Abstract
Ninety-three Malaysian extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae isolates were investigated for ciprofloxacin resistance. Two mismatch amplification mutation (MAMA) assays were developed and used to facilitate rapid detection of gyrA and parC mutations. The isolates were also screened for plasmid-mediated quinolone resistance (PMQR) genes including aac(6′)-Ib-cr, qepA, and qnr. Ciprofloxacin resistance (MICs 4– ≥ 32 μg/mL) was noted in 34 (37%) isolates, of which 33 isolates had multiple mutations either in gyrA alone (n = 1) or in both gyrA and parC regions (n = 32). aac(6′)-Ib-cr was the most common PMQR gene detected in this study (n = 61), followed by qnrB and qnrS (n = 55 and 1, resp.). Low-level ciprofloxacin resistance (MICs 1-2 μg/mL) was noted in 40 (43%) isolates carrying qnrB accompanied by either aac(6′)-Ib-cr (n = 34) or a single gyrA 83 mutation (n = 6). Ciprofloxacin resistance was significantly associated with the presence of multiple mutations in gyrA and parC regions. While the isolates harbouring gyrA and/or parC alteration were distributed into 11 PFGE clusters, no specific clusters were associated with isolates carrying PMQR genes. The high prevalence of ciprofloxacin resistance amongst the Malaysian ESBL-producing K. pneumoniae isolates suggests the need for more effective infection control measures to limit the spread of these resistant organisms in the hospital.
1. Introduction
The emergence and spread of extended-spectrum β-lactamase (ESBL)-producing organisms have posed a great challenge to clinicians worldwide. As ESBL-producing organisms are usually resistant to many other antimicrobial agents, limited therapeutic options are available for treatment of infections caused by these organisms [1]. Ciprofloxacin is one of the therapeutic choices for infections caused by bacteria belonging to the family Enterobacteriaceae. This antibiotic acts by inhibiting bacterial DNA gyrase and topoisomerase IV which are required for replication [2].
Resistance to fluoroquinolones (FQ) is now common in many ESBL-producing Gram-negative bacteria including Klebsiella pneumoniae [3, 4]. FQ resistance has been linked to specific amino acid substitutions in the chromosomal quinolone resistance determining regions (QRDRs) in GyrA and B subunits of DNA gyrase and ParC and E subunits of topoisomerase IV [5]. Mutations at Ser83 and Asp87 codons of GyrA subunit and Ser80 and Glu84 codons of ParC subunit have been commonly reported in FQ resistant K. pneumoniae isolates worldwide [6–8]. DNA sequencing is the gold standard method for the detection of these mutations; however, this method is expensive, laborious, and time consuming. Hence, cheaper, simpler, and rapid methods are required to facilitate mutation detection. A few assays have been developed for rapid detection of mutations in gyrA and/or parC genes of Campylobacter jejuni [9], Escherichia coli [10], and Neisseria gonorrhoeae [11] using mismatch amplification mutation assay (MAMA), a modified polymerase chain reaction that permits discriminatory amplification of specific allele sequences at QRDRs [12].
Plasmid-mediated quinolone resistance (PMQR) genes, including qnr, aac(6′)-Ib-cr and efflux pumps, are known to confer low-level FQ resistance [13]. Quinolone target protection by Qnr proteins are widely distributed in Enterobacteriaceae worldwide [14]. Until now, six Qnr families, namely, Qnr A, B, C, D, S, and VC, have been identified (http://www.lahey.org/qnrStudies/). While qnrA, B, and S genes are commonly detected at variable rates in K. pneumoniae worldwide [3, 13, 15], qnrC and D have been reported at low rates amongst K. pneumoniae isolates in China [16]. Moreover, a variant of aminoglycoside acetyltransferase (AAC(6′)-Ib-cr) with the ability to modify and inactivate ciprofloxacin has been widely spread in K. pneumoniae isolates from Asia [17, 18] and worldwide [3, 14].
FQ resistance may also arise as a result of reduced intracellular drug accumulation caused by porin loss or active efflux pump [5]. QepA, a quinolone-specific efflux pump, has been identified in Escherichia coli isolates from several Asian countries such as Japan, Korea, and China [19–21] but was rarely detected in K. pneumoniae [22, 23].
There is a paucity of data on the prevalence and the genetic determinants associated with ciprofloxacin resistance in Malaysian K. pneumoniae isolates. Hence, this study was conducted to identify chromosomal as well as plasmid-mediated mechanisms of ciprofloxacin resistance in a group of Malaysian ESBL-producing K. pneumoniae isolates. To facilitate rapid detection of gyrA and parC mutations, two mismatch amplification mutation assays (MAMA-PCR) were developed and validated in this study.
2. Materials and Methods
2.1. Bacterial Isolates
A group of 93 nonduplicate ESBL-producing K. pneumoniae isolates from patients attending to University of Malaya Medical Centre and a private hospital in Kuala Lumpur, Malaysia, in 2010–2012 were investigated in this study. The isolates were confirmed as K. pneumoniae using a PCR assay targeting the internal transcribed spacer unit of the bacteria [24]. Confirmation of ESBL production was performed using Cefpodoxime Combination Disc Kit (Oxoid, UK).
2.2. Antibiotic Susceptibility Testing
Minimum inhibitory concentration (MIC) of ciprofloxacin was determined by E-test strips (bioMéreiux, Marcy L'Etoile, France) on Mueller-Hinton agar (Oxoid, UK) in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines [25]. MIC values ≥4, 2, and ≤1 μg/mL were used to define resistance, intermediate susceptibility, and susceptibility to ciprofloxacin, respectively.
2.3. Development of MAMA-PCR for gyrA and parC Mutations Detection
Two duplex PCR assays (gyrA83 + parC80 assay and gyrA87 + parC84 assay) were developed for the simultaneous detection of mutations in Ser83 codon of GyrA subunit and Ser80 codon of ParC subunit, and Asp87 codon of GyrA subunit and Glu84 codon of ParC subunit, respectively.
Universal gyrA and parC forward primers [16] were used together with the reverse primers (MAMA primers) designed in this study for the amplification of gyrA (Ser83 and Asp87) and parC (Ser80 and Glu84) genetic regions (Figure 1). MAMA primer design was performed using NCBI/Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The reverse primers were complementary to the wild-type alleles of gyrA and parC sequences of K. pneumoniae strain ATCC 13883 (GenBank accession numbers: DQ673325 and AF303641, resp.), except for a mismatch at the antepenultimate (−3) nucleotide of the 3′ end of each MAMA primer, which was included to improve allele discrimination. The MAMA primer:template mismatches included in this study were C:C (in gyrA83 and 87), A:G (in parC80), G:A (in parC84). The selection of the mismatches was based on previous observations of their effects on the overall PCR yield [26]. The presence of a single primer:template mismatch has minimal effect on the PCR yield; thus the wild-type gene can be amplified efficiently. In case of mutation(s), PCR efficiency will be extremely reduced due to the presence of additional mismatch(es) at the 3′ end of the MAMA primer which will not bind to the template; thus, amplification of the target gene is failed [27].
Figure 1.
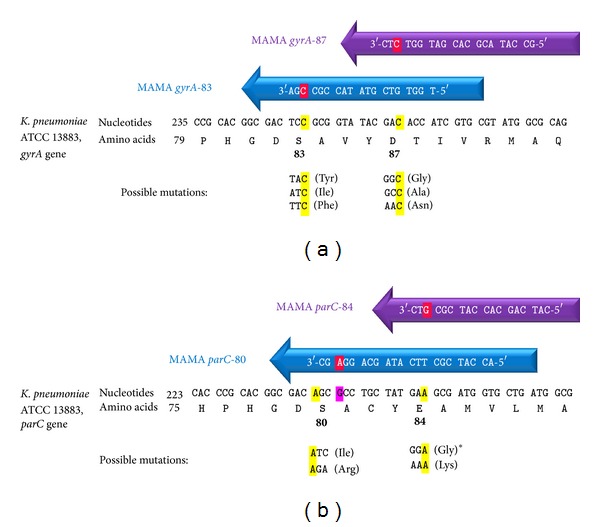
MAMA-PCR primers for gyrA (a) and parC (b) mutation detection. Red highlighted nucleotides are the mismatched nucleotides at the 3′ end of each MAMA primer. Mismatches were positioned at the conserved nucleotides of each codon (highlighted by yellow) located at the 3rd nucleotide from the 3′ end of each primer, except for parC80 primer where the conserved nucleotide (1st nucleotide in the parC80 codon) was excluded from the MAMA primer and the alteration was situated at a nucleotide outside the coding region (pink highlighted nucleotide). Quality control strains with the expected mutations shown in the figure were used for the assay development and optimization except the mutation with ∗ which was not available.
The performance of the primers was first evaluated using monoplex PCR prior to use in the duplex PCR assays which were finally optimized for the simultaneous detection of mutations in Ser83 codon of GyrA subunit with Ser80 codon of ParC subunit and in Asp87 codon of GyrA subunit with Glu84 codon of ParC subunit.
For the first duplex PCR assay, the concentrations of primers were optimized to 0.4 μM for each parC80 MAMA primer and parC universal forward primer, in addition to 0.25 μM for each gyrA83 MAMA primer and gyrA universal forward primer. For the second assay, the concentrations of primers were optimized to 0.45 μM for each parC84 MAMA primer and parC universal forward primer, in addition to 0.2 μM for each of gyrA87 MAMA primer and gyrA universal forward primer. The primer mixtures were added to a final PCR reaction volume of 20 μL containing 4 μL of 5x HOT FIREPol Blend Master Mix (Solis BioDyne, Estonia) which was comprised of 200 μM of each dNTP, 0.05 U/μL of DNA polymerase, and 1.5 mM MgCl2. Finally, 1 μL (<100 ng) of boiled bacterial extract [28] was added to each reaction. Amplification was carried out on a Veriti 96-well thermal cycler (Applied Biosystems, USA) programmed as follows: initial denaturation at 95°C for 10 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 40 s and extension at 72°C for 50 s, and a final extension step at 72°C for 7 min. PCR products were analysed on a 2% agarose gel prestained with 0.5 μg/mL ethidium bromide in 0.5x TBE buffer.
K. pneumoniae strains with known mutations in gyrA (45 isolates) and parC (10 isolates) were used as quality control strains for the assay optimization and validation [29].
2.4. Detection of gyrA and parC Mutations by MAMA-PCR
Following validation of the MAMA-PCR assays, the 93 clinical isolates investigated in this study were tested for alterations in gyrA and/or parC regions. For confirmation purpose, amplification and sequence analysis of the entire coding regions of gyrA and parC for 25 randomly selected isolates were performed as described previously [16]. The nucleotide sequences and deduced proteins were analyzed by NCBI tools and BioEdit software (version 7) and were compared with those of GyrA and ParC subunits of K. pneumoniae strain ATCC 13883 (GenBank accession numbers: DQ673325 and AF303641, resp.).
2.5. Detection of Plasmid-Mediated Quinolone Resistance (PMQR) Genes
All isolates were subjected to screening using a multiplex PCR assay for the detection of qnr types (A, B, C, and S) [13] and a monoplex PCR assay for the detection of qnrD type [30]. Detection of efflux pump (qepA) and the aminoglycoside acetyltransferase (aac(6′)-Ib) was performed using a multiplex PCR assay [13]. Allele-specific PCR assay was used to identify the cr mutation in aac(6′)-Ib [31]. To confirm the PCR results, representative amplicons of each PMQR gene were sequenced. Additionally, nine isolates from different susceptibility categories (resistant, intermediately susceptible, and susceptible to ciprofloxacin) were selected for sequence determination of the entire qnrB gene, as described previously [32]. The nucleotide sequences and deduced proteins were compared to the reference sequences in Lahey website (http://www.lahey.org/qnrStudies/) and GenBank database using BLAST search engine (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.6. Pulsed-Field Gel Electrophoresis (PFGE)
PFGE was used to determine the genetic relationship of the isolates as described previously [33]. Fragments generated by restriction with XbaI enzyme (New England Biolabs, USA) were separated by the CHEF-DR II system (Bio-Rad Laboratories, USA). The resultant banding patterns were analysed with BioNumerics software, version 7.1 (Applied Maths, Belgium), by the unweighted pair group method with arithmetic mean (UPGMA) algorithm. Cluster designation was based on isolates showing ≥80% relatedness.
2.7. Statistical Analyses
Categorical variables were compared by the Chi-square or Fisher's exact test and continuous variables were compared by Mann-Whitney U test. The relationship between ciprofloxacin MIC values with the number of gyrA and/or parC mutations and with the total number of quinolone resistance determinants was assessed by calculating Spearman's correlation coefficient. The total number of quinolone resistance determinants was calculated by adding the number of PMQR genes and the number of mutations in Ser83 and/or Asp87 codons of GyrA subunit and in Ser80 and/or Glu84 codons of ParC subunit. All tests were two-tailed and a P value < 0.05 was considered statistically significant. All statistical analyses were performed by PASW software version 18 (SPSS, Chicago, IL, USA).
3. Results
Reduced susceptibility to ciprofloxacin (resistance and intermediate susceptibility) was observed in 66 (71%) isolates investigated in this study. Ciprofloxacin MIC values of the isolates ranged from 0.032 to ≥32 μg/mL with MIC90 and MIC50 equal to ≥32 and 2 μg/mL, respectively. Based on ciprofloxacin MICs, the 93 ESBL-producing K. pneumoniae isolates were grouped into three susceptibility categories. The MIC values with quinolone resistance determinants in each category are shown in Table 1.
Table 1.
Ciprofloxacin susceptibility patterns and fluoroquinolone (FQ) resistance determinants detected in the 93 K. pneumoniae isolates investigated in this study.
| Ciprofloxacin susceptibility | MIC (μg/mL) | Number of isolates | FQ resistance determinants | Total number of gyrA and/or parC alterations | Total number of FQ resistance determinants | |
|---|---|---|---|---|---|---|
| PMQR* genes (n) | gyrA and/or parC alterations (n) | |||||
| Susceptible (n = 27, 29%) |
0.032–0.047 | 2 | None (2) | None (2) | None | None |
| 0.094–0.38 | 12 | aac(6′)-Ib-cr (12) | None (12) | None | 1 | |
| 0.5–0.75 | 5 |
qnrB (2), qnrS (1) None (2) |
None (3) gyrA83 (2) |
None 1 |
1 1 |
|
| 1 | 8 |
qnrB + aac(6′)-Ib-cr (5) qnrB (3) |
None (5) gyrA83 (3) |
None 1 |
2 2 |
|
|
| ||||||
| Intermediately susceptible (n = 32, 34%) |
2 | 32 |
qnrB + aac(6′)-Ib-cr (29) qnrB (3) |
None (29) gyrA83 (3) |
None 1 |
2 2 |
|
| ||||||
|
Resistant (n = 34, 37%) |
4–6 | 4 | None (4) | gyrA83 + parC80 (4) | 2 | 2 |
| ≥32 | 30 |
qnrB + aac(6′)-Ib-cr (8) qnrB + aac(6′)-Ib-cr (2) qnrB + aac(6′)-Ib-cr (1) qnrB + aac(6′)-Ib-cr (1), aac(6′)-Ib-cr (2) qnrB + aac(6′)-Ib-cr (1) |
gyrA83 + parC80 (10) gyrA83 + parC84 (2) gyrA83 + gyrA87 (1) gyrA83 + gyrA87 + parC80 (16) None (1) |
2 2 2 3 None |
2 or 4 4 4 3–5 2 |
|
*PMQR: Plasmid-mediated quinolone resistance.
3.1. Development and Validation of MAMA-PCR
The universal forward and MAMA reverse primers generated a PCR product from the wild-type gene in the absence of mutation(s). On the other hand, PCR was inhibited in the presence of mutation(s) (two or more mismatches at the 3′ end of the MAMA primer); therefore, negative PCR result was an indication of mutation in the corresponding genetic region. MAMA primers were able to distinguish wild types from mutations for all of the quality control strains (45 isolates for gyrA and 10 isolates for parC). The four MAMA monoplex PCR assays were then combined into two duplex assays (gyrA83 + parC80 assay and gyrA87 + parC84 assay). Figure 2 shows the results of MAMA duplex assays for some of the isolates investigated in this study.
Figure 2.
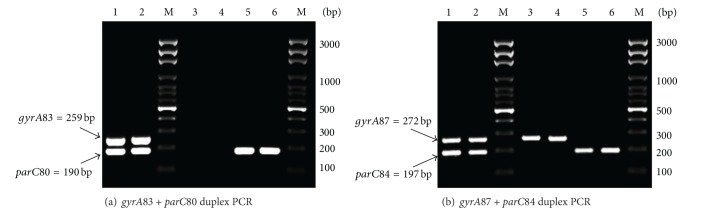
Agarose gel electrophoresis image of PCR products generated from duplex MAMA-PCR assays (a) gyrA83 + parC80 and (b) gyrA87 + parC84. Identification of each target was based on the expected product size. Lanes 1-2 represent PCR products generated in the presence of the wild-type alleles. Lanes 3–6 are examples of products generated in case of mutations in one gene or in both target genes. M: DNA molecular size marker (100 bp DNA Ladder, Solis BioDyne, Estonia).
3.2. Detection of gyrA and parC Mutations by MAMA-PCR Assays
Alterations in gyrA and/or parC genetic regions were detected in 41 of the 93 K. pneumoniae clinical isolates by MAMA method as shown in Table 2. Mutations in gyrA were detected in 44% (n = 41) of the isolates, of which 17 isolates had both Ser83 and Asp87 alterations, and 24 isolates had Ser83 mutation detected either alone in ciprofloxacin susceptible (n = 5) or intermediately susceptible isolates (n = 3) or coupled with parC mutations in ciprofloxacin resistant isolates (n = 16).
Table 2.
Alterations in gyrA and parC genes detected by MAMA-PCR and confirmed by sequence analysis for selected isolates.
| Total number of mutations | Number of isolates | Alterations detected by MAMA-PCR | Confirmation by sequencing (n) | ||||
|---|---|---|---|---|---|---|---|
| gyrA | parC | ||||||
| 83 | 87 | 80 | 84 | gyrA | parC | ||
| None | 52 | None | None | None | None | Wild type (7) | Wild type (7) |
| 1 | 8 | Mutation | None | None | None | Ser83Tyr (4) | ND |
| 2 | 1 | Mutation | Mutation | None | None | ND | ND |
| 14 | Mutation | None | Mutation | None | Ser83Ile (8) | Ser80Ile (9), Ser80Arg (1) | |
| 2 | Mutation | None | None | Mutation | Ser83Ile (2) | Glu84Lys (2) | |
| 3 | 16 | Mutation | Mutation | Mutation | None | Ser83Phe + Asp87Ala (4) | Ser80Ile (6) |
ND: not done.
Alterations in ParC subunit of DNA topoisomerase IV were detected in 34.4% (n = 32) of the isolates, of which 30 isolates had Ser80 mutation and two isolates had Glu84 alteration. Mutations in parC were detected in ciprofloxacin resistant isolates which had single or multiple gyrA mutations.
MAMA findings were confirmed by sequence analysis of the entire coding regions of gyrA and parC for selected isolates (seven isolates with the wild type of gyrA and parC and 18 isolates with MAMA results indicative of gyrA and/or parC alterations). Detected amino acid substitutions in GyrA were Ser83Ile (n = 10), Ser83Tyr (n = 4), and Ser83Phe + Asp87Ala (n = 4), whilst substitutions in ParC were Ser80Ile (n = 15), Ser80Arg (n = 1), and Glu84Lys (n = 2) (Table 2).
3.3. PMQR Genes
aac(6′)-Ib gene was detected in 74 (79.6%) of the isolates, of which 61 (65.6%) carried the cr variant. qnr genes were detected from 56 isolates (60.2%), of which 55 carried qnrB (59.1%) and one carried qnrS (1.1%). Sequence analysis of qnrB gene in nine randomly selected isolates revealed 100% nucleotide sequence identity with qnrB1 (n = 4), qnrB6 (n = 3), and qnrB7 (n = 2) (GenBank accession numbers DQ351241, GQ914054, and EU043311, resp.). The deduced proteins (223 amino acids) also exhibited 100% amino acid identity with QnrB1, QnrB6, and QnrB7 (GenBank accession numbers DQ351241, ADH03417, and ABW03156, resp.).
aac(6′)-Ib-cr and qnr genes were detected from isolates which were susceptible (17 and 11 isolates, resp.), intermediately susceptible (29 and 32 isolates, resp.), and resistant to ciprofloxacin (15 and 13 isolates, resp.). Interestingly, 47 isolates (50.2%) harbored both qnrB and aac(6′)-Ib-cr genes; thus the association between both genes was statistically significant (P < 0.001). Neither qepA efflux pump nor qnrA, C, and D genes were detected in this study.
3.4. The Relationship between Ciprofloxacin MIC and FQ Resistance Determinants
Table 1 shows the increase in the ciprofloxacin MICs of our isolates which was accompanied by a stepwise accumulation of FQ resistance determinants. The increase in ciprofloxacin MICs is correlated strongly with the increase in the total number of FQ resistance determinants (both gyrA and/or parC mutations and PMQR genes) (Spearman's correlation coefficient = 0.918; P < 0.001).
The lowest MIC values were noted in the isolates lacking any FQ resistance determinants (0.032–0.047 μg/mL). For the isolates with one FQ resistance determinant, MICs of isolates expressing aac(6′)-Ib-cr alone were significantly lower (0.094–0.38 μg/mL) than those of isolates expressing qnr gene alone or having a single gyrA83 mutation (0.5–0.75 μg/mL) (P = 0.001). MICs of isolates expressing two FQ resistance determinants (qnr gene accompanied by either aac(6′)-Ib-cr or a single gyrA83 mutation) were significantly higher (1-2 μg/mL) compared to those of isolates expressing one of the above mentioned genes alone (P < 0.001).
Of note, 34 out of 40 isolates which demonstrated reduced susceptibility to ciprofloxacin (MIC = 1-2 μg/mL) harbored both qnrB and aac(6′)-Ib-cr genes; thus both genes were significantly associated with low-level ciprofloxacin resistance (P < 0.001).
There is significant association between the resistance phenotype and the presence of more than one mutation in gyrA and/or parC (P < 0.001), as most (33 out of 34) ciprofloxacin resistant isolates harbored 2-3 mutations in gyrA and/or parC codons. These isolates demonstrated significantly higher MIC values (4–≥32 μg/mL) compared to those of isolates harboring single gyrA83 mutation with and without qnrB gene (MICs 0.5–2 μg/mL, P < 0.001) and those harboring qnr and/or aac(6′)-Ib-cr genes without any alterations in QRDRs (MICs 0.094–2 μg/mL, P < 0.001). Thus, the increase in ciprofloxacin MICs is correlated strongly with the increase in the total number of mutations in gyrA and/or parC subunits (Spearman's correlation coefficient = 0.78; P < 0.001).
Notably, aac(6′)-Ib-cr and qnrB were detected in some of the ciprofloxacin resistant isolates (15 and 13 isolates, resp.). The real contribution of PMQR on ciprofloxacin MIC is not clear in the ciprofloxacin resistant isolates as there is nonsignificant difference in ciprofloxacin MICs of the resistant isolates with and without aac(6′)-Ib-cr and qnrB genes (P > 0.05).
3.5. PFGE
The 93 isolates investigated in this study were differentiated into 41 PFGE clusters (Figure 3). The isolates harboring gyrA and/or parC alteration were distributed into 11 clusters, of which six clusters (X1–X6) were composed of 2–14 genetically related isolates, whereas the remaining five clusters were comprised of only one isolate each. Identical gyrA and/or parC mutations were found amongst isolates within the same cluster, with the only exception of two ciprofloxacin resistant isolates in cluster X4. In this cluster, two highly related isolates (92.3%) harboring gyrA83 and parC84 mutations were genetically related (less than 89%) to another two isolates without any gyrA and parC mutations (one was sensitive and the other was intermediately susceptible to ciprofloxacin). While gyrA and/or parC alteration were limited to isolates within 11 clusters, PMQR genes were detected in isolates distributed into 38 clusters. The FQ resistance determinants of the isolates in different clusters were presented in the supplementary data file (Supplementary Material available online at http://dx.doi.org/10.1155/2014/601630).
Figure 3.
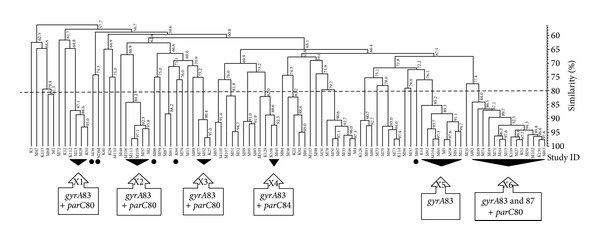
PFGE dendrogram of the 93 K. pneumoniae isolates investigated in this study. Alterations in GyrA and/or ParC subunits were detected in the isolates which belong to 11 clusters. Black triangles represent clusters with multiple isolates possessing the same gyrA and/or parC mutations. Black circles represent monoisolate clusters with gyrA and/or parC mutations. The dashed line represents the similarity level (80%) used in the clusters definition.
4. Discussion
A big proportion of our isolates (71%) were nonsusceptible (resistant and intermediately susceptible) to ciprofloxacin, which is a common finding in ESBL-producing isolates as reported in several countries such as Taiwan (59.1%) [34], France (60.3%) [3], and UK (62.3%) [35]. According to the latest study of antimicrobial resistance trends (SMART) in the Asia-Pacific region, ciprofloxacin nonsusceptibility in K. pneumoniae was much higher in the ESBL-producing isolates (65.8%) compared to the non-ESBL-producing isolates (7.4%) [36]. This may explain the reason why our ciprofloxacin nonsusceptibility rate (71%) was higher than the rate reported in a previous Malaysian study (18%) because the isolates investigated in that study were a mixture of ESBL and non-ESBL-producing K. pneumoniae isolates [37].
In this study, two multiplex MAMA-PCR assays have been successfully developed for the detection of mutations in gyrA83 + parC80 and in gyrA87 + parC84 codons of quinolone resistance determining regions in K. pneumoniae. To the best of our knowledge, no specific assay has been developed previously for the detection of alterations in the QRDRs of K. pneumoniae. A few monoplex PCR assays using MAMA method have been developed to detect alterations in bacterial QRDRs. For C. jejuni and N. gonorrhoeae, MAMA primers were designed to amplify particular mutations in the gyrA codon and not the wild type [9, 11]. On the contrary, the primers used in the E. coli MAMA-PCR method were designed in a way that gyrA or parC wild types were amplified and no PCR product would be obtained if mutations were present [10]. A similar approach was used in designing the MAMA primers in this study. Additionally, instead of using four monoplex MAMA-PCR assays as described in the E. coli study, two duplex assays were designed for simultaneous amplification of the gyrA and parC genetic regions. The duplex assay strategy is expected to facilitate rapid detection of mutations in the gyrA and parC since it shortens the time and the steps involved. The MAMA method developed in this study is rapid and cost effective compared to DNA sequencing approach and is useful for screening of a large number of K. pneumoniae isolates. Isolates with results suggestive of mutations can be selected for sequence analysis in order to define the amino acid at the mutation site.
DNA sequence analysis for selected isolates revealed several types of amino acid substitutions in GyrA (Ser83Ile, Ser83Tyr, Ser83Phe, and Asp87Ala) and ParC (Ser80Ile, Ser80Arg, and Glu84Lys). Similar amino acid substitutions were detected in GyrA regions of K. pneumoniae isolates from Malaysia [29] and other Asian countries [16, 38]. No data is available on ParC alterations in the Malaysian K. pneumoniae isolates; however, the amino acid substitutions detected in our study (Ser80Ile, Ser80Arg, and Glu84Lys) have been reported previously in K. pneumoniae isolates from other Asian countries such as Singapore [39], Japan [6], and Taiwan [38].
In agreement with previous reports [6, 40], multiple alterations in GyrA and/or ParC have been associated with ciprofloxacin resistance. Isolates with single alterations in gyrA83 exhibited reduced susceptibility to ciprofloxacin (MIC = 0.5–2 μg/mL). This observation has been reported previously [5] and is considered as the first step for the development of full resistance to ciprofloxacin. Ciprofloxacin MICs of our isolates increased with the acquisition of additional mutations in gyrA and/or parC genetic regions. This was expected as previous studies have shown that the progression from FQ susceptible towards resistant phenotype is a gradual process, starting from mutations in gyrA, the primary target of FQ, and followed by parC alterations which has a complementary role in the development of higher resistance [2, 41].
Surprisingly, we had one ciprofloxacin resistant isolate (MIC ≥ 32 μg/mL) which lacked any mutation in gyrA and parC genes but harbored both qnrB and aac(6′)-Ib-cr genes. Similar observations have been reported in isolates from other geographical regions [39, 42, 43]. The possible involvement of resistance mechanisms such as the reduction of bacterial drug uptake due to active efflux system (for instance, OqxAB) and/or membrane impermeability due to porin loss, or mutation in other genetic regions such as gyrB or parE, is yet to be explored.
The high prevalence of PMQR genes (65.6% for aac(6′)-Ib-cr and 60.2% for qnr) in our ESBL-producing K. pneumoniae isolates is in agreement with previous findings from other parts of the world [34, 44]. This high prevalence can be attributed to the coexistence of ESBL and PMQR genes on the same plasmid, as reported previously [34, 45]. Similarly, the simultaneous detection of both qnrB and aac(6′)-Ib-cr genes in 50.2% of the isolates is an indication that they are most likely located on the same plasmid, as shown in previous studies [17, 45].
aac(6′)-Ib-cr was the most common PMQR gene detected in this study. No information is available on the prevalence of this gene in the Malaysian Enterobacteriaceae isolates; however, studies from other Asian countries such as China [18], Korea [17], and Thailand [44] confirmed the emergence and spread of this gene amongst K. pneumoniae isolates.
qnrB was the predominant qnr gene identified in this study, in agreement with a previous report from our hospital [29] and reports from other Asian countries [15, 46]. qnrB1, qnrB6, and qnrB7 were detected by sequence analysis in some of our isolates. Both qnrB1 and qnrB6 have been previously detected in K. pneumoniae isolates from Malaysia [29] and other parts of Southeast Asia [38, 44], whereas qnrB7 has only been reported in two K. pneumoniae isolates from Norway and Sweden [47].
qnrS was detected at a very low frequency in our isolates, in contrast to a recent Thai report whereby this gene was the dominant qnr type in K. pneumoniae isolates [44]. Other qnr types including qnrA, C, and D were not detected in this study. The prevalence of qnr genes is variable from time to time and in different geographical locations [13]. This phenomenon has been attributed to the variations in the qnr-carrying plasmids which can also possess multiple antibiotic resistance genes. As a result of antibiotic selective pressure, some plasmids may dominate in certain clinical settings [48]. qepA efflux pump gene was not detected in our isolates. This was expected as the prevalence of this gene is generally low worldwide [22, 23].
In this study, the progressive increase in ciprofloxacin MICs of our isolates is correlated with the stepwise accumulation of FQ resistance determinants. Isolates with single FQ resistance determinant (a single PMQR gene (qnr or aac(6′)-Ib-cr) or a single gyrA83 mutation) demonstrated ciprofloxacin MICs (0.094–0.75 μg/mL) lower than those of isolates expressing two FQ resistance determinants (1-2 μg/mL) including two PMQR genes (qnrB and aac(6′)-Ib-cr) or qnrB with a single gyrA83 mutation (Table 1). Our results indicate that the effect of different FQ resistance determinants on ciprofloxacin MIC is cumulative, as reported previously [14].
Although the expression of qnr and aac(6′)-Ib-cr confers low-level ciprofloxacin resistance, it may have a negative impact on the therapeutic efficacy of ciprofloxacin as observed in rat animal models of experimental infection with qnr producing K. pneumoniae [49, 50]. Moreover, the expression of these genes can increase the mutant prevention concentration, which is the lowest antimicrobial concentration required to prevent the emergence of resistant mutants; thus, resistant mutants can be selected under ciprofloxacin therapeutic levels [51, 52].
The PFGE results in this study show the evidence of the spread of ciprofloxacin resistant isolates harboring GyrA and/or ParC alterations by clonal expansion, as identical mutations in gyrA and/or parC were detected from genetically related isolates. Similar findings have also been reported previously by other investigators [35]. Interestingly, both gyrA83 and parC84 mutations were detected in two highly related isolates (92.3%), which were genetically related to another two isolates without any gyrA and parC mutations. All the four isolates were hospital-associated (data not shown); therefore, it is possible that they have originated from a common ancestor in the hospital environment. Ciprofloxacin resistance in two of the four isolates was probably caused by de novo mutations in gyrA83 and parC84 induced by ciprofloxacin selective pressure, as previously observed in E. coli [53]. This assumption was supported by the finding of both qnrB and aac(6′)-Ib-cr in the two ciprofloxacin resistant isolates as both genes have the ability to enhance the selection of chromosomal mutations [2].
PMQR genes were widely distributed into isolates within different PFGE clusters. Horizontal dissemination of the plasmids carrying PMQR genes is probably responsible for the high prevalence of PMQR genes in our isolates due to their wide distribution amongst genetically unrelated isolates [44].
5. Conclusions
A high prevalence of ciprofloxacin resistance was reported amongst the Malaysian ESBL-producing K. pneumoniae isolates investigated in this study. The current scenario can complicate the clinical management of patients because very few antimicrobial agents are active against these bacteria. This study also identified chromosomal and plasmid-mediated genetic determinants associated with ciprofloxacin resistance. The MAMA method developed in this study is simple, cost effective, and rapid for detection of gyrA and parC mutations. It is important for epidemiologic investigations particularly when a large number of bacterial isolates need to be screened. The high prevalence of PMQR genes in our isolates is alarming as these genes can be widely spread via plasmids, confer low-level ciprofloxacin resistance, and facilitate the selection of chromosomal mutations implicated in higher level ciprofloxacin resistance. Regular surveillance of ciprofloxacin resistance determinants and molecular epidemiologic investigations are essential in order to follow up the progress of resistance development and spread in our hospital settings.
Supplementary Material
The distribution of 93 K. pneumoniae isolates into 41 PFGE clusters. The ciprofloxacin MIC and detection of plasmid-mediated quinolone resistance (PMQR) genes and chromosomal mutations in gyrA and/or parC gene regions for each isolate are shown.
Acknowledgments
This study was supported by the University of Malaya Postgraduate Research Grant (PV037-2012A) and the University of Malaya High Impact Research Grant (HIR/E000013-20001). PFGE analysis was performed using a fully functional temporary evaluation license of BioNumerics software (version 7.1). Permission to publish the PFGE results was obtained from Applied Maths, Belgium. The authors thank Ms. Amal S. Saiful Anuar for her kind assistance in this study.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clinical Infectious Diseases. 2001;32(8):1162–1171. doi: 10.1086/319757. [DOI] [PubMed] [Google Scholar]
- 2.Drlica K, Zhao X, Malik M, Salz T, Kerns R. Fluoroquinolone resistance: mechanisms, restrictive dosing, and anti-mutant screening strategies for new compounds. In: Dougherty TJ, Pucci MJ, editors. Antibiotic Discovery and Development. New York, NY, USA: Springer; 2012. pp. 485–514. [Google Scholar]
- 3.Crémet L, Caroff N, Dauvergne S, Reynaud A, Lepelletier D, Corvec S. Prevalence of plasmid-mediated quinolone resistance determinants in ESBL Enterobacteriaceae clinical isolates over a 1-year period in a French hospital. Pathologie Biologie. 2011;59(3):151–156. doi: 10.1016/j.patbio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Paterson DL, Mulazimoglu L, Casellas JM, et al. Epidemiology of ciprofloxacin resistance and its relationship, to extended-spectrum β-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clinical Infectious Diseases. 2000;30(3):473–478. doi: 10.1086/313719. [DOI] [PubMed] [Google Scholar]
- 5.Jacoby GA. Mechanisms of resistance to quinolones. Clinical Infectious Diseases. 2005;41(2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 6.Deguchi T, Fukuoka A, Yasuda M, et al. Alterations in the gyrA subunit of DNA gyrase and the parC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae . Antimicrobial Agents and Chemotherapy. 1997;41(3):699–701. doi: 10.1128/aac.41.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, Guo L, Xu Y, et al. Alteration of GyrA amino acid required for ciprofloxacin resistance in Klebsiella pneumoniae isolates in China. Antimicrobial Agents and Chemotherapy. 2008;52(8):2980–2983. doi: 10.1128/AAC.00151-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y, Zhang W, Wang H, et al. Specific patterns of gyrA mutations determine the resistance difference to ciprofloxacin and levofloxacin in Klebsiella pneumoniae and Escherichia coli . BMC Infectious Diseases. 2013;13(1, article 8) doi: 10.1186/1471-2334-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zirnstein G, Li Y, Swaminathan B, Angulo F. Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. Journal of Clinical Microbiology. 1999;37(10):3276–3280. doi: 10.1128/jcm.37.10.3276-3280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiang YZ, Qin T, Fu W, Cheng WP, Li YS, Yi G. Use of a rapid mismatch PCR method to detect gyrA and parC mutations in ciprofloxacin-resistant clinical isolates of Escherichia coli . Journal of Antimicrobial Chemotherapy. 2002;49(3):549–552. doi: 10.1093/jac/49.3.549. [DOI] [PubMed] [Google Scholar]
- 11.Sultan Z, Nahar S, Wretlind B, Lindback E, Rahman M. Comparison of mismatch amplification mutation assay with DNA sequencing for characterization of fluoroquinolone resistance in Neisseria gonorrhoeae . Journal of Clinical Microbiology. 2004;42(2):591–594. doi: 10.1128/JCM.42.2.591-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birdsell DN, Pearson T, Price EP, et al. Melt analysis of mismatch amplification mutation assays (Melt-MAMA): a functional study of a cost-effective SNP genotyping assay in bacterial models. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0032866.e32866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong BK, Chi HP, Chung JK, Kim EC, Jacoby GA, Hooper DC. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrobial Agents and Chemotherapy. 2009;53(2):639–645. doi: 10.1128/AAC.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz J, Pons MJ, Gomes C. Transferable mechanisms of quinolone resistance. International Journal of Antimicrobial Agents. 2012;40(3):196–203. doi: 10.1016/j.ijantimicag.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Wang A, Yang Y, Lu Q, et al. Presence of qnr gene in Escherichia coli and Klebsiella pneumoniae resistant to ciprofloxacin isolated from pediatric patients in China. BMC Infectious Diseases. 2008;8(1, article 68) doi: 10.1186/1471-2334-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Yi Y, Wang Q, et al. Analysis of drug resistance determinants in Klebsiella pneumoniae isolates from a tertiary-care hospital in Beijing, China. PLOS ONE. 2012;7(7) doi: 10.1371/journal.pone.0042280.e42280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin SY, Kwon KC, Park JW, et al. Characteristics of aac(6′)-Ib-cr gene in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from Chungnam area. Korean Journal of Laboratory Medicine. 2009;29(6):541–550. doi: 10.3343/kjlm.2009.29.6.541. [DOI] [PubMed] [Google Scholar]
- 18.Yang H, Chen H, Yang Q, Chen M, Wang H. High prevalence of plasmid-mediated quinolone resistance genes qnr and aac(6′)-Ib-cr in clinical isolates of Enterobacteriaceae from nine teaching hospitals in China. Antimicrobial Agents and Chemotherapy. 2008;52(12):4268–4273. doi: 10.1128/AAC.00830-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamane K, Wachino JI, Suzuki S, et al. New plasmid-mediated fluoroquinolone efflux pump, qepA, found in an Escherichia coli clinical isolate. Antimicrobial Agents and Chemotherapy. 2007;51(9):3354–3360. doi: 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim ES, Jeong JY, Choi SH, et al. Plasmid-mediated fluoroquinolone efflux pump gene, qepA, in Escherichia coli clinical isolates in Korea. Diagnostic Microbiology and Infectious Disease. 2009;65(3):335–338. doi: 10.1016/j.diagmicrobio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Liu JH, Deng YT, Zeng ZL, et al. Coprevalence of plasmid-mediated quinolone resistance determinants qepA, Qnr, and AAC(6′)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrobial Agents and Chemotherapy. 2008;52(8):2992–2993. doi: 10.1128/AAC.01686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J, Zeng Z, Chen Z, et al. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrobial Agents and Chemotherapy. 2009;53(2):519–524. doi: 10.1128/AAC.00886-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Y, Yang J, Zhang Y, Ye L, Wang L, Guo L. Prevalence of β-lactamases and 16S rRNA methylase genes amongst clinical Klebsiella pneumoniae isolates carrying plasmid-mediated quinolone resistance determinants. International Journal of Antimicrobial Agents. 2011;37(4):352–355. doi: 10.1016/j.ijantimicag.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Liu C, Zheng W, et al. PCR detection of Klebsiella pneumoniae in infant formula based on 16S-23S internal transcribed spacer. International Journal of Food Microbiology. 2008;125(3):230–235. doi: 10.1016/j.ijfoodmicro.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Performance Standards for Antimicrobial Susceptibility Testing. Wayne, Pa, USA: Clinical and Laboratory Standards Institute document M100-S23; 2013. [Google Scholar]
- 26.Kwok S, Kellogg DE, McKinney N, et al. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Research. 1990;18(4):999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons BL, Heflich RH. Genotypic selection methods for the direct analysis of point mutations. Mutation Research. 1997;387(2):97–121. doi: 10.1016/s1383-5742(97)00026-4. [DOI] [PubMed] [Google Scholar]
- 28.Huang TM, Chang YF, Chang CF. Detection of mutations in the gyrA gene and class I integron from quinolone-resistant Salmonella enterica serovar Choleraesuis isolates in Taiwan. Veterinary Microbiology. 2004;100(3-4):247–254. doi: 10.1016/j.vetmic.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Saiful Anuar AS, Mohd Yusof MY, Tay ST. Prevalence of plasmid-mediated qnr determinants and gyrase alteration in Klebsiella pneumoniae isolated from a university teaching hospital in Malaysia. European Review for Medical and Pharmacological Sciences. 2013;17(13):1744–1747. [PubMed] [Google Scholar]
- 30.Cavaco LM, Hasman H, Xia S, Aarestrup FM. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrobial Agents and Chemotherapy. 2009;53(2):603–608. doi: 10.1128/AAC.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wareham DW, Umoren I, Khanna P, Gordon NC. Allele-specific polymerase chain reaction (PCR) for rapid detection of the aac(6′)-Ib-cr quinolone resistance gene. International Journal of Antimicrobial Agents. 2010;36(5):476–477. doi: 10.1016/j.ijantimicag.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Tamang MD, Seol SY, Oh JY, et al. Plasmid-mediated quinolone resistance determinants qnrA, qnrB, and qnrS among clinical isolates of Enterobacteriaceae in a Korean hospital. Antimicrobial Agents and Chemotherapy. 2008;52(11):4159–4162. doi: 10.1128/AAC.01633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gautom RK. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. Journal of Clinical Microbiology. 1997;35(11):2977–2980. doi: 10.1128/jcm.35.11.2977-2980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin CJ, Siu LK, Ma L, Chang YT, Lu PL. Molecular epidemiology of ciprofloxacin-resistant extended-spectrum β-lactamase-producing Klebsiella pneumoniae in Taiwan. Microbial Drug Resistance. 2012;18(1):52–58. doi: 10.1089/mdr.2011.0060. [DOI] [PubMed] [Google Scholar]
- 35.Dashti AA, Paton R, Amyes SGB. Linkage of ciprofloxacin resistance with a single genotypic cluster of Klebsiella pneumoniae . International Journal of Antimicrobial Agents. 2006;27(1):73–76. doi: 10.1016/j.ijantimicag.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 36.Hsueh PR. Study for Monitoring Antimicrobial Resistance Trends (SMART) in the Asia-Pacific region, 2002–2010. International Journal of Antimicrobial Agents. 2012;40(supplement 1):S1–S3. doi: 10.1016/S0924-8579(12)00244-0. [DOI] [PubMed] [Google Scholar]
- 37.Lim KT, Yeo CC, Yasin RM, Balan G, Thong KL. Characterization of multidrug-resistant and extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains from Malaysian hospitals. Journal of Medical Microbiology. 2009;58(11):1463–1469. doi: 10.1099/jmm.0.011114-0. [DOI] [PubMed] [Google Scholar]
- 38.Liao CH, Hsueh PR, Jacoby GA, Hooper DC. Risk factors and clinical characteristics of patients with qnr-positive Klebsiella pneumoniae bacteraemia. Journal of Antimicrobial Chemotherapy. 2013;68(12):2907–2914. doi: 10.1093/jac/dkt295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneiders T, Amyes SGB, Levy SB. Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrobial Agents and Chemotherapy. 2003;47(9):2831–2837. doi: 10.1128/AAC.47.9.2831-2837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bansal S, Tandon V. Contribution of mutations in DNA gyrase and topoisomerase IV genes to ciprofloxacin resistance in Escherichia coli clinical isolates. International Journal of Antimicrobial Agents. 2011;37(3):253–255. doi: 10.1016/j.ijantimicag.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 41.Drago L, Nicola L, Mattina R, de Vecchi E. In vitro selection of resistance in Escherichia coli and Klebsiella spp. at in vivo fluoroquinolone concentrations. BMC Microbiology. 2010;10(1, article 119) doi: 10.1186/1471-2180-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazzariol A, Zuliani J, Cornaglia G, Rossolini GM, Fontana R. AcrAB efflux system: expression and contribution to fluoroquinolone resistance in Klebsiella spp. Antimicrobial Agents and Chemotherapy. 2002;46(12):3984–3986. doi: 10.1128/AAC.46.12.3984-3986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. Journal of Antimicrobial Chemotherapy. 2003;51(5):1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 44.Pasom W, Chanawong A, Lulitanond A, et al. Plasmid-mediated quinolone resistance genes, aac(6’)-Ib-cr, qnrS, qnrB, and qnrA, in urinary isolates of Escherichia coli and Klebsiella pneumoniae at a Teaching Hospital, Thailand. Japanese Journal of Infectious Diseases. 2013;66(5):428–432. doi: 10.7883/yoken.66.428. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y, Zhou Z, Qian Y, et al. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. Journal of Antimicrobial Chemotherapy. 2008;61(5):1003–1006. doi: 10.1093/jac/dkn063. [DOI] [PubMed] [Google Scholar]
- 46.Teo JWP, Ng KY, Lin RTP. Detection and genetic characterisation of qnrB in hospital isolates of Klebsiella pneumoniae in Singapore. International Journal of Antimicrobial Agents. 2009;33(2):177–180. doi: 10.1016/j.ijantimicag.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 47.Karah N, Poirel L, Bengtsson S, et al. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in Escherichia coli and Klebsiella spp. from Norway and Sweden. Diagnostic Microbiology and Infectious Disease. 2010;66(4):425–431. doi: 10.1016/j.diagmicrobio.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Pai H, Seo MR, Choi TY. Association of QnrB determinants and production of extended-spectrum β-lactamases or plasmid-mediated AmpC β-lactamases in clinical isolates of Klebsiella pneumoniae . Antimicrobial Agents and Chemotherapy. 2007;51(1):366–368. doi: 10.1128/AAC.00841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuursted K, Schumacher H. Significance of low-level resistance to ciprofloxacin in Klebsiella pneumoniae and the effect of increased dosage of ciprofloxacin in vivo using the rat granuloma pouch model. Journal of Antimicrobial Chemotherapy. 2002;50(3):421–424. doi: 10.1093/jac/dkf148. [DOI] [PubMed] [Google Scholar]
- 50.Rodríguez-Martínez JM, Pichardo C, García I, et al. Activity of ciprofloxacin and levofloxacin in experimental pneumonia caused by Klebsiella pneumoniae deficient in porins, expressing active efflux and producing QnrA1. Clinical Microbiology and Infection. 2008;14(7):691–697. doi: 10.1111/j.1469-0691.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- 51.Briales A, Rodríguez-Martínez JM, Velasco C, et al. In vitro effect of qnrA1, qnrB1, and qnrS1 genes on fluoroquinolone activity against isogenic Escherichia coli isolates with mutations in gyrA and parC . Antimicrobial Agents and Chemotherapy. 2011;55(3):1266–1269. doi: 10.1128/AAC.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodríguez-Martínez JM, Velasco C, García I, Cano ME, Martínez-Martínez L, Pascual A. Mutant prevention concentrations of fluoroquinolones for Enterobacteriaceae expressing the plasmid-carried quinolone resistance determinant qnrA1 . Antimicrobial Agents and Chemotherapy. 2007;51(6):2236–2239. doi: 10.1128/AAC.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hees BC, Tersmette M, Willems RJL, de Jong B, Biesma D, van Hannen EJ. Molecular analysis of ciprofloxacin resistance and clonal relatedness of clinical Escherichia coli isolates from haematology patients receiving ciprofloxacin prophylaxis. Journal of Antimicrobial Chemotherapy. 2011;66(8):1739–1744. doi: 10.1093/jac/dkr216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The distribution of 93 K. pneumoniae isolates into 41 PFGE clusters. The ciprofloxacin MIC and detection of plasmid-mediated quinolone resistance (PMQR) genes and chromosomal mutations in gyrA and/or parC gene regions for each isolate are shown.


