Abstract
Aim
Metacommunity theories attribute different relative degrees of importance to dispersal, environmental filtering, biotic interactions and stochastic processes in community assembly, but the role of spatial scale remains uncertain. Here we used two complementary statistical tools to test: (1) whether or not the patterns of community structure and environmental influences are consistent across resolutions; and (2) whether and how the joint use of two fundamentally different statistical approaches provides a complementary interpretation of results.
Location
Grassland plants in the French Alps.
Methods
We used two approaches across five spatial resolutions (ranging from 1 km × 1 km to 30 km × 30 km): variance partitioning, and analysis of metacommunity structure on the site-by-species incidence matrices. Both methods allow the testing of expected patterns resulting from environmental filtering, but variance partitioning allows the role of dispersal and environmental gradients to be studied, while analysis of the site-by-species metacommunity structure informs an understanding of how environmental filtering occurs and whether or not patterns differ from chance expectation. We also used spatial regressions on species richness to identify relevant environmental factors at each scale and to link results from the two approaches.
Results
Major environmental drivers of richness included growing degree-days, temperature, moisture and spatial or temporal heterogeneity. Variance partitioning pointed to an increase in the role of dispersal at coarser resolutions, while metacommunity structure analysis pointed to environmental filtering having an important role at all resolutions through a Clementsian assembly process (i.e. groups of species having similar range boundaries and co-occurring in similar environments).
Main conclusions
The combination of methods used here allows a better understanding of the forces structuring ecological communities than either one of them used separately. A key aspect in this complementarity is that variance partitioning can detect effects of dispersal whereas metacommunity structure analysis cannot. Moreover, the latter can distinguish between different forms of environmental filtering (e.g. individualistic versus group species responses to environmental gradients).
Keywords: Alps, community assembly, France, incidence matrix, metacommunity structure, plant communities, site-by-species, variance partitioning
INTRODUCTION
The concept of metacommunity, defined as a set of communities connected through dispersal, has recently gained much popularity in ecology (Logue et al., 2011). This success stems in part from the need to integrate ecological knowledge at different spatial scales and to consider the interplay between the regional pool of species and local biological forces driving community assembly (Leibold et al., 2004; Holyoak et al., 2005; Harrison & Cornell, 2008), a necessary step in ultimately developing a mechanistic understanding of biodiversity distribution (Lavergne et al., 2010). Four main metacommunity theories are distinguished on the basis of the importance each one gives to the four main processes identified as shaping the structure of metacommunities: dispersal, stochastic events of colonization and extinction, environmental filtering, and biological interactions (Leibold et al., 2004; Holyoak et al., 2005; Logue et al., 2011). Although theoretical and experimental work is still progressing with regard to how these processes interact to produce testable predictions, empirical work has already made some valuable contributions to the subject (Logue et al., 2011). The four metacommunity frameworks combine these processes in different ways: (1) the neutral model focuses on the effects of stochastic dispersal events and competition among ecologically equivalent species; (2) the patch dynamics framework is dominated by colonization–extinction trade-offs in patchy environments (e.g. a lower dispersal ability requires a higher competitive ability for long-term survival); (3) species sorting focuses on environmental filtering (i.e. species occupy sites according to their environmental preferences); and (4) mass effects are the interaction between environmental filtering and dispersal (Leibold et al., 2004; Holyoak et al., 2005). Although directly testing the relative effects of these processes in real communities is often very difficult, owing to the large scales and number of species involved, many efforts to link contrasting predictions to the observed diversity patterns have shown some success (Logue et al., 2011). These analyses have generally found that environmental filtering or a combination of it with dispersal dominates temperate communities at large spatial scales (e.g. Gilbert & Lechowicz, 2004; Cottenie, 2005; Meynard & Quinn, 2008), whereas studies based on tropical systems tend to find more support for neutral processes (e.g. Tuomisto et al., 2003; Cottenie, 2005; Keppel et al., 2010). Note, however, that results have been fairly mixed in some cases. One of the main impediments to the development of a mechanistic view of community assembly comes from the challenge to distinguish empirically the effects of dispersal from those related to environmental filtering because the two processes may generate similar patterns of spatial autocorrelation in species diversity and composition (Cottenie, 2005; Münkemüller et al., 2012). Recent studies have therefore suggested the need to use multiple approaches and multiple scales in metacommunity analysis (Giladi et al., 2011; Logue et al., 2011; Münkemüller et al., 2012).
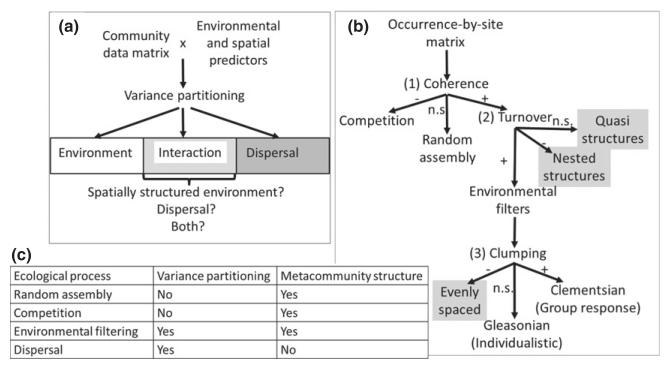
Three empirical tests, which have usually been applied separately, have been proposed within a metacommunity framework to relate ecological patterns and theory (Logue et al., 2011). The first strategy is targeted at revealing the importance of neutral processes (Hubbell, 2001); that is, the effects of stochastic processes under the assumption that species competing for similar resources are equivalent. We will not deal with the neutral approach here but will focus instead on the other two strategies, which consider the relative roles of the four processes (environmental filtering, biological interactions, dispersal limitation and stochastic events) in community assembly. The second approach, variance partitioning, is used to tease apart the roles of spatial structure and environmental filtering in community data (Cottenie, 2005). Spatial structure and environmental influences are decomposed using partial redundancy analysis (Fig. 1a), which is equivalent to partial regression analysis but using a multivariate response (a community matrix) (Borcard et al., 1992). The part of variance explained that can be linked solely to environmental variables is usually attributed to environmental filtering; the part that is linked to spatial structure and is non-environmentally driven is usually attributed to dispersal limitations; and the interaction term between environment and spatial structure represents covariation between environmental and spatial factors that are difficult to tease apart (Borcard et al., 1992; Cottenie, 2005) (Fig. 1a). This approach allows the study of different processes at different spatial scales, but suffers from the problem that the spatial component is difficult to interpret (e.g. Meynard et al., 2011). Indeed, the spatial structure that is independent of the environment could always be justified by a lack of knowledge regarding the relevant environmental variables, one or several of which could have been left out of the analysis (Borcard et al., 1992; Meynard et al., 2011). Conversely, the variance that is attributed to environmentally correlated spatial structure could arise from dispersal limitations that happen to occur in a spatially structured environment (Fig. 1a). Finally, the third approach to link ecological patterns and metacommunity theory involves the use of a site-by-species incidence matrix to test for specific elements of metacommunity structure (Leibold & Mikkelson, 2002).
Figure 1. The two metacommunity analyses used in this study of grassland plant communities within the French Alps.
(a) Variance partitioning allows the identification of relevant environmental predictors structuring species composition and separation of the effects of environmental filtering from that of dispersal. However, the interaction term between spatial structure and environmental predictors as well as the unexplained variance in the models give ambiguous results with respect to the four metacommunity processes (dispersal, environmental filtering, biological interactions, and stochastic colonization–extinction dynamics). (b) Analysis of metacommunity structure through coherence, range turnover and boundary clumping using the site-by-species incidence matrix allows some observed patterns to be linked to their possible causal processes: random assembly (non-significant coherence), competitive exclusion (negative coherence) and environmental filtering (Gleasonian or Clementsian responses to environmental gradients). However, some other results cannot be clearly linked to the four metacommunity processes (grey boxes in the figure), and it may be possible that competition and environmental filtering may explain some of them as well. (c) Summary of the processes that can be distinguished by using one or the other type of analysis, showing that their combination opens new avenues in metacommunity analyses. n.s., non-significant.
Randomization tests on these matrices as well as turnover and nestedness analyses allow us to test whether observed elements of metacommunity structure are different from chance expectation, and whether species replace each other along consistent environmental gradients (Fig. 1b). This approach was recently proposed as an integrated framework for the study of metacommunities (Presley et al., 2010). More specifically, a combination of statistical tools allows us to determine, for example, whether or not species show individualistic responses to environmental gradients (i.e. Gleasonian view) or whether communities are actually changing more or less consistently through groups of species that respond in a similar way to environmental gradients (i.e. Clementsian view). Although this approach is appealing because of its links to ecological theory, to our knowledge it has not been tested on the same data set as the partial regression approach. Moreover, some of the potential outcomes in the analysis remain without ecological interpretation (Fig. 1), making the translation between pattern and theory uncertain. Because the two approaches provide complementary tests regarding the relative roles of the main driving processes in metacommunity theory, using them together may lead to a better understanding of metacommunity driving processes.
Here, we used variance partitioning and analysis of metacommunity structure on the site-by-species incidence matrices to test (1) whether or not the patterns of community structure and environmental influences are consistent across resolutions, and (2) whether the joint use of the two statistical approaches mentioned above provides a consistent and complementary interpretation of results. We also used spatial regressions to identify relevant environmental predictors for species richness at different resolutions and to link regression and metacommunity structure results. We used a comprehensive database of alpine grassland plants, including more than 2600 plant species, 30 years of exhaustive community plot surveys in 12,000 sites, plus more than 1 million presence-only records across the French Alps, and an exhaustive environmental database including climate and soil characteristics over a 30-year period. We show below how variance partitioning and the analysis of metacommunity structure can provide complementary results, improving our understanding of community assembly across resolutions.
MATERIALS AND METHODS
Plant community data
Species composition data were provided by the French National Alpine Botanic Conservatory (CBNA: http://www.cbn-alpin.fr/, data downloaded October 2010; see also Boulangeat et al., 2012). The database contains information on almost 12,000 community plots over the French Alps (Fig. 2), and records for more than 2600 plant species. The community plots include exhaustive species lists for plots surveyed between 1980 and 2009, which we term here ‘community data’. We filtered these data for standardization purposes: we considered only grassland plots of known size classes, and checked for spatial accuracy (< 200 m) and the botanical expertise of the observers (see also Boulangeat et al., 2012). The database was also validated through multiple consultations with taxonomic experts and field biologists. This resulted in 2544 community plots of grassland habitat of intermediate size (10–1000 m2) available for the analysis.
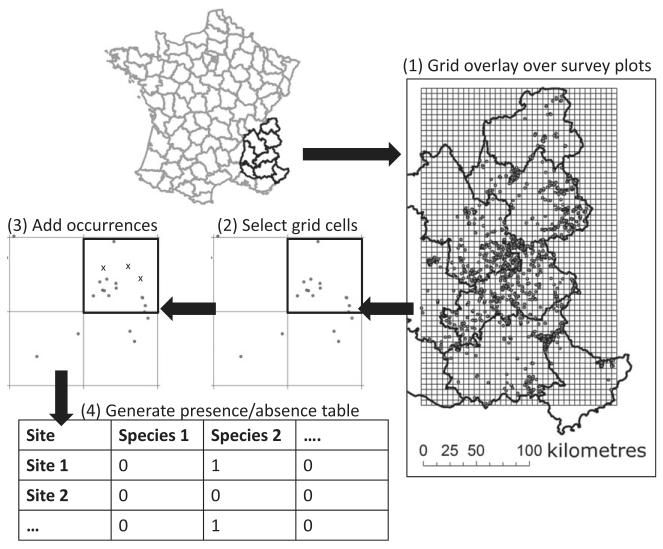
Figure 2. Study area and spatial aggregation strategy.
The French Alps are located in south-eastern France. (1) Regular grids of cells of different resolutions (1, 5, 10, 20 and 30 km) were overlaid over exhaustive vegetation survey plots from the CBNA community plot database. (2) Grid cells containing a minimum number of community plots (3, 5, 10, 20 and 30 community plots, respectively) were selected for further analyses (and the rest discarded) to ensure good sampling coverage of each grid cell considered in the analysis. In this example, only the upper right grid cell (thicker black contour) was selected, given that it was the only one with more than five community plots (represented here as grey dots). (3) Grid cells remaining in the analysis were further complemented with additional occurrence data (i.e. presence-only data, not coming from exhaustive community plots) from the CBNA occurrence-only database, and are represented here as crosses. (4) A table considering only grid cells selected in the previous steps (here called ‘sites’) was generated, in which each species received a presence (1) or absence (0) code, regardless of the number of occurrences.
When aggregating data at different spatial scales (see below) we also added occurrence records from the CBNA database, and checked for consistency and accuracy as described above (here termed ‘occurrence data’). These are presence-only records of species across the French Alps over the same 30-year period but that were not necessarily part of an exhaustive community survey. This database includes more than 1 million records of species occurrences distributed across the entire region of the French Alps.
Because of the completeness of this database and the extent of the study area (Fig. 2), which includes an important elevational gradient and relevant environmental gradients, this data set provides a unique opportunity to test ecological patterns at intermediate scales (i.e. larger than a single community, < 1000 km), at which the metacommunity provides a relevant theoretical framework (Holyoak et al., 2005).
Environmental data
Given the abundant literature on the potential drivers of species richness (e.g. Currie, 1991; Thuiller et al., 2006; Anderson et al., 2007), we included variables related to habitat heterogeneity (topography and climatic variability), energy (moisture index) and favourableness (soil, mean annual temperature). More specifically, we considered climate temporal mean and standard deviation summarized over a period of 30 years, soil properties such as percentage calcareous soil, depth and moisture index, topography, and synthetic biological variables such as growing degree-days (see Appendix S1 in Supporting Information). These variables have previously been shown to shape species distributions and affect community structure in Alpine plants (e.g. Körner, 1999; Dullinger et al., 2007, 2012).
Spatial scales
We used equal-area square grids to aggregate plant community plots and calculate species richness or build site-by-species incidence matrices at various resolutions (Fig. 2). Five spatial aggregation grains (here grain and resolution are used interchangeably) were considered: 1, 5, 10, 20 and 30 km (therefore the area varied from 1 to 900 km2). Multiple-occurrence data (coming from community plots or from occurrence records) that fell within one single grid cell were aggregated as a single-occurrence entry (Fig. 2). Because this strategy resulted in a different sampling effort between different grid cells, we carried out a simple standardization based on (1) eliminating from the analysis the grid cells that contained too few community plots (the minimum number of plots depended on the resolution, going from 3 plots at 1-km resolution to 30 plots at 30-km resolution), and (2) randomly selecting the same number of community plots for all remaining grid cells for the statistical analysis. This ensured that the grid cells considered for the analysis were surveyed thoroughly and that all grid cells had the same sampling effort. Additional occurrence data were overlaid on those grid cells to reduce the chances of false absences (Fig. 2). The environmental information for all community plots within each grid cell was aggregated using the mean and standard deviation of the environmental conditions of the community plots.
Environmental factors explaining species richness
We calculated the number of species for each grid cell at each resolution, and modelled species richness as the response variable in all regressions. Spatial autoregressive (SAR) models were used to incorporate the effects of spatial structure because they allow the modelling of spatial effects as well as the incorporation of environmental predictors into the analysis. As in other regression analyses, the response variable is modelled as a function of explanatory variables plus an error term. However, in an SAR model, the error term (ei) is modelled as a function of space:
| (1) |
where bij represents the spatial dependence between sites and is used to model the spatially dependent error, and εi represents the independently distributed residual error (assumed to be normally distributed). SAR models were first built here by incorporating the effects of the spatial autocorrelation in the absence of other (environmental) predictors. We fitted several SAR models by using two different shapes of the spatially dependent component (1/x and 1/x2, where x represents the distance between sites) and several maximum distances (from 50 to 200 km, in 50-km intervals) to consider in the spatial autocorrelations. As recommended in Kissling & Carl (2008), the spatial model (combination of shape of autocorrelation as a function of distance and maximal distance considered) was chosen to minimize the Akaike information criterion (AIC), therefore imposing a penalty for models with too many parameters. The environmental predictors were then added to this spatial model using a forward selection strategy. Initially we considered a total of 27 environmental variables (Appendix S1) as potential predictors in the statistical analysis. To select a subset of relevant variables, we started with a minimal model including the spatially dependent term and one environmental predictor. The predictor chosen to stay in the model was the one that maximized the Nagelkerke pseudo-R2 (Nagelkerke, 1991), which is the estimate of variance explained provided with the SAR models. Its calculation is based on log-likelihoods rather than on residual variance, but its interpretation is equivalent to the unadjusted R2 in classical linear regressions. Once a variable entered the model, all other variables that were strongly correlated with it (∣Pearson’s r∣ > 0.8) were excluded from further consideration. Then a second predictor was chosen to maximize the model R2. The process went on until the variable added became non-significant in the model (P-value > 0.05) or until the variable added did not significantly increase the predictive power of the model (Crawley, 2007). SAR models were built using the errorsarlm function within the package spdep in R 2.13.1 (R Development Core Team, 2011).
Variance partitioning on community data
Variance partitioning applied to the study of community structure allows the effects of the spatial structure that are independent of the environmental gradients (and therefore attributed to dispersal) to be isolated from the environmental effects that are independent of that spatial structure (and therefore attributed to environmental filtering) (Fig. 1a) (Legendre & Legendre, 1998; Cottenie, 2005; Tuomisto & Ruokolainen, 2006; Meynard & Quinn, 2008). However, this partitioning always leaves some variation that is shared between environment and spatial structure and that is difficult to attribute to either one of the two processes (Fig. 1a). This interaction could represent, for example, a dispersal effect that is correlated with topography, or the joint effect of several environmental factors that have a similar spatial structure. However, variance partitioning does not allow the presence of stochastic assembly or of biological interactions to be tested directly (Fig. 1a,c). Here, we used the function varpart within the package vegan, which applies a partial redundancy analysis (RDA) to partition variance between spatial and environmental components (Borcard et al., 1992; Cottenie, 2005). The environmental effects were represented by the variables selected at each resolution in the regression analyses described above, and the spatial effect was represented by a third-degree polynomial of geographical coordinates (Borcard et al., 1992). The total variance explained can thus be partitioned between the effects that are exclusive of environmental factors, those that are exclusive of spatial structure (i.e. dispersal) and those that result from the interactions between spatial and environmental structure (Legendre & Legendre, 1998) (Fig. 1b).
Elements of metacommunity structure using site-by-species incidence matrices
The second analysis was aimed at studying elements of metacommunity structure along environmental gradients using the metacommunity framework originally proposed by Lei-bold & Mikkelson (2002) and subsequently modified by Presley et al. (2010). Here we interpreted results according to Presley et al. (2010), and used matlab scripts made available by the authors at http://faculty.tarleton.edu/higgins/metacommunity-structure.html (accessed 18 November 2010). Three types of patterns of metacommunity structure were analysed in the data: (1) coherence, which corresponds to the level to which different species are structured and respond to the same environmental gradient; (2) species range turnover, which corresponds to how often species ranges replace each other; and (3) boundary clumping, which corresponds to how often multiple species have their range limits in the same sites (Presley et al., 2010). The first step in the analysis consists of ordering the site-by-species matrix using reciprocal averaging (RA, i.e. a regular canonical correspondence analysis using only the site-by-species incidence matrix without environmental predictors). This first step allows the identification of one or more RA axes that produce structure among communities. This produces an ordered site-by-species incidence matrix that is compared with random expectations through randomization, which allows coherence to be characterized as non-significant (random community assembly), significantly negative (checkerboard pattern where some species avoid each other, reflecting competitive exclusion) or significantly positive (communities are structured along environmental gradients, either individualistically or by groups of species that respond similarly to the environment) (Fig. 1b). In most cases, coherence is positive and the use of the other two indices helps in determining whether individualistic (Gleasonian) or synchronous (Clementsian) species turnover is important (Presley et al., 2010), and whether or not there are nested subsets of species along the environmental gradients (Fig. 1b). Note, however, that while some elements of community structure provide a clear link to one of the four processes associated with metacommunity theory, other results do not (e.g. nested and evenly spaced structures in Fig. 1b). Moreover, the fact that competitive exclusion produces checkerboard patterns, for example, does not preclude the possibility that other forms of competition could produce other metacommunity structures.
We carried out the same analysis using the first and second axes of an RA analysis on the site-by-species incidence matrix (Presley et al., 2009) at each resolution. We used default settings for all other parameters (see Presley et al., 2010 for details, and references therein). We also calculated Spearman’s rank correlations between the RA axes and richness as well as to the environmental predictors available to relate metacommunity structures to the environmental gradients present in the study region (Presley et al., 2009, 2010). The effects of dispersal cannot, however, be teased apart using this approach (Fig. 1b,c).
RESULTS
Environmental factors explaining species richness
The total variance explained increased at coarser resolutions, with R2 = 0.26 at 1-km resolution and R2 = 0.63 at 30-km resolution (Table 1). The environmental variables selected varied across resolutions (Table 1). For instance, at 1-km resolution the selected variables were mean values of temperature, slope and elevation, while at 30-km resolution the most important variables were related to spatial heterogeneity (summer moisture and temperature of coldest months; Table 1). At least one variable reflecting environmental heterogeneity (temporal or spatial) was selected in each model, although the relationship to species richness could be positive as well as negative depending on the variable and scale (Table 1). For example, the annual standard deviation of temperature of the coldest month had a negative effect on species richness at 1-km resolution, but a positive effect at 30-km resolution. However, at 30-km resolution, the only other variable selected – the spatial standard deviation on the summer moisture index – had a negative effect on richness (Table 1).
Table 1. Results from a spatial autoregressive (SAR) modelling on species richness in grassland plant communities within the French Alps.
Only the environmental variables selected on a forward stepwise selection are shown at each resolution. Values in each column represent the estimated coefficient value for each variable, with the corresponding standard error and significance level (P-value). At each resolution we also show the number of grid cells in the analysis (n), the number of community plots per grid cell, and the total explained variance (R2).
| Estimate ± SE | P-value | |
|---|---|---|
| 1 km (n = 284, 3 plots per cell, R2 = 0.26) | ||
| Intercept | 27.86 ± 0.34 | <0.001 |
| Temperature of coldest month | 3.22 ± 0.33 | <0.001 |
| Slope | 1.73 ± 0.23 | <0.001 |
| Elevation | −1.55 ± 0.27 | <0.001 |
| YSD temperature of coldest month | −1.63 ± 0.36 | <0.001 |
| 5 km (n = 121, 5 plots per cell, R2 = 0.29) | ||
| Intercept | 364.47 ± 13.85 | <0.001 |
| SSD growing degree-days | 24.39 ± 8.96 | 0.006 |
| Percentage calcareous soil | 27.99 ± 10.77 | 0.009 |
| Topographic wetness index | −33.45 ± 10.04 | <0.001 |
| YSD annual precipitation | −37.23 ± 12.42 | 0.003 |
| 10 km (n = 69, 10 plots per cell, R2 = 0.60) | ||
| Intercept | −239.65 ± 360.62 | 0.51 |
| SSD growing degree-days | 0.04 ± 0.01 | 0.001 |
| YSD annual temperature | 165.54 ± 56.14 | 0.003 |
| SSD percentage calcareous soil | 2.23 ± 0.59 | <0.001 |
| Elevation | 0.20 ± 0.08 | 0.010 |
| YSD annual precipitation | −0.87 ± 0.26 | <0.001 |
| Percentage calcareous soil | 0.80 ± 0.27 | 0.003 |
| SSD topographic wetness index | −96.18 ± 41.05 | 0.019 |
| 20 km (n = 37 grid cells, 20 plots per cell, R2 = 0.47) | ||
| Intercept | 834.12 ± 45.18 | <0.001 |
| YSD annual temperature | 74.38 ± 20.18 | <0.001 |
| SSD percentage calcareous soil | 52.24 ± 18.30 | 0.004 |
| 30 km (n = 25 grid cells, 30 plots per cell, R2 = 0.63) | ||
| Intercept | 962.39 ± 49.14 | <0.001 |
| SSD temperature of coldest month | 55.60 ± 15.02 | <0.001 |
| SSD summer moisture index | −47.95 ± 17.93 | 0.007 |
SE, standard error; SSD, spatial standard deviation; YSD, yearly standard deviation.
Variance partitioning on community data
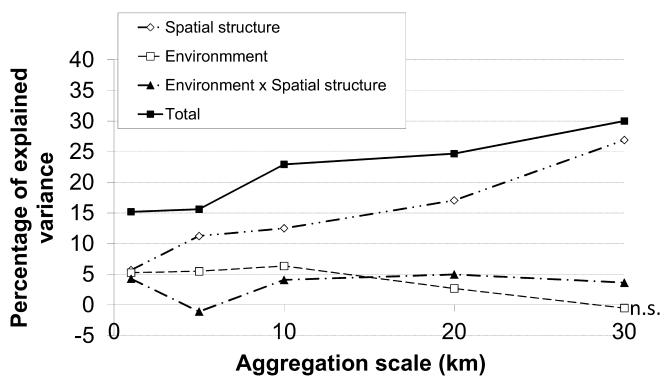
As with species richness, the variance explained on community data at coarser resolution was higher than at lower resolution (Fig. 3, black squares). The role of the spatial component after controlling for environment (i.e. the role of dispersal, white diamonds in Fig. 3) increased from 5% at 1-km resolution to 27% at 30-km resolution, while the role of the environment decreased from 5% to 0 (Fig. 3, white squares). The interaction term remained low as compared with the purely spatial effect (Fig. 3, black triangles), and the effect of the environment (after controlling for spatial structure) became non-significant at 30-km resolution (ANOVA on 200 permutations, P-value = 0.5).
Figure 3. Variance partitioning of species composition at various spatial scales in grassland plant communities within the French Alps.
While the total variance explained (black squares) reflects the R2 for a redundancy analysis (RDA) including the spatial as well as the environmental components, the other curves represent the results from a variance partitioning: the R2 that can be attributed to the environment after controlling for spatial structure (white squares), the R2 that can be attributed to spatial structure (i.e. dispersal) after controlling for the environment (white diamonds), and the interaction between spatial structure and environment (black triangles), which can be attributed to either environmental influences that are spatially structured or to dispersal effects that are environmentally structured. n.s., non-significant (ANOVA permutation test, P-value > 0.05).
Elements of metacommunity structure using site-by-species incidence matrices
The elements of metacommunity structure on the first and second reciprocal averaging (RA) axes showed very consistent results across scales: positive coherence (P-value < 0.001); positive turnover (P-value < 0.001); and significant clumping (P-value < 0.001) at all scales. In other words, communities are responding to consistent environmental gradients, here represented by the two RA axes; groups of species have coincident species range boundaries; and species composition changes consistently in similar places of the same environmental clines (Clementsian structure, see Fig. 1).
This analysis revealed that at least two RA axes reflect different metacommunity structures, both supporting a Clementsian response to common latent environmental gradients. Because reciprocal averaging groups sites by similarity in species composition, and groups species by their similarity in occurrence patterns (Presley et al., 2010; López-González et al., 2012), we found it useful to look at the correlations between the RA axes site scores and richness, and between the RA axes and environmental predictors. While the first axis showed significant positive correlations with richness at all resolutions (Table 2), the second axis showed a significant negative correlation with richness only at 1-km resolution (results not shown). This makes it more difficult to interpret the second RA axis in relation to the regression results, and we therefore focus on the first RA axis from now on. Correlations between RA axes and the various environmental predictors were numerous and did not allow us to single out a few environmental drivers among the large set of predictors available (Table 3). However, the strongest correlations were fairly consistent across scales. For example, positive relationships can be observed with mean annual temperature, growing degree-days and mean temperature of the coldest month across all scales. Negative correlations included spring, summer, autumn and annual moisture index, and temporal standard deviation on autumn moisture index (Table 3). Different measures of temporal and spatial standard deviation showed negative as well as positive correlations with the first axis at all scales. However, the overall spatial heterogeneity seemed to have a positive effect at all scales, whereas temporal variability on some factors (annual precipitation and autumn moisture index) seemed to have a negative impact on the first axis (Table 3).
Table 2. Correlation values between richness and the first axis of the reciprocal averaging (RA) calculated within a metacommunity structure analysis based on the incidence-by-site matrix in grassland plant communities within the French Alps.
Values represent Pearson correlations and the corresponding P-values for each resolution studied.
| Resolution | Correlation | P-value |
|---|---|---|
| 1 km | 0.42 | 0.037 |
| 5 km | 0.44 | 0.007 |
| 10 km | 0.35 | 0.003 |
| 20 km | 0.40 | < 0.001 |
| 30 km | 0.35 | < 0.001 |
Table 3. Summary of correlations between the first axis of a reciprocal averaging on the occurrence-by-site matrix and environmental variables in grassland plant communities within the French Alps.
Values are the result of Spearman’s rank correlations for each resolution; n.s., non-significant (P-value > 0.05); SD, standard deviation. For simplicity, we omitted variables that did not show significant correlations at three or more aggregation scales, and we only show significant correlations.
| Resolution |
||||||
|---|---|---|---|---|---|---|
| Variable | 1 km | 5 km | 10 km | 20 km | 30 km | |
| Mean values | Growing degree-days | 0.83 | 0.86 | 0.85 | 0.85 | 0.66 |
| Annual temperature | 0.83 | 0.86 | 0.83 | 0.82 | 0.64 | |
| Annual moisture index | −0.42 | −0.60 | −0.52 | −0.43 | −0.82 | |
| Spring moisture index | −0.48 | −0.70 | −0.65 | −0.59 | −0.82 | |
| Summer moisture index | −0.51 | −0.66 | −0.62 | −0.53 | −0.86 | |
| Autumn moisture index | −0.33 | −0.52 | −0.45 | −0.37 | −0.68 | |
| Temperature coldest month | 0.81 | 0.81 | 0.76 | 0.75 | 0.58 | |
| Summer precipitation | n.s. | −0.33 | −0.25 | n.s. | −0.58 | |
| Water-holding capacity | 0.24 | 0.30 | 0.26 | n.s. | n.s. | |
| Percentage calcareous soils | 0.20 | 0.21 | n.s. | 0.35 | n.s. | |
| Solar radiation | 0.25 | 0.26 | 0.42 | 0.49 | 0.51 | |
| Temporal SD | Growing degree-days | 0.81 | 0.85 | 0.87 | 0.87 | 0.65 |
| Annual moisture index | 0.47 | 0.45 | 0.52 | 0.50 | n.s. | |
| Autumn moisture index | −0.20 | −0.43 | −0.37 | −0.37 | −0.57 | |
| Temperature coldest month | 0.35 | 0.24 | 0.37 | 0.45 | n.s. | |
| Annual precipitation | −0.22 | −0.39 | −0.31 | n.s. | −0.63 | |
| Annual temperature | 0.59 | 0.68 | 0.69 | 0.75 | 0.83 | |
| Spatial SD | Growing degree-days | n.s. | 0.22 | 0.39 | 0.69 | 0.42 |
| Annual moisture index | 0.16 | 0.25 | 0.49 | 0.65 | n.s. | |
| Spring moisture index | 0.22 | 0.28 | 0.48 | 0.64 | n.s. | |
| Autumn moisture index | n.s. | 0.28 | 0.50 | 0.63 | n.s. | |
DISCUSSION
Our study is unique in bringing together two types of statistical analysis that have traditionally been used separately to infer metacommunity driving forces. By using them systematically on the same data set and at different spatial resolutions, we are able to show their complementarity in relating the four main processes proposed within the metacommunity framework (dispersal, biological interactions, environmental filtering and stochastic events) to the observed patterns. However, the interpretation of results from the two strategies is not straightforward, and the regression analysis on richness, which is not strictly necessary in this context, will help us to interpret and discuss the links between variance partitioning and community structure analysis.
Certainly one of the most puzzling results in our study is the fact that variance partitioning points to a strong role of dispersal that increases at coarser resolution (Fig. 3), while metacommunity structure analysis based on site-by-species incidence matrices points to a Clementsian community assembly that is very consistent across scales, and therefore to a strong role for environmental filtering. Although this difference may appear at first to be a contradiction, it actually shows the complementarity between the two approaches. Indeed, metacommunity structure analysis based on site-by-species incidence matrices cannot detect the effects of dispersal, even if they exist (Fig. 1b). Presley et al. (2010) recognized that this was a shortcoming of the method. The only way dispersal could potentially be detected in this type of analysis would be if dispersal effects were correlated with the latent environmental gradient identified in the RA axes as structuring the metacommunity. However, this would require a strong interaction between the relevant environmental gradient and the dispersal effects. Here, the variance partitioning analysis showed a strong effect of dispersal that is independent of environmental gradients, and an interaction term between environment and spatial structure that remains small compared with the spatial effect, especially at coarser resolution (Fig. 3). Therefore our results suggest that dispersal effects need to be analysed separately in order to be detected, and variance partitioning provides a useful tool to complement analyses based on metacommunity structure in this direction.
One of the disadvantages of the metacommunity structure analysis is the difficulty in interpreting the structuring axes that result from the reciprocal averaging and their correlation with environmental gradients. By optimizing site and species order in the site-by-species incidence matrix, reciprocal averaging will result in a first axis that would be correlated with species richness when the site-by-species matrix is nested. This correlation is not universal, however, and should not be significant in random assembly scenarios nor in loosely nested metacommunities. Here we found a significant positive correlation between the first RA axis and species richness across resolutions, despite the fact that the system is not nested (positive turnover at all scales). This correlation makes it possible to match the results from regressions based on richness to the results from the site-by-species incidence analysis in our study. However, in the absence of such a correlation this matching would not be possible. Second, by using univariate correlations between RA axes and environmental factors we may be looking at superfluous relationships. In a multivariate regression, we control the effect of one variable with another. For example, at 1-km resolution the effect of mean temperature during the coldest month on richness is positive and significant. However, the effect of mean temperature is also controlled for the effects of elevation, slope and temporal temperature variability (Table 1). In the analysis of metacommunity structure, correlations of the RA axes with all environmental variables may be significantly positive or negative (Table 3) but they do not account for the confounding effects of other variables. This also requires the interpretation of multiple correlations simultaneously, adding more complexity to the interpretation of results. Although in this case the strongest correlations are highly consistent across scales, the value of those correlations varies, and the fact that we cannot single out a few variables along the environmental gradient may be problematic. López-González et al. (2012) partly avoided this problem by carrying out a correspondence analysis, this time considering the incidence matrix and a reduced set of environmental variables. Although this is a valid alternative, the resulting axes are correlated with the RA axes but are not exactly the same. In the end, we still need to interpret multiple axes and multiple environmental variables, where the effects of a few relevant variables may be diluted by the noise introduced by others.
By contrast, variance partitioning introduces a different problem, namely that of variable selection. Here we have chosen to use environmental predictors based on regressions between richness and environmental factors within a forward stepwise selection strategy. However, we note that using all environmental predictors available without variable selection produced very similar results (not shown), suggesting that the same predictors capture fundamental variations in environmental gradients.
Our results do not reveal any effects of stochastic events or competitive exclusion at the finest resolutions studied here. On the contrary, Clementsian patterns are consistent across spatial resolutions. Spatial scale is usually described by two components: spatial grain or resolution, and spatial extent (Fortin & Dale, 2006). Presley & Willig (2010) studied bat metacommunity structure in the Caribbean Basin, and found that at the smallest spatial extents the metacommunity structure could differ between biogeographical regions, but that at larger extents structures were all Clementsian. Similar results were shown by Lewinsohn et al. (2006), wherein nestedness could only be revealed at the largest extents. Here we only varied grain, which could explain the consistency of patterns across scales. Presumably, the inclusion of several biogeographical regions could result in different structuring forces. However, it is particularly striking that environmental affinity has such an important role in structuring alpine grassland plant communities at all spatial resolutions. Tamme et al. (2010) suggested that resolution may be more important to detect mechanisms of coexistence because this aspect of scale determines how ‘diluted’ species interactions are. Paradoxically, our results along with the previous studies mentioned above suggest that extent, and not resolution, is more important in determining the resulting community patterns detected. In other words, different patterns found in previous studies may well reflect varying driving processes when including different biogeographical regions, rather than variations due to aggregating the data at different resolutions. This point certainly needs further attention and would need to be complemented with similar analyses across taxonomic groups. However, this also reveals the importance of clearly defining grain and extent when addressing spatial scale issues in ecology (Fortin & Dale, 2006). The consistency of the Clementsian pattern of species turnover suggests that environmental filtering remains a structuring force throughout scales and has important evolutionary implications. These results suggest that, as individualistic as the species responses may be, there must be some physiological or evolutionary trade-offs associated with important environmental thresholds, which will translate into similar species occurring and disappearing at the same locations along environmental gradients (Dahlgren & Ehrlen, 2011). This, along with the increasing role of dispersal at coarser resolutions (Fig. 3), fits well within the mass-effect metacommunity framework (Shmida & Wilson, 1985; Mouquet & Loreau, 2002; Leibold et al., 2004), where community composition is highly dependent on environmental filtering, but where dispersal may also have an important influence on it. A deeper understanding of adaptive forces that are related to environmental filtering processes would be necessary to better apprehend the relationship between dispersal and environmental filtering in this context.
A last point that is worth discussing is the role of environmental heterogeneity in structuring diversity at large spatial scales. It has been previously recognized that heterogeneity is important in several ways (Anderson et al., 2007; Lundholm, 2009; Tamme et al., 2010). Having very heterogeneous landscapes close together could increase turnover between sites, because species occupying different types of habitats should be different (Rahbek et al., 2007). In mountain ranges, for example, sites above and below the tree line may be very close to each other but support very different vegetation covers and different faunas. The overall species diversity of such areas could therefore be significantly higher than that in areas taken exclusively below or above the tree line. However, the effects of temporal heterogeneity may be different, as it has been argued that more stable environments form cradles of diversity over evolutionary time-scales, and species inhabiting more variable environments throughout the year should also be adapted to cope with those different environmental conditions (Fjeldså et al., 1999). Furthermore, the effects of environmental heterogeneity on species richness should be positive at some scales and negative at others (Tamme et al., 2010; Giladi et al., 2011). Our results show mostly positive relationships of species richness with spatial variability, but negative relationships with the temporal variability of some factors at all spatial resolutions (Table 3), suggesting that the effects of spatial and temporal variability depend on the environmental variable considered. We suggest that those environmental variables that impose important physiological stress would probably limit species richness, because fewer species will be adapted to such conditions, whereas those that promote habitat diversity or environmental diversity without imposing physiological constraints could have the opposite effect, for example through a storage effect over time (Chesson, 1994).
Overall, the recent literature indicates that the use of several analyses in conjunction, together with a combination of indices or of statistical approaches, can give us better insights into community assembly (e.g. Münkemüller et al., 2012). Recent efforts to incorporate phylogenetic and functional considerations into these types of analyses may provide further insights into these processes (Cavender-Bares et al., 2009; Mouquet et al., 2012). We showed here that this is also true for variance partitioning and metacommunity structure analyses, both of them providing complementary information regarding the processes that are behind diversity patterns (Fig. 1). Most notably, while metacommunity structure analysis based on site-by-species incidence matrices allows us to distinguish patterns from random expectations and between different modes of environmental filtering (e.g. individualistic species responses versus groups of species with similar environmental responses), it would not allow any roles for dispersal to be distinguished directly. On the contrary, variance partitioning suggests that dispersal does play a preponderant role, and that this role increases at coarser resolution. However, the interpretation of results is not straightforward, and complementary analysis on richness provides insights into metacommunity structure by providing links between the two approaches. This analysis also provides strong support for a Clementsian community structure, where species change consistently across environmental gradients, suggesting that there are important environmental thresholds (i.e. places along the environmental clines where many species have their distribution limit) that determine community composition across resolutions. Finally, our work also points to the importance of distinguishing between the two axes of spatial scale, namely resolution and extent, in ecological studies (Fortin & Dale, 2006). Comparison of our results with previous studies suggests that this distinction is crucial for understanding the role of spatial scale in community assembly processes.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded preliminarily by the French ‘Agence Nationale de la Recherche’ through the project DIVERS-ITALP (ANR-07-BDIV-014). C.N.M. and N.M. were supported by a University Montpellier II research grant. I.B. was funded by the French ‘Agence Nationale de la Recherche’ through the project SCION (ANR-08-PEXT-03). W.T. and S.L. received funding from the European Research Council under the European Community’s Seven Framework Programme FP7/2007–2013 Grant Agreement no. 281422 (TEEMBIO). Three anonymous referees provided useful comments on previous versions of this paper.
BIOSKETCH
Christine N. Meynard is an ecologist interested in diversity patterns at local to regional scales, including the modelling of species distributions and understanding metacommunity dynamics. Her recent research has focused on incorporating phylogenetic and functional diversity into these fields, as well as on understanding spatial patterns of diversity in relation to environmental gradients.
REFERENCES
- Anderson TM, Metzger KL, McNaughton SJ. Multi-scale analysis of plant species richness in Serengeti grasslands. Journal of Biogeography. 2007;34:313–323. [Google Scholar]
- Borcard D, Legendre P, Drapeau P. Partialling out the spatial component of ecological variation. Ecology. 1992;73:1045–1055. [Google Scholar]
- Boulangeat I, Lavergne S, Van Es J, Garraud L, Thuiller W. Niche breath, rarity and ecological characteristics within a regional flora spanning large environmental gradients. Journal of Biogeography. 2012;39:204–214. [Google Scholar]
- Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. The merging of community ecology and phylogenetic biology. Ecology Letters. 2009;12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- Chesson P. Multispecies competition in variable environments. Theoretical Population Biology. 1994;45:227–276. [Google Scholar]
- Cottenie K. Integrating environmental and spatial processes in ecological community dynamics. Ecology Letters. 2005;8:1175–1182. doi: 10.1111/j.1461-0248.2005.00820.x. [DOI] [PubMed] [Google Scholar]
- Crawley M. The R book. John Wiley & Sons Ltd; Chichester, UK: 2007. [Google Scholar]
- Currie DJ. Energy and large-scale patterns of animal-species and plant-species richness. The American Naturalist. 1991;137:27–49. [Google Scholar]
- Dahlgren JP, Ehrlen J. Incorporating environmental change over succession in an integral projection model of population dynamics of a forest herb. Oikos. 2011;120:1183–1190. [Google Scholar]
- Dullinger S, Kleinbauer I, Pauli H, et al. Weak and variable relationships between environmental severity and small-scale co-occurrence in alpine plant communities. Journal of Ecology. 2007;95:1284–1295. [Google Scholar]
- Dullinger S, Gattringer A, Thuiller W, et al. Extinction debt of high-mountain plants under twenty-first-century climate change. Nature Climate Change. 2012;2:619–622. [Google Scholar]
- Fjeldså J, Lambin E, Mertens B. Correlation between endemism and local ecoclimatic stability documented by comparing Andean bird distributions and remotely sensed land surface data. Ecography. 1999;22:63–78. [Google Scholar]
- Fortin MJ, Dale M. Spatial analysis: a guide for ecologists. Cambridge University Press; Cambridge: 2006. [Google Scholar]
- Giladi I, Ziv Y, May F, Jeltsch F. Scale-dependent determinants of plant species richness in a semi-arid fragmented agro-ecosystem. Journal of Vegetation Science. 2011;22:983–996. [Google Scholar]
- Gilbert B, Lechowicz MJ. Neutrality, niches, and dispersal in a temperate forest understory. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7651–7656. doi: 10.1073/pnas.0400814101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S, Cornell H. Toward a better understanding of the regional causes of local community richness. Ecology Letters. 2008;11:969–979. doi: 10.1111/j.1461-0248.2008.01210.x. [DOI] [PubMed] [Google Scholar]
- Holyoak M, Leibold MA, Mouquet NM, Holt RD, Hoopes MF. Metacommunities: a framework for large-scale community ecology. In: Holyoak M, Leibold MA, Holt RD, editors. Metacommunities: spatial dynamics and ecological communities. University of Chicago Press; Chicago: 2005. pp. 1–31. [Google Scholar]
- Hubbell SP. The unified neutral theory of biodiversity and biogeography. Princeton University Press; Princeton, NJ: 2001. [DOI] [PubMed] [Google Scholar]
- Keppel G, Buckley YM, Possingham HP. Drivers of lowland rain forest community assembly, species diversity and forest structure on islands in the tropical South Pacific. Journal of Ecology. 2010;98:87–95. [Google Scholar]
- Kissling WD, Carl G. Spatial autocorrelation and the selection of simultaneous autoregressive models. Global Ecology and Biogeography. 2008;17:59–71. [Google Scholar]
- Körner C. Alpine plant life. Springer-Verlag; Berlin: 1999. [Google Scholar]
- Lavergne S, Mouquet N, Thuiller W, Ronce O. Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Annual Review of Ecology, Evolution, and Systematics. 2010;41:321–350. [Google Scholar]
- Legendre P, Legendre L. Numerical ecology. 2nd edn Elsevier Science; Amsterdam: 1998. [Google Scholar]
- Leibold MA, Mikkelson GM. Coherence, species turnover, and boundary clumping: elements of metacommunity structure. Oikos. 2002;97:237–250. [Google Scholar]
- Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, Holt RD, Shurin JB, Law R, Tilman D, Loreau M, Gonzalez A. The metacommunity concept: a framework for multi-scale community ecology. Ecology Letters. 2004;7:601–613. [Google Scholar]
- Lewinsohn TM, Prado PI, Jordano P, Bascompte J, Olesen JM. Structure in plant–animal interaction assemblages. Oikos. 2006;113:174–184. [Google Scholar]
- Logue JB, Mouquet N, Peter H, Hillebrand H, The Metacommunity Working Group Empirical approaches to metacommunities: a review and comparison with theory. Trends in Ecology and Evolution. 2011;26:482–491. doi: 10.1016/j.tree.2011.04.009. [DOI] [PubMed] [Google Scholar]
- López-González C, Presley SJ, Lozano A, Stevens RD, Higgins CL. Metacommunity analysis of Mexican bats: environmentally mediated structure in an area of high geographic and environmental complexity. Journal of Biogeography. 2012;39:177–192. [Google Scholar]
- Lundholm JT. Plant species diversity and environmental heterogeneity: spatial scale and competing hypotheses. Journal of Vegetation Science. 2009;20:377–391. [Google Scholar]
- Meynard CN, Quinn JF. Bird metacommunities in the temperate forests of South America: direct and indirect effects of vegetation structure, area and climate. Ecology. 2008;89:981–990. doi: 10.1890/07-0350.1. [DOI] [PubMed] [Google Scholar]
- Meynard CN, Devictor V, Mouillot D, Thuiller W, Jiguet F, Mouquet N. Beyond taxonomic diversity patterns: how do α, β and υ components of bird functional and phylogenetic diversity respond to environmental gradients across France? Global Ecology and Biogeography. 2011;20:893–903. [Google Scholar]
- Mouquet N, Loreau M. Coexistence in metacommunities: the regional similarity hypothesis. The American Naturalist. 2002;159:420–426. doi: 10.1086/338996. [DOI] [PubMed] [Google Scholar]
- Mouquet N, Devictor V, Meynard CN, et al. Ecophylogenetics: advances and perspectives. Biological Reviews. 2012;87:769–785. doi: 10.1111/j.1469-185X.2012.00224.x. [DOI] [PubMed] [Google Scholar]
- Münkemüller T, de Bello F, Meynard CN, Gravel D, Lavergne S, Mouillot D, Mouquet N, Thuiller W. From diversity indices to community assembly processes: a test with simulated data. Ecography. 2012;35:468–480. [Google Scholar]
- Nagelkerke NJD. A note on a general definition of the coefficient of determination. Biometrika. 1991;78:691–692. [Google Scholar]
- Presley SJ, Willig MR. Bat metacommunity structure on Caribbean islands and the role of endemics. Global Ecology and Biogeography. 2010;19:185–199. [Google Scholar]
- Presley SJ, Higgins CL, López-González C, Stevens RD. Elements of metacommunity structure of Paraguayan bats: multiple gradients require analysis of multiple ordination axes. Oecologia. 2009;160:781–793. doi: 10.1007/s00442-009-1341-x. [DOI] [PubMed] [Google Scholar]
- Presley SJ, Higgins CL, Willig MR. A comprehensive framework for the evaluation of metacommunity structure. Oikos. 2010;119:908–917. [Google Scholar]
- R Development Core Team . R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2011. [Google Scholar]
- Rahbek C, Gotelli NJ, Colwell RK, Entsminger GL, Rangel T, Graves GR. Predicting continental-scale patterns of bird species richness with spatially explicit models. Proceedings of the Royal Society B: Biological Sciences. 2007;274:165–174. doi: 10.1098/rspb.2006.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmida A, Wilson MV. Biological determinants of species diversity. Journal of Biogeography. 1985;12:1–20. [Google Scholar]
- Tamme R, Hiiesalu I, Laanisto L, Szava-Kovats R, Partel M. Environmental heterogeneity, species diversity and co-existence at different spatial scales. Journal of Vegetation Science. 2010;21:796–801. [Google Scholar]
- Thuiller W, Midgley GF, Rouget M, Cowling RM. Predicting patterns of plant species richness in megadiverse South Africa. Ecography. 2006;29:733–744. [Google Scholar]
- Tuomisto H, Ruokolainen K. Analyzing or explaining beta diversity? Understanding the targets of different methods of analysis. Ecology. 2006;87:2697–2708. doi: 10.1890/0012-9658(2006)87[2697:aoebdu]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Tuomisto H, Ruokolainen K, Yli-Halla M. Dispersal, environment, and floristic variation of western Amazonian forests. Science. 2003;299:241–244. doi: 10.1126/science.1078037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.