Significance
The folding of outer membrane proteins (OMPs) in biological membranes in vivo requires an evolutionarily conserved and essential protein, β-barrel assembly machinery subunit A (BamA). By investigating the folding kinetics of OMPs into membranes composed of Escherichia coli lipids, we discovered that periplasmic lipid head groups impose a kinetic barrier to OMP folding. This barrier serves as a negative selection to prevent OMPs from folding into inner bacterial membranes where they would dissipate the proton motive force and kill the cell. We showed that BamA enables OMPs to overcome this barrier at bacterial outer membranes by enhancing their observed folding rates. We propose BamA does so by creating bilayer defects, which are known to accelerate the intrinsic folding reaction.
Keywords: membrane protein folding, beta-barrel transmembrane protein
Abstract
Outer membrane β-barrel proteins (OMPs) are crucial for numerous cellular processes in prokaryotes and eukaryotes. Despite extensive studies on OMP biogenesis, it is unclear why OMPs require assembly machineries to fold into their native outer membranes, as they are capable of folding quickly and efficiently through an intrinsic folding pathway in vitro. By investigating the folding of several bacterial OMPs using membranes with naturally occurring Escherichia coli lipids, we show that phosphoethanolamine and phosphoglycerol head groups impose a kinetic barrier to OMP folding. The kinetic retardation of OMP folding places a strong negative pressure against spontaneous incorporation of OMPs into inner bacterial membranes, which would dissipate the proton motive force and undoubtedly kill bacteria. We further show that prefolded β-barrel assembly machinery subunit A (BamA), the evolutionarily conserved, central subunit of the BAM complex, accelerates OMP folding by lowering the kinetic barrier imposed by phosphoethanolamine head groups. Our results suggest that OMP assembly machineries are required in vivo to enable physical control over the spontaneously occurring OMP folding reaction in the periplasm. Mechanistic studies further allowed us to derive a model for BamA function, which explains how OMP assembly can be conserved between prokaryotes and eukaryotes.
Outer membrane β-barrel proteins (OMPs) are found in the outer membranes of Gram-negative bacteria, mitochondria, and chloroplasts (1). The functions of OMPs are versatile and often essential as they include transport of metabolites and toxins as well as membrane biogenesis (2). Alterations of outer membranes and outer membrane proteins can lead to the development of antibiotic-resistance in pathogenic bacteria, and dysfunction of OMPs in outer membranes of mitochondria plays a role in diabetes and neurodegenerative diseases, among other life-threatening illnesses (3–7). How OMPs attain their native fold in their natural lipid environment is therefore a fundamental question in biological and biomedical research.
The biological assembly of outer membrane proteins into bacterial outer membranes requires a functionally conserved protein complex, termed β-barrel assembly machinery (BAM) (8, 9). Previous work suggested that the main subunit of the BAM complex, the OMP BamA, carries out its essential function by providing a structural basis for OMP folding (10–12). However, it has been shown many times that OMPs are capable of spontaneously folding to their native state in model membranes in vitro through an intrinsic folding pathway in the absence of BamA (13–17). Neither the folding in vivo nor in vitro requires an external energy source such as ATP or a redox potential (18, 19).
The observation that OMPs can fold to their native states in vitro raises the important question of why OMPs require assembly machineries such as BAM to fold into their cellular outer membranes. To address this question, we developed an experimental strategy that enabled us to monitor the folding kinetics of bacterial OMPs in the absence and presence of prefolded BamA under membrane conditions that mimicked the periplasmic lipid environment. We discovered that native lipid head groups impose a kinetic barrier to folding that is relieved by the catalytic action of BamA. Our findings explain many in vivo observations and allowed us to derive a biophysical model of OMP sorting to the correct cellular membrane followed by its folding into bacterial outer membranes.
Results
Native Lipids of Escherichia coli Outer Membranes Support Poor Folding Efficiencies in Vitro for Outer Membrane Proteins.
To obtain generally applicable insights into the biological folding of outer membrane β-barrels, we investigated the folding of eight distinct OMPs: full-length outer membrane protein A (OmpA325), OmpT, OmpX, OmpW, PhoP/PhoQ-activated gene product (PagP), outer membrane phospholipase (OmpLA), the long-chain fatty acid transport protein (FadL), and the β-barrel assembly machinery subunit A (BamA) from E. coli. We chose these OMPs for several reasons: (i) These proteins all derive from the same biological membrane and have experienced similar evolutionary pressures with respect to their native lipid environments; (ii) they have distinct sequences with no significant pairwise sequence similarity; (iii) their folding has been extensively studied in vitro and in vivo (15); and (iv) their structures are solved at high atomic resolution, which shows that the set is structurally diverse within the β-barrel fold (20). All of these proteins will fold into synthetic lipid vesicles composed of phosphocholine head groups. In contrast, SI Appendix, Fig. S1 shows the generally poor folding we observed using membranes composed of native lipids purified from E. coli. Despite an extensive search for productive folding conditions, we only ever observed moderate folding for OmpT, BamA, and OmpA, an insignificant amount of folding for PagP and OmpX, and no folding for OmpLA, FadL, and OmpW after a 20-h incubation. These results demonstrate that bacterial OMPs have limited abilities to attain their native state in a membrane prepared from native lipids in the absence of the BAM complex. The observed folding efficiencies are extremely low in comparison with those carried out using membranes composed of nonnative lipids (15). Based on these results, we hypothesize that the E. coli outer membrane lipids represent a physical environment that slows intrinsic OMP folding.
Native Lipid Head Groups Impose a Kinetic Barrier to the Intrinsic OMP Folding Reaction.
The vast majority of in vitro folding experiments have been conducted using lipids with phosphocholine (PC) head groups. This lipid environment supports fast and efficient folding, but PC is not a naturally occurring head group in E. coli (15). In contrast, the outer membrane, inner leaflet surface encountered by a folding OMP in vivo completely lacks PC and is instead enriched in phosphatidylethanolamine and contains phosphatidylglycerol as well as a small fraction of cardiolipin (21, 22). To quantify the impact of these native head groups on folding, we developed a lipid “host” and “native-guest” head group strategy in which the host head group was PC and the native-guest head groups were phosphoethanolamine (PE) and/or phosphoglycerol (PG), and lipids were mixed in different mole ratios. We collected kinetic traces for OMP folding into various lipid compositions using a representative set of the initial eight OMPs, namely, OmpA, OmpLA, OmpX, and OmpA171 (a truncated version of OmpA containing only the TM β-barrel domain; ref. 23).
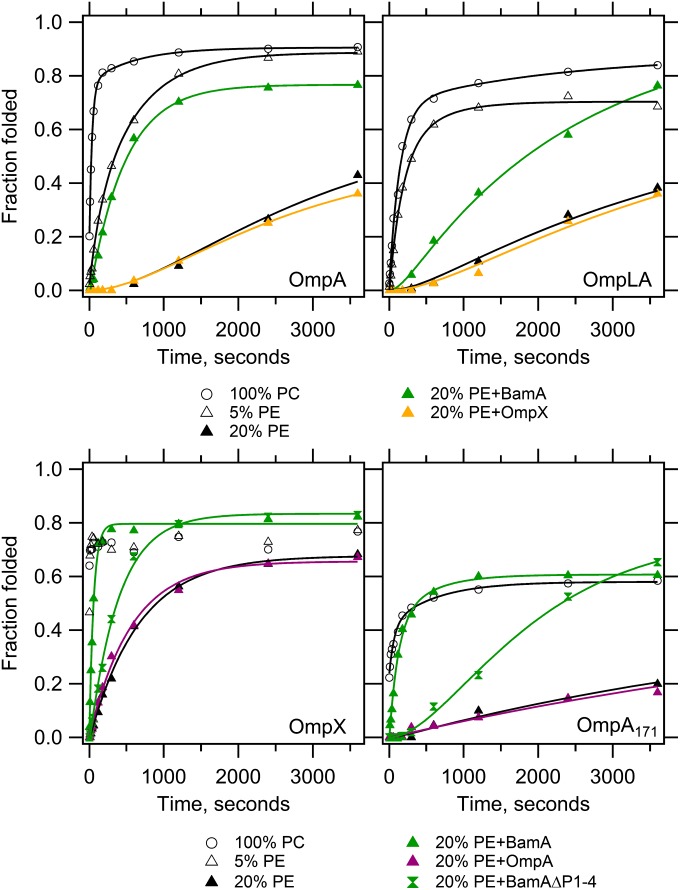
Fig. 1 shows that the introduction of a guest PE head group slows the observed folding rates of all four OMPs. Further, the PE-induced kinetic retardation demonstrates a dose-dependence as evidenced by the finding that the observed folding rates of OmpX, OmpA, and OmpLA are even slower in 20% PE as compared with 5% PE, which are already slowed in comparison with 100% PC lipids. In contrast to the observation that 20% of OmpA171 folds in the burst phase into 100% PC lipids, the introduction of 20% PE induces a lag time in the appearance of the folded protein. Such a lag is also evident for both OmpLA and OmpA in 20% PE. The impact of this PE-induced slower folding on the folded efficiency is dramatic: Although 50–80% of the OMPs are folded in 100% PC after 1 min, this same folding efficiency was not reached even after 1 h in membranes containing 20% PE guest lipid. This result is completely compatible with the poor folding observed using native E. coli lipids where the PE fraction is much higher, and our findings for OmpA agree well with a previous observation that PE slows the folding of OmpA (24). Our study using multiple OMPs suggests PE has a general effect on OMP folding.
Fig. 1.
BamA lowers the kinetic barrier to OMP folding imposed by PE head groups. Unfolded OMPs were folded into LUVs containing 100% PC or 95% PC + 5% PE or 80% PC + 20% PE in the absence or presence of one of the following prefolded OMPs as indicated: BamA, BamAΔP1-4, OmpA or OmpX. The client OMP is indicated in each panel, and its total concentration was 4 or 2 μM without or with prefolded OMP, respectively, at a constant lipid:protein mole ratio of 800:1. Samples were taken at indicated times and quenched with SDS/PAGE loading buffer. SDS/PAGE gels (SI Appendix, Fig. S2) were analyzed by densitometry and fraction folded was plotted as the ratio of the folded band intensity divided by the boiled band intensity. Lines indicate fitted curves; black, no prefolded OMP; green, prefolded BamA or BamAΔP1-4; orange, prefolded OmpX; magenta, prefolded OmpA. Each experiment was conducted in triplicate, and one representative data set is shown for each condition. SI Appendix, Fig. S3 shows representative data that includes the extended time points collected for the slower kinetic data, demonstrating that folding was collected to saturation. The average fit values with SDs are reported in SI Appendix, Tables S1–S4 and Fig. S4.
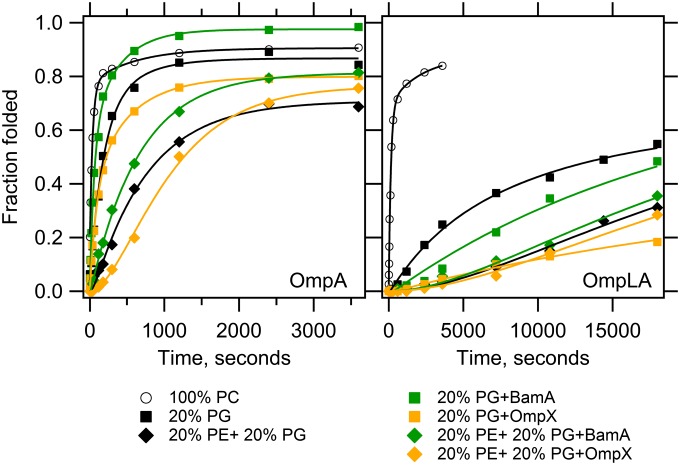
Fig. 2 shows that the introduction of 20% PG guest lipid also kinetically retards the folding of OmpA and OmpLA, although to different extents. Compared with 100% PC, the kinetic retardation of 20% PG on OmpLA severely affected its observed folding rates, reducing both the fast and slow rates by an order of magnitude. The consequential effect on final folding efficiency was a reduction of 18%, which is similar to the effect on efficiency of 20% PE (−15% compared with 100% PC). However, because 20% PE induced the formation of a lag phase for OmpLA, higher amounts of folded protein appeared at earlier time points in 20% PG compared with 20% PE. The inclusion of 20% PG slowed the fast and slow observed rates for OmpA folding by 12-fold and threefold, respectively. Although the appearance of folded OmpA was slowed, the final efficiency was not affected by 20% PG compared with 100% PC. Similarly, the combination of 20% PE + 20% PG (henceforth PE+PG) displayed OMP-specific effects. Although the presence of PG relieved some of the inhibitory effect of PE lipids on OmpA folding, the PE+PG combination had an additive inhibitory effect on OmpLA folding. Our results therefore disagree with previous suggestions that folding of bacterial OMPs is overall favorable in phospholipids with glycerol head groups (25). Unlike the general trend of PE lipids slowing OMP folding, we demonstrated that PG lipids can either kinetically promote or inhibit folding of individual OMPs in the presence of PE. Overall, our results suggest that the periplasmic lipid head groups of PE and PG kinetically modulate the intrinsic OMP folding reaction with the general effect of imposing a kinetic barrier to folding.
Fig. 2.
BamA functionality is modulated by PE and PG head groups. Unfolded OmpA and OmpLA were folded into PC LUVs composed of 20% PG or 20% PE + 20% PG with or without prefolded BamA or OmpX. The client OMP is indicated in each panel, and its total concentration was 4 or 2 μM without or with prefolded OMP, respectively, at a constant lipid:protein ratio of 800:1. Samples were recorded, analyzed and fitted as described in Fig. 1. Kinetic traces of OMP folding into 100% PC LUVs containing no prefolded OMP are replotted from Fig. 1 for comparison. Lines indicate fitted curves; black, no prefolded OMP; green, prefolded BamA; orange, prefolded OmpX. Each experiment was conducted in triplicate, and one representative data set is shown for each condition. Average fit values with SDs are reported in SI Appendix, Tables S1 and S2 and Fig. S4. Longer time points for the slow OmpLA transients are shown in SI Appendix, Fig. S5.
BamA Catalyzes OMP Folding by Lowering the Kinetic Barrier Imposed by PE Head Groups.
To enable cells to overcome the lipid-imposed kinetic barrier to folding at the periplasmic surface of bacterial outer membranes, we hypothesized that the BAM complex must function to reduce the activation barrier to folding. Because the central component, BamA, is evolutionarily conserved, we tested its ability to accelerate the folding of client OMPs in our host-guest membrane system by prefolding BamA into the various lipid compositions. As a negative control, we folded client OMPs into vesicles containing prefolded OmpA or OmpX, neither of which should function to accelerate the folding of a client OMP (SI Appendix, Fig. S2). Fig. 1 shows that prefolded BamA accelerates the folding for all OMPs tested in 20% PE; BamA, therefore, lowers the kinetic barrier imposed by PE head groups. The acceleration is significant: for example, OmpX folded ∼10x faster in the presence of prefolded BamA in 20% PE guest membranes in comparison with the absence of BamA (Fig. 1 and SI Appendix, Fig. S4).
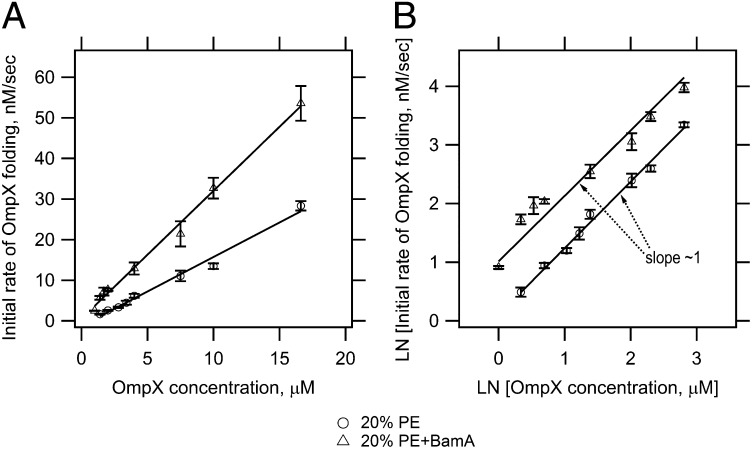
This type of reaction acceleration is suggestive of an enzymatic function for BamA in OMP biogenesis. To test this hypothesis, we sought to determine a Vmax and Km for BamA using OmpX as substrate by carrying out experiments in which we increased the OmpX concentration from 1 to 16.6 μM while keeping BamA and the lipid concentration constant (Fig. 3 and SI Appendix, Fig. S6). In both the presence and absence of prefolded BamA, the initial rates of the OmpX folding reaction followed a first order rate law (Fig. 3). Additionally, the observed OmpX folding rates increased as the OmpX substrate concentration was increased, as expected for Michaelis–Menten kinetics. However, we were unable to observe a saturation point for BamA catalysis, even by going as high as 16.6 μM OmpX. This result may indicate a high Km for BamA with respect to a protein substrate. Higher substrate concentrations could not be tested due to increased self-association of unfolded OmpX and decreased OMP folding rates as a consequence of low lipid to protein ratios of <90:1 (26–28). For these reasons, full enzymatic parameters could not be determined for BamA-catalyzed folding under these conditions. Another possibility is that the interactions between BamA and a client OMP are not saturable.
Fig. 3.
BamA catalyzes the folding reaction of OmpX. (A) Initial rates of OmpX folding as a function of OmpX concentration. (B) Double logarithmic plot of initial OmpX folding rates as a function of OmpX concentration. OmpX was folded into PC LUVs containing 20% PE with or without prefolded BamA at different final OmpX concentrations. The lipid concentration was 1.6 mM, and the final concentration of prefolded BamA was ∼1.6 μM. SI Appendix, Fig. S6 shows representative kinetic traces of OmpX folding with and without prefolded BamA that were used to determine the initial OmpX folding rates. Experiments were conducted in triplicate and samples were recorded, analyzed and fitted as described in Fig. 1.
The Periplasmic Head Group PG Displays Mixed Effects on BamA-Catalyzed Folding of Client OMPs.
We further tested whether BamA activity was affected by lipids with a PG head group and found that its effect is more limited than that of PE. Fig. 2 shows that prefolded BamA accelerated the folding of OmpA in lipids containing either 20% PG or the combination PE+PG compared with OmpA folding in the absence of prefolded BamA. In contrast, prefolded BamA slightly inhibited the folding of OmpLA in lipids with 20% PG. Moreover, the effects of PE and PG on BamA did not display an additive effect on OmpLA folding because the PE+PG combination did not accelerate OmpLA folding kinetics (Fig. 2), despite the fact that 20% PE alone does allow BamA to accelerate OmpLA folding (Fig. 1). Negative controls confirm that these kinetic effects were specific to BamA, because replacement of prefolded BamA with prefolded OmpA or prefolded OmpX did not show these trends.
The Catalytic Activity of BamA Is Located at the Membrane and Depends on the C-Terminal Phenylalanine of the Client OMP β-Signal.
Bacterial OMPs have been shown to contain a conserved sequence of about ten amino acid residues that form a β-strand at the C-terminus, termed the “OMP β-signal” (29). This signature sequence is crucial for the assembly of OMPs in vivo, specifically the highly conserved C-terminal phenylalanine (C-Phe) (30, 31). To investigate the function of the OMP β-signal in more detail, we deleted the C-Phe from OmpA171 and OmpLA and folded the proteins into large unilamellar vesicles (LUVs) containing 0 or 20% PE with or without prefolded BamA. The experiments in 100% PC lipids allowed us to demonstrate that disruption of the OMP β-signal did not perturb the intrinsic folding of OMPs and to distinguish this possibility from BamA-catalyzed folding acceleration of OMPs. Fig. 4 and SI Appendix, Fig. S7 show that deletion of the C-Phe in both client OMPs caused a significant delay in BamA-catalyzed folding in comparison with the wild-type sequences of both proteins. As expected, we observed no statistically significant differences in the absence of prefolded BamA. Taken together, these results demonstrate that defects in OMP assembly upon deletion of the C-Phe are not the consequence of altered intrinsic folding kinetics. Rather, the lack of the C-Phe negatively impacts the ability of BamA to lower the kinetic barrier imposed by PE lipids. Optimal catalytic activity of BamA therefore depends on the C-Phe of the β-signal in a client OMP.
Fig. 4.
The C-terminal phenylalanine of the OMP β-signal is required for folding via BamA. Unfolded OmpA171ΔPhe and OmpLAΔPhe were folded into PC LUVs with or without prefolded BamA. The total concentration of the target OMP was 4 or 2 μM without or with prefolded BamA, respectively, at a constant lipid:protein ratio of 800:1. Data were recorded, analyzed, and fitted as described in Fig. 1. Kinetic traces for the corresponding wild-type OMP are replotted from Fig, 1. Fit values are reported in SI Appendix, Tables S2 and S4 and Fig. S7. Lines indicate fitted curves; black, no prefolded OMP; green, prefolded BamA. Longer time points for the slow transients are shown in SI Appendix, Fig. S8.
BamA consists of five periplasmic N-terminal polypeptide-transport-associated (POTRA) domains and a C-terminal transmembrane β-barrel domain (10, 32). To test whether the BamA POTRA domains are required for OMP folding acceleration, we folded OmpA171 and OmpX into 20% PE LUVs containing a prefolded BamA variant lacking POTRA domains 1–4 (BamAΔP1-4). Fig. 1 shows that BamAΔP1-4 retained the ability to enhance OMP folding kinetics in PE lipids. Nevertheless, the folding kinetics of OmpA171 and OmpX in the presence of prefolded BamAΔP1-4 were not accelerated to the same extent in comparison with wild-type prefolded BamA. We rationalize that this decrease in catalytic activity is partly due to the fact that BamAΔP1-4 exhibits around 50% less folding efficiency than wild-type BamA under the same folding conditions (SI Appendix, Fig. S2). As expected for an enzymatically catalyzed reaction, a decrease in the amount of enzyme would lead to diminishment in observed activity. Indeed, it has been observed that the acceleration in OMP folding kinetics increases with increasing amounts of folded BamA (24). However, because the apparent rate is diminished fivefold by a twofold reduction in enzyme concentration, a reduced specific activity of this truncated form could also contribute to this result. Despite this latter possibility, our experiments support the idea that the BamA mechanistic abilities are located at the membrane and only require the transmembrane β-barrel domain of BamA and POTRA domain 5.
Discussion
Outer Membrane β-Barrel Folding Is Physically Controlled by the Periplasmic Lipid Head Groups and BamA.
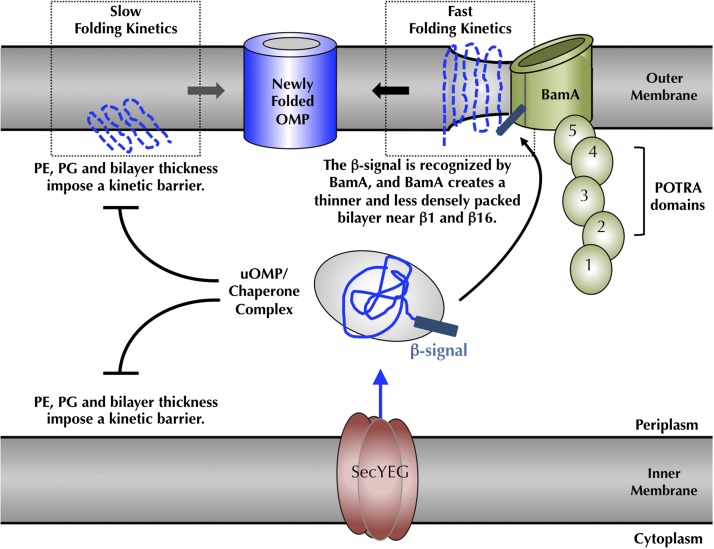
Our experiments demonstrate that only a subset of OMPs are capable of adopting a native fold in membranes of native lipid compositions without the “assistance” of the BAM complex. Moreover, even when some folding occurs, these intrinsic folding reactions into native E. coli membranes are inefficient and too slow to be biologically relevant. Using a lipid “host–native guest” system, we showed that the presence of either PE or PG head groups kinetically retards OMP folding. Biologically, this is initially a paradoxical finding: Why should native head groups slow the folding of proteins that are eventually destined to be incorporated into those membranes? This apparent contradiction can be reconciled by recognizing that the periplasmic membrane surfaces of both the inner and outer membranes have similar lipid compositions and would pose similar kinetic barriers (33). We reason that this physical property of the periplasmic lipid head groups is crucial for cell viability, because it enables negative control over a spontaneously occurring cellular reaction (13–15), namely intrinsic OMP folding. Because many OMPs function as porins, which create holes in membranes (34), it is conceivable that the folding of even one OMP into the bacterial inner membrane could dissipate the proton motive force and kill the cell. Therefore, slow and negligible spontaneous folding into a native lipid membrane would function to kinetically partition OMPs away from inner membranes and into periplasmic chaperone complexes that deliver unfolded OMPs to bacterial outer membranes (Fig. 5). Wu et al. (35) have shown that unfolded OMPs bind to chaperones with half times on the millisecond time range; thus, the kinetic retardation we observe with native head groups is more than sufficient to partition unfolded OMPs away from inner membranes and into chaperone complexes that presumably direct unfolded OMPs to the BAM complex in outer membranes.
Fig. 5.
Cellular consequences of the lipid-induced kinetic folding barrier and model for BamA-catalyzed folding. Folding of bacterial OMPs into the outer membrane of E. coli is kinetically prevented by the bilayer thickness and a densely packed membrane surface, caused by PE and PG head groups (15, 37–39). At the outer membrane, BamA accelerates client OMP folding by the creation of local defects in and thinning of the membrane bilayer (32), which promote faster intrinsic folding kinetics (15). β-signals of client OMPs are recognized by BamA, which localizes clients to the site of the membrane defect. A high Km value of BamA for client OMPs together with favorable OMP folding free energies (49) drive the client OMPs to dissociate from BamA and move away into the membrane. This mechanistic view indicates how bacterial and mitochondrial OMPs can be recognized and folded by the eukaryotic BamA homologs (44, 45). POTRA, polypeptide-transport-associated.
Although this physical control mechanism preserves the integrity of the inner membrane, kinetic retardation creates a challenge for the cell at the periplasmic membrane surface of the bacterial outer membrane. We propose that BamA, the central component of the outer membrane BAM complex, functions to overcome this biological obstacle by reducing the lipid-imposed activation barrier to OMP folding at the site of the correct biological membrane. It is noteworthy that BamA is itself a transmembrane β-barrel, and this physical control mechanism also prevents its incorporation into bacterial inner membranes. The fact that this catalytic activity does not require the remaining BAM subunits is consistent with the observation that BamA is the only evolutionarily conserved subunit of the BAM complex (36).
Mechanistic Interpretations of BamA Function.
An intriguing question concerns how BamA mechanistically carries out its function. We and others previously determined that a reduction of bilayer thickness as well as higher curvature in membranes both accelerated the intrinsic folding kinetics of bacterial OMPs (15). These findings lead to the idea that bilayer defects, distortion of the membrane surface, and thinning of the bilayer decrease the kinetic barrier to intrinsic OMP folding. It is also notable that PC lipid membranes, which support fast and efficient folding in vitro, are less densely packed and have different lateral pressures than membranes containing PE and/or PG lipids (37–39). To accelerate folding in vivo, creating perturbations in the membrane surface and thinning the bilayer should therefore be physical mechanisms used in catalysis. Remarkably, the recently solved crystal structure of BamA showed exactly that result: microsecond simulations of membranes composed of PE lipids showed bilayers were ∼16 Å thinner and less densely packed around BamA β-strands 1 and 16 in contrast to the opposite side of the BamA transmembrane β-barrel (32). The creation of this type of local membrane defect, a thinner membrane with decreased lipid packing and lower lateral pressure, would facilitate fast intrinsic OMP folding kinetics without the requirement to provide any conformational folding instructions, consistent with the Anfinsen hypothesis and the many in vitro folding studies demonstrating that the protein sequence of an OMP itself contains all of the information to specify the fold. In accordance with the idea that BamA should function to create a local membrane defect, we demonstrated that the catalytic activity of BamA is located at the membrane. The fact that BamA can so profoundly affect the membrane structure by stabilizing an excited state “membrane-defect” conformation also highlights the idea that the membrane is a second substrate of the BamA enzymatic activity. Because this membrane-defect stabilization is so pronounced, a high Km as we observe toward the other substrate, an OMP folding client, may be all that is required for efficient folding catalysis.
This mechanistic hypothesis leaves the important question of how OMPs are localized to the membrane defect created by BamA. Aside from chaperones, which are certainly thought to play a role in delivering client OMPs to outer membranes (40), previous in vitro studies demonstrated that BamA interacts with the client OMP β-signal, specifically the C-Phe residue, and this recognition caused alterations in the BamA transmembrane β-barrel domain (41). Upon inspection of the E. coli BamA structural model, we noticed that β-strand 16 of BamA is a mirror image of the E. coli OMP β-signal (SI Appendix, Fig. S9) with the conserved C-Phe residue of a client OMP corresponding to Phe802 in strand 16 of the E. coli BamA sequence. Intriguingly, the molecular dynamics simulations suggested that this same region of the protein, β-strands 1 and 16, are not only the site of a BamA-induced membrane defect, but also form a lateral gate to open the transmembrane β-barrel domain of BamA (32). It is tempting to speculate that BamA Phe802 may play a role in BamA substrate recognition of a client OMP β-signal, especially if the client β-signal C-Phe replaces a BamA Phe802 ground state intraprotein interaction during catalysis.
Although the molecular details of catalysis remain to be determined, our results nevertheless show that BamA-catalyzed OMP folding is slowed upon disruption of the client OMP β-signal in a manner that cannot be dismissed as a defect in the intrinsic folding abilities of OMPs. In excellent agreement with our findings in vitro, the lack of β-signal C-Phe resulted in the accumulation of the E. coli OMP PhoE in the periplasm of living bacteria, and reducing the PhoE expression levels restored its assembly into the outer membrane (30, 31). It has also been observed that β-signal peptides, when added in trans, inhibited binding of a client OMP to Sam50/Tob55, the mitochondrial homolog of BamA (42). Adding scrambled peptide sequences of identical composition had no effect. In a separate study, deletion of the β-signal C-Phe prevented formation of the native Tom40 β-barrel by Sam50/Tob55 (43).
In conclusion, our suggested catalytic function for BamA-mediated OMP folding in the cell (Fig. 5) is founded upon physical folding principles. Our data indicate that both the membrane and the client OMP are substrates acted upon by BamA in the catalytic reaction. Accelerating folding by stabilizing excited states of both substrates implies that bacterial OMPs can be recognized and folded by the homologous eukaryotic assembly machineries and vice versa, in agreement with the current literature (44, 45).
Materials and Methods
Mutagenesis of Outer Membrane Proteins.
Plasmids encoding mature wild-type OMPs were constructed previously (15, 23). Variants lacking the C-terminal phenylalanine for the truncated OmpA171 (OmpA171ΔPhe) and OmpLA (OmpLAΔPhe) were PCR amplified from full-length genes and subsequently cloned into the popinE expression vector (Oxford Protein Production Facility) (46). The BamA mutant lacking POTRA domains 1–4 (BamAΔP1-4) was generated by ligation-independent cloning directly into a popinE plasmid. The methods are described in greater details in SI Appendix, SI Text.
Production of Denatured Outer Membrane Proteins.
Expression, isolation, and purification of denatured OMPs were performed according to refs. 15 and 23.
Growth and Extraction of E. coli Outer Membrane Lipids.
The E. coli mutant strain WBB06 was obtained from the Yale E. coli genetic stick center. Extraction of outer membrane lipids was performed according to ref. 47 and is described in greater detail in the SI Appendix, SI Text.
OMP Folding into SUVs Made from Extracted E. coli Outer Membrane Lipids.
Preparation of small unilamellar vesicles (SUVs) is described in the SI Appendix, SI Text. Folding was initiated by rapid dilution of denatured OMPs into folding buffer containing SUVs, while stirring, to a final concentration of 4 μM protein and 1.2 mM lipids in 1 M urea, 20 mM borate pH 8. Folding temperature was controlled using a rotating incubator (Hybridization Incubator Model 400, Robbins Scientific) and the folding samples were incubated for 20 h at 47 °C and 6 rpm.
OMP Folding into Synthetic LUVs and Analysis of Folding Kinetics.
Preparation of LUVs is described in the SI Appendix, SI Text. Folding was initiated by rapid dilution of denatured OMPs into folding buffer to a final concentration of 1–16.6 μM uOMP, 3.2 mM lipids, 1 M urea, 2 mM EDTA, 20 mM borate pH 10 with constant stirring at 35 °C (10 Sample Thermoelectric Temperature Incubator, Model T-10, Aviv Biomedical). Time points were taken by removing small aliquots and quenching the reaction with 5× SDS gel-loading buffer (LB) to a final concentration of 1× SDS gel LB. Additional samples were taken in the first minute and at the end of the experiment termed “pre” and “postsample,” respectively. These samples were boiled for 5 min at 95 °C.
Folding of denatured OMPs into host PC LUVs containing 20% PE, PG, or 20% PE + 20% PG with prefolded OMPs, BamA/BamAΔP1-4/OmpA/OmpX, was conducted as follows: BamA, BamAΔP1-4, OmpA, or OmpX were folded as described above to a final folding mixture of 4 μM OMP and 3.2 mM lipids. For all lipid compositions, the folding solution was incubated with stirring at 35 °C for 2 h for BamA and OmpX and overnight for BamAΔP1-4 and OmpA. Folding kinetics were then measured for a particular client OMP by rapidly adding that OMP to the folding mixture containing the prefolded OMP. Final concentrations were 1–16.6 μM OMP client, 2 μM total prefolded OMP, 1.6 mM lipid, 1 M urea, 2 mM EDTA, 20 mM borate pH 10. Prefolded BamA, OmpX, BamAΔP1-4, and OmpA exhibited a folding efficiency of 79 ± 7%, 62 ± 4%, 43 ± 1%, and 63 ± 2%, which accounts for ∼1.6 μM, ∼1.2 μM, ∼0.9 μM, and ∼1.3 μM folded, respectively. Samples were taken at indicated time points and immediately quenched with SDS gel-loading buffer as described above.
Quenched samples were stored for no longer than 5 h at room temperature (RT) and subsequently run on 10 or 12% precast gels (Mini-PROTEAN TGX, Bio-Rad) at a constant voltage of 150 mV for 55 min at RT. Gels were stained with Coomassie Blue, scanned, and analyzed using ImageJ (NIH). Fraction folded was calculated as the ratio of the folded band intensity divided by the “Pre”boiled band intensity. Kinetic traces were fitted using IgorPro (WaveMetrics) according to refs. 15 and 48 and are described in more details in the SI Appendix, SI Text.
Supplementary Material
Acknowledgments
We thank Sarah Kempka and Dr. Patrick Fleming for critical discussion of the experiments and the manuscript and the late Professor Christian R. H. Raetz (Duke University) for advice and protocols on purification of E. coli lipids. This work was supported by National Science Foundation Grant MCB0919868 and National Institutes of Health Grants R01 GM079440 and T32 GM008403.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322473111/-/DCSupplemental.
References
- 1.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666(1-2):250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13(4):404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33(11):1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajaj H, et al. Antibiotic uptake through membrane channels: Role of Providencia stuartii OmpPst1 porin in carbapenem resistance. Biochemistry. 2012;51(51):10244–10249. doi: 10.1021/bi301398j. [DOI] [PubMed] [Google Scholar]
- 5.Manczak M, Reddy PH. Abnormal interaction of VDAC1 with amyloid beta and phosphorylated tau causes mitochondrial dysfunction in Alzheimer’s disease. Hum Mol Genet. 2012;21(23):5131–5146. doi: 10.1093/hmg/dds360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki K, et al. VDAC: Old protein with new roles in diabetes. Am J Physiol Cell Physiol. 2012;303(10):C1055–C1060. doi: 10.1152/ajpcell.00087.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender A, et al. TOM40 mediates mitochondrial dysfunction induced by α-synuclein accumulation in Parkinson’s disease. PLoS ONE. 2013;8(4):e62277. doi: 10.1371/journal.pone.0062277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299(5604):262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 9.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121(2):235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317(5840):961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 11.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: Conformational flexibility and substrate recognition. Structure. 2008;16(12):1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knowles TJ, et al. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68(5):1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 13.Kleinschmidt JH, den Blaauwen T, Driessen AJ, Tamm LK. Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry. 1999;38(16):5006–5016. doi: 10.1021/bi982465w. [DOI] [PubMed] [Google Scholar]
- 14.Huysmans GH, Radford SE, Brockwell DJ, Baldwin SA. The N-terminal helix is a post-assembly clamp in the bacterial outer membrane protein PagP. J Mol Biol. 2007;373(3):529–540. doi: 10.1016/j.jmb.2007.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283(39):26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley AM, Fleming KG. The process of folding proteins into membranes: Challenges and progress. Arch Biochem Biophys. 2008;469(1):46–66. doi: 10.1016/j.abb.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 17.Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc Natl Acad Sci USA. 2011;108(25):10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rigel NW, Ricci DP, Silhavy TJ. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc Natl Acad Sci USA. 2013;110(13):5151–5156. doi: 10.1073/pnas.1302662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagan CL, Kahne D. The reconstituted Escherichia coli Bam complex catalyzes multiple rounds of β-barrel assembly. Biochemistry. 2011;50(35):7444–7446. doi: 10.1021/bi2010784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeth K, Thein M. Porins in prokaryotes and eukaryotes: Common themes and variations. Biochem J. 2010;431(1):13–22. doi: 10.1042/BJ20100371. [DOI] [PubMed] [Google Scholar]
- 21.White DA, Lennarz WJ, Schnaitman CA. Distribution of lipids in the wall and cytoplasmic membrane subfractions of the cell envelope of Escherichia coli. J Bacteriol. 1972;109(2):686–690. doi: 10.1128/jb.109.2.686-690.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lugtenberg EJ, Peters R. Distribution of lipids in cytoplasmic and outer membranes of Escherichia coli K12. Biochim Biophys Acta. 1976;441(1):38–47. doi: 10.1016/0005-2760(76)90279-4. [DOI] [PubMed] [Google Scholar]
- 23.Danoff EJ, Fleming KG. The soluble, periplasmic domain of OmpA folds as an independent unit and displays chaperone activity by reducing the self-association propensity of the unfolded OmpA transmembrane β-barrel. Biophys Chem. 2011;159(1):194–204. doi: 10.1016/j.bpc.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel GJ, Kleinschmidt JH. The lipid bilayer-inserted membrane protein BamA of Escherichia coli facilitates insertion and folding of outer membrane protein A from its complex with Skp. Biochemistry. 2013;52(23):3974–3986. doi: 10.1021/bi400103t. [DOI] [PubMed] [Google Scholar]
- 25.Patel GJ, Behrens-Kneip S, Holst O, Kleinschmidt JH. The periplasmic chaperone Skp facilitates targeting, insertion, and folding of OmpA into lipid membranes with a negative membrane surface potential. Biochemistry. 2009;48(43):10235–10245. doi: 10.1021/bi901403c. [DOI] [PubMed] [Google Scholar]
- 26.Kleinschmidt JH, Tamm LK. Secondary and tertiary structure formation of the beta-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J Mol Biol. 2002;324(2):319–330. doi: 10.1016/s0022-2836(02)01071-9. [DOI] [PubMed] [Google Scholar]
- 27.Ebie Tan A, Burgess NK, DeAndrade DS, Marold JD, Fleming KG. Self-association of unfolded outer membrane proteins. Macromol Biosci. 2010;10(7):763–767. doi: 10.1002/mabi.200900479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huysmans GH, Radford SE, Baldwin SA, Brockwell DJ. Malleability of the folding mechanism of the outer membrane protein PagP: Parallel pathways and the effect of membrane elasticity. J Mol Biol. 2012;416(3):453–464. doi: 10.1016/j.jmb.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tommassen J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology. 2010;156(Pt 9):2587–2596. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- 30.de Cock H, Struyvé M, Kleerebezem M, van der Krift T, Tommassen J. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein PhoE of Escherichia coli K-12. J Mol Biol. 1997;269(4):473–478. doi: 10.1006/jmbi.1997.1069. [DOI] [PubMed] [Google Scholar]
- 31.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218(1):141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 32.Noinaj N, et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature. 2013;501(7467):385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harwood JL, Russell NJ. Lipids in Plants and Microbes. London: George Allen & Unwin; 1984. [Google Scholar]
- 34.Cowan SW, et al. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 35.Wu S, et al. Interaction between bacterial outer membrane proteins and periplasmic quality control factors: A kinetic partitioning mechanism. Biochem J. 2011;438(3):505–511. doi: 10.1042/BJ20110264. [DOI] [PubMed] [Google Scholar]
- 36.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164(1):19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boggs JM. Lipid intermolecular hydrogen bonding: Influence on structural organization and membrane function. Biochim Biophys Acta. 1987;906(3):353–404. doi: 10.1016/0304-4157(87)90017-7. [DOI] [PubMed] [Google Scholar]
- 38.Zhang YP, Lewis RN, McElhaney RN. Calorimetric and spectroscopic studies of the thermotropic phase behavior of the n-saturated 1,2-diacylphosphatidylglycerols. Biophys J. 1997;72(2 Pt 1):779–793. doi: 10.1016/s0006-3495(97)78712-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murzyn K, Róg T, Pasenkiewicz-Gierula M. Phosphatidylethanolamine-phosphatidylglycerol bilayer as a model of the inner bacterial membrane. Biophys J. 2005;88(2):1091–1103. doi: 10.1529/biophysj.104.048835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4(1):57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 41.Robert V, et al. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4(11):e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kutik S, et al. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell. 2008;132(6):1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 43.Qiu J, et al. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell. 2013;154(3):596–608. doi: 10.1016/j.cell.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 44.Walther DM, Papic D, Bos MP, Tommassen J, Rapaport D. Signals in bacterial beta-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc Natl Acad Sci USA. 2009;106(8):2531–2536. doi: 10.1073/pnas.0807830106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walther DM, Bos MP, Rapaport D, Tommassen J. The mitochondrial porin, VDAC, has retained the ability to be assembled in the bacterial outer membrane. Mol Biol Evol. 2010;27(4):887–895. doi: 10.1093/molbev/msp294. [DOI] [PubMed] [Google Scholar]
- 46.Berrow NS, et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35(6):e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raetz CR, et al. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res. 2006;47(5):1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- 48.Nölting B. Protein Folding Kinetics, Biophysical Methods. 2nd Ed. Berlin: Springer; 2006. [Google Scholar]
- 49.Moon CP, Zaccai NR, Fleming PJ, Gessmann D, Fleming KG. Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proc Natl Acad Sci USA. 2013;110(11):4285–4290. doi: 10.1073/pnas.1212527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.