Significance
During translation, ribosomes decode mRNAs in a sequential fashion. In this paper, we report the discovery of more than 80 translational bypassing elements (byps) 27–55 nt long in mitochondrial protein-coding regions of the yeast Magnusiomyces capitatus. We demonstrate experimentally that byps are retained in mRNA but not translated into protein. Byps somewhat resemble the single bypass element in bacteriophage T4 but also display unique features. We further discovered byp-like sequences in other yeast species, indicating that these inserts are mobile genetic elements. In contrast to byps, byp-like sequences are not bypassed during translation. When inserted in variable protein regions, they have the potential to drive the evolutionary diversification of protein structure and function.
Keywords: ribosome hopping, mitochondrial genome, proteome analysis, heterologous expression
Abstract
Programmed translational bypassing is a process whereby ribosomes “ignore” a substantial interval of mRNA sequence. Although discovered 25 y ago, the only experimentally confirmed example of this puzzling phenomenon is expression of the bacteriophage T4 gene 60. Bypassing requires translational blockage at a “takeoff codon” immediately upstream of a stop codon followed by a hairpin, which causes peptidyl-tRNA dissociation and reassociation with a matching “landing triplet” 50 nt downstream, where translation resumes. Here, we report 81 translational bypassing elements (byps) in mitochondria of the yeast Magnusiomyces capitatus and demonstrate in three cases, by transcript analysis and proteomics, that byps are retained in mitochondrial mRNAs but not translated. Although mitochondrial byps resemble the bypass sequence in the T4 gene 60, they utilize unused codons instead of stops for translational blockage and have relaxed matching rules for takeoff/landing sites. We detected byp-like sequences also in mtDNAs of several Saccharomycetales, indicating that byps are mobile genetic elements. These byp-like sequences lack bypassing activity and are tolerated when inserted in-frame in variable protein regions. We hypothesize that byp-like elements have the potential to contribute to evolutionary diversification of proteins by adding new domains that allow exploration of new structures and functions.
The traditional view of translation is that mRNA is read sequentially, one codon at a time. However, low-level nonprogrammed translational bypassing (i.e., the occasional skipping of a few nucleotides) can be triggered by various factors, including tRNA paucity, unusual codons, and homo-polymer sequence tracts (1). In addition, programmed translational bypassing of 50 nt has been demonstrated for the gene 60 transcript of bacteriophage T4 (2–4). In vitro mutagenesis experiments showed that efficient translational “jumping” or “hopping” in T4 requires matching takeoff and landing codons (most effective is the wild-type GGA), a stop codon, and both a hairpin RNA secondary structure directly downstream of the takeoff site, and a Shine–Dalgarno (SD) sequence a few nucleotides upstream of the landing codon. Finally, a particular amino acid sequence in the nascent peptide encoded upstream of the takeoff site confers highest jumping efficiency. Additional cases of programmed bypassing have been postulated but currently lack supporting evidence (e.g., ref. 5), making the T4 gene 60 expression the only confirmed instance.
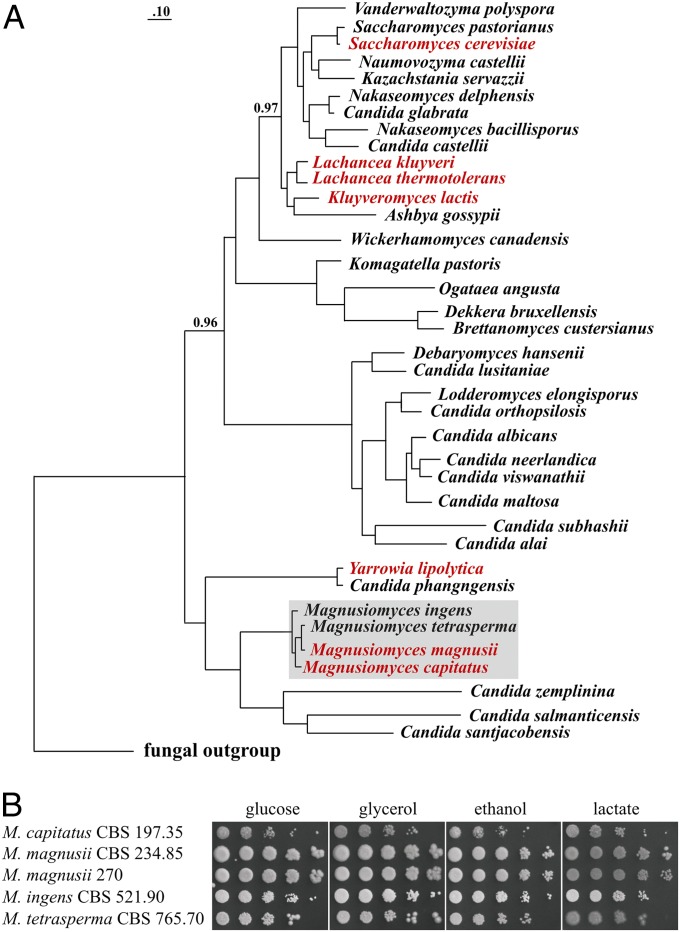
Here, we report the massive occurrence of translational bypassing elements in mitochondria of the opportunistic human pathogen Magnusiomyces (also known as Blastoschizomyces or Geotrichum) capitatus (6), which belongs to a deeply branching lineage of Saccharomycetales (Fig. 1A). Our findings suggest that translational bypassing might be more widespread than previously thought.
Fig. 1.
Magnusiomyces growth on nonfermentable carbon sources and phylogeny. (A) Phylogeny of yeast species based on mitochondrial protein sequences. The tree (PhyloBayes and the CAT/GTR model) was built using 13 mtDNA-encoded proteins. All divergence points are supported by posterior probability values of 1.0, except where indicated. The fungal outgroup consists of Rhizopus oryzae, Aspergillus niger, Podospora anserina, Fusarium oxysporum, Cantharellus cibarius, and Ustilago maydis. Species containing byps or byp-like insertion elements (ins) analyzed in this study are colored red. Note that GC clusters in mtDNAs of many more yeast species may be byp-related. Species for which mtDNAs were sequenced in the context of this work are labeled by gray shading. (B) Yeast cells were cultivated on synthetic medium with glucose, glycerol, ethanol, or lactate as the sole carbon source. See Materials and Methods for details.
Results
Protein-Coding Genes Interrupted by Dozens of Insertions.
We sequenced mitochondrial DNAs (mtDNAs) from five Magnusiomyces strains (M. capitatus, M. ingens, M. magnusii (2 isolates), and M. tetrasperma (Table S1). The identified genes specify a standard set of highly conserved proteins involved in oxidative phosphorylation (atp6, -8, and -9; cob; cox1, -2, and -3; and nad1, -2, -3, -4, -4L, -5, and -6) and translation (rps3), two ribosomal RNAs (rnl and rns), and 25 transfer RNAs (trnA-Y). During comparative genome analysis, we identified 57 short insertions (30–55 bp) in protein-coding regions of otherwise typical genes in M. capitatus mtDNA (Fig. 2). Using covariance models for computational searches, we detected 24 additional inserts (27–54 bp) in intron ORFs of this mtDNA (Table 1 and Table S2). The most “infested” gene is nad5 with 12 inserts, increasing the overall gene size by more than 20%. In total, these inserts make up 7.2% of the M. capitatus mitochondrial genome sequence. The four other magnusiomycete mtDNAs lack these particular insertions, which therefore allowed their precise positioning by sequence alignment. All but 4 of the 57 ORF inserts introduce frameshifts and/or internal stop codons (Table S2). If translated, the inserts would abolish oxidative phosphorylation in this obligate aerobic yeast; yet, M. capitatus grows well on nonfermentable carbon sources (Fig. 1B).
Fig. 2.
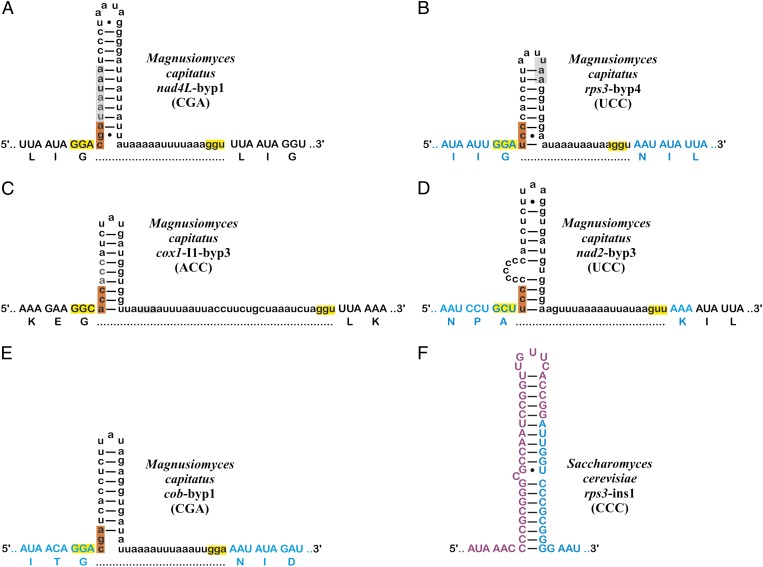
Representative byps and byp-like elements. Upper- and lowercase nucleotides, putatively translated and untranslated sequence, respectively. Dotted lines, continuity of protein sequence. Blue and purple, translation demonstrated by proteomics data. Highlighting in yellow, takeoff and landing sites; orange, unused codon following takeoff site; gray, in-frame stop codons. (A–E) Byps in M. capitatus mtDNA. (A–C) Byps representing the three groups, CGA, UCC, and ACC, (A and B) in standard protein-coding genes and (C) in an intron ORF. (B, D, and E) Byps for which translational bypassing has been demonstrated experimentally. (F) Byp-like element in mitochondrion-encoded rps3 of S. cerevisiae that, in contrast to A–E, is translated. The two tryptic peptides encoded by the purple and blue sequence, respectively, are MLNNNNMNPAGANPVVHR and IGPAGNINNK (our data and data at http://gpmdb.thegpm.org/seq.html) (43).
Table 1.
Byps in mitochondrial genes and intron ORFs of Magnusiomyces capitatus
| Gene/intron ORF | No. of byps | Byp group* | |||
| CGA | UCC | ACC | Other | ||
| atp6 | 1 | — | 1 | — | — |
| cob | 1 | 1 | — | — | — |
| cox1 | 2 | 1 | — | — | 1 |
| cox3 | 3 | 2 | 1 | — | — |
| nad1 | 3 | — | 3 | — | — |
| nad2 | 11 | 4 | 6 | — | 1 |
| nad3 | 2 | 1 | 1 | — | — |
| nad4 | 7 | 4 | 2 | — | 1 |
| nad4L | 3 | 3 | — | — | — |
| nad5 | 12 | 3 | 9 | — | — |
| nad6 | 5 | 3 | 1 | — | 1 |
| rps3 | 7 | 4 | 3 | — | — |
| cob-I1 | 1 | — | 1 | — | — |
| cob-I2 | 5 | — | 4 | — | 1 |
| cox1-I1 | 3 | — | 2 | 1 | — |
| cox1-I2 | 5 | — | 5 | — | — |
| cox1-I3 | 4 | — | 3 | 1 | — |
| cox1-I4 | 5 | — | 3 | 2 | — |
| cox1-I6 | 1 | — | — | 1 | — |
Groups are defined according to the unused codon. Other, unclassified.
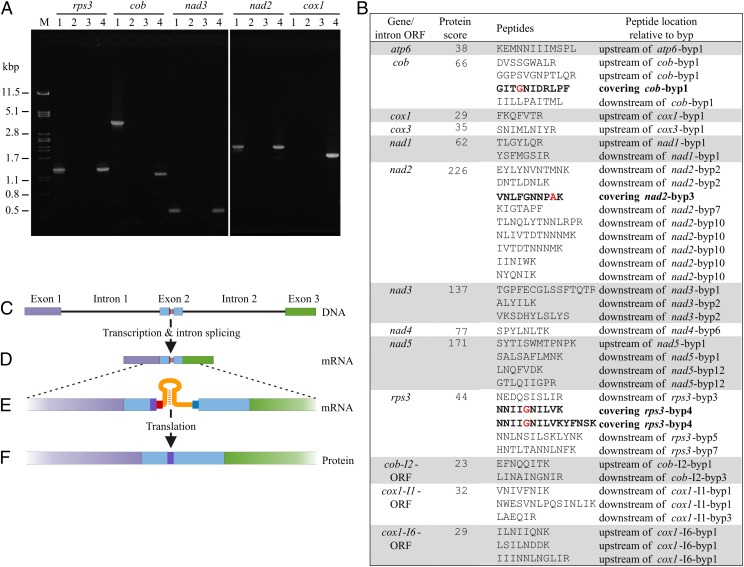
To produce functional proteins, the inserts must be either removed from the RNA or ignored during translation by a programmed translational bypass. These two possibilities can be readily distinguished by comparing mRNA and protein sequences. Analysis of reverse transcriptase PCR (RT-PCR) products of cob, cox1, nad2, nad3, and rps3 transcripts shows that the inserts are included in mRNAs (Fig. 3A) whereas mass spectrometry analysis of mitochondrial proteins demonstrates that amino acid sequences corresponding to the inserts are absent from proteins. Specifically, we found peptides for 10 insert-containing genes and three insert-containing endonuclease-encoding intron ORFs (Fig. 3B). Many of the peptides originate from regions preceded by a stop codon- and/or frameshift-bearing insert, and none of the observed peptides corresponds to a translated insertion sequence. Therefore, inserts must have been circumvented, with potential mechanisms including nonsense suppression, single-nucleotide frameshifts, or translational bypassing. Notably, four distinct peptides bridge three insertion points (Fig. 3B and Fig. S1 A–C), providing compelling evidence for effective and precise translational jumping. We posit that all insertion elements in M. capitatus, which we term “byps,” are translationally bypassed. This conjecture is based on the structural similarities shared by all byps and their similarity with the phage T4 hop element (see Mechanism of Translational Bypassing).
Fig. 3.
Transcript processing and translation of byp-containing genes in M. capitatus. (A) Analysis of mitochondrial transcripts indicates that mRNAs contain byps. PCR reactions were carried out using primers specific for M. capitatus genes rps3, cob, nad3, nad2, and cox1 and the following templates: lane 1, genomic DNA; lane 2, RNA extracted from purified mitochondria; lane 3, cDNA synthesized from RNase A-treated mitochondrial RNA; and lane 4, cDNA synthesized from mitochondrial RNA (for details see Materials and Methods). Amplification products were separated by electrophoresis. As molecular weight marker (M) served λ DNA digested with PstI endonuclease. The sizes of PCR and RT-PCR products are identical for rps3, nad3, and nad2 but differ for cob and cox1, which contain introns removed from the mRNA by splicing. The retention of byps and elimination of introns from cob and cox1 cDNAs was confirmed by DNA sequencing. The cox1 gene fragment was not amplified from genomic DNA due to its large size (∼10.1 kbp). (B) List of peptides corresponding to byp-containing genes and intron ORFs. Peptides spanning byps are indicated in bold, and amino acids in red correspond to the takeoff codon. Mass spectrometry profiles of peptides spanning byps are shown in Fig. S1. (C–F) Diagram of the translational bypassing process in M. capitatus mitochondria. Purple-, sky blue-, and mint green-filled boxes indicate individual exons at the gene and transcript level and the corresponding portions in the protein. (C) Gene structure of the cob gene. The multicolored narrow box within exon 2 represents the byp element. (D) Mature cob transcript after intron removal by RNA splicing. (E) Messenger RNA magnified in size. Dark purple rectangle, takeoff codon; red rectangle, unused codon; orange hairpin, folded RNA sequence; bright blue rectangle, landing codon. (F) Protein sequence of apocytochrome b that contains an amino acid corresponding to the takeoff codon whereas the other components of byps are not translated.
Byps share several structural features, which apparently instruct the mitochondrial translation machinery to bypass a precise mRNA interval (Fig. 3 C–F). (i) The 5′ portion of these elements consists of a G+C-rich stretch able to fold into a hairpin structure with a variable-length A+U-rich loop. (ii) In nearly all instances, the codon preceding byps and the triplet at the 3′ end of byps (putative takeoff and landing sites, respectively) are identical or differ only in the third position (e.g., GGA and GGU in Table S2). (iii) The three nucleotides at the 5′ end of byps, most frequently CGA or UCC, represent codons not used in M. capitatus coding regions (for codon frequencies, see Tables S3–S8). Avoidance of UCC codons is a common feature of mitochondria, with a general bias against codons ending in C or G (7). CGA, in contrast, is probably not decoded in M. capitatus mitochondria, because the only mitochondrion-encoded tRNAArg in this system has an ACG anticodon, which will not decode CGA (unless the A in the anticodon’s wobble position is converted to inosine posttranscriptionally).
Most byps fall into two groups referred to as CGA and UCC (Fig. 2 A and B and Table 1), based on sequence similarity and a shared unused codon following the takeoff site. Using sequence comparison and covariance models (8) (Materials and Methods), we found 24 additional byps in seven out of eight intron ORFs (Table 1). Interestingly, none of the intron ORF-included byps belongs to the CGA group, and five are members of the intron-only ACC group. No significant match was detected in the nuclear protein-coding genes of M. capitatus.
Mechanism of Translational Bypassing.
In vitro mutagenesis experiments in T4 showed that translational jumping or hopping requires matching takeoff and landing codons (most effective is the wild-type GGA), a stop codon, and a hairpin RNA secondary structure both directly downstream of the takeoff site, and a Shine–Dalgarno (SD) sequence a few nucleotides upstream of the landing codon. Further, a particular amino acid sequence in the nascent peptide encoded upstream of the takeoff site confers highest jumping efficiency across the single bypassing element (“hop”) in T4 gene 60 (2–4, 9). We posit that translational bypassing in M. capitatus mitochondrial mRNAs follows a similar mechanism as T4 gene 60 expression. Mito-ribosomes will stall at a takeoff codon immediately upstream of an untranslatable codon; then the peptidyl–tRNA complex will dissociate from mRNA and reassociate further downstream at the next available cognate (landing) codon. The byp landing site is rarely identical to the takeoff but typically belongs to the same codon family according to the extended superwobble rules of tRNA recognition (10). Exceptions are cox1-I3-byp3 and nad2-byp3, where takeoff and landing sites (GCU and GUU, respectively) belong to different codon families (Fig. 2D and Table S2). Mass-spectrometry data for Nad2 show that the corresponding peptide has an alanine at the position of the takeoff codon, followed by a lysine that corresponds to the predicted codon immediately downstream of the landing site (Fig. 3B and Fig. S1A). This result strongly suggests that the takeoff GCU is decoded by tRNAAla(ugc) and translated accordingly as alanine whereas the landing codon (GUU, corresponding to valine) is ignored during translation. We infer that the anticodon of tRNAAla(ugc) associates well enough with the GUU landing site by forming a G-U (instead of a G-C) pair in the second position of the codon–anticodon interaction.
There are additional differences between byps in M. capitatus mitochondria and the hop element in the T4 gene 60. In T4, the codon immediately downstream of the takeoff site is a stop, and the hairpin has a UUCG tetraloop (tightly packed at the 3D level) rather than a variable A+U-rich region. Further, the T4 element has an SD motif upstream of the landing site whereas byps do not have recognizable sequence conservation in that region [note that most mitochondria lack bacterial SD motifs except jakobid flagellates (11) and that yeast mitochondria use long untranslated leader sequences for gene-specific translation initiation instead (12)]. Moreover, the T4 element requires for effective bypassing a sequence motif (MKKY) in the nascent peptide at positions −33 to −30 (4). In M. capitatus, however, upstream sequences may be as short as 10 aa (e.g., rps3-byp1) and do not share significant sequence similarity among each other (Fig. S2), nor do they have numerous positively charged amino acids known to delay translation (13), which may increase translational bypassing. Finally, translational bypassing in M. capitatus must be by far more efficient than in the T4 gene 60 transcript where it occurs successfully only half the time (14). With suboptimal performance, the multiple byps per gene (e.g., 12 in nad5) would lead to insufficient amounts of functional protein plus presumably intolerable quantities of abortive polypeptides.
Because of the similarities between translational jumping in the T4 gene 60 case and the mitochondrial system, we tested whether Escherichia coli ribosomes are able to bypass M. capitatus byps, including variants carrying T4 gene-60 features (1), such as a stop codon immediately downstream of the takeoff codon, a UUCG tetraloop, an SD motif, and the nascent peptide sequence of the host mitochondrial gene. However, the number of E. coli transformants was extremely low, and the few clones obtained had the byp hairpin partly or fully deleted (Table S9). This finding indicates that the expression in E. coli is toxic, presumably due to extensive ribosome stalling. Apparently, bypassing elements have coevolved with their particular translation machinery and are not interchangeable between different systems.
Distribution of Byp-Like Elements Suggests Mobility.
GC clusters are frequent in intergenic mtDNA regions of a broad range of eukaryotes, predominantly fungi (15–18). GC clusters in several Saccharomycetales mtDNAs are reminiscent of byps in that they have flanking ≥3-nt-long repeats and an internal hairpin (Figs. S3–S6). These byp-like elements are mostly inserted in intron ORFs: for example, in the cox3 intron 1 ORF of one out of two analyzed M. magnusii strains (Fig. S3), the cox1 intron ORF of three out of six examined Lachancea species (Fig. S4), and in the cob intron 2 ORF of one among three species of the Yarrowia clade (Fig. S5) (19, 20). The only “regular” mitochondrial gene harboring byp-like elements in Saccharomycetales is rps3, a gene previously designated var1 due to its variable size and poor sequence conservation (Fig. S6). These byp-like elements in rps3 do not disrupt the reading frame in conceptual translation and have thermodynamically more stable hairpins than M. capitatus byps. Mass spectrometry data from S. cerevisiae show that, in contrast to byps, rps3-ins1 is fully translated (Fig. 2F).
Byps and byp-like elements are likely mobile, spreading within the same mtDNA and across species boundaries. Intragenome proliferation is exemplified by two identical copies of an insertion element, one in rps3 and the other in an intergenic region of S. cerevisiae isolate FY1679 (Fig. S6 A and B), and two almost identical copies in the cob intron 2 ORF of Yarrowia (Fig. S5 A and B) (19). Horizontal transfer is most convincingly illustrated by the presence of highly similar byp-like elements in Kluyveromyces lactis and Saccharomyces cerevisiae mtDNA (Fig. S6C).
In phage T4, the hop element, together with a homing endonuclease ORF, constitutes a mobile DNA cassette that is able to invade related bacteriophages. Bonocora et al. (21) show that the hop element protects the ORF against self-cleavage by its own gene product, providing a rationale for horizontal (co)transfer of hop. In contrast, mitochondrial byps and byp-like elements probably transpose by themselves, because they do not carry along additional sequence, and the flanking short direct repeats may have arisen by target-site duplication typical for transposons (Figs. S3A, S4A and S5 A and B). However, the mobility of byp and byp-like elements may also be enabled or at least facilitated in trans by group I intron-ORF encoded LAGLIDADG_1 endonucleases, which make up the overwhelming majority of intron ORFs in Saccharomycetales.
Although byp-like elements are distributed broadly across Saccharomycetales mtDNAs, translational bypassing seems confined to M. capitatus. Essential mitochondrial protein-coding genes in other yeasts appear to tolerate elements only if they are inserted in highly variable protein regions, do not shift the reading frame, or do not contain stops or unused codons. Therefore, byp-like elements have the potential to contribute to evolutionary diversification of proteins by adding new domains with accelerated sequence change, as, for instance, byp-like elements in rps3 (Fig. S6C) and intronic ORFs in M. magnusii or Lachancea sp. (Figs. S3A and S4A).
Discussion
We discovered a plethora of bypassing elements in M. capitatus mitochondria, suggesting that programmed translational jumping is more frequent than previously thought. Given the apparent mobility of byps and byp-like elements, it is conceivable that they also occur in mtDNAs outside fungi and in nuclear genomes. We posit that the mitochondrial translation system is particularly “predestined” for acquiring translational jumping, owing to relaxed codon:tRNA-anticodon recognition. A weaker interaction between the ribosomal P-site and tRNAs aids dissociation of the peptidyl-tRNA from the takeoff codon and its reassociation with a landing site that may even belong to a different codon family. Another presumed requirement is that certain codons are unused in mitochondrial translation so that this codon can be used in new, creative ways. In M. capitatus, untranslated codons are exploited as a translation barrier for bypassing undesired sequences. In other instances, such codons are reassigned to new amino acid identities by a codon capture mechanism [e.g., CUN (leucine) to CUN (threonine) in baker’s yeast (22)]. It will be interesting to decipher the particular adaptations of the mitochondrial translation system in M. capitatus that enable such astoundingly effective programmed translational bypassing as reported here. It will also be interesting to unravel the mechanisms involved in horizontal transfer and integration of these “selfish” elements into genomic DNA.
Materials and Methods
Detailed descriptions of materials and methods can be found in SI Materials and Methods.
Yeast Strains and Culture Conditions.
Yeast strains (Table S1) were obtained from C. P. Kurtzman and J. Swezey (National Center for Agricultural Utilization Research, Peoria, IL) and R. A. Zvyagilskaya (Bach Institute of Biochemistry, Moscow, Russia). Yeast cells were cultivated at 28 °C in synthetic medium containing either glucose, glycerol, ethanol, or lactate as the sole carbon source.
DNA Purification and Sequencing.
Mitochondrial DNA from the Magnusiomyces magnusii strain 270 was prepared as described previously (23), used for random library construction, and sequenced by the dideoxy-chain termination method (24). For the other magnusiomycete strains, total cellular DNA was isolated as described earlier (25). Mitochondrial DNA was sequenced along with nuclear DNA using the Illumina HiSeq 2000 and paired-end (2 × 100 nt) technology, and assembled with Velvet (version 1.1.06) (26).
Mitochondrial Genome Annotation.
Gene annotation of Magnusiomyces mtDNAs was performed with the automated tool MFannot, developed in-house. MFannot predicts genes for structural RNAs and proteins and group I and II introns by using the search engines Erpin (27), Exonerate (28) and HMMER (29). Translation table 4 was used for conceptual translation. Termini and precise exon-intron boundaries of structural RNA genes were predicted manually using comparative structure modeling.
In Silico Search for Byps.
Byp insertions in M. capitatus were initially identified with TFASTA (30), by comparison with protein sequences from other Magnusiomyces strains without byps. For in silico searches, byp sequences were aligned with Muscle version 3.6 (31), and the resulting multiple alignment was manually curated for optimal fit of primary sequence and secondary structure predicted by RNAalifold (32). Byps were searched for with covariance models (8), based on the alignment of UCC and CGA byps in mitochondrial protein genes of M. capitatus. Only confidently aligned nucleotides were used for model building by applying the –hand option. Searches were performed with a cutoff E-value of 0.01.
Mapping of rRNA Termini in M. magnusii.
The termini of mitochondrial large and small rRNAs of the M. magnusii strain 270 were mapped using an RNA circularization procedure (33). The regions containing ligated rRNA termini were amplified by RT-PCR, cloned, and sequenced. Sequences of the identified rRNA termini are conserved across the analyzed magnusiomycetes.
Mitochondrial RNA Isolation and Transcript Analysis by RT-PCR.
Purified mitochondria were isolated as described earlier (34–36), and their RNA was extracted with the RNeasy Mini kit (Qiagen), treated with DNase I, and used for cDNA synthesis primed with random hexamers. The cDNA was used as a template in PCR reactions using gene-specific primer pairs. Control PCR reactions were performed using the following templates: (i) genomic DNA (positive control), (ii) mitochondrial RNA treated with DNase I, and (iii) cDNA prepared from mitochondrial RNA treated with DNase I and RNase A (negative controls). Amplicons were separated by electrophoresis in a 1% agarose gel, and selected products were sequenced using the dideoxy-chain termination method.
Identification of Mitochondrial Proteins by Mass Spectrometry.
Isolated mitochondria were solubilized with digitonin and homogenized, and the remaining membranes were removed by centrifugation. The supernatant was electrophoretically separated on a nondenaturating poly-acrylamide gel (37), individual bands were cut out and analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) (38, 39), and peptides were annotated by Mascot (40).
LacZ Constructs of Byps and Expression in E. coli.
For LacZ constructs of byps and expression in E. coli., see SI Materials and Methods.
Phylogeny of Yeast Species Based on mtDNA-Encoded Protein Sequences.
Thirteen inferred protein sequences (Cob, Cox1, -2, and -3, Atp6 and -9, and Nad1, -2, -3, -4, -4L, -5, and -6) were aligned with FSA (41). Columns with posterior probability <0.9 were discarded, and alignments were concatenated. The final dataset includes 44 species and 3,254 positions. Sequences except those for Magnusiomyces species were downloaded from the Organelle Genome Resources at National Center for Biotechnology Information. The phylogenetic tree was constructed with PhyloBayes (42), the CAT/GTR model, six discrete categories, four independent chains, 14,000 cycles (corresponding to ∼770, 000 generations), and the –dc parameter to remove constant sites. The first 10,000 cycles were discarded as burn-in.
Supplementary Material
Acknowledgments
We thank M. W. Gray (Dalhousie University), D. Morse (Université de Montréal), J. Ling (Yale University), and L. Kovac and J. Kolarov (Comenius University) for insightful discussion and comments. This work was supported by the Natural Sciences and Engineering Research Council (Canada) and the Canadian Research Chair Program (B.F.L.), Canadian Institutes of Health Research Grant MOP-79309 (to G.B.), Slovak Research and Development Agency Grants APVV 0123-10 (to J.N.) and 0035-11 (to L.T.), and Slovak Grant Agency Grants VEGA 1/0405/11 (to J.N.), 1/0311/12 (to L.T.), 1/1085/12 (to B.B.), and 1/0719/14 (to T.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KJ459950 (M. ingens), KJ459951 (M. tetrasperma), KJ459952 (M. capitatus), KJ459953 (M. magnusii 270), and JQ236859 (M. magnusii CBS 234.85)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322190111/-/DCSupplemental.
References
- 1. Wills NM (2010) Translational bypassing: Peptidyl-tRNA repairing at non-overlapping sites. Recoding: Expansion of Decoding Rules Enriches Gene Expression, Nucleic Acids and Molecular Biology, eds Atkins JF, Gesteland RF (Springer, New York), Vol 24, pp 365–380.
- 2.Herr AJ, Atkins JF, Gesteland RF. Coupling of open reading frames by translational bypassing. Annu Rev Biochem. 2000;69:343–372. doi: 10.1146/annurev.biochem.69.1.343. [DOI] [PubMed] [Google Scholar]
- 3.Huang WM, et al. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science. 1988;239(4843):1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- 4.Weiss RB, Huang WM, Dunn DM. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990;62(1):117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manch-Citron JN, Dey A, Schneider R, Nguyen NY. The translational hop junction and the 5′ transcriptional start site for the Prevotella loescheii adhesin encoded by plaA. Curr Microbiol. 1999;38(1):22–26. doi: 10.1007/pl00006766. [DOI] [PubMed] [Google Scholar]
- 6.Birrenbach T, et al. Emergence of Blastoschizomyces capitatus yeast infections, Central Europe. Emerg Infect Dis. 2012;18(1):98–101. doi: 10.3201/eid1801.111192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight RD, Landweber LF, Yarus M. How mitochondria redefine the code. J Mol Evol. 2001;53(4-5):299–313. doi: 10.1007/s002390010220. [DOI] [PubMed] [Google Scholar]
- 8.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: Inference of RNA alignments. Bioinformatics. 2009;25(10):1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wills NM, et al. Translational bypassing without peptidyl-tRNA anticodon scanning of coding gap mRNA. EMBO J. 2008;27(19):2533–2544. doi: 10.1038/emboj.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heckman JE, Sarnoff J, Alzner-DeWeerd B, Yin S, RajBhandary UL. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci USA. 1980;77(6):3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5(2):418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green-Willms NS, Fox TD, Costanzo MC. Functional interactions between yeast mitochondrial ribosomes and mRNA 5′ untranslated leaders. Mol Cell Biol. 1998;18(4):1826–1834. doi: 10.1128/mcb.18.4.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charneski CA, Hurst LD. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013;11(3):e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maldonado R, Herr AJ. Efficiency of T4 gene 60 translational bypassing. J Bacteriol. 1998;180(7):1822–1830. doi: 10.1128/jb.180.7.1822-1830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin S, Heckman J, RajBhandary UL. Highly conserved GC-rich palindromic DNA sequences flank tRNA genes in Neurospora crassa mitochondria. Cell. 1981;26(3 Pt 1):325–332. doi: 10.1016/0092-8674(81)90201-4. [DOI] [PubMed] [Google Scholar]
- 16.Paquin B, Forget L, Roewer I, Lang BF. Molecular phylogeny of Allomyces macrogynus: Congruency between nuclear ribosomal RNA- and mitochondrial protein-based trees. J Mol Evol. 1995;41(5):657–665. doi: 10.1007/BF00175824. [DOI] [PubMed] [Google Scholar]
- 17.Sor F, Fukuhara H. Nature of an inserted sequence in the mitochondrial gene coding for the 15S ribosomal RNA of yeast. Nucleic Acids Res. 1982;10(5):1625–1633. doi: 10.1093/nar/10.5.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullerwell CE, Leigh J, Forget L, Lang BF. A comparison of three fission yeast mitochondrial genomes. Nucleic Acids Res. 2003;31(2):759–768. doi: 10.1093/nar/gkg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerscher S, Durstewitz G, Casaregola S, Gaillardin C, Brandt U. The complete mitochondrial genome of Yarrowia lipolytica. Comp Funct Genomics. 2001;2(2):80–90. doi: 10.1002/cfg.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaillardin C, Neuvéglise C, Kerscher S, Nicaud JM. Mitochondrial genomes of yeasts of the Yarrowia clade. FEMS Yeast Res. 2012;12(3):317–331. doi: 10.1111/j.1567-1364.2011.00782.x. [DOI] [PubMed] [Google Scholar]
- 21.Bonocora RP, Zeng Q, Abel EV, Shub DA. A homing endonuclease and the 50-nt ribosomal bypass sequence of phage T4 constitute a mobile DNA cassette. Proc Natl Acad Sci USA. 2011;108(39):16351–16356. doi: 10.1073/pnas.1107633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su D, et al. An unusual tRNAThr derived from tRNAHis reassigns in yeast mitochondria the CUN codons to threonine. Nucleic Acids Res. 2011;39(11):4866–4874. doi: 10.1093/nar/gkr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griac P, Nosek J. Mitochondrial DNA of Endomyces (Dipodascus) magnusii. Curr Genet. 1993;23(5-6):549–552. doi: 10.1007/BF00312651. [DOI] [PubMed] [Google Scholar]
- 24.Burger G, Lavrov DV, Forget L, Lang BF. Sequencing complete mitochondrial and plastid genomes. Nat Protoc. 2007;2(3):603–614. doi: 10.1038/nprot.2007.59. [DOI] [PubMed] [Google Scholar]
- 25.Philippsen P, Stotz A, Scherf C. DNA of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- 26.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gautheret D, Lambert A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J Mol Biol. 2001;313(5):1003–1011. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- 28.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eddy SR (2008) HMMER: Biological Sequence Analysis Using Profile Hidden Markov Models, Version 3.0. Available at http://hmmer.janelia.org.
- 30.Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: Improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokobori S, Pääbo S. Transfer RNA editing in land snail mitochondria. Proc Natl Acad Sci USA. 1995;92(22):10432–10435. doi: 10.1073/pnas.92.22.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomáska L, Nosek J, Fukuhara H. Identification of a putative mitochondrial telomere-binding protein of the yeast Candida parapsilosis. J Biol Chem. 1997;272(5):3049–3056. doi: 10.1074/jbc.272.5.3049. [DOI] [PubMed] [Google Scholar]
- 35.Lambowitz AM. Preparation and analysis of mitochondrial ribosomes. Methods Enzymol. 1979;59:421–433. doi: 10.1016/0076-6879(79)59103-4. [DOI] [PubMed] [Google Scholar]
- 36.Newman SM, Zelenaya-Troitskaya O, Perlman PS, Butow RA. Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiae that lacks the mitochondrial HMG box protein Abf2p. Nucleic Acids Res. 1996;24(2):386–393. doi: 10.1093/nar/24.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daoud R, Forget L, Lang BF. Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res. 2012;40(4):1728–1736. doi: 10.1093/nar/gkr941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wessels HJ, et al. LC-MS/MS as an alternative for SDS-PAGE in blue native analysis of protein complexes. Proteomics. 2009;9(17):4221–4228. doi: 10.1002/pmic.200900157. [DOI] [PubMed] [Google Scholar]
- 39.Fandiño AS, et al. LC-nanospray-MS/MS analysis of hydrophobic proteins from membrane protein complexes isolated by blue-native electrophoresis. J Mass Spectrom. 2005;40(9):1223–1231. doi: 10.1002/jms.903. [DOI] [PubMed] [Google Scholar]
- 40.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Bradley RK, et al. Fast statistical alignment. PLOS Comput Biol. 2009;5(5):e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21(6):1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 43.Meisinger C, Sickmann A, Pfanner N. The mitochondrial proteome: From inventory to function. Cell. 2008;134(1):22–24. doi: 10.1016/j.cell.2008.06.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.