Significance
The power of adaptive immunity in humans is realized through the hypervariable molecules: the T-cell receptors (TCRs). Each of those is built from genetically encoded parts with the addition of random nucleotides finally forming individual TCR repertoire. Despite that the individual TCR repertoire potentially can include 1011–1014 different variants, substantially less molecules are found in a single individual. The particular genetic impact on the final set of TCR molecules is still poorly understood. In this study, for the first time to the best of our knowledge, we compare deep TCR repertoires of genetically identical twins. We found that, although TCR repertoires of any pair of individuals have the same amount of identical receptors, twin repertoires share certain specific features.
Keywords: immunogenetics, TCR repertoire analysis, twin studies
Abstract
Adaptive immunity in humans is provided by hypervariable Ig-like molecules on the surface of B and T cells. The final set of these molecules in each organism is formed under the influence of two forces: individual genetic traits and the environment, which includes the diverse spectra of alien and self-antigens. Here we assess the impact of individual genetic factors on the formation of the adaptive immunity by analyzing the T-cell receptor (TCR) repertoires of three pairs of monozygous twins by next-generation sequencing. Surprisingly, we found that an overlap between the TCR repertoires of monozygous twins is similar to an overlap between the TCR repertoires of nonrelated individuals. However, the number of identical complementary determining region 3 sequences in two individuals is significantly increased for twin pairs in the fraction of highly abundant TCR molecules, which is enriched by the antigen-experienced T cells. We found that the initial recruitment of particular TCR V genes for recombination and subsequent selection in the thymus is strictly determined by individual genetic factors. J genes of TCRs are selected randomly for recombination; however, the subsequent selection in the thymus gives preference to some α but not β J segments. These findings provide a deeper insight into the mechanism of TCR repertoire generation.
Adaptive immunity is provided by B and T cells bearing B-cell receptors (BCRs) and Ig-like T-cell receptors (TCRs), respectively. These hypervariable molecules are the key part of the adaptive immune system as they can potentially recognize any alien agent and drive specific immune responses. The α/β TCRs recognize short peptides in the complex with major histocompatibility complex (MHC) molecules and play the key role in the targeted immune response. The total diversity of TCR molecules in an individual human organism is initially formed via genomic recombination with subsequent positive and negative selection at several stages of maturation and activation. The maximal theoretical diversity of TCRβ chain’s amino acid sequences in humans is estimated between 5 × 1011 (1) and 1014 (2), whereas the maximal number of α/β pairs reaches 1018 (3). This huge number of variants is probably never achieved: the whole TCRβ chain repertoire size in a single human organism is estimated at 1–5 × 106 (1, 4–6), although this is only a lower bound estimate. Two driving forces shape the final face of individual TCR repertoire: the individual genetics and the complexity of environmental factors. The genes coding for proteins involved in VDJ recombination, antigen processing and presentation, and products of genes participating in the immune response signaling belong to the first type of the repertoire-forming factors. The spectrum of the organism’s self-peptides presented in the thymus also depends on the individual’s set of the MHC molecules. Moreover, this spectrum of peptides is determined by the amino acid sequences of the organism’s proteins, which thus can also be considered a genetic factor. Furthermore, TCRs arising to the same alien antigenic peptides are known to be MHC restricted (7). The environmental factors include the whole range of pathogens met by the individual including disease-causing bacteria and viruses, as well as vaccines, symbionts, etc. The genetic component can potentially have a major impact on the initial recombination and selection in the thymus forming the naïve TCR repertoire, whereas the subsequent interference with antigens provides the selective expansion of some TCRs and forms the final repertoire structure. However, the particular impact of genetic factors on TCR repertoire structure and diversity is unknown.
All genes of monozygous (MZ) twins are identical (including those responsible for the TCR repertoire formation), and therefore, MZ twins are widely used in the studies where the genetic impact is evaluated. Several studies of TCR repertoires were performed mainly focusing on diseases concordant and discordant MZ twins and using complementarity determining region 3 (CDR3) spectra-typing and/or low depth sequencing (8–11). Some of these studies reported the common use of particular V genes and common clonotypes. In recent years, the high-throughput sequencing technologies paved the way to whole-repertoire studies of individual TCRs that led to new findings in the field of adaptive immunity (1, 5, 6, 12–22). In this study, for the first time to the best of our knowledge, we obtain and compare the α and β chain TCR repertoires of three pairs of MZ twins using next-generation sequencing (NGS).
Results
Library Preparation and Sequencing.
RNA samples from three female pairs of monozygous twins (A, C, and D) were studied in this work. All individuals were HLA (MHC I) typed, and we found that each of the three MZ pairs had the same MHC I alleles profile (Table S1). TCR repertoires (for α and β chains) were obtained for all three pairs of monozygous twins. Sequencing libraries were prepared as described previously (12, 15, 23) with several modifications (Fig. S1 and SI Materials and Methods). To avoid any potential cross-contamination during library preparation and sequencing, we introduced sample-specific barcodes on both ends of each TCR cDNA library in the course of cDNA synthesis and amplification. In total, we obtained around 50,000,000 sequencing reads for 12 samples (6 α and 6 β) using the Illumina sequencing platform. After removing the reads with improper index combinations, TCR clonesets were generated for each library using our MiTCR software package (24) (see Table S2 for reads and clones distribution). Briefly, MiTCR extracts the CDR3 sequence from each read, identifies V, D, and J gene segments, merges and counts sequences with the identical CDR3 to form clonotypes, filters out or rescues low-quality reads, and provides advanced correction of PCR and sequencing errors. MiTCR CDR3 extraction yielded 2.2–3.6 million reads for each of the 12 libraries (Table S2). The identified number of clonotypes was different for each individual, varying from ∼100,000 to nearly 500,000. The resulting clonotypes are available at http://labcfg.ibch.ru/tcr.html#MZTwins and http://mitcr.milaboratory.com/datasets/twins2013/ in MiTCR format (24). A portion of each library was comprised by the out-of-frame clonotypes representing the nonfunctional TCR sequences formed during the recombination step. The percentage of such sequences was different for α and β TCR libraries, varying in most cases from 12.5% to 14.5% and from 2.9% to 4.1% for α and β libraries, respectively (Table S2).
V Gene Usage but Not J Gene Usage Is Similar in Twins Before the T-Cell Selection in Thymus.
Proteins involved in TCR gene recombination could affect the initial TCR repertoire formed at the very early stage before selection in the thymus. MZ twins have identical genes coding for the TCR recombination machinery; thus, their TCR repertoires are expected to be more similar at this step. In course of T-cell maturation, TCR locus recombination events can produce nonfunctional TCRs with frameshifts or stop codons (25). In this case, the T cell tries to arrange the second allele, and if the successful (in-frame) TCR formation occurs, the T cell carries both functional and nonfunctional TCR genes (26). The latter allele expression is usually down-regulated by the nonsense-mediated mRNA decay (NMD). However, the corresponding RNA is still present in the T cell at some concentration.
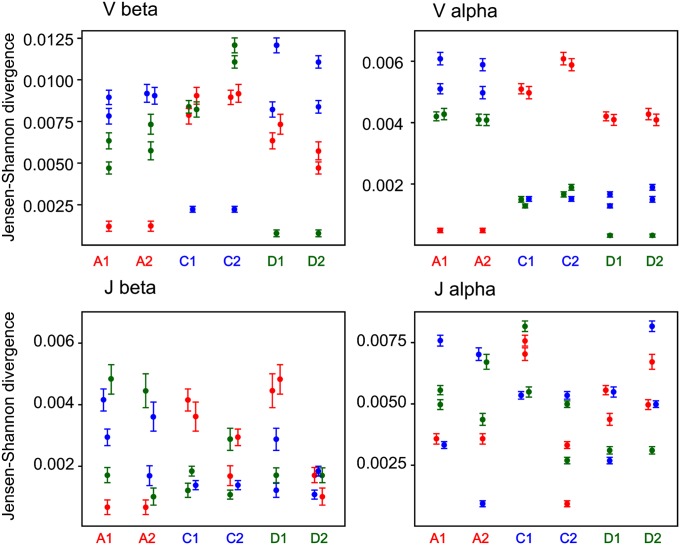
To analyze the nonfunctional TCR repertoire, we first removed artificial (carrying PCR or sequencing errors) out-of-frame clonotypes by mapping them to the in-frame sequences with one indel nucleotide allowed in the CDR3. The remaining genuine out-of-frame sequences were not affected by any type of selection (either in the thymus or on the periphery) and thus were used to characterize the initial repertoire formed by recombination itself (1, 2, 20). We then calculated the Jensen–Shannon (JS) divergence divided by mean entropy (SI Materials and Methods) for V and J gene usage in all possible pairs formed by six individuals for α and β out-of-frame TCR clonotypes, irrespective of each clonotype frequency (Fig. 1). The more similar distribution of V or J segments between a pair of individuals is characterized by the lower JS divergence (distance) value. V segment usage for TCRβ out-of-frame clonotypes was more similar (4–10 times lower JS divergence) in each twin pair compared with other individuals (Fig. 1, Vβ). For TCRα out-of-frame clonotypes the lowest JS distance was also observed between the individual and her twin except for C1 (Fig. 1, Vα). In contrast, JS divergence for J gene usage was distributed in a nearly stochastic manner, indicating no significant skew in J segment use in twin pairs for both α and β TCR chains (Fig. 1, Jα and Jβ).
Fig. 1.
JS divergence for Vβ, Jβ and Vα, Jα segment usage of out-of-frame clonotypes. Each column indicates the JS divergence divided by the mean entropy (for each pair) between an individual and five other individuals. Error bars indicate SD calculated using the bootstrap analysis. Red circles, individuals A1 or A2; blue circles, individuals C1 or C2; green circles, individuals D1 or D2.
J Gene Usage Is Affected by Selection in the Thymus.
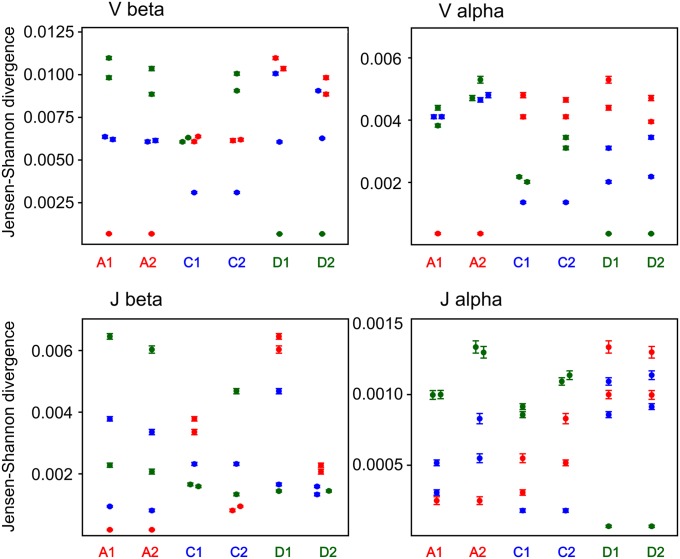
The next step of TCR repertoire formation includes the selection of T cells with functional CDR3 sequences against the individual’s specific repertoire of MHC molecules in the thymus. After maturation in the thymus, a pool of naïve T cells able to recognize MHC and at the same time not to recognize auto-antigens is formed. At this step, individual genetics is the main driving force as the MHC molecules and self-antigenic peptides are both genetically encoded. As the high-throughput sequencing provides deep TCR repertoire profiling, the main diversity of the obtained repertoire is generated by the naïve T cells. At the same time, antigen-experienced expanded clones represented by a large number of sequencing reads have a minor influence on the total clonotype diversity [for example, Robins et al. (1) report a mean of 420,000 unique CDR3 sequences in FACS-sorted naïve samples compared with 69,000 unique sequences in a memory pool]. Thus, we suggested that the entire set of the in-frame TCR clonotypes sequenced is dominated by a naïve T-cell repertoire. We calculated the JS divergence for V and J gene usage for α and β in-frame TCR clonotypes of all possible pairs of individuals (Fig. 2).
Fig. 2.
JS divergence for Vβ, Jβ and Vα, Jα segment usage of in-frame clonotypes. Each column indicates the JS divergence divided by the mean entropy (for each pair) between an individual and five other individuals. Error bars indicate SD calculated using the bootstrap analysis. Red circles, individuals A1 or A2; blue circles, individuals C1 or C2; green circles, individuals D1 or D2.
V gene usage for both α and β in-frame TCR clonotypes was obviously closer for all three twin pairs compared with any pair of unrelated individuals (Fig. 2, Vβ and Vα). J gene usage for the TCRβ in-frame clonotypes demonstrated diverse patterns of JS divergence: for individuals A1 and A2, the shortest JS distance was to the twin individual, whereas for individuals D1/D2 and C1/C2, the distribution of JS distance was stochastic (Fig. 2, Jβ).
A significant change in the pattern of JS divergence of Jα usage was observed for the in-frame clonotypes compared with the out-of-frame clonotypes. Jα usage for the in-frame clonotypes was much more similar for all three twin pairs than for random pairs of individuals, whereas for the out-of-frame TCRα repertoires, J genes demonstrated stochastic distribution independent of kinship (Figs. 1, Jα, and 2, Jα). Additionally average JS distance for Jα in-frames was considerably (5–10 times) shorter compared with out-of-frames between any pair of individuals. This finding suggests that TCRs’ J segments are initially selected for recombination in a relatively random manner in all humans, but during the subsequent selection, the usage of Jα segments becomes more universal for all individuals. Twin pairs use even a more similar pattern of Jα segments after selection compared with unrelated individuals.
MZ Twins Have More Shared Clonotypes Among the Most Abundant T-Cell Clones.
The final individual repertoire of TCRs results from three sequential processes: the recombination of the germ-line TCR gene locus, selection in the thymus, and interaction with different antigens (including self-antigens). Therefore, as MZ twins bear identical genes coding for proteins involved in TCR recombination and identical sets of genes involved in antigen presentation, they are expected to have more similar sets of TCR molecules. Indeed, this fact is clearly observed from the similar usage of V gene segments (see Fig. 2) coding for CDR1 and CDR2 responsible for interaction with MHC. Moreover, twins reared together are exposed to a similar set of antigens processed in the same way for a long period of their life. Thus, we expected the set of TCR CDR3 sequences to have more similarity between twins than between unrelated individuals.
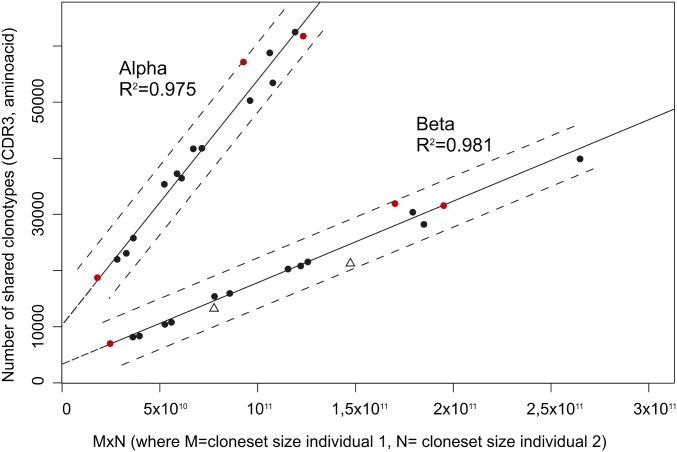
We analyzed the number of amino acid CDR3 clonotypes (i.e., having an identical amino acid sequence of CDR3 independently of their nucleotide CDR3 sequence) shared between each pair of individuals (Table S3) for α and β TCR repertoires. The minimal number of shared CDR3β clonotypes (6,906) was observed for individuals A1 and A2, both having the lowest number of identified clonotypes (hereafter “cloneset size”; 228,772 and 102,989 individual clonotypes, respectively), whereas the maximal number (39,479) was determined for C1 and D2 having the largest clonotype sets (around 527,000 and 500,000, respectively). The same pattern was observed for overlap of the α chain CDR3 repertoires. The number of shared clonotypes for all possible pairs of individuals and the product of the intersected clonesets sizes is shown in Fig. 3. We observed a linear correlation between the number of shared CDR3 clonotypes and the product of the cloneset sizes for both α and β TCR chains. R2 was 0.9752 and 0.9814 (for α and β regression lines, respectively). Thus, the number of shared clonotypes between TCR repertoires of two individuals is mainly determined by the intersected cloneset sizes. The impact of the identical TCR recombination machinery and MHC turns out not to be substantial as at least two of the three twin pairs exactly fit the regression line (Fig. 3, red circles). Using the data for all pairs, we calculated 98% prediction intervals for both lines. We also added the data on the number of shared CDR3 clonotypes obtained by Warren et al. (6) for two pairs of individuals (Fig. 3, empty triangles). These clonesets were generated using the similar RNA-based TCR library preparation method. Data on them fell into our model’s prediction interval despite that the TCR extraction and cloneset generation methods were different. Thus, the number of CDR3 clonotypes shared between TCR repertoires of two individuals can be predicted from the intersected clonesets sizes quite precisely. The normalized number of shared CDR3 clonotypes (i.e., the number of shared CDR3 clonotypes divided by the product of the intersected cloneset sizes) can be approximated as a cloneset size-independent measure of CDR3 repertoire similarity between two individuals.
Fig. 3.
Correlation between the number of shared clonotypes in each pair of individuals and the product of intersected cloneset sizes. Red circles correspond to twin pairs, and black circles correspond to unrelated pairs. Empty triangles correspond to the number of shared clonotypes obtained for two pairs of individuals by Warren et al. (6). Dash lines indicate the 98% prediction intervals for both regression lines.
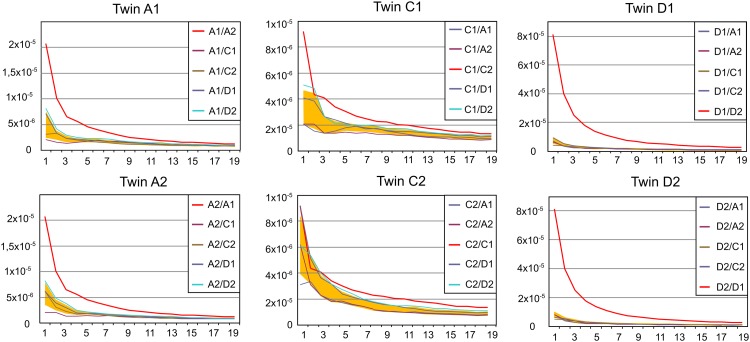
The abundant clonotypes (i.e., clonotypes with a higher number of sequencing reads) are enriched by antigen experienced T-cell clones, whereas the clonotypes with a low number of reads are to a great extent represented by the naïve T cells [it should be mentioned, however, that some naïve T lymphocytes can be abundant (27) and some antigen experienced T-cell clones are quite rare]. Fig. 4 shows the relationship between the number of shared CDR3 clonotypes and the abundance of the selected clonotypes in the datasets for each possible pair of individuals (1,000 most abundant clonotypes in individual 1 vs. 1,000 most abundant clonotypes in individual 2; 2,000 most abundant clonotypes in individual 1 vs. 2,000 most abundant clonotypes in individual 2, etc.). The number of shared CDR3 clonotypes was divided by the product of the intersected cloneset sizes (1,000 × 1,000, 2,000 × 2,000, etc.) for normalization. Previous observations suggest that if we randomly pick a limited number (1,000, 2,000, etc.) of clonotypes from each individual cloneset (independent of each clonotype read count), the normalized number of shared clonotypes remains constant.
Fig. 4.
The normalized number of identical TCRβ CDR3 amino acid sequences for each possible pair of individuals among 1,000 most abundant clonotypes, 2,000 most abundant clonotypes, etc. x axis, the number of the most abundant clonotypes (×1,000) intersected; y axis, the normalized number of shared clonotypes. The shaded area (orange) indicates the mean ± SD calculated for pairs formed by twin with each unrelated individual.
We found that the normalized number of shared clonotypes is significantly higher among the most abundant TCRβ CDR3 clonotypes for any pair of individuals. Notably, each of the six individuals had an even more increased number of the most abundant CDR3β clonotypes shared with her twin (Fig. 4). Similarly, more shared CDR3 amino acid sequences were observed among abundant α clonotypes. However, the difference between the twin and unrelated pairs was not obvious. Although individuals C1, C2, D1, and D2 shared the highest number of common clonotypes with their twins, the difference with the unrelated individuals was minimal, whereas no difference was observed for the individuals A1 and A2 (Fig. S2).
Shared Clonotypes Features Are Different for Twins and Unrelated Individuals.
The antigen specificity of the T-cell receptors to the peptide-MHC complex is provided by three complement determining regions (CDRs). One of them (CDR3) is hypervariable (i.e., can consist of potentially any combination of amino acids), whereas CDR1 and CDR2 are selected from a set of about 50 possible variants (genetically encoded V genes). Thus, T-cell receptors having identical amino acid sequences of all three CDRs most probably recognize the same antigen-MHC complex in different individuals.
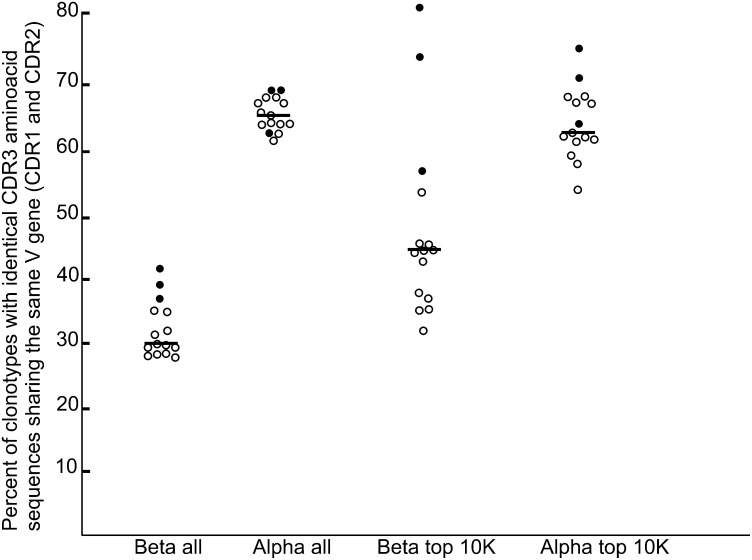
We analyzed the percentage of TCR clonotypes having the same V gene among clonotypes with identical amino acid CDR3s for each possible pair of six individuals for both α and β chains (Fig. 5). For the whole set of identified clonotypes the percentage of identical V genes among TCRs with identical CDR3 was similar for any pair of individuals for both α and β chains. Twenty-eight percent to 42% of clonotypes with identical CDR3β additionally have the same V gene, wherein three monozygous twin pairs have the highest percent (P = 0.0088, Mann–Whitney U test; Fig. 5, β all). For the TCRα clonotypes, this percent lied between 62% and 69% with no significant deviation for the twin pairs (P = 0.233, Mann–Whitney U test; Fig. 5, α all). We also calculated the percent of clonotypes with identical V genes among the carriers of identical CDR3s taken from the top 10,000 most abundant clonotypes for each possible pair of individuals for α and β TCRs (Fig. 5, β top 10K and α top 10K). We observed a significant increase of this value among the most abundant TCRβ sequences for all pairs (mean, 47%) compared with all clonotypes (mean, 33%). Interestingly, this percent was significantly (P = 0.0044, Mann–Whitney U test) higher for all three twin pairs (from 57% for C1/C2 to 81% for D1/D2). In contrast, there was no significant difference between the percentage of identical V genes among all and the top 10,000 most abundant TCRα clonotypes with identical CDR3 (means were 65% and 64%, respectively). This percentage for twin pairs among the 10,000 most abundant TCRα was slightly higher (P = 0.048, Mann–Whitney U test).
Fig. 5.
The percentage of clonotypes with identical CDR3 amino acid sequences sharing the same V gene for all possible pairs of individuals. Black circles indicate twin pairs, empty circles indicate all other pairs, and lines indicate median values. β all, among all identified TCRβ clonotypes; α all, among all identified TCRα clonotypes, β top 10K, among 10,000 most abundant TCRβ clonotypes; α top 10K, among 10,000 most abundant TCRα clonotypes.
To further characterize the shared TCRs pool, we analyzed the number of random nucleotides added at the junction between the V-D and D-J segments for the β chain and V-J for the α chain. The average number of added nucleotides in the individual repertoire was 7.4 (sum for both junctions) for the β chain and 6.2 (for the single junction) for the α chain. In the shared TCR pools, we observed a significant decrease in the number of added nucleotides (3.3 and 3.8 for β and α, respectively), with no difference between twin pairs and unrelated individual pairs. This decrease is not surprising as TCRs with a lower number of inserted nucleotides have a higher chance for independent convergent generation in different individuals [as indicated previously (18, 20, 21)] and thus a higher chance to match in two or more humans. In contrast, when we intersected the most abundant clonotypes (top 10,000), the number of inserted nucleotides for matched TCRs differed between twin pairs and unrelated pairs (2.9 vs. 1.9, respectively, P < 0.00001, two-sided t test) for β chains. For α chains, the number of inserted nucleotides was very close for twin and unrelated pairs for the most abundant shared clonotypes (2.07 and 1.90, respectively, P = 0.22, two-sided t test).
Clonotypes shared by twins and by unrelated individuals can represent T cells fighting common viruses like CMV and Epstein–Barr virus (EBV). We stained peripheral blood mononuclear cells (PBMCs) from donor A2 with the HLA-A*02 multimer with CMV peptide NLVPMVATV(NLV), FACS sorted the stained cells, generated the TCR libraries, and sequenced them by Illumina. We identified 25 β and 21 α distinct clonotypes and searched for their exact CDR3 amino acid matches in other individual datasets. Nine β and 12 α matched clonotypes were found in at least one other individual (Table S4). Interestingly, the matched clonotypes were found in the datasets of C1/C2 twins despite that they do not have the HLA-A*02 alleles. When comparing the V segments of matched CMV-NLV-specific donor A2 clonotypes, we found that individuals A1, D1, and D2 in many cases have the same V gene in addition to the same CDR3β (5/6 for A1, 4/6 for D1, and 3/5 for D2). In contrast, none of the matched CDR3β sequences from C1 and C2 had the same V segment. Thus, we can suggest that these clonotypes recognize the same CMV peptide (as they have identical CDR3) but presented by another MHC molecule (as they have different V segments coding for the CDR1/CDR2 that recognize MHC) (28). No such observation was made for the α chain where identical/different V segments among matched CMV-positive CDR3s are distributed quite randomly between individuals. For donor A2, we calculated the portion of HLA-A02-NLV-specific CDR3 clonotypes among all of the clonotypes shared with each other individual for several abundance categories (top 10,000 most abundant clonotypes in both donors, top 30,000, etc.; Fig. S3A). These data suggest that more identical CMV-specific T cells are present among abundant clonotypes. However, the absolute numbers of such clonotypes are too low to make confident conclusions. To get deeper insight into this issue, we generated a list of 257 TCRβ CDR3 sequences known to recognize different CMV or EBV peptides in complex with a number of MHC I variants (HLA-A*02; HLA-B*07, B*08; at least one allele is present in every individual; Table S1) from the previously published works (SI Materials and Methods). Then we compared the list with the clonesets of all six individuals from this work. Sixty-nine of 257 CMV/EBV-specific amino acid CDR3 sequences were found in at least one individual, and 6 sequences were found in all six individuals. Next, we calculated the percent of CMV/EBV-specific CDR3s among clonotypes shared by each pair of six individuals (Fig. S3B). The portion of such CDR3 sequences is again higher in the fraction of abundant clonotypes. The higher percentage of CMV/EBV-specific clonotypes suggests that a significant portion of abundant clonotypes in the human organism may be represented by common virus-specific TCRs. More generally, the higher proportion of shared clones among abundant clonotypes could be a consequence of peripheral selection and expansion of antigen-specific clonotypes driven by persistent pathogens. We observed no significant difference between twins and nonrelated individuals for this parameter.
Discussion
To summarize the results of the current study, we conclude the following.
V gene segment selection for TCR β and α chains is very similar in twin pairs. This result could be expected because twins have identical sets of MHC molecules and V segments coding for MHC recognizing motifs (29, 30). However, we found that not only the TCRs of cells affected by thymic selection (in-frame TCRs), but also the TCRs formed before selection (out-of-frame TCRs) are strictly dependent on individual genetic factors. Each of the analyzed twin pairs has a much closer distribution of V gene usage in the out-of-frame clonesets than any pair of random individuals, as evaluated by JS divergence. The same conclusion is valid for V gene segments usage in α chain TCRs. These findings indicate that the selection of variable segments for the mature TCRs during recombination is determined by the genes coding for the TCR recombination machinery, as it was suggested previously (31).
In contrast, the selection of J gene segments for both α and β TCR chains at the initial recombination step (as evaluated from the out-of-frame clonesets) is not different in twin pairs and random pairs of individuals. This finding indicates that the selection of J gene segments for recombination is not strictly determined by genetic factors. The pattern of JS divergence of J segment distributions is dramatically changed for the in-frame α TCRs after thymic selection. For each of the six individuals, her twin becomes the closest one in α segment usage leaving behind all unrelated individuals. Moreover, we observe that JS divergences fall down for all pairs of individuals when moving from the out-of-frame to the in-frame Jα segment usage. At the same time, Jβ segment usage remains generally independent on the individuals’ kinship. This finding most probably suggests that the part of the TCRα chain receptor encoded by the J gene segment is involved in the recognition of MHC molecules at the selection stage in the thymus, whereas the same part of the β chain molecule is not. However, this suggestion has to be validated by more independent studies.
An accurate estimation of the shared clonotypes amount when comparing large individual TCR repertoires appears to be quite a challenging task: missing particular points when performing such calculations can lead to incorrect conclusions and false discoveries. First, the compared datasets should be generated using the same protocol of sample preparation, comparable depth of sequencing, and TCR sequence extraction algorithms. Additionally, many PCR cycles and millions of sequencing reads inevitably generate a huge artificial TCR diversity and lead to an incorrect evaluation of the cloneset size. This artificial diversity can be partially avoided by using proper error correction algorithms (12, 24). Second, possible cross-contamination between individual TCR datasets should be avoided by standard precautions (PCR boxes, clean rooms, etc.), as well as by the use of additional platform-specific strategies preventing molecular exchange in course of library preparation and sequencing (e.g., double sample barcoding; SI Materials and Methods and Fig. S1). Finally, the number of shared clonotypes between a pair of individuals can be compared correctly only if the comparable amount of starting material (T cells and isolated RNA or DNA) and the similar sequencing depth are achieved. For example, if a total of 10,000 clonotypes is identified for samples 1 and 2, they are mainly comprised by the expanded T-cell clones and have a higher proportion of public (common) clonotypes, whereas 200,000 sample 3 clonotypes would to a great extent consist of the naïve T cells and contain a lower portion of public TCRs. In this example the normalized (divided by the product of cloneset sizes) number of shared clonotypes will be higher for the sample pair 1/2 than for the pairs 1/3 and 2/3.
In this study, we clearly demonstrate that the number of CDR3 clonotypes shared between any pair of individuals is strictly dependent on the product of the intersected cloneset sizes for both α and β TCR chains. This finding is in agreement with the observation made for the out-of-frame (generated) repertoire by Murugan et al. (2). Surprisingly, MZ twins do not deviate from this trend. The lines depicting the linear relationship are approximated with p = b1 × MN + b0, where p is the number of identical CDR3s in a pair of individuals; M and N are the sizes of the two intersected clonesets; and b1 and b0 are the slope and the intercept, respectively. Different values of b1 specifying the slope of lines for β (1.472 × 10−7) and for α chains (4.351 × 10−7) indicate different potential diversity and thus different numbers of shared clonotypes for α and β chains. The more diverse β chain potentially has less shared clonotypes in a pair of individual repertoires because amino acid sequence convergence is less probable. An extrapolation of the line to the zero intersected cloneset sizes indicates that every pair of individuals will inevitably have some shared clonotypes and their number is reflected by the b0 coefficient. Presence of such shared clonotypes can be partially explained by the existence of natural killer T (NKT) and mucosa-associated invariant T (MAIT) cells having the identical TCRα chain sequences in all humans (32, 33). Indeed, we found all of the CDR3 sequences originated from MAIT cells and 8 of 12 CDR3 sequences originated from NKT cells (32) in our datasets. Moreover, the majority of MAIT CDR3 sequences were present in all individuals within 10,000 most abundant clonotypes. The frequently formed public clonotypes against common persisting viruses like CMV and EBV can also contribute to the preexisting shared sequences. Thus, the real behavior of the line near zero is more complex: when a small number of clonotypes (less than ∼10,000–20,000) is detected the chance to pick public clonotypes is higher, thus increasing the actual portion of shared clonotypes in a pair of individuals (as reflected in Fig. 4 and Fig. S2).
In general, deep TCR repertoires (mainly composed by low abundant T cells) of MZ twins have the same amount of shared CDR3 clonotypes compared with unrelated individuals. The shared clonotypes in all people are characterized by a lower number of nucleotides added at the germ-line segment junctions compared with individual repertoires, as shown previously (20, 34, 35). Interestingly, individual TCR repertoires of mice (nearly genetically identical within the same strain) share at least 10 times more CDR3 clonotypes (36) than human pairs. This fact is probably explained by the overall lower number of added nucleotides in mice TCRs [around 4.0 (34)] providing a higher chance to generate identical CDR3 independently. The observed percentage of identical V segments among TCRs with identical CDR3 shared by twins is also similar to unrelated pairs.
In contrast, the most abundant clonotypes (supposed to mainly reflect the antigen experienced T cells) shared by identical twins show some characteristics of β chain that clearly distinguish them from the ones shared by random individuals. First, the proportion of shared CDR3 sequences among abundant β clonotypes is significantly higher in twin pairs. Second, abundant clonotypes shared by twins have a higher number of added nucleotides compared with unrelated humans. Finally, the most abundant clonotypes with identical CDR3s share the same Vβ segments in twin pairs for β TCR chains more often. Most probably, these clonotypes in twins have an initial higher probability to rearrange and then are activated by the same antigens presented in the same MHC context. The results of the experiments with CMV-specific T cells sorting also favor this suggestion. However, it is not possible to exclude the influence of environmental factors: all three pairs of twins in this study were reared together and most probably met the same infectious agents, vaccines, etc. To further clarify this point deep TCR profiling of twins reared apart is needed. At the same time, we did not observe any significant difference in characteristics of most abundant TCRα chain clonotypes shared between the twins and between the unrelated individuals. This finding suggests that α chain CDR3 is less involved in specific antigenic peptides recognition.
Materials and Methods
A detailed description of experimental procedures is given in SI Materials and Methods and Fig. S4. Briefly, RNA was isolated from PBMCs of three female pairs of healthy MZ twins and used for TCR cDNA library preparation (see Table S5 for oligonucleotides used). cDNA libraries were sequences by Illumina. TCR clonotypes were generated using MiTCR software.
Supplementary Material
Acknowledgments
This work was supported by the Molecular and Cellular Biology Program Russian Academy of Sciences, Russian Foundation for Basic Research (14-04-01062, 14-04-01823, 13-04-01124, and 14-04-01247), European Regional Development Fund (CZ.1.05/1.1.00/02.0068), Council of the President of the Russian Federation for Young Scientists (MD-3044.2014.4 and SP-2039.2012.4), and the Dmitry Zimin Dynasty Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI sequence read archive database (accession no. SRP028752).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319389111/-/DCSupplemental.
References
- 1.Robins HS, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2(47):47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murugan A, Mora T, Walczak AM, Callan CG., Jr Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proc Natl Acad Sci USA. 2012;109(40):16161–16166. doi: 10.1073/pnas.1212755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334(6181):395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 4.Arstila TP, et al. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286(5441):958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 5.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren RL, et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011;21(5):790–797. doi: 10.1101/gr.115428.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miles JJ, Douek DC, Price DA. Bias in the αβ T-cell repertoire: Implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011;89(3):375–387. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 8.Fozza C, et al. T-cell receptor repertoire analysis in monozygotic twins concordant and discordant for type 1 diabetes. Immunobiology. 2012;217(9):920–925. doi: 10.1016/j.imbio.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Gulwani-Akolkar B, Shalon L, Akolkar PN, Fisher SE, Silver J. Analysis of the peripheral blood T-cell receptor (TCR) repertoire in monozygotic twins discordant for Crohn’s disease. Autoimmunity. 1994;17(3):241–248. doi: 10.3109/08916939409010660. [DOI] [PubMed] [Google Scholar]
- 10.Somma P, et al. Characterization of CD8+ T cell repertoire in identical twins discordant and concordant for multiple sclerosis. J Leukoc Biol. 2007;81(3):696–710. doi: 10.1189/jlb.0906584. [DOI] [PubMed] [Google Scholar]
- 11.Utz U, et al. Skewed T-cell receptor repertoire in genetically identical twins correlates with multiple sclerosis. Nature. 1993;364(6434):243–247. doi: 10.1038/364243a0. [DOI] [PubMed] [Google Scholar]
- 12.Bolotin DA, et al. Next generation sequencing for TCR repertoire profiling: Platform-specific features and correction algorithms. Eur J Immunol. 2012;42(11):3073–3083. doi: 10.1002/eji.201242517. [DOI] [PubMed] [Google Scholar]
- 13.Britanova OV, et al. First autologous hematopoietic SCT for ankylosing spondylitis: A case report and clues to understanding the therapy. Bone Marrow Transplant. 2012;47(11):1479–1481. doi: 10.1038/bmt.2012.44. [DOI] [PubMed] [Google Scholar]
- 14.Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009;19(10):1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mamedov IZ, et al. Quantitative tracking of T cell clones after haematopoietic stem cell transplantation. EMBO Mol Med. 2011;3(4):201–207. doi: 10.1002/emmm.201100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen P, et al. Identification of errors introduced during high throughput sequencing of the T cell receptor repertoire. BMC Genomics. 2011;12:106. doi: 10.1186/1471-2164-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherwood AM, et al. Deep sequencing of the human TCRγ and TCRβ repertoires suggests that TCRβ rearranges after αβ and γδ T cell commitment. Sci Transl Med. 2011;3(90):90ra61. doi: 10.1126/scitranslmed.3002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venturi V, et al. A mechanism for TCR sharing between T cell subsets and individuals revealed by pyrosequencing. J Immunol. 2011;186(7):4285–4294. doi: 10.4049/jimmunol.1003898. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. 2010;107(4):1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putintseva EV, et al. Mother and child T cell receptor repertoires: Deep profiling study. Front Immunol. 2013;4:463. doi: 10.3389/fimmu.2013.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shugay M, et al. Huge Overlap of Individual TCR Beta Repertoires. Front Immunol. 2013;4:466. doi: 10.3389/fimmu.2013.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turchaninova MA, et al. Pairing of T-cell receptor chains via emulsion PCR. Eur J Immunol. 2013;43(9):2507–2515. doi: 10.1002/eji.201343453. [DOI] [PubMed] [Google Scholar]
- 23.Mamedov IZ, et al. Preparing unbiased T-cell receptor and antibody cDNA libraries for the deep next generation sequencing profiling. Front Immunol. 2013;4:456. doi: 10.3389/fimmu.2013.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolotin DA, et al. MiTCR: Software for T-cell receptor sequencing data analysis. Nat Methods. 2013;10(9):813–814. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
- 25.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21(2):133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weischenfeldt J, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22(10):1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quigley MF, et al. Convergent recombination shapes the clonotypic landscape of the naive T-cell repertoire. Proc Natl Acad Sci USA. 2010;107(45):19414–19419. doi: 10.1073/pnas.1010586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koning D, et al. CD8+ TCR repertoire formation is guided primarily by the peptide component of the antigenic complex. J Immunol. 2013;190(3):931–939. doi: 10.4049/jimmunol.1202466. [DOI] [PubMed] [Google Scholar]
- 29.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10(2):143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 31.Melenhorst JJ, et al. Contribution of TCR-beta locus and HLA to the shape of the mature human Vbeta repertoire. J Immunol. 2008;180(10):6484–6489. doi: 10.4049/jimmunol.180.10.6484. [DOI] [PubMed] [Google Scholar]
- 32.Greenaway HY, et al. NKT and MAIT invariant TCRα sequences can be produced efficiently by VJ gene recombination. Immunobiology. 2013;218(2):213–224. doi: 10.1016/j.imbio.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Venturi V, Rudd BD, Davenport MP. Specificity, promiscuity, and precursor frequency in immunoreceptors. Curr Opin Immunol. 2013;25(5):639–645. doi: 10.1016/j.coi.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Li H, et al. Recombinatorial biases and convergent recombination determine interindividual TCRβ sharing in murine thymocytes. J Immunol. 2012;189(5):2404–2413. doi: 10.4049/jimmunol.1102087. [DOI] [PubMed] [Google Scholar]
- 35.Venturi V, et al. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc Natl Acad Sci USA. 2006;103(49):18691–18696. doi: 10.1073/pnas.0608907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ndifon W, et al. Chromatin conformation governs T-cell receptor Jβ gene segment usage. Proc Natl Acad Sci USA. 2012;109(39):15865–15870. doi: 10.1073/pnas.1203916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.