Significance
Microbes commonly form dense communities that are central to many diseases and bioremediation. Here we demonstrate a simple and general principle of living in dense communities: microbes will commonly compete to reach nutrients at the community edge, akin to plants competing for light. We follow the evolution of a highly competitive genotype in colonies of a soil bacterium. We show that the genotype gains its advantage not from its intrinsic growth rate but by positioning itself at the surface of the community, where it gains preferential access to oxygen. A large-scale genetic analysis reveals striking parallel evolution and evidence of strong natural selection. Our work suggests that positioning is a major basis for evolutionary competition in dense microbial communities.
Keywords: bacteria, biofilm, experimental evolution, social interaction
Abstract
Microbes commonly live in dense surface-attached communities where cells layer on top of one another such that only those at the edges have unimpeded access to limiting nutrients and space. Theory predicts that this simple spatial effect, akin to plants competing for light in a forest, generates strong natural selection on microbial phenotypes. However, we require direct empirical tests of the importance of this spatial structuring. Here we show that spontaneous mutants repeatedly arise, push their way to the surface, and dominate colonies of the bacterium Pseudomonas fluorescens Pf0-1. Microscopy and modeling suggests that these mutants use secretions to expand and push themselves up to the growth surface to gain the best access to oxygen. Physically mixing the cells in the colony, or introducing space limitations, largely removes the mutant’s advantage, showing a key link between fitness and the ability of the cells to position themselves in the colony. We next follow over 500 independent adaptation events and show that all occur through mutation of a single repressor of secretions, RsmE, but that the mutants differ in competitiveness. This process allows us to map the genetic basis of their adaptation at high molecular resolution and we show how evolutionary competitiveness is explained by the specific effects of each mutation. By combining population level and molecular analyses, we demonstrate how living in dense microbial communities can generate strong natural selection to reach the growing edge.
Microbes commonly live in dense surface-attached communities where cell division results in layer upon layer of cells lying on top of one another (1–4). Key examples include biofilms and the closely related colony mode of growth (5). These types of structured communities are not only common but also central to many of the ways that bacteria affect us, which includes an important role in health and disease via commensal gut communities and chronic infections (6, 7). In addition, these microbial communities are central to bioremediation, such as in waste-water treatment, but also biofouling in industry and shipping (8).
Why do microbes form these structured surface-associated communities? A key explanation is that the process allows cells to establish themselves in the best environment for growth and survival. This theory is well illustrated by the adaptive radiation of Pseudomonas fluorescens SBW25 in undisturbed liquid cultures (9). Here, specific mutants arise that colonize the glass surface of test tubes and generate a mat-like biofilm across the air–liquid interface, which subsequently collapses via the re-emergence of non–mat-forming strains. From such examples, we can understand the processes driving the formation and breakdown of biofilm communities. However, many such communities persist for long periods, over many generations (8, 10, 11). Understanding microbial phenotypes, therefore, also requires us to study the processes that affect cells within these dense communities.
A key feature of growth within a biofilm or colony is an extremely high cell density relative to liquid growth. This high cell density readily generates localized nutrient limitation and, particularly, the existence of microgradients, whereby the availability of nutrients decreases the further one goes into the cell mass and away from the nutrient source (12). There is good evidence that these gradients commonly occur in a range of systems, both when nutrients come from the surface to which cells attach, and when nutrients diffuse in from the environment (13–17). Moreover, there is good reason to believe that these microgradients will be central to understanding microbial phenotypes.
A growing body of theory emphasizes how strong nutrient gradients mean that cells near the nutrient source are able to divide but cells in the center of a colony or biofilm often cannot (18–20). In evolutionary terms, the difference between the two positions can be stark: division versus dormancy or death. A key corollary is that cells will benefit from investing in phenotypes that solely act to allow them to reach the growing edge ahead of a competitor. An analogy from macroscopic biology is the evolution of woody tissue in plants to grow tall and gain better access to light than competitors. In microbes, candidate mechanisms from the theory that may enable competition for the growing edge include secreted polymers, which allow a genotype to expand to reach growth surfaces, and cell motility (18, 20). In sum, given that microbes commonly live with both spatial structure and nutrient limitation, natural selection to gain the best position within biofilm-type communities should be central to understanding microbial phenotypes. However, we need direct tests of this hypothesis.
Colonies have long been considered a model laboratory system to study spatially structured microbial communities (12, 21–23), and are one of the main experimental approaches in studying biofilms (5). Here we use experimental evolution in laboratory colonies to study competition in microbial groups. Although artificial, our goal is to use a defined and tractable system to dissect key principles responsible for evolutionary success within dense microbial communities. We show that mutants arise repeatedly de novo within colonies of the bacterium P. fluorescens Pf0-1 and outcompete the parent. Importantly, we find their evolution occurs not because of intrinsically faster growth but rather the ability to expand as a group and eventually dominate the surface of the colony. We also follow the process at the genetic level, which reveals striking parallel evolution that is consistent with very strong natural selection to reach the surface of the colonies. Accordingly, the study has two main components: (i) the characterization of a bacterial phenotype that results from strong natural selection on cells to position themselves at the growth surfaces, and (ii) the genetic analysis of how this strong natural selection is manifest at the molecular scale.
Results and Discussion
Mucoid Variants Repeatedly Emerge as a Dominant Genotype.
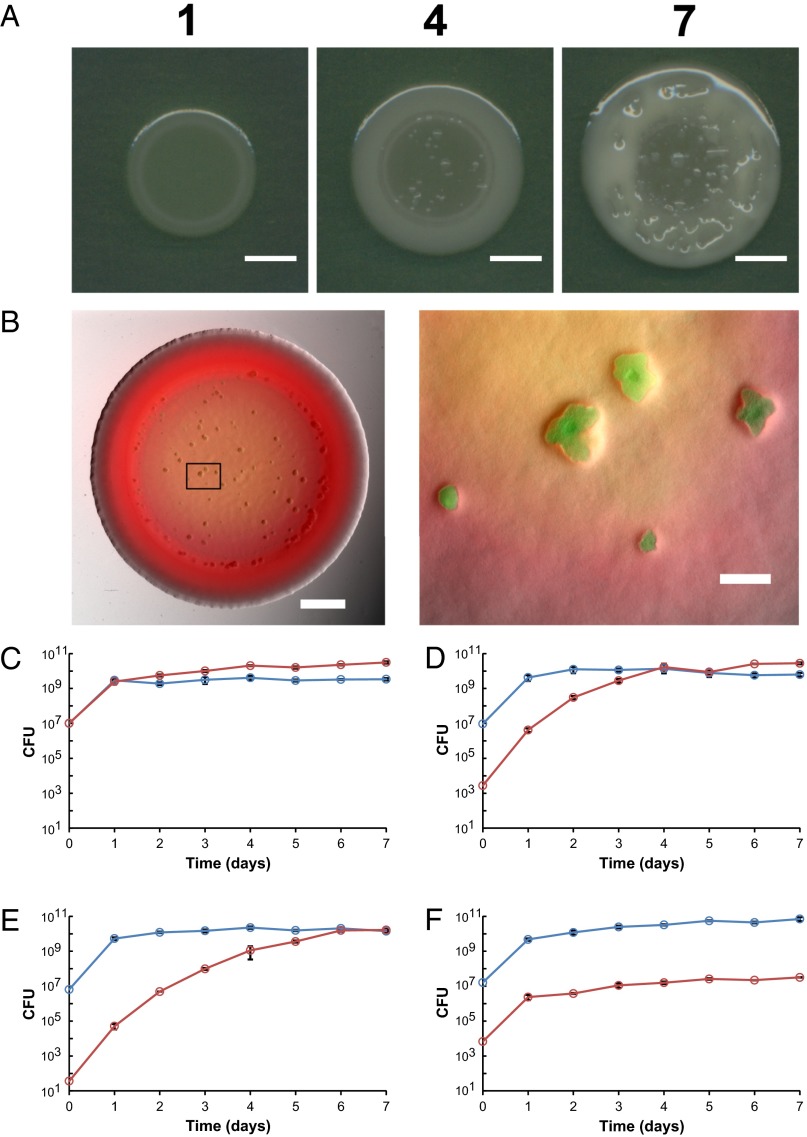
Mucoid variants reliably arise in colonies of the soil bacterium P. fluorescens Pf0-1. These variants emerge initially as distinct spots in colonies before spreading into patches and finally emerging and spreading across the surface of the colony (Fig. 1 A and B and Fig. S1A). Isolating a subset of the variants and replating them multiple times as pure colonies revealed a stable phenotype consistent with a genetic mutation. We therefore labeled a representative strain of the variant, herein mucoid variant (MV), and the wild-type (WT), with neutral antibiotic-resistance markers to track their frequency in mixed colonies. MV cells were seeded at various initial frequencies in mixed colonies with the WT and the frequency of the two genotypes was tracked over time by destructively sampling colonies each day (Fig. 1 C–F). This process revealed that MV cells are under strong positive selection in mixed colonies and rapidly increase in frequency as the colony grows. This effect is strongest when MV cells are seeded at very low frequencies but, even when MV cells initially vastly outnumber the WT cells (Fig. S1 B–D), they retain their fitness advantage.
Fig. 1.
Emergence and reproductive dominance of the mucoid variants. (A) Propagation of a WT colony consistently leads to the emergence of mucoid variants that appear to overtake the WT population over time. Indicated above each panel is the number of days postinoculation. (Scale bars, 5 mm.) (B) Discrete patches of tagged MV (GFP) emerge to the surface when introduced concurrently with WT (DsRed-Express) (Left). The mixed population was initially seeded at MV:WT ratio of 10−5:1, and visualized by fluorescence microscopy. (Scale bar, 2 mm.) (Right) Magnification of the section of the colony bound by the rectangle. (Scale bar, 0.2 mm.) (C–E) Results of competitions between MV and WT commenced at different starting ratios (MV:WT): (C) 1:1, (D) 10−3:1, and (E) 10−5:1. MV and WT were tagged with kanamycin and streptomycin resistance cassettes, respectively. The graphs show the mean population size of MV (red circle) and WT (blue circle) in CFU obtained from destructively sampling three independent populations at each interval over a period of 7 d. (F) Results of competition between MV tagged with kanamycin resistance (red) and MV tagged with streptomycin resistance (blue). Error bars represent the 95% confidence interval.
MV’s dominance over the WT could simply be a result of acquiring the ability to grow faster than its counterpart within the given environment. We thus compared the growth rate of the two strains when alone as single genotypes. In strong contrast to the fitness difference seen in mixed colonies, we could detect no differences in the growth rate of the two strains when alone, either over timescales of hours or days, in liquid or on plates (Fig. S1 E and F). The fitness advantage of the MV, therefore, rests upon the interactions between cells of the two genotypes.
The fact that mucoid variants readily emerge from any given WT colony indicates that they could already be present and perhaps even selected for in the WT population before plate inoculation. However, we found that mucoid variants also emerge within colonies commenced from a single WT cell (Fig. S1A). We provide additional evidence that the causal mutation occurs de novo in each experiment after the WT populations are seeded on the plates in SI Text, Estimation of the Timeline of rsmE Mutations.
A Model for the Evolutionary Advantage of the MV.
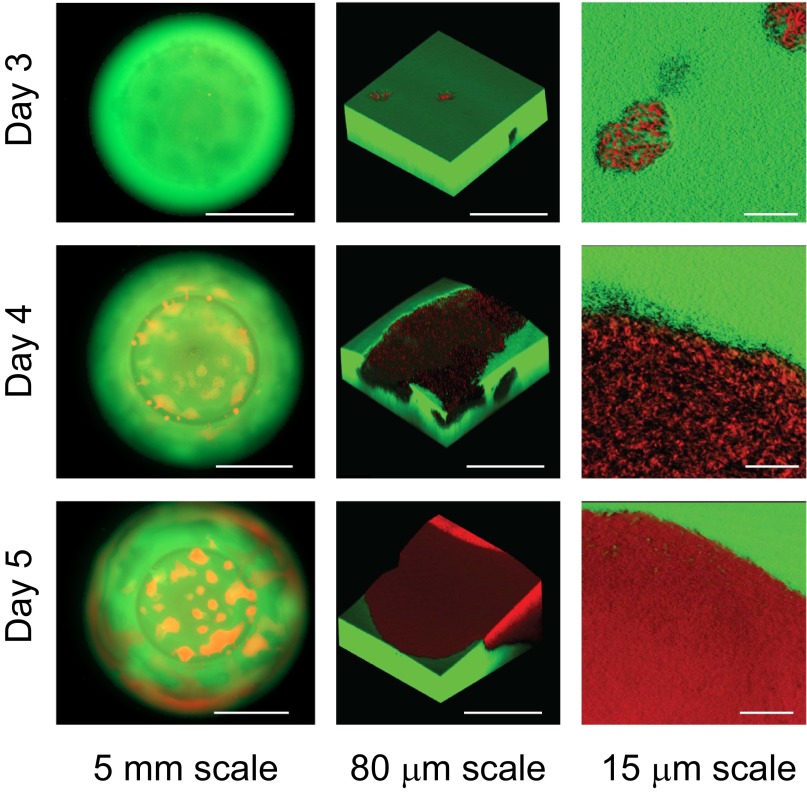
The fact that the MV does not grow faster than WT when alone suggests that the MV does not gain its advantage from a simple growth-promoting secretion, like an enzyme that provides nutrients. This aspect and its mucoid appearance suggest a model where the MV uses secretions to reduce cell density and expand outwards and upwards. Moving to the surface will allow cells to gain the best access to oxygen, which is strongly growth-limiting within colonies (13–16, 21). We used confocal imaging to assess the spatial patterning of the MV within WT colonies. Importantly, imaging was done noninvasively without a coverslip added to the colony, but we still achieve single-cell resolution. Consistent with our model, the MV does indeed display a much lower cell density than the WT and, moreover, it forms expanding patches at the surface of the colony that push up and out (Figs. 2 and 3E and Fig. S2).
Fig. 2.
Confocal imaging of colony competitions reveals that MV’s density is initially much lower than WT. Mixed populations of fluorescently tagged ΔrsmE (DsRed-Express) and WT (GFP) were seeded at the initial ratio of 10−5:1 (ΔrsmE:WT). Independent colonies were visualized over time by fluorescence microscopy (Left) and confocal microscopy (Center and Right, rendered in 3D). The images also show how MV comes to dominate the surface of the colony.
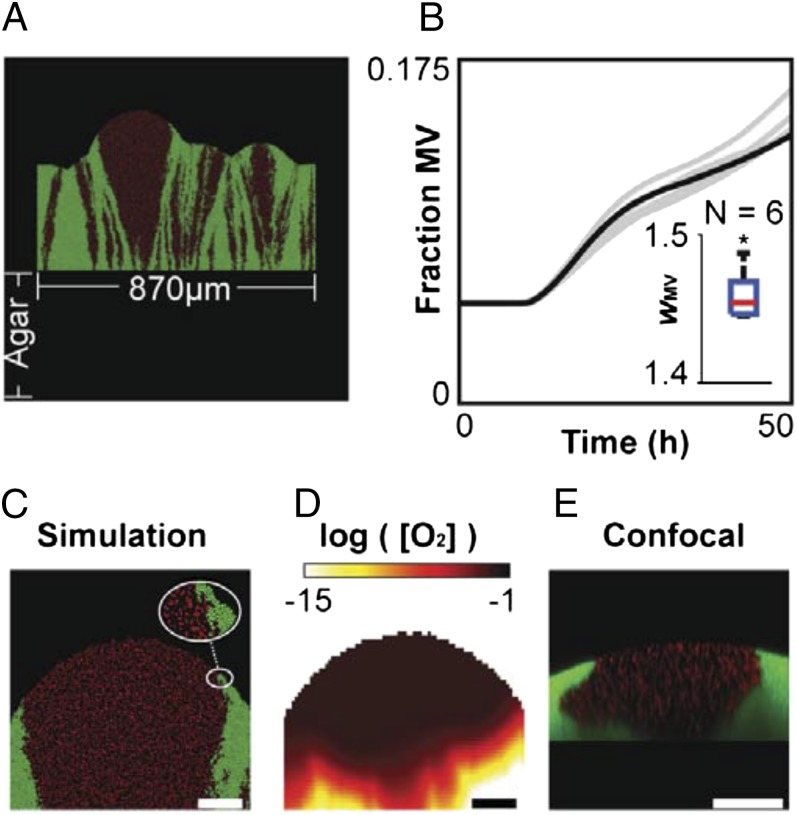
Fig. 3.
Individual-based simulations. (A) Snapshot from a 2D simulation of an 870-µm-wide cross-section of a colony growing on agar; MV in red, WT in green. (B) Fraction of the total mass belonging to the MV over 50 h in six independent simulations (black line: simulation shown in A, C, and D); initial fraction 0.05. Inset is a boxplot showing the relative fitness (W) of the mucoid variants at t = 50 h; the asterisk (*) means results are significantly different from equal fitness (W = 1), Wilcoxon signed-rank test (P = 0.0313). (C) Close-up of a region from the simulated colony. Because of the secretion of polymers, mucoid variant cells are less densely packed than WT cells. (D) Oxygen concentration profile in the simulation of the region shown in C). More oxygen is available in the region of mucoid variant cells because of the lower local cell density. (E) Confocal microscopy image of a colony of MV cells expressing DsRed-Express and WT cells expressing GFP. (Scale bars, 50 µm.)
The model is also consistent with the theoretical work discussed in the introduction that suggests that volume expansion by polymers can allow cells to push themselves to the edge of a submerged biofilm (18–20). We therefore adapted these models for colonies rather than submerged biofilms to show the same processes play out (SI Text, Individual-Based Simulation of WT-MV Competitions, and Fig. 3 and Fig. S3). Finally, general support for the idea that polymer secretion can provide a competitive advantage to a secreting strain comes from Nadell and Bassler, who showed a polymer-secreting genotype of Vibrio cholerae makes more robust biofilms than a nonproducer genotype, even when the two genotypes are mixed in direct competition (24). However, in this case the primary benefit came from better attachment to the substratum in a flow environment rather than competition for the growing edge.
MV Cells’ Fitness Rests upon Positioning Themselves in the Colony.
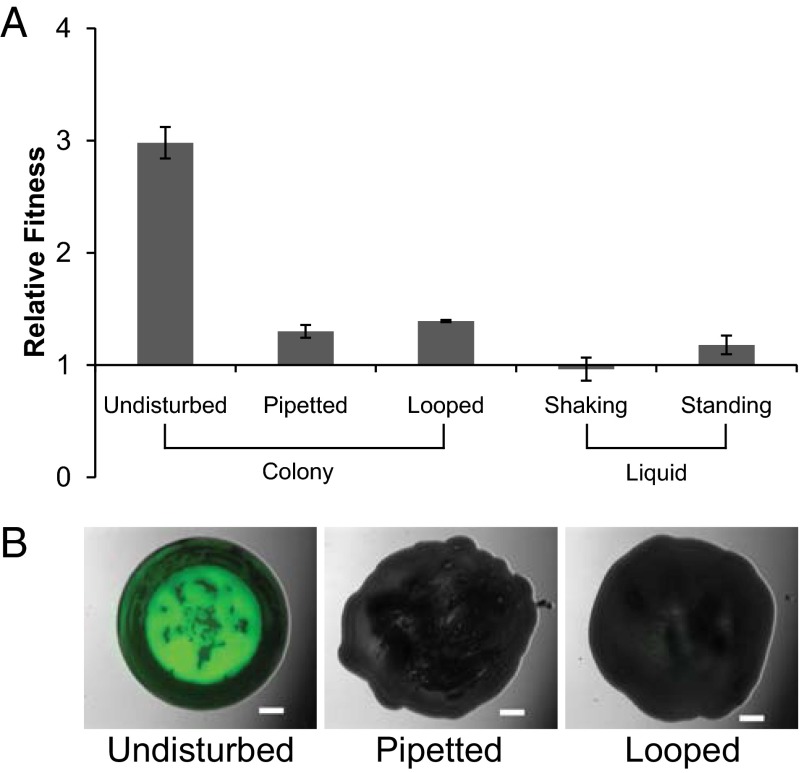
Our model for the evolutionary advantage of MV cells is that they use secretion to push themselves up and out and dominate growth surfaces (Figs. 1B and 2 and Fig. S2A). Specifically, it suggests that the ability of MV cells to gain a preferential position in the colony is what leads to their evolutionary advantage (Fig. 2) (18, 19). If correct, this model predicts that disrupting spatial structure will prevent MV cells from preferentially colonizing growth surfaces in mixed genotype populations. We tested this theory by having the two genotypes compete in shaking and standing liquid culture, where spatial structure is absent or modest. As predicted, we observe little or no fitness difference between the two genotypes under these conditions (Fig. 4A).
Fig. 4.
Spatiogenetic structure is essential for the fitness of MVs. (A) Effect of physical mixing of colonies and liquid cultures on the relative fitness of ΔrsmE compared with WT after 4 d of incubation. WT and ΔrsmE were tagged with streptomycin or kanamycin resistance cassettes, respectively. All competitions were seeded at the starting ratio of 10−5:1 (ΔrsmE:WT). Colonies were either left undisturbed or mixed daily with a pipette tip or sterile loop, and liquid cultures were incubated either standing or constantly shaking. The datapoints represent the mean (ΔrsmE over WT) from three independent populations, and the error bars represent the SD. The undisturbed colony treatment was found to be significantly different against all other treatments (two-tailed t test; P < 0.0001). (B) Visual assessment of physical mixing on competitions between the ΔrsmE mutant and WT by fluorescence microscopy (ΔrsmE tagged with GFP, WT untagged) after 4 d of incubation. Competitions were seeded at the starting ratio of 10−5:1 (ΔrsmE:WT) and either left undisturbed or mixed daily with a pipette tip or sterile loop. (Scale bars, 2 mm.)
Although the liquid experiments support our predictions, liquid culture and colonies differ in many ways and it is not clear that the MV’s ability to gain a better position in a colony is the causal factor explaining the fitness of the MV in colonies. We, therefore, performed a second assay to more directly test the importance of cell position for fitness. In addition to our normal experimental design, we added new treatments where each day we physically mixed up the colony using a pipette tip or a microbiological loop (Fig. 4). Although such a coarse manipulation will not fully disturb all patches of MV cells, this had a strong effect on the fitness of MV cells. MV fitness was greatly reduced close to the level observed in liquid competitions. Disrupting spatial structure in this way likely has two effects. First, it prevents MV cells from preferentially localizing themselves near the growing edge at the top of the colony. Second, mixing cells means that the secretions provided by MV cells could also help WT cells. Both effects may be important in reducing the performance of MV cells in mixed colonies. We, therefore, designed a complementary experiment to test the idea that the ability of MV cells to position themselves at the top of the colony is specifically necessary for their evolutionary advantage. Here, we had the two genotypes compete in colonies that were grown under an agar pad, which prevents upwards spreading and is intended to allow all cells equal access to nutrients and oxygen (Fig. S4); this again removed the fitness advantage of the MV. Together, these experiments suggest that the ability to position themselves in the colony is necessary for the strong natural selection on MV cells.
MV Selection Results from a Mutation Within the rsmE Locus.
To further understand the MV phenotype, we next sought to identify the underlying genetic changes. We used whole-genome pyrosequencing to identify potential causal mutations in one evolved isolate. This process revealed a deletion of a single nucleotide at the 126th position within the coding sequence of rsmE, which causes a frameshift, leading to a premature termination codon and the probable loss of protein function. RsmE functions as a specific repressing clamp in the GacA/S regulon by binding to regulatory small-RNAs and mRNA. The Gac system and its homologs have been a focal point of research for many years, as they regulate the production of various factors implicated in metabolism, host-colonization, and pathogenesis in a wide range of organisms (25). Finding mutations in the Gac regulon is consistent with other data from Pseudomonas spp., which suggests that the Gac pathway is a common mutational target, which includes evidence that gac genes undergo reversible mutations (26) and gac mutants frequently evolve during rhizosphere colonization (27).
RsmE is one of three Rsm proteins in the Pf0-1 genome. Among the pseudomonads, Rsm proteins are known to promote motility (28) and suppress various secreted products, such as protease, extracellular polysaccharides, and a quorum-sensing system that regulates phenazine biosynthesis (25, 29–31). Given the nature of such components, the Gac/Rsm regulatory cascade has previously been proposed as a key modulator of bacterial social behavior (25). Pf0-1 also appears to have a suppressed Gac regulon (32), suggesting that our WT strain has a low activity of the Gac pathway relative to other strains of P. fluorescens. Accordingly, mutation in rsmE is a way to activate the corresponding component of the Gac regulon and generate a more social phenotype. To confirm that the identified mutation was directly responsible for the MV phenotype, we constructed a strain from the WT harboring the same single-nucleotide deletion, and a second strain by deleting the entire rsmE locus. These strains appear phenotypically identical to the MV (Fig. 5A) and displayed the same strong evolutionary advantage in competitions with the WT (Fig. 5B). This finding supports the notion that the original mutation in the MV results in loss of RsmE function and is sufficient to cause the MV phenotype. Note that RsmE is a global repressor, so although the mutations cause a loss of function at the level of RsmE, the dominant phenotypic outcome is predicted to be an increase in multiple types of secretion under the control of the Gac/Rsm regulatory cascade (25, 29–31).
Fig. 5.
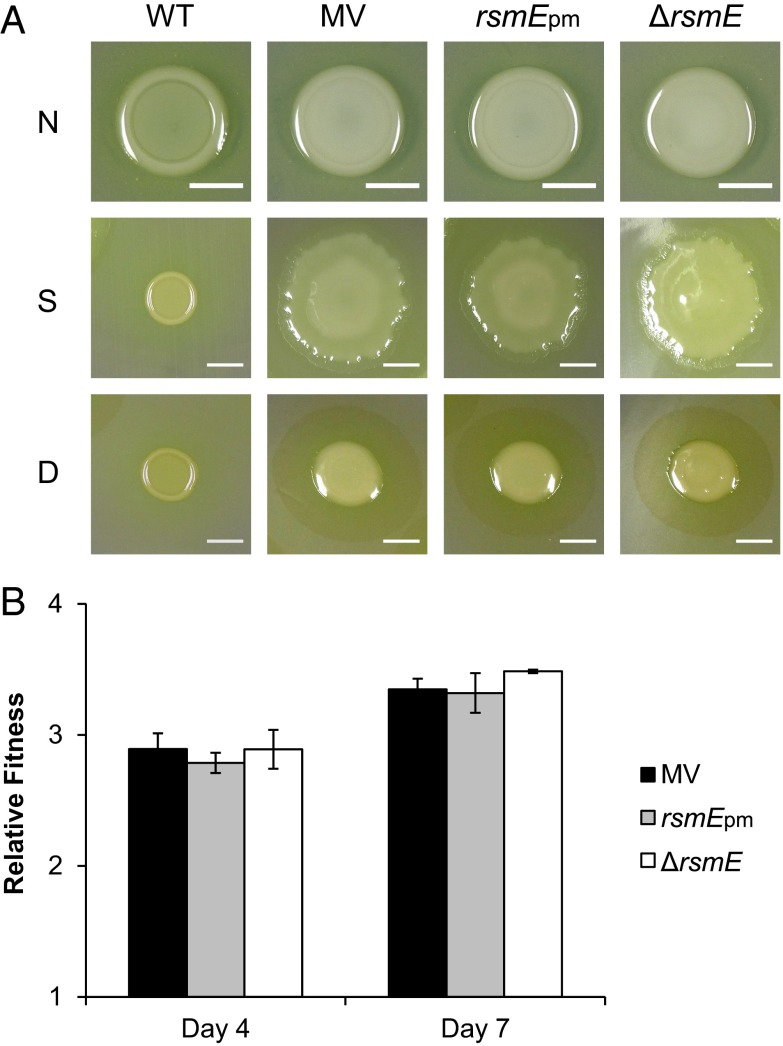
Mutation in rsmE triggers the secretion of products leading to the MV phenotype. (A) Phenotypic comparison of WT, MV, and engineered rsmE mutant colonies following 2 d of growth: “N” shows the relative mucoidy of colonies without polycarbonate membrane, “S” shows colony spreading on the shiny side of the membrane, and “D” shows biosurfactant production on the dull side of the membrane. (Scale bars, 5 mm.) (B) Results of competitions between WT and mutants in colony. Competitions were initiated at the mutant:WT ratio of 10−5:1. WT was tagged with streptomycin resistance and all mutants were tagged with kanamycin resistance. Error bars represent the SD of the mean relative fitness (mutant over WT) calculated from destructively sampling three independent populations after 4 and 7 d of incubation. There were no significant differences among the mutants on either day (Kruskal–Wallis, P = 0.4296 and 0.0665, respectively).
Strong Natural Selection Drives Parallel Evolution at the rsmE Locus.
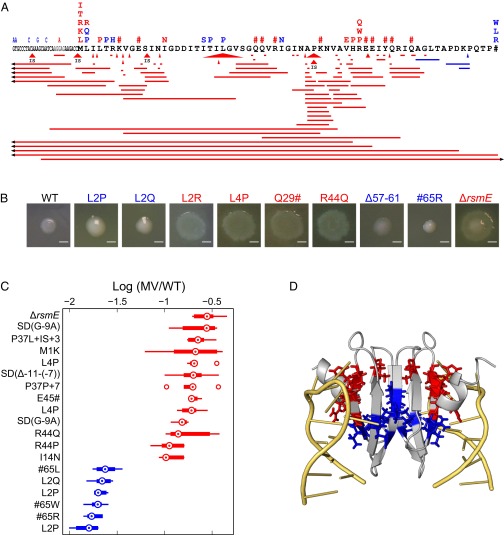
Our data are consistent with strong natural selection on cells to gain the best position within a bacterial community. If correct, we should also see evidence of this selection at the genetic level. Strong natural selection for a particular phenotype is often associated with parallel evolution whereby the same phenotype, and potentially genotype, arises reliably whenever an organism experiences the environmental conditions of interest. A familiar example from bacteria is the evolution of antibiotic resistance (33, 34). Consistent with parallel evolution, preliminary work suggested that rsmE mutants were a common occurrence in our experimental set-up. We therefore isolated and sequenced the rsmE locus in 565 independently derived mucoid variants to determine how many of the variants contained a mutation to a new rsmE allele (Fig. 6A and Table S1). Every MV strain harbored a single instance of mutated rsmE allele with changes either in the coding (534 mutants) or the upstream regulatory sequence (31 mutants), an example of perfectly repeatable evolution at the level of the gene. Cataloguing the mutations by type reveals a diverse array throughout the length of the rsmE locus: 322 base pair substitutions (282 missense and 40 nonsense), 190 deletions, and 22 insertions within the coding sequence, and 24 base pair substitutions, 6 deletions, and 1 insertion within the upstream sequence. Given their nature and the physical organization of the rsmE gene, these mutations are unlikely to exert any polar effects. Few exceptions may be the insertion sequence element insertions and the large deletions that extend beyond the coding sequence (Fig. 6A).
Fig. 6.
Molecular map and competitive phenotypes of individually derived rsmE mutations. (A) A schematic of mutations identified in each MV emerging from independent WT populations. Each mutant is classified into two categories based on the assay shown in B: strong competitor phenotype similar to ΔrsmE (red) and weak competitor phenotype closer to WT (blue). Mapped to the amino acid sequence of RsmE are missense mutations (denoted by corresponding substitutions), nonsense mutations (#), insertions (triangles denoting insertion site and size; IS denotes insertion sequence element), and deletions (horizontal bars showing size). Substitution mutations in the 5′ UTR are denoted by the actual nucleotide substitutions, and the Shine-Dalgarno sequence is shown in gray. Arrowhead denotes deletions that extend beyond the range. (B) Comparison of the colony spreading phenotype of representative mutant strains on the shiny side of the polycarbonate membrane. Δ57–61 denotes the amino acid residues that are deleted, which is listed under the Δ171–181 genotype in Table S1. (Scale bars, 2 mm.) (C) All mutants outcompete the WT, but not to the same extent. Comparison of competition outcome after 4 d between WT and select MVs representing different classes of mutations. MVs were seeded at 10−5 frequency against WT. Boxplots illustrate the distribution (medians, upper and lower quartiles, and outliers) of the relative frequency of mutant over WT among six competition replicates. Multiple occurrences of the same genotype indicate independently isolated mutants. According to a nonparametric Kruskal–Wallis test (n = 6, P < 0.05), all strains in red were similar to the knock-out control case (ΔrsmE vs. WT) and six strains were significantly weaker (blue). (D) Structural representation of the RsmE dimer (gray) bound to two cognate mRNA molecules (yellow) as determined previously by NMR (39). Missense mutations have either a large (red) or small (blue) effect on RsmE activity as shown in the other panels of the figure. Image was rendered by PyMOL (PDB accession ID 2JPP).
In addition to strong natural selection, parallel evolution can occur as a result of mutational bias if certain genes—in our case rsmE—mutate more often than is typical. Inspection of the rsmE sequence data, however, does not suggest a raised mutation rate. Specifically, we do not see any synonymous substitutions in rsmE (Table S2) and we find only one loss-of-function mutation per clone (additional such mutations would be neutral). To further evaluate the possible role of mutational bias in our system, we estimated the mutation rate at the rsmE locus using a modified form of the methodology of Lang and Murray (35). The analysis revealed that the mutation rate at rsmE is well within the estimates of the genome average and effective target size of rsmE (SI Text, Comparison of Rates Between the Emergence of Mucoid Variants and Mutations in rsmE). In addition, the great majority of mutations in rsmE, if not all, occur de novo in each experiment after the cells have been plated (SI Text, Estimation of the Timeline of rsmE Mutations, and Fig. S1H). We, therefore, conclude that our identifications of mutations in rsmE is the product of strong natural selection, not an elevated mutation rate.
We can then always link the emergence of any one mucoid variant to a single mutation in the same gene, rsmE. This approach raises a rare opportunity to explore in fine detail how the idiosyncrasies of different mutations impact upon evolutionary competitiveness. We therefore explored this by comparing a representative subset of the mutants in competition with the WT in two ways. In the first assay, we mixed each mutant at the ratio of 1:105 in pairwise competition with the WT to see how well they proliferate during colony growth (Fig. 6C). The second assay mimicked our mutant isolation process by seeding a small number of mutant cells with a large number of WT and then counting how many of the mutants successfully emerged to the top of the colony (Fig. S5; see SI Text, Comparison of Emergence Rates Between Mucoid Variants for detailed analysis). Both assays showed that all mutants outcompete the WT, but that the mutants differed in their competitive abilities against the WT.
The competition assays are difficult to do on a large scale. We therefore sought a proxy phenotype that would allow us to assess the competitive ability in all of our mutants. One major difference was revealed when the strains were propagated on top of polycarbonate membranes (16). The two sides of these membranes have different properties. One side is visibly dull and the other side is shiny (Materials and Methods). Here, we found that the MV and the constructed rsmE mutants could spread across the shiny surface, whereas WT could not, leading to a clear diagnostic phenotype (Fig. 5A). Moreover, none of the strains could spread across the dull side; however, a zone of transparent biosurfactant-like substance was visible around the MV genotypes but not the WT. We posit that this RsmE-regulated secreted product reduces surface tension and allows colonies to spread (36) across the shiny polycarbonate membrane. The dull side physically hinders expansion of the colony but not that of the secreted product.
The spreading phenotype of the mutants mapped predictably onto the two measures of competitive ability. In particular, mutants that appeared more like the WT performed relatively poorly in competition, whereas mutants that appeared like the ΔrsmE strain performed relatively well against the WT (Figs. 5A and 6 A and B). There exists a possibility that secondary mutations could account for the observed variations in competitiveness. However, this is unlikely given that independent strains that share the same mutation fall into the same measure of competitive ability and most fit mutants are predicted to be knockouts (Fig. 6C). In addition, as we show in the next section, the competitive ability of the mutants also makes sense in terms of precisely where the rsmE mutations are found.
The difference in the ability to expand on membranes might indicate a role for flagella-driven motility rather than secretion in the competitive ability of the MV. However, the introduction of a flagellin mutation in the MV had no effect in competitions against WT (Fig. S6). Although active motility appears unimportant, we do not exclude a role for passive motility in the fitness advantage of the MV over WT. In our simulations (Fig. 3) secretions act by carrying cells along with them, which means that secreting cells move further than nonsecretors. A related observation has been made in Pseudomonas aeruginosa, whereby cells slide on a surface independent of flagella, specifically when the type-iv pili are absent (37). Our particular WT strain does not exhibit twitching motility (38), thus a similar mechanism may aid MV cells to reach the growing edge of the colony.
Competitiveness Can Be Explained in Terms of Molecular Structure and Function.
We find then that all mutants outcompete the WT, but they differ in their relative ability to compete. The variability among mutants is consistent with differences in the production of the secretions that help groups of MV cells to gain preferential access to the growing edge in the colony in competition with the WT. We next sought to understand the molecular basis for this variability. Using the spreading phenotype as a proxy for competitive ability against WT, we divided the different mutants into two classes: strong competitors (more secretion) and weak competitors (less secretion). Fig. 6A summarizes the position of the different mutations found among the 565 mutants (full details in Tables S1 and S2). For many positions, we found the same mutation multiple times, up to a maximum of 69 cases of a particular single nucleotide substitution (Arg44). Moreover, we found 7 of 7 possible loss of start codon mutations and 11 of 14 possible nonsense mutations in rsmE, where two of the three missing nonsense mutations are expected to be silent as they occur at the nonfunctional tail end of the protein (39, 40). Because we know which of these classes of mutations lead to a loss of function, we can then estimate that we have found more than 95% (21 of 22) of the other loss-of-function mutations (35). Although approximate only, this calculation suggests that we have a detailed molecular map of the possible routes to the origin of the MV phenotype.
We used this map to evaluate how well one can translate the degree of adaptation at the population (colony) level into processes at the molecular level. We first considered insertion and deletion mutations (indels), which are expected to lead to a nonfunctional protein because of frameshift. Consistent with this result, we found that most indels cause a strong competitor phenotype, including multiple cases of insertion sequence element insertion (Fig. 6A). The tail end of the protein, however, has indels with both the strong and the weak competitor phenotype. Here, the observed deletion within the Gly54 codon leads to an immediate truncation of the protein at the next position and the strong competitor phenotype. This finding fits with the prediction that the tail end of the protein has little functional importance but from position 55 onwards (39, 40). However, if the tail end of the protein is not functionally important, why do we also see weak competitor phenotypes in this region? All of the weak competitor phenotypes in this region result from frameshift that add 59–66 amino acids to the carboxyl-terminus of RsmE, which is a significant bulk to a protein that is normally only 64 amino acids in length. We also found clear patterns for the single-nucleotide substitutions. At the carboxyl terminus, there were again a number of weak competitors that resulted from the loss of the stop codon and the addition of 18 amino acids to the protein. At the other end of the locus, the 5′ UTR is associated with weak competitor mutations with a single exception in the Shine-Dalgarno sequence.
All nonsense mutations that we found led to the strong competitor phenotype. In contrast, the missense mutation spectrum showed that only specific amino acids in specific positions lead to the MV adaptation (Table S2). This finding is of course expected but it does appear that RsmE is a robust protein (41), in the sense that very few missense mutations lead to a complete loss of function (see Table S2 for robustness calculation). Moreover, in contrast to nonsense mutations, the missense mutations that we found caused both weak and strong competitors. At first glance, the distribution of the two phenotypes caused by a missense mutation appeared to be arbitrary across the linear sequence (Fig. 6A). However, there are detailed structure-function predictions for the binding of RsmE, and its closely related homolog CsrA, to its cognate mRNA (39, 40). These studies immediately provide insight on our data as we found many cases of mutation at the two residues, Leu4 and Arg44 (Fig. 6A), that were subjected to additional in vivo and in vitro functional studies on the basis of their predicted importance (39). However, we sought to further test the explanatory power of the structure-function data in our evolutionary experiment. We, therefore, plotted the positions of the missense mutations on the RsmE-mRNA structure by competitive class. It is striking that the strong and weak competitor mutations each fall out into discrete regions within the structure. Strong competitor residues cluster at the interface of RsmE-mRNA, and weak competitor residues cluster more at the interface between the two dimers of RsmE (Fig. 6D). The distinction between the two classes of competitive adaptation by missense mutation, therefore, appears to arise from the modulation of different classes of molecular interaction.
What Maintains rsmE Expression in Nature?
We observed the reliable mutation of rsmE in our experiments. What then maintains rsmE expression under natural conditions? We speculate that two key factors are at play. The first factor is that there may be hidden fitness costs to the loss of rsmE that are not seen in the laboratory. We observed no fitness cost associated with mutation in rsmE under laboratory conditions, even though it is associated with the expression of multiple secretions. The lack of costs may partly result from rsmE being expressed primarily at stationary phase rather than during exponential growth (29). Loss of rsmE, therefore, will tend to derepress secretions when cells are not growing at their fastest, which can limit the fitness costs of the secretions (36). Nevertheless, loss of rsmE may have fitness costs under some natural condition that we are unable to recapitulate in the laboratory. The second factor is that our WT strain (Pf0-1) may be more likely than some natural isolates to lose rsmE function. The production of numerous extracellular secretions appears to correspond to the relative activity of the Gac system. Activity in the Gac pathway is variable in the pseudomonads (26, 27) and Pf-01 is known to have a suppressed Gac system (32). This finding suggests that Pf-01 has a relatively low level of secretions to begin with and, under conditions that favor secretion, it may be more likely to lose rsmE function than a natural isolate that is already a strong secretor. In the end, experimental evolution studies are limited by the fact that what happens in the laboratory may not reflect precisely what happens in natural systems. However, there is natural variation in Rsm homolog copy number in the pseudomonads (25, 28, 42), so the gain and loss of Rsm proteins is part of the evolutionary trajectory of natural systems.
Conclusion
Microbes growing on a solid surface, such as submerged biofilms and colonies, commonly form dense communities that rapidly deplete incoming nutrients. This general effect is predicted to exert strong natural selection on cells to gain the best access to nutrients, in a similar way to plants competing to gain the best access to light. Here we have described a series of simple evolutionary experiments that suggest that pushing to the surface can provide large fitness benefits. MV cells spontaneously arise within colonies and use secretions to collectively expand and push themselves to the surface of WT colonies. Notably, the MV cells show no growth rate advantage when in single genotype colonies; the evolutionary process demonstrably rests upon social interaction between MV and WT cells within a colony. In addition, we find that moving cells around or limiting their ability to form thick-layered colonies removes the evolutionary advantage of MV cells. We also find evidence of the importance of this competition at the genetic level: it leads to strong natural selection and parallel evolution comparable to the clearest known examples, such as the evolution of antibiotic resistance (33, 34). We used this evidence to generate a fine-scale map of the mutations that cause the MV phenotype and explain their competitiveness in terms of molecular structure and function. Our work suggests that strategies that allow microbes to reach the edge of dense communities will often be under strong natural selection.
Materials and Methods
An extended version of the materials and methods used can be found in SI Materials and Methods.
Bacterial Strains.
P. fluorescens Pf0-1 is a natural strain (43) that was directly isolated from soil by S.B.L. All mucoid variants described in this study were derived from individually isolated or spotted colonies of Pf0-1 on Difco Pseudomonas agar F (PAF), which is a commercial formulation of King’s Medium B (KMB) (44). The evolution of mucoid variants is observed in minimal and complex media supplemented with glycerol or glucose as carbon source. One mucoid variant, MV, was designated as the prototype for detailed analyses.
Estimation of Frequency in Single and Mixed Genotype Populations.
Competition experiments in colonies were carried out by mixing strains and spotting in 20-μL volumes on PAF plates. Individual strains were tagged with either a kanamycin or streptomycin resistance cassette, which are neutral in P. fluorescens Pf0-1 (45) (Fig. 1F). Colonies were harvested and the frequency of individual strains was estimated by serial dilutions and plating on selective media. Individual strains were also tagged with cassettes encoding GFP, YFP, or DsRedExpress proteins (46), and competition was visualized by fluorescence and confocal laser scanning microscopy. For competition experiments in liquid, 20 μL of the mixture was inoculated into test tubes containing 2 mL of KMB or PAF without agar. The tubes were incubated either shaking or left standing undisturbed, and frequency was estimated as above. The outcome of each competition was analyzed by comparing both the raw CFU data and calculating the relative fitness (W) (47), or as noted otherwise.
Genome Sequencing, Identification, and Confirmation of the Causal Mutation.
Whole-genome sequencing (454 FLX) was carried out by the Washington Genome Sequencing Center (St. Louis, MO), to compare MV to its parent strain. A single nucleotide (A) deletion at the 126th position of the coding DNA sequence of the rsmE gene was confirmed to be the causal mutation, resulting in the mucoid phenotype by introducing the same single-nucleotide deletion (rsmEpm) or deleting the entire rsmE locus (ΔrsmE) in the parent strain.
Mutant Construction and Tagging.
Mutants were constructed using the gene splicing by overlap extension method (48) and homologous recombination as previously outlined (49). Primers complimentary to regions flanking the targeted gene were used to monitor the proper replacement with the mutant constructs by PCR, and confirmed by sequencing both template strands. The miniTn7 system was used to tag the chromosomes of the strains used in this study using established procedures (46).
Colony and Biosurfactant Spreading Assay on Polycarbonate Membranes.
Nuclepore polycarbonate membrane (Whatman) was laid on top of PAF plates using sterile forceps and spotted with overnight cultures. The smooth and shiny side of the membrane was spotted to compare the colony spreading phenotype and the matted and dull side was spotted for visualizing biosurfactant production.
Individual-Based Simulations.
An extension of an established framework was used for the computer simulation of bacterial growth (18, 19, 50–52) to model a cross-section of a mixed colony of WT and MV cells. In the simulations, cells are considered to be spheres that metabolize diffusing nutrients that grow and eventually divide. Through this activity, local concentration gradients of nutrients arise. Multigrid solvers were used to calculate the steady-state solution of the 2D diffusion reaction equations that return these gradients in each iteration. Both cell types have the same growth rate but only the MV secretes polymers. Differences in the relative fitness (W) were calculated as described for experimental competitions. See Table S3 for a summary of the parameters used in the simulations.
Parallel Evolution Experiments.
Overnight WT cultures (20 µL) were spotted on PAF plates and incubated for 4 d at room temperature until mucoid variants became clearly visible. A single variant was randomly isolated from each single WT colony, with one exception being that three spatially separated patches of variants were isolated from a common WT colony. Each variant was purified and phenotype confirmed on fresh PAF plates. The mutation in each variant was identified by sequencing PCR products amplified with primers specific to regions flanking the rsmE locus.
Statistical Analyses.
Given that the sample sizes were too small (n = 3) for the Mann–Whitney test, a two-tailed t test was used to compare the relative fitness differences between any two given strains. A Kruskal–Wallis test was applied to compare the relative fitness of the constructed rsmE mutants to the MV. The Kruskal–Wallis test, corrected for multiple comparisons (Tukey’s honestly significant difference criterion), was used to compare CFU ratios of different mucoid variants to the WT. A two-tailed Mann–Whitney test was applied to compare the emergence ratios of different mucoid variants. The Wilcoxon signed-rank test was applied to compare relative fitness in the simulations. Bonferoni correction was applied when making multiple pairwise comparisons, and the relevant values for the n and α parameters are indicated for each test where appropriate. All statistical tests were conducted using Matlab.
Imaging.
Still pictures of colonies were generated using the CanoScanLiDE 200 flatbed scanner (Canon) or the EOS 30D DSLR camera (Canon), and images were scaled to calibrated dimensions using the ImageJ software (53). Fluorescently tagged strains were imaged using the Typhoon 9400 scanner (GE Healthcare) and the associated ImageQuant TL software as described elsewhere (36), the SteREO Lumar.V12 microscope (Zeiss) under the NeoLumar S 0.8× objective lens and the associated AxioVision software, or the Axio Zoom.V16 microscope (Zeiss) under the PlanApo Z 0.5× objective lens and the associated Zen software. Confocal imaging was carried out on the LSM 700 laser scanning microscope (Zeiss) using the 20× and 50× objectives and the associated Zen software. A square piece of agar containing the entire colony was cut out and placed on slides without a coverslip for confocal imaging. For all other imaging procedures, entire plates were imaged without disturbing the agar surface.
Supplementary Material
Acknowledgments
We thank M. Cant, L. Keller, G. Lang, M. Laub, A. Murray, C. Nadell, B. Stern, K. Verstrepen, S. West, and J. Xavier for providing comments, and S. Mitri for assistance on statistical analyses. These studies were funded by Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (to W.K.), US Department of Agriculture Grants 2006-35604-16673 and 2010-04952 (to S.B.L.), National Institute of General Medical Sciences Center of Excellence Grant 5P50 GM 068763 (to K.R.F.), and European Research Council Grant 242670 (to K.R.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323632111/-/DCSupplemental.
References
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Kolter R, Greenberg EP. Microbial sciences: The superficial life of microbes. Nature. 2006;441(7091):300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 3.Monds RD, O’Toole GA. The developmental model of microbial biofilms: Ten years of a paradigm up for review. Trends Microbiol. 2009;17(2):73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33(1):206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 5.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13(1):20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292(2):107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 7.Costerton JW, Montanaro L, Arciola CR. Biofilm in implant infections: Its production and regulation. Int J Artif Organs. 2005;28(11):1062–1068. doi: 10.1177/039139880502801103. [DOI] [PubMed] [Google Scholar]
- 8.Jass J, Walker J. In: Industrial Biofouling: Detection, Prevention and Control. Walker J, Surman S, Jass J, editors. New York: John Wiley & Sons; 2000. [Google Scholar]
- 9.Rainey PB, Rainey K. Evolution of cooperation and conflict in experimental bacterial populations. Nature. 2003;425(6953):72–74. doi: 10.1038/nature01906. [DOI] [PubMed] [Google Scholar]
- 10.Gellatly SL, Hancock REW. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog Dis. 2013;67(3):159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 11.Des Marais DJ. Microbial mats and the early evolution of life. Trends Ecol Evol. 1990;5(5):140–144. doi: 10.1016/0169-5347(90)90219-4. [DOI] [PubMed] [Google Scholar]
- 12.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 13.Pirt SJ. A kinetic study of the mode of growth of surface colonies of bacteria and fungi. J Gen Microbiol. 1967;47(2):181–197. doi: 10.1099/00221287-47-2-181. [DOI] [PubMed] [Google Scholar]
- 14.Wimpenny JW, Lewis MW. The growth and respiration of bacterial colonies. J Gen Microbiol. 1977;103(1):9–18. doi: 10.1099/00221287-103-1-9. [DOI] [PubMed] [Google Scholar]
- 15.Peters AC, Wimpenny JW, Coombs JP. Oxygen profiles in, and in the agar beneath, colonies of Bacillus cereus, Staphylococcus albus and Escherichia coli. J Gen Microbiol. 1987;133(5):1257–1263. doi: 10.1099/00221287-133-5-1257. [DOI] [PubMed] [Google Scholar]
- 16.Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47(1):317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64(10):4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA. 2007;104(3):876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadell CD, Xavier JB, Levin SA, Foster KR. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008;6(1):e14. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xavier JB, Martinez-Garcia E, Foster KR. Social evolution of spatial patterns in bacterial biofilms: When conflict drives disorder. Am Nat. 2009;174(1):1–12. doi: 10.1086/599297. [DOI] [PubMed] [Google Scholar]
- 21.Rani SA, et al. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol. 2007;189(11):4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro JA. Pattern and control in bacterial colony development. Sci Prog. 1992;76(301–302 Pt 3–4):399–424. [PubMed] [Google Scholar]
- 23.López D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009;74(3):609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci USA. 2011;108(34):14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapouge K, Schubert M, Allain FH-T, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: From RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67(2):241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 26.van den Broek D, Chin-A-Woeng TFC, Bloemberg GV, Lugtenberg BJJ. Molecular nature of spontaneous modifications in gacS which cause colony phase variation in Pseudomonas sp. strain PCL1171. J Bacteriol. 2005;187(2):593–600. doi: 10.1128/JB.187.2.593-600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez-Granero F, Rivilla R, Martín M. Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl Environ Microbiol. 2006;72(5):3429–3434. doi: 10.1128/AEM.72.5.3429-3434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Granero F, et al. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS ONE. 2012;7(2):e31765. doi: 10.1371/journal.pone.0031765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reimmann C, Valverde C, Kay E, Haas D. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J Bacteriol. 2005;187(1):276–285. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irie Y, et al. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78(1):158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, et al. Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30-84. Microbiologyopen. 2013;2(3):505–524. doi: 10.1002/mbo3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loper JE, et al. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8(7):e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandgren A, et al. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6(2):e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacLean RC, Buckling A. The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genet. 2009;5(3):e1000406. doi: 10.1371/journal.pgen.1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178(1):67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xavier JB, Kim W, Foster KR. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol Microbiol. 2011;79(1):166–179. doi: 10.1111/j.1365-2958.2010.07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray TS, Kazmierczak BI. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol. 2008;190(8):2700–2708. doi: 10.1128/JB.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barton MD, Petronio M, Giarrizzo JG, Bowling BV, Barton HA. The genome of Pseudomonas fluorescens strain R124 demonstrates phenotypic adaptation to the mineral environment. J Bacteriol. 2013;195(21):4793–4803. doi: 10.1128/JB.00825-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schubert M, et al. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol. 2007;14(9):807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 40.Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem. 2006;281(42):31832–31842. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- 41.Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc Natl Acad Sci USA. 2004;101(25):9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris ER, et al. Structural rearrangement in an RsmA/CsrA ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure. 2013;21(9):1659–1671. doi: 10.1016/j.str.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Compeau G, Al-Achi BJ, Platsouka E, Levy SB. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl Environ Microbiol. 1988;54(10):2432–2438. doi: 10.1128/aem.54.10.2432-2438.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44(2):301–307. [PubMed] [Google Scholar]
- 45.Silby MW, Nicoll JS, Levy SB. Requirement of polyphosphate by Pseudomonas fluorescens Pf0-1 for competitive fitness and heat tolerance in laboratory media and sterile soil. Appl Environ Microbiol. 2009;75(12):3872–3881. doi: 10.1128/AEM.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;6(7):726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- 47.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138(6):1315–1341. [Google Scholar]
- 48.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene. 1989;77(1):61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 49.Silby MW, Levy SB. Use of in vivo expression technology to identify genes important in growth and survival of Pseudomonas fluorescens Pf0-1 in soil: Discovery of expressed sequences with novel genetic organization. J Bacteriol. 2004;186(21):7411–7419. doi: 10.1128/JB.186.21.7411-7419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nadell CD, Foster KR, Xavier JB. Emergence of spatial structure in cell groups and the evolution of cooperation. PLOS Comput Biol. 2010;6(3):e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10(11):e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.