CR1gfp/+ mice (red line) or CD11c+CX3CR1+ cells from Rbpj+/+:CX3CR1gfp/+ mice (black line) were stained with anti-CD40, CD80, or CD86 for evaluation by flow cytometry. Fluorescence-conjugated streptavidin or isotype-matched control antibody (gray) was used as the negative control. (D) Expression of Il10 mRNA from sorted R1–R4 populations from Rbpj+/+ mice or from the R5 population from Rbpj−/− mice was evaluated by real-time PCR and is shown as the relative mRNA expression levels of Il10 versus Actb. The data are shown as means ± SE. **P < 0.01. The gating strategy is the same as in Fig. S3A. (E) Alexa Fluor 647-conjugated OVA (black line) or unconjugated OVA (gray) was injected through the duodenum in Rbpj−/−:CX3CR1gfp/+ mice or Rbpj+/+:CX3CR1gfp/+ mice. Five hours after injection, expression of Alexa Fluor 647 in CD11clowCX3CR1+ cells and CD11c+CX3CR1+ cells was evaluated by flow cytometry. The data shown in these figures are representative of four independent experiments.
We next compared the expression of cell-surface molecules on CD11clowCX3CR1+ and CD11c+CX3CR1+ cells (Fig. 1C). CD11clowCX3CR1+ cells expressed CD40 and CD80 at levels equivalent to CD11c+CX3CR1+ cells (Fig. 1C). In contrast, the expression of CD86 (Fig. 1C) and CD68 (Fig. 1A) was higher in CD11clowCX3CR1+ cells than in CD11c+CX3CR1+ cells, and the expression of CD4 was lower in CD11clowCX3CR1+ cells than in CD11c+CX3CR1+ cells (Fig. S1A). The expression pattern of cell-surface molecules on CD11clowCX3CR1+ cells in Rbpj−/−:CX3CR1gfp/+ mice was similar to that in Rbpj+/+:CX3CR1gfp/+ mice (Fig. S1B). The absence of the Cx3cr1 does not affect the development of CD11c+CX3CR1+ cells (Fig. S2), indicating that CX3CR1 is not involved in the development of Rbpj-mediated differentiation of CD11c+CX3CR1+ cells.
Previous studies categorized intestinal antigen-presenting cells into four groups (R1–R4) based on their expression of CD11c and CD11b (Fig. S3A) (5). The R1, R2, R3, and R4 populations consist of CD11b−CD103+ DCs, CD11b+CD103+ DCs, macrophages, and eosinophils, respectively (5). The R3 population expresses MHC class II (Fig. S3B), and about 90% of the R3 population is positive for CX3CR1 and F4/80 (Fig. S3C), indicating that the R3 population is almost identical to CD11c+CX3CR1+ cells. Therefore, when analyzing mice that do not have CX3CR1-gfp allele, we used R3 as a population that corresponds to CD11c+CX3CR1+ cells. By analyzing CD11c and CD11b in Rbpjf/f-CD11c mice, we found an increased proportion of CD11clowCD11b+ cells that we named “R5” (Fig. S3A). CD11clowCD11b+ cells were positive for MHC class II (Fig. S3B), CX3CR1, and F4/80 (Fig. S3C), indicating that the R5 population is almost identical to CD11clowCX3CR1+ cells. A previous study revealed that the R3 population secretes IL-10, which is involved in the low T-cell–stimulatory activity of this population (25, 26). We tested the mRNA expression of Il10 in R1–R4 populations from control mice and in the R5 population from Rbpj−/− mice. As previously reported, the R3 (CD11c+CX3CR1+) population expressed Il10 at a high level, and the R5 (CD11clowCX3CR1+) population also expressed substantial levels of Il10 mRNA (Fig. 1D).
To compare the antigen-capturing activity of CD11c+CX3CR1+ and CD11clowCX3CR1+ cells, Alexa Fluor 647-conjugated ovalbumin (OVA) was injected through the duodenum, and OVA uptake was evaluated 5 h later. The CD11clowCX3CR1+ population in Rbpj−/−:CX3CR1gfp/+ mice and the CD11clowCX3CR1+-like population in Rbpj+/+:CX3CR1gfp/+ mice took up more OVA than did the CD11c+CX3CR1+ population (Fig. 1E), indicating that CD11clowCX3CR1+ cells have a greater ability than CD11c+CX3CR1+ cells to take up exogenous antigens.
We assessed functional difference in intestinal immune responses between Rbpj+/+ and Rbpj−/− mice by using a dextran sodium sulfate (DSS)-induced colitis model. However, severity as evaluated by the body weight loss was comparable between two groups (Fig. S4).
Rbpj−/− Mice Have Few CD11c+Cells in the Small Intestine.
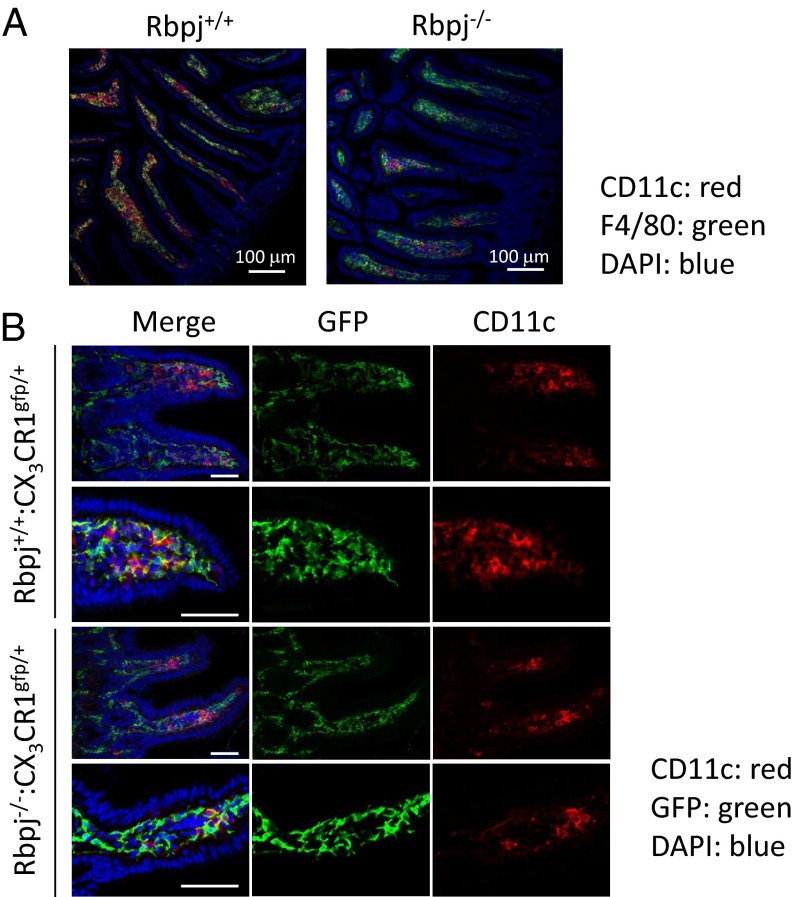
We analyzed the expression of F4/80 and CD11c in the small intestine of Rbpj−/− and Rbpj+/+ mice. The numbers and distributions of F4/80+ cells were comparable in Rbpj+/+ mice and Rbpj−/− mice (Fig. 2A). CD11c+ cells were restricted to the LP region in Rbpj+/+ mice but were scarcely detected in Rbpj−/− mice (Fig. 2A). Those results are consistent with findings that Rbpj−/− mice have few CD11c+CX3CR1+ cells. We next compared the intestinal sites in which CD11c+CX3CR1+ cells and CD11clowCX3CR1+ cells were found. The number and distribution of CX3CR1+ cells were comparable in Rbpj+/+:CX3CR1gfp/+ and Rbpj−/−:CX3CR1gfp/+ mice (Fig. 2B). Those data suggest that CD11c-dependent Rbpj deficiency skews the development of CD11c+CX3CR1+ cells to CD11clowCX3CR1+ cells, but the distributions of those two populations in the tissue were similar.
Fig. 2.
Rbpj deficiency in CD11c+ cells disturbs the development of CD11c+CX3CR1+ cells in the small intestine. (A) Sections of ileum of Rbpj+/+ or Rbpj−/− mice were stained with anti-CD11c (red) and F4/80 (green) antibodies. The nuclei were stained with DAPI (blue). Sections were evaluated by confocal microscopy. (Original magnification: 200×.) (Scale bar: 100 μm.) (B) Sections of small intestine of Rbpj+/+:CX3CR1gfp/+ mice or Rbpj−/−:CX3CR1gfp/+ mice were stained with anti-CD11c (red) and GFP (green) antibodies. The nuclei were stained with DAPI (blue). Sections were evaluated by confocal microscopy. (Original magnification 400×.) (Scale bar: 50 μm.) The data shown in these figures are representative of four independent experiments.
Notch1 and Notch2 Are Required for the Differentiation of CD11c+CX3CR1+ Cells.
We analyzed the expression of Notch receptors on CD11c+CD11b+ (R3: CD11c+CX3CR1+), CD11c+CD11b− (R1: CD11b−CD103+ DCs), and CD11chighCD11bhigh (R2: CD11b+CD103+ DCs) populations. R2 (CD11b+CD103+ DCs) populations expressed Notch2 and Notch3 at low levels. R3 (CD11c+CX3CR1+) populations expressed Notch1, 2, and 3, and the expression of Notch 3 was relatively higher than in the R1 (CD11b−CD103+ DCs) and R2 (CD11b+CD103+ DCs) populations (Fig. S5A). To determine if the R3 (CD11c+CX3CR1+) population received Notch signals, the expression of Hes1 (a Notch target gene) was evaluated by real-time PCR (Fig. S5B). The R3 (CD11c+CX3CR1+) population expressed higher amounts of Hes1 than did the R1 (CD11b−CD103+ DCs) and R2 (CD11b+CD103+ DCs) populations (Fig. S5B), suggesting that the R3 (CD11c+CX3CR1+) population received stronger Notch signaling than other types of intestinal antigen-presenting cells.
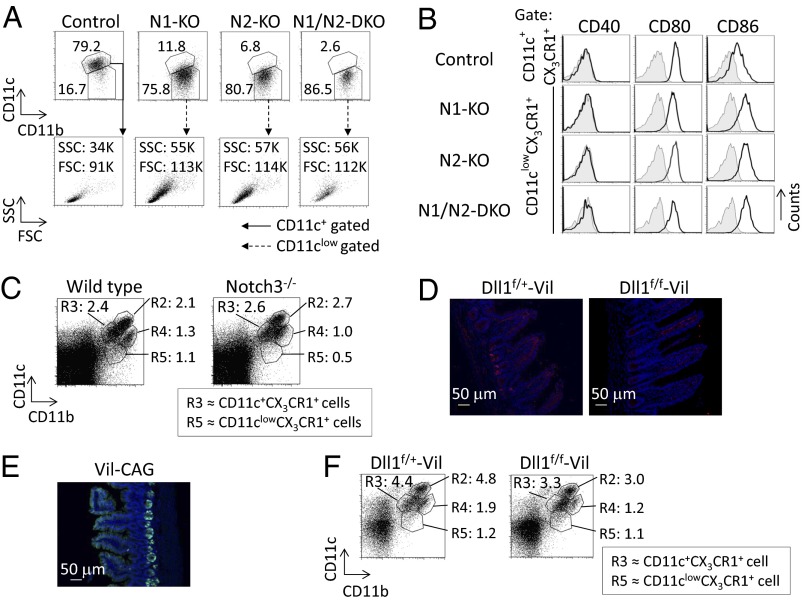
To examine which Notch receptors are required for the differentiation of CD11c+CX3CR1+ cells, we crossed Notch1flox/flox mice and Notch2flox/flox mice with CD11c promoter-driven Cre transgenic mice and with Cx3cr1 knockin mice (N1-KO or N2-KO, respectively). Mice deficient in both N1 and N2 were termed “N1/N2-DKO mice.” N1-KO mice and N2-KO mice had reduced numbers of CD11c+CX3CR1+ cells. N1/N2-DKO mice had markedly reduced numbers of CD11c+CX3CR1+ cells (Fig. 3A). The N1-KO mice, the N2-KO mice, and N1/N2-DKO mice had increased numbers of CD11clowCX3CR1+ cells with higher FSC and SSC (Fig. 3A) that expressed higher levels of CD86 than control cells (Fig. 3B). In contrast, the R3 (CD11c+CX3CR1+) population in Notch3-deficient mice was comparable to that in wild-type mice (Fig. 3C and Fig. S6A). A previous report (21) showed that Notch2 controls the development of CD11b+CD103+ DCs. We also found that numbers of CD11b+CD103+ DCs in small intestine were reduced in N2-KO and N1/N2-DKO mice (Fig. S6B). In contrast, the number of CD11b+CD103+ DCs is partially decreased in Rbpj−/−:CX3CR1 mice (Fig. S3A and Fig. S6B), and there was no significant reduction of CD11b+CD103+ DCs in N1-KO mice (Fig. S6B). Those data indicate that both Notch1 and Notch2 contribute to the differentiation of CD11c+CX3CR1+ cells and that Notch2, but not Notch1, controls the development of CD11b+CD103+ DCs. Although Notch3-deficient mice did not show any impaired differentiation of R3 (CD11c+CX3CR1+) cells, it would be necessary to assess the contribution of Notch3 to the development of R3 (CD11c+CX3CR1+) on a Notch1- and Notch2-deficient background.
Fig. 3.
Notch1 and Notch2 are required for the differentiation of CD11c+CX3CR1+ cells. LP cells from the small intestine were isolated from four mouse strains that have the CD11c-Cre transgene and are heterozygous for the CX3CR1-gfp allele: N1-KO, N2-KO, N1/N2-DKO, and control mice. (A) Cells were stained for evaluation with antibodies against F4/80, MHC class II, CD11c, and CD11b. CD11b and CD11c expression (Upper; numbers indicate the percentage of cells) or FSC and SSC (Lower; numbers show the value of FSC and SSC) were analyzed by gating on the 7AAD−F4/80+CX3CR1+MHC class II+ population. The gating strategy is the same as in Fig. 1A. (B) Expression of CD40, CD80, and CD86 in CD11c+CX3CR1+ cells or CD11clowCX3CR1+ cells was evaluated by flow cytometry. Fluorescence-conjugated streptavidin or isotype-matched control antibody (gray area) was used as the negative control. (C) LP cells from the small intestine were isolated from Notch3-deficient mice. Cells were stained with antibodies against CD11c and CD11b, and their expression was evaluated. The gating strategy was the same as in Fig. S3A. (D) Sections of ileum from Dll1f/+-Vil or Dll1f/f-Vil mice were stained with anti-Dll1 (red) antibody. The nuclei were stained with DAPI (blue). Sections were evaluated by confocal microscopy. (Original magnification: 200×.) (Scale bar: 50 μm.) (E) GFP expression in the sections of ileum in Vil-CAG mice was evaluated by confocal microscopy. (Original magnification: 200×.) (Scale bar: 50 μm.) (F) LP cells from the small intestine were isolated from Dll1f/+-Vil and Dll1f/f-Vil mice. Cells were stained with antibodies against CD11c and CD11b, and their expression was evaluated. The gating strategy is the same as in Fig. S3A. The data shown in these figures are representative of four independent experiments.
We sought to assess the Notch ligands that regulated the development of CD11c+CX3CR1+ cells. We first assessed the expression of Delta-like 1 (Dll1) in the small intestine. To assess the expression of Dll1, we used mice in which Dll1 was deleted by Vil1-Cre (Dll1f/f-Vil mice) and control mice. We found that Dll1 is expressed in the crypt base where precursors of intestinal epithelial cells are present in control Dll1f/+-Vil mice (Fig. 3D). This result was confirmed by the absence of Dll1 staining in the small intestine from Dll1f/f-Vil mice (Fig. 3D). To evaluate the expression pattern of Vil1-Cre, Vil1-Cre mice were crossed with CAG-CAT-GFP (Vil-CAG) mice, in which GFP expression was directed upon Cre-mediated excision of the loxP-flanked chloramphenicol acetyltransferase gene located between the CAG promoter and Gfp. The expression of Vil1-Cre was restricted to the crypt base and intestinal epithelial cells (Fig. 3E). However, the numbers of the R3 (CD11c+CX3CR1+) population in Dll1f/f-Vil mice were comparable to those in control mice (Fig. 3F). These results demonstrate that Dll1 expression on intestinal epithelial cells is not required for the development or differentiation of CD11c+CX3CR1+ cells. The numbers of the R3 (CD11c+CX3CR1+) population in Jagged1f/f-Vil mice, in which Jagged1 was deleted by Vil1-Cre, also were comparable to those in control mice (Fig. S6C).
Intrinsic Notch Signaling Is Required for CD11b+CD11c+ Development.
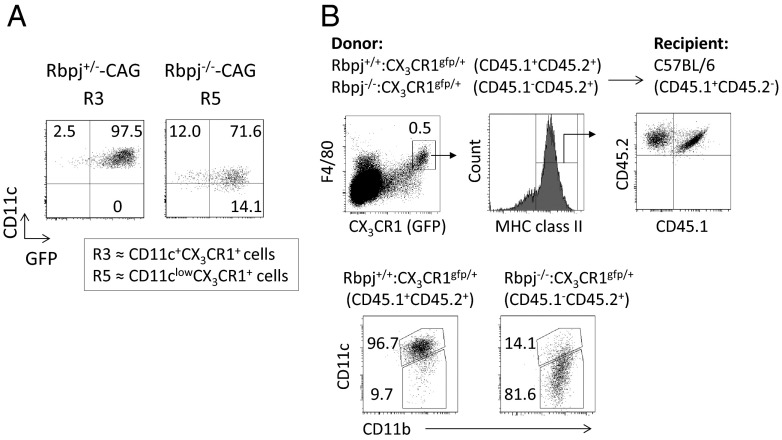
We asked whether the deletion of Rbpj from CD11c+ cells is intrinsically involved in the defective development of the R3 (CD11c+CX3CR1+) and R5 (CD11clowCX3CR1+) populations. Therefore, we crossed Rbpj+/−or Rbpj−/− mice with CAG-CAT-GFP mice (Rbpj+/−-CAG or Rbpj−/−-CAG, respectively). About 90% of the R3 (CD11c+CX3CR1+) population from Rbpj+/−-CAG mice and the R5 (CD11clowCX3CR1+) population from Rbpj−/−-CAG mice expressed GFP (Fig. 4A), indicating that Rbpj is deleted efficiently in both the R3 (CD11c+CX3CR1+) and R5 (CD11clowCX3CR1+) populations. To exclude the possibility that Rbpj deficiency in CD11c+CX3CR1+ cells affect neighboring cells and so contribute to the differentiation of CD11clowCX3CR1+ cells, we transplanted bone marrow cells from Rbpj+/+:CX3CR1gfp/+ mice (CD45.1/CD45.2) or from Rbpj−/−:CX3CR1gfp/+ (CD45.2) mice into lethally irradiated C57BL/6 (CD45.1) mice. Four weeks after transplantation, we analyzed the development of CD11c+CX3CR1+ cells in the small intestine (Fig. 4B). CD11c+CX3CR1+ cells were more numerous in the CD45.1+CD45.2+ fraction, whereas large numbers of CD11clowCX3CR1+ cells differentiated in CD45.2+ cells (Fig. 4B). These data demonstrate that the lack of intrinsic Rbpj-mediated signaling was responsible for the differentiation of CD11clowCX3CR1+ cells.
Fig. 4.
Intrinsic Rbpj is required for the differentiation of CD11c+CX3CR1+ cells. (A) LP cells from the small intestine were isolated from Rbpj+/−-CAG or Rbpj−/−-CAG mice. Cells were stained with antibodies against CD11c and CD11b, and R3 (CD11c+CX3CR1+) and R5 (CD11clowCX3CR1+) populations were evaluated as in Fig. S3A. (B) Lethally irradiated C57BL/6 (CD45.1) mice were reconstituted with bone marrow cells from Rbpj+/+:CX3CR1gfp/+ (CD45.1/CD45.2) or from Rbpj−/−:CX3CR1gfp/+ (CD45.2) mice (1:1). Four weeks after transplantation, the expression of CD11b and CD11c by CX3CR1+F4/80+MHC class II+CD45.1+CD45.2+ cells or CX3CR1+F4/80+MHC class II+CD45.1−CD45.2+ cells was evaluated by flow cytometry. The data shown in these figures are representative of four independent experiments.
Notch Alters the Differentiation of CD11c+CX3CR1+ Cells.
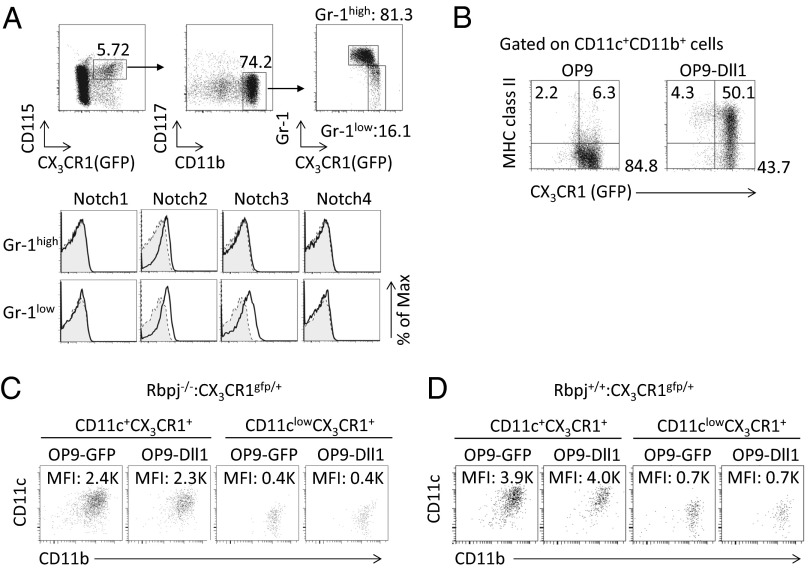
CD11c+CX3CR1+ cells are derived from monocyte precursors (27), and CX3CR1+CD115+CD11bhighCD117−Gr1high cells (Gr1high cells) differentiate toward CX3CR1+CD115+CD11bhighCD117−Gr1low cells (Gr1low cells). We examined the expression of Notch receptors on Gr1high cells and Gr1low cells. The Gr1high cells expressed Notch2, and Gr1low cells expressed Notch2 and 3 (Fig. 5A). Notch1 was not expressed by either cell type. We sought to determine whether Gr1low cells could differentiate toward CD11c+CX3CR1+ cells on OP9 or OP9-Dll1 cells in which Dll1 was overexpressed. Gr1low cells could differentiate to CD11c+CX3CR1+ cells on OP9 cells but could not differentiate toward CD11c+CX3CR1+MHC class II+ cells (Fig. 5B). In contrast, Gr1low cells could differentiate toward CD11c+CX3CR1+MHC class II+ cells in vitro on OP9-Dll1 (Fig. 5B), supporting the notion that Gr1low cells are progenitors for CD11c+CX3CR1+ (MHC class II+) cells and that Notch signaling is crucial for the differentiation of CD11c+CX3CR1+ (MHC class II+) cells.
Fig. 5.
CD11clowCX3CR1+ cells represent a lineage distinct from that of CD11c+CX3CR1+ cells. (A) Expression of Notch1–Notch4 by the Gr1low or Gr1high fraction from CX3CR1gfp/+ mouse bone marrow cells present in the CD115+CX3CR1+CD11bhighCD117− gate. Fluorescence-conjugated streptavidin or isotype-matched control antibody (gray). (B) CD115+CX3CR1+CD11bhighCD117−Gr1low cells from CX3CR1gfp/+ mice were cultured on OP9 or OP9-Dll1 cells for 5 d in the presence of macrophage colony-stimulating factor (M-CSF). Expression of CX3CR1 and MHC class II was evaluated by flow cytometry by gating on both CD11c+ and CD11b+ cells. (C) CD11c+CX3CR1+ cells or CD11clowCX3CR1+ cells were sorted from Rbpj−/−:CX3CR1gfp/+ mice and subsequently were cultured on OP9 or OP9-Dll1 cells with M-CSF, granulocyte macrophage colony-stimulating factor (GM-CSF), and Flt3L for 5 d. Expression of CD11b and CD11c was evaluated by flow cytometry by gating on MHC class II+ cells. (D) CD11c+CX3CR1+ cells and CD11clowCX3CR1+ cells were sorted from Rbpj+/+:CX3CR1gfp/+ mice and subsequently were cultured on OP9 or OP9-Dll1 cells with M-CSF, GM-CSF, and Flt3L for 5 d. Expression of CD11b and CD11c was evaluated by flow cytometry by gating on MHC class II+ cells. The data shown in these figures are representative of four independent experiments.
Using this culture system, we evaluated the differentiation of CD11clowCX3CR1+ cells and CD11c+CX3CR1+ cells in vitro. The increase of CD11clowCX3CR1+ cells in Rbpj−/−:CX3CR1gfp/+ mice allowed us to evaluate whether CD11clowCX3CR1+ cells are precursors of CD11c+CX3CR1+ cells or can change to CD11c+CX3CR1+ cells. CD11c+CX3CR1+ cells and CD11clowCX3CR1+ cells were sorted from Rbpj−/−:CX3CR1gfp/+ mice and subsequently were cultured on OP9 or OP9-Dll1 cells (Fig. 5C). CD11c+CX3CR1+ cells did not differentiate to CD11clowCX3CR1+ cells; moreover, CD11clowCX3CR1+ cells could not differentiate toward CD11c+CX3CR1+ cells. Furthermore, CD11c+CX3CR1+ cells from Rbpj+/+:CX3CR1gfp/+ mice were unable to give rise to CD11clowCX3CR1+ cells; similarly, CD11clowCX3CR1+ cells from Rbpj+/+:CX3CR1gfp/+ mice did not produce CD11c+CX3CR1+ cells (Fig. 5D). Those data suggest that CD11clowCX3CR1+ cells are not precursors of CD11c+CX3CR1+ cells, nor can they change to CD11c+CX3CR1+ cells.
Discussion
The intestine is in continuous contact with both pathological and beneficial commensal bacteria (2, 4, 28). Epithelial cell recognition of such microorganisms is required for the maintenance of immune homeostasis in the gut (4, 28). CD11c+CX3CR1+ cells contribute to the sampling of intestinal antigens (7–9), and they are crucial for the trafficking of bacteria to lymph nodes under dysbiotic condition (10). However, the molecular mechanisms that regulate the differentiation or development of CD11c+CX3CR1+ cells remain unclear. Our present studies show that Notch signaling in CD11c+ cells is essential for the differentiation of intestinal CD11c+CX3CR1+ cells. Moreover, the lack of Notch1 and Notch2 signaling skews the cell lineage toward CD11clowCX3CR1+ cells that have stronger antigen uptake activity, differential expression of cell-surface markers, and distinct morphology as compared with CD11c+CX3CR1+ cells. Those data suggest that Notch is crucial for fixing the CD11c+CX3CR1+ cell lineage among intestinal antigen-presenting cells.
CX3CR1+CD115+CD11bhighCD117−Gr1high cells in the bone marrow differentiate to CX3CR1+CD115+CD11bhighCD117−Gr1low cells that are precursors for CD11c+CX3CR1+ cells (27). Here, we found that CX3CR1+CD115+CD11bhighCD117−Gr1low cells differentiate to CD11c+CX3CR1+MHC class II+ cells on OP9-Dll1 cells. In contrast, in the OP9 culture system, CD11clowCX3CR1+ cells could not differentiate toward CD11c+CX3CR1+ cells. Moreover, CD11c+CX3CR1+ cells could not develop into CD11clowCX3CR1+ cells. Those results demonstrate that Notch signaling is required for the differentiation of CD11c+CX3CR1+ cells and that CD11clowCX3CR1+ cells are not precursors for CD11c+CX3CR1+ cells or vice versa. Rather, the data suggest that Notch is required for lineage fixation of CD11c+CX3CR1+ cells and that the lack of Notch signaling skews the cell lineage from CD11c+CX3CR1+ to CD11clowCX3CR1+ cells. However, we have not eliminated the possibility that CD11clowCX3CR1+ cells and CD11c+CX3CR1+ cells are derived from distinct precursors or that the role of Notch in each precursor is to promote or inhibit their differentiation. This issue could be clarified by identifying the precursor cells for each population.
CD11c-Cre–mediated deletion of Notch1 or Notch2 disturbs the differentiation of CD11c+CX3CR1+ cells. The Notch3-deficient population was normal in size, although Notch3 was highly expressed on the R3 population. The deficiency of both Notch1 and Notch2 leads to defects of CD11c+CX3CR1+ cell differentiation similar to those seen in Rbpj-deficient cells, suggesting that Notch1 and Notch 2 are major receptors controlling CD11c+CX3CR1+ cell differentiation. Previous studies reported that Notch2 signaling regulates CD11c+ESAM+ DCs in the spleen and CD11b+CD103+ DCs in the intestine (20–22), as confirmed in the present study, and also suggested that CD11b+CD103+ DCs are a major producer of IL-23, controlling Citrobacter rodentium infection (22). However, those studies did not find a defect in intestinal CD11c+CX3CR1+ cell differentiation caused by Notch2 deficiency. Those studies did not use CX3CR1 as a marker or analyze CD11c and CD11b by gating in MHC class II+ cells. Those approaches could explain why those reports could not detect the defect in CD11c+CX3CR1+ cell differentiation caused by Notch2 deficiency. Therefore, the alteration of the intestinal immune system of mice in which Notch2 was deleted by CD11c-Cre is attributable to defective differentiation of both CD11b+CD103+ DCs and CD11c+CX3CR1+ cells, together with an increased number of CD11clowCX3CR1+ cells. In any case, Notch signaling plays key roles in establishing the repertoire of antigen-presenting cells in the intestine.
Five canonical Notch ligands that regulate CD11c+CX3CR1+ cells are present in mice (12), and Vil1-Cre–mediated deletion of Jagged1 or Dll1 did not affect the development of CD11c+CX3CR1+ cells. BecauseVil1-Cre is expressed only in intestinal epithelial cells, it is unlikely that the expression of Jagged1 or Dll1 in intestinal epithelial cells is involved in the differentiation of CD11c+CX3CR1+ cells. The expression of Dll1 in the intestine is restricted in the intestinal epithelial cells; however, the expression pattern of other Notch ligands has not been studied. Therefore, it would be necessary to determine the expression pattern of Notch ligands in the intestine and bone marrow to identify the crucial Notch ligands that support CD11c+CX3CR1+ cell differentiation.
The current results demonstrate that Notch signaling is indispensable for lineage fixation of CD11c+CX3CR1+ cells. Moreover, the lack of Notch signaling in CD11c+ cells increases the number of CD11clowCX3CR1+ cells. Although we did not observe any difference between control and Rbpj−/− mice in the severity of DSS-induced colitis, the deletion of Rbpj by CD11c-Cre also affects the development of CD11b+CD103+ dendritic cells, as previously reported and shown here, and thereby might affect the immunological responses in the small intestine (21). Therefore, to analyze the functional role of Rbpj in CD11c+CX3CR1+ cells precisely in future studies, it will be necessary to find more a specific marker that deletes only Rbpj in CD11c+CX3CR1+ cells. Nevertheless, these results provide molecular insights into the differentiation of CD11c+CX3CR1+ cells. CD11c+CX3CR1+ cells are involved not only in sampling luminal antigens but also in conveying bacteria to lymph nodes under dysbiotic conditions. Therefore, we suggest that the identification of Notch signaling as the essential pathway for their differentiation may open new ways to modulate CD11c+CX3CR1+ cells in dysbiosis-mediated pathological bacterial infections and oral tolerance by targeted inhibition of Notch signaling.
Methods
Mice.
C57BL/6 mice (6- to 8-wk-old) were purchased from Japan SLC. Notch1flox/flox, Cx3cr1gfp/gfp, Notch1flox/flox, Vil1-Cre, and CD11c-Cre transgenic mice were purchased from Jackson Laboratory. Notch2flox/flox (29), Rbpjflox/flox (30), Notch3-deficient (31, 32), and CAG-CAT-GFP (33) mice were provided by S. Chiba (University of Tsukuba, Tsukuba, Japan), T. Honjo (Kyoto University, Kyoto, Japan), R. Kopan (University of Cincinnati College of Medicine, Cincinnati), and J. Miyazaki (Osaka University, Osaka, Japan), respectively. Dll1flox/flox (34) and Jagged1flox/flox (35) mice were previously described. All mice were maintained under specific pathogen-free conditions in the animal facilities at the University of Tokushima, Japan. All experiments were performed in accordance with institutional guidelines for animal care at the University of Tokushima.
Statistical Analysis.
For all experiments, the significant differences between groups were calculated using the Mann–Whitney u test for unpaired data. Differences were considered significant when P < 0.05.
Other methods are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Dr. S. Uematsu for discussions and technical help in isolating intestinal macrophages, Drs. S. Chiba, J. Miyazaki, P. Kopan, T. Honjo, and K. Tanigaki for providing mice, and Ms. C. Kinouchi and C. Tomari for their technical and editorial assistance. This work was supported by a Grant-in-Aid for Young Scientists (S) from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401671111/-/DCSupplemental.
References
- 1.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 2.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: Friends or foes? Nat Rev Immunol. 2010;10(10):735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 3.Papatriantafyllou M. Tolerance: The origins of colonic TReg cells. Nat Rev Immunol. 2013;13(6):394. doi: 10.1038/nri3468. [DOI] [PubMed] [Google Scholar]
- 4.Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 5.Uematsu S, et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol. 2008;9(7):769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- 6.Persson EK, et al. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38(5):958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Niess JH, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307(5707):254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 8.Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203(13):2841–2852. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hapfelmeier S, et al. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in DeltainvG S. Typhimurium colitis. J Exp Med. 2008;205(2):437–450. doi: 10.1084/jem.20070633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl GE, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494(7435):116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral Tolerance Can Be Established via Gap Junction Transfer of Fed Antigens from CX3CR1(+) Macrophages to CD103(+) Dendritic Cells. Immunity. 2014;40(2):248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13(6):427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 13.Wolfer A, et al. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat Immunol. 2001;2(3):235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- 14.Maekawa Y, et al. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity. 2003;19(4):549–559. doi: 10.1016/s1074-7613(03)00270-x. [DOI] [PubMed] [Google Scholar]
- 15.Amsen D, et al. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117(4):515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 16.Amsen D, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27(1):89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang TC, et al. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27(1):100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maekawa Y, et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol. 2008;9(10):1140–1147. doi: 10.1038/ni.1649. [DOI] [PubMed] [Google Scholar]
- 19.Bailis W, et al. Notch simultaneously orchestrates multiple helper T cell programs independently of cytokine signals. Immunity. 2013;39(1):148–159. doi: 10.1016/j.immuni.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204(7):1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis KL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35(5):780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satpathy AT, et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14(9):937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez-Cabezas B, et al. Ligation of Notch receptors in human conventional and plasmacytoid dendritic cells differentially regulates cytokine and chemokine secretion and modulates Th cell polarization. J Immunol. 2011;186(12):7006–7015. doi: 10.4049/jimmunol.1100203. [DOI] [PubMed] [Google Scholar]
- 24.Xu H, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13(7):642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8(10):1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 26.Chang SY, et al. Circulatory antigen processing by mucosal dendritic cells controls CD8(+) T cell activation. Immunity. 2013;38(1):153–165. doi: 10.1016/j.immuni.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varol C, et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med. 2007;204(1):171–180. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leavy O. Mucosal immunology: Inflammasome activation in the gut. Nat Rev Immunol. 2010;10(5):293. doi: 10.1038/nri2776. [DOI] [PubMed] [Google Scholar]
- 29.Saito T, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18(5):675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 30.Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14(6):637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 31.Skarnes WC, Moss JE, Hurtley SM, Beddington RS. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc Natl Acad Sci USA. 1995;92(14):6592–6596. doi: 10.1073/pnas.92.14.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu K, et al. Lunatic Fringe-mediated Notch signaling is required for lung alveogenesis. Am J Physiol Lung Cell Mol Physiol. 2010;298(1):L45–L56. doi: 10.1152/ajplung.90550.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamoto S, et al. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 2000;470(3):263–268. doi: 10.1016/s0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- 34.Hozumi K, et al. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5(6):638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 35.Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133(7):1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.