Abstract
Claudin proteins are a major component of the tight junctions. Dysregulation of claudin protein expression has been described in a number of malignancies. Gene expression profiling has stratified breast cancers into distinct molecular subtypes: luminal, HER2+ and basal-like. Recently, a novel claudin-low molecular subtype has been described. In this study we correlated the expression patterns of claudins with the molecular subtypes of breast cancer. On the basis of immunohistochemical expression 226 grade 3 invasive ductal carcinomas were stratified into 65 luminal (ER+), 65 HER2 positive (HER2+), 86 basal-like, including 14 metaplastic carcinomas (ER−, HER2−, CK5/6 and /or EGFR+), and 10 unclassified. Tissue microarrays were analyzed for expression of claudins 1, 3, 4, 7 and 8 by immunohistochemistry and scored semiquantitatively. High levels of expression were detected in 17% of all cases for claudin 1, 32% claudin 3, 41% claudin 4, 44% claudin 7, and 40% claudin 8. Luminal cancers exhibited increased claudins 7 and 8; basal-like tumors demonstrated increased claudins 1 and 4 expression. Low expression of all five claudins was detected in 30 of 226 cases (13%) and this group was designated “claudin-low”. The majority of the claudin-low subgroup were basal-like cancers (23 of 30, 77%). In contrast, only 1 of 30 (3%) claudin-low tumors were of the luminal phenotype and 6 of 30 cases (20%) were HER2+ (P<0.001). Within the basal-like subgroup, 64% of the metaplastic and 19% of the non-metaplastic tumors were claudin-low. The claudin-low group was strongly associated with disease recurrence (P=0.0093). In conclusion, this study is the first to comprehensively examine the differential expression of claudins 1, 3, 4, 7 and 8 in the molecular subtypes of high grade breast cancer. Claudin-low subtype is a frequent phenomenon in metaplastic and basal-like breast cancer and appears to be a strong predictor of disease recurrence.
Keywords: Breast, Carcinoma, Claudin
Introduction
Breast cancer is the most common type of female cancer and represents 14% of cancer-related deaths in women (1). It has been estimated that there will be about 226,870 new cases of breast cancer in United States in 2012, and about 39,510 women will die from this disease (1). Breast cancer is a genetically heterogeneous group of tumors with a variety of morphologic features. Global gene expression profiling studies have produced a new molecular classification of breast cancer with 4 distinct subtypes : lumunal, HER2, basal-like, and unclassified (2, 3). Each of these subtypes has unique biologic and prognostic features. As microarray gene expression analysis is not routinely available, immunohistochemical surrogates have been developed which approximate the molecular subtypes of breast cancer (4, 5). Based on an immunohistochemical panel of 4 markers (estrogen receptor (ER), HER2, epidermal growth factor receptor (EGFR), and cytokeratin 5/6), breast cancers may be stratified into ER+/HER2− (luminal), HER2+/any ER (HER2 enriched), ER−/HER2−/EGFR+ and/or cytokeratin 5/6+ (basal like), and ER−/HER2−/EGFR−/cytokeratin 5/6− (unclassified) (4). Recent gene expression studies have identified a novel claudin-low subtype of breast cancer (6). This claudin-low subgroup has been proposed to be related to metaplastic cancer, epithelial-mesenchymal transition and poor prognosis (7–9).
Claudins are members of a large family of tight junction proteins which regulate cellular adhesion, polarity, and glandular differentiation. There are currently at least 24 different claudins known to exist in humans, and the expression of each appears to be tissue specific (10, 11). Claudin expression is frequently altered in several cancers (12–14). Because of the high specificity of claudin expression patterns in cancer tissue, it has been proposed that claudins may represent useful diagnostic and prognostic markers in gastrointestinal, genitourinary, and pulmonary neoplasms (15–21).
Comparatively few studies have examined the expression of claudins in breast cancer. Claudin 7 has been reported to be down-regulated in high grade invasive ductal carcinomas, in the ER negative and basal-like molecular subtypes (22–24). In contrast, claudins 3, and 4 have been found to be overexpressed in breast cancer at both the mRNA and protein level (25–28).
In this study we examined the expression of claudins 1, 3, 4, 7, and 8 in a cohort of patients with grade 3 breast carcinoma representing the different molecular subtypes of breast cancer in order to identify and better characterize the claudin-low subgroup.
Materials and Methods
Tissue selection
Tissue samples of grade 3 invasive ductal carcinoma from 226 consecutive patients aged 27 to 96 years were collected between the years 1996 and 2009 from the archives of the Departments of Pathology at the Rhode Island Hospital and The Miriam Hospital. The cohort of grade 3 tumors was selected based on the fact that claudin-low breast carcinomas are mainly histologically grade 3 tumors (9). None of these patients received neoadjuvant chemotherapy or radiotherapy prior to surgery. After surgery, 100 (44%) of patients received chemotherapy alone, 20 (9%) received hormonal therapy alone, and 36 (16%) received both chemotherapy and hormonal therapy. This study was approved by the institutional review board at the Rhode Island Hospital. The histologic grade was previously determined according to the Nottingham modification of the Bloom-Richardson scoring system (29) and confirmed independently by 2 pathologists (E. Y. and K. S.). Stage of disease was defined according to the American Joint Committee on Cancer (30). Based on the immunohistochemical expression of ER, HER2, EGFR, and cytokeratin 5/6, the tumors were stratified into 65 luminal (ER+/HER2−/any EGFR and/or cytokeratin 5/6), 65 HER2 enriched (HER2+/ any ER), 86 basal like (ER−, HER2−, CK5/6, and /or EGFR+), and 10 unclassified (ER−/HER2−/EGFR−/cytokeratin 5/6−) according to Nielsen et al (4). The basal-like group included 14 metaplastic carcinomas, of which 6 had a squamous component, 5 spindle sarcomatous areas and 7 with heterologous elements (5 chondroid and 2 osseous). Four tumors contained two or more components.
Tissue microarray construction
Paraffin blocks containing representative tumor areas were identified on corresponding hematoxylin-eosin–stained sections. Areas of interest were identified and marked on the source block. The source block was cored, and a 1-mm core was transferred to the recipient “master block” using the Beecher Tissue Microarrayer (Beecher Instruments, Silver Spring, MD). Five representative cores of tumor and 2 cores of normal breast tissue were arrayed per specimen.
Immunohistochemical staining
Immunohistochemical staining was performed according to the following protocol. Sections from paraffin-embedded tissue microarrays were cut at 4 µm, deparaffinized, and rehydrated with xylene and graded alcohols. Microwave epitope retrieval was performed in target retrieval pH 6.0 (DAKO, Carpinteria, CA) for ER, and HER2; high pH target retrieval for cytokeratin 5/6 (DAKO); or 10 mM citrate buffer (pH 6.0) for 10 minutes followed by cooling for 15 minutes at room temperature for claudins. The following primary antibodies were used: clone ER1D5 against ER (1:300 dilution; DAKO), clone CB11 against HER2 (1:150 dilution; Vector Laboratories), clone D5/16B4 against cytokeratin 5/6 (1:40 dilution; Cell Marque, Rocklin, CA), rabbit polyclonal antibodies against claudin-1 (1:50 dilution, NeoMarkers, Lab Vision, Fremont, CA, USA), claudin-3 (1:1000, NeoMarkers), claudin-4 (1:1000, NeoMarkers), claudin-7 (1:500, NeoMarkers), and claudin-8 (1:100, GeneTex Inc., San Antonio, TX, USA). Appropriate positive and negative controls were used simultaneously with test slides. Positive controls consisted of normal colonic mucosa (15). Normal rabbit serum (sc-2338; Santa Cruz Biotechnology, Inc, Santa Cruz, CA) was used as the negative control for anti-claudin antibodies.
Immunohistochemical staining for ER and HER2 was performed using the DAKO Autostainer Plus and EnVision Dual Link detection reagent (DAKO) with DAB (DAKO). Immunohistochemical staining for claudins 3, 4, 7, and 8 was performed using the Envision Plus kit (Dako) and for claudin-1 using the Ventana Discovery automated staining system employing the Ventana Basic DAB detection kit and endogenous biotin-blocking kit (Ventana Medical Systems, Tucson, AZ). EGFR was stained using the PharmDX kit (DAKO) according to manufacturer's instructions. For HER2 fluorescent in situ hybridization assay, slides were hybridized with probes to LSI HER2/neu and CEP 17 with the PathVysion HER-2 DNA Probe Kit (Abbott Molecular, Inc, Des Plaines, IL, USA) according to the manufacturer's instructions. Slides were counterstained with 4',6-diamidino-2-phenylindole and visualized on a Zeiss Axioplan epifluorescent microscope (Zeiss, Baden-Wurttemberg, Germany).
Staining results were assessed by two pathologists (E. Y. and S. L.) in a blinded fashion. ER stains were considered positive if immunostaining was seen in more than 1% of tumor nuclei. EGFR stains were considered positive if any (weak or strong) membranous invasive carcinoma cell staining was observed. CK5/6 stains were scored positive if any (weak or strong) cytoplasmic and/or membranous staining was detected in the tumor cells. For HER2 status, tumors were considered positive if scored as 3+ according to the guidelines of the American Society of Clinical Oncology/College of American Pathologists (31), and fluorescent in situ hybridization with amplification ratio 2.2 or more was used to segregate immunohistochemically equivocal (2+) results. Claudin immunoreactivity was assessed based on a combined score of the extent and intensity of staining. Scores 0–3 were assigned according to the percentage of positive tumor cells (0=0%; 1=<25%; 2=25–50%; 3=>51%) and the intensity of staining in tumor (0=0; 1=1+; 2=2+; 3=3+) as previously described (18). The two scores were multiplied to give an overall score of 0–9, of which 0 was considered negative, 1–2 was considered weak, 3–6 moderate, and 9 strong staining (16). Negative and weak expression was considered as low, whereas moderate and strong as high. Tumors with low expression of all five claudins were defined as claudin-low. Any discordant scores were reviewed together by both scorers to obtain a consensus score.
Statistical methods
The χ2 analysis was used to assess the associations between the expression of claudins and molecular subtypes. Hierarchical clustering was used to detect the association between claudins and ER, HER2, EGFR, and CK5/6 expression by centroid method with data standardization. Complete method with data standardization was used for hierarchical clustering of histologic features of claudin-low breast cancer. Overall survival (OS) time was calculated from the time of diagnosis until the time of death. The method of Kaplan-Meier was used to generate OS curves, and curves were compared using a log-rank test. The prognostic significance of claudin expression and the other clinical variables including tumor size, lymph node status, presence of metastasis, and American Joint Committee on Cancer (AJCC) stage was determined using a univariate Cox proportional hazards model. Multivariate analysis was done using a multivariate Cox proportional hazards model including the following variables: age, claudin expression, tumor molecular subtype, AJCC stage, and systemic adjuvant therapy. All tests were 2 sided with 0.05 as the threshold to be considered statistically significant. All analyses were performed using SAS software, JMP Base version 8.0.0 (SAS, Cary, NC, USA).
Results
Clinicopathologic characteristics
The clinicopathological characteristics of the patients with grade 3 invasive ductal carcinomas are summarized in Table 1. One hundred two patients (45%) presented with T2 or larger tumors; 71 (31%), with nodal involvement; and 6 (3%), with metastatic disease. Twenty six patients (12%) had advanced stage (3–4) disease at diagnosis (defined according to the AJCC). Clinical follow-up was available for all cases. Median follow-up was 52.5 months (range, 1–211.6 months).
Table 1.
Clinicopathologic Characteristics of Patients with Different Molecular Subtypes of Breast Carcinoma
| Parameter | Total | Molecular subtype | |||
|---|---|---|---|---|---|
| Luminal | HER2 | Basal like | Unclassified | ||
| Sample size (%) | 226 | 65 (29%) | 65 (29%) | 86 (38%) | 10 (4%) |
| Mean age (y) ± SD | 61 ± 15 | 64 ± 16 | 61 ± 15 | 59 ± 15 | 65 ± 12 |
| Race | |||||
| White | 202 (90%) | 62 (95%) | 59 (92%) | 73 (85%) | 9 (90%) |
| African American | 24 (10%) | 3 (5%) | 5 (8%) | 13 (15%) | 1 (10%) |
| AJCC stage | |||||
| Stage 1 | 91 (41%) | 20 (31%) | 30 (46%) | 35 (41%) | 6 (60%) |
| Stage 2 | 95 (42%) | 31 (470%) | 24 (37%) | 37 (43%) | 3 (30%) |
| Stage 3 | 19 (9%) | 7 (11%) | 4 (6%) | 7 (8%) | 1 (10%) |
| Stage 4 | 7 (3%) | 3 (5%) | 1 (2%) | 3 (3%) | 0 |
| Tumor status | |||||
| T1 | 120 (53%) | 26 (40%) | 38 (58%) | 48 (56%) | 8 (80%) |
| T2 | 84 (37%) | 29 (45%) | 21 (32%) | 33 (38%) | 1 (10%) |
| T3 | 9 (4%) | 3 (5%) | 2 (3%) | 3 (4%) | 1 (10%) |
| T4 | 9 (4%) | 6 (8%) | 1 (2%) | 2 (2%) | 0 |
| Node status | |||||
| N0 | 117 (52%) | 28 (44%) | 36 (58%) | 46 (53%) | 7 (70%) |
| N1 | 55 (24%) | 15 (23%) | 18 (27%) | 20 (23%) | 2 (20%) |
| N2 | 11 (5%) | 4 (6%) | 1 (2%) | 5 (6%) | 1 (10%) |
| N3 | 5 (2%) | 2 (3%) | 2 (3%) | 1 (1%) | 0 |
| Nx | 38 (17%) | 16 (25%) | 8 (12%) | 14 (16%) | 0 |
| Metastasis status | |||||
| M0 | 215 (95%) | 62 (95%) | 63 (96%) | 80 (93%) | 8 (80%) |
| M1 | 6 (3%) | 3 (5%) | 1 (2%) | 2 (2%) | 0 |
| Mx | 5 (2%) | 0 | 1 (2%) | 4 (5%) | 2 (20%) |
Expression of claudins in breast cancer
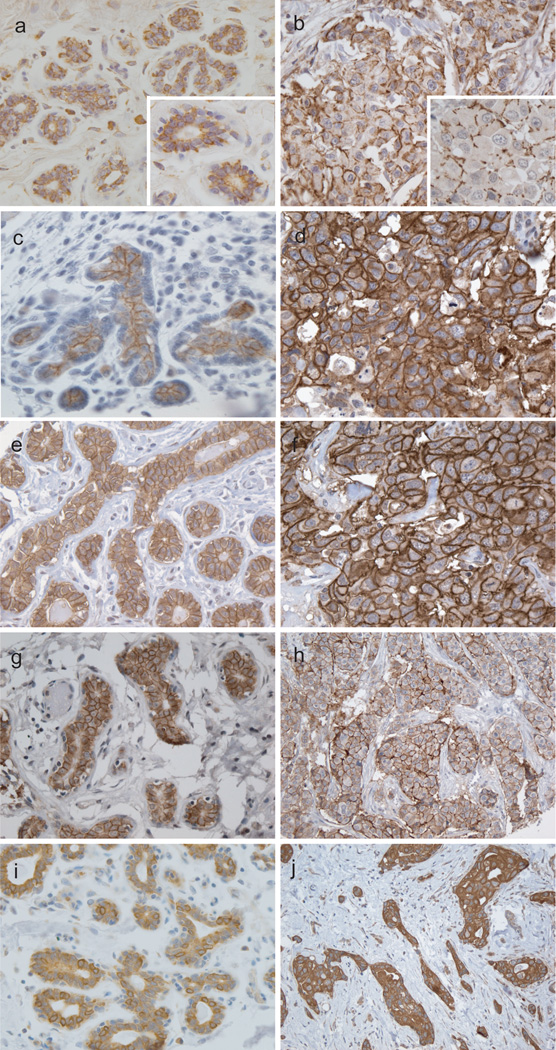
The normal breast ductal and lobular epithelium expressed claudins 1, 3, 4, 7 and 8 (Figure 1). In normal breast tissue all of the claudins exhibited predominantly an apicolateral membranous staining pattern of luminal cells with no expression by myoepithelial cells. In invasive ductal carcinoma cases claudin-1 demonstrated a predominantly punctate membranous staining pattern, whereas claudins 3, 4, 7, and 8 staining varied from a punctate to complete circumferential membranous staining with no preservation of the apicolateral polarity seen in normal breast epithelium.
Figure 1.
Expression of claudins in normal breast (A, C, E, G, and I) and invasive ductal carcinoma (B, D, F, H and J). Claudin 1 is expressed in the majority of luminal cells of terminal ductal lobular unit (A) and has scattered pattern in a basal-like carcinoma (B). Claudin 3 is expressed in the apicolateral side of the normal luminal epithelium (C) and the polarity is lost in a basal-like cancer (D). Predominantly apicolateral Claudin 4 expression in normal breast (E) and complete circumferential membranous staining in a basal-like carcinoma (F). Claudin 7 and 8 is also apicolateral in normal gland (G, I) and circumferential membranous in luminal subtype tumors (H, J).
High levels of expression were detected in 17% of all tumors for claudin 1, 32% claudin 3, 41% claudin 4, 44% claudin 7, and 40% for claudin 8. The frequency of high-level claudin 1 expression was approximately 50% that of other claudin types. Based on the protein expression of claudins, ER, HER2, CK5/6, and EGFR, a hierarchical clustering analysis was performed and the results provided a global overview of the relationship among these markers (Figure 2). Cases with high expression of claudins 1, 3 and 4 clustered in one group with cases positive for basal markers CK5/6 and EGFR. In contrast, tumors with high claudin 7 and 8 expression clustered with ER+ neoplasms. HER2+ tumors did not cluster with any of claudins.
Figure 2.
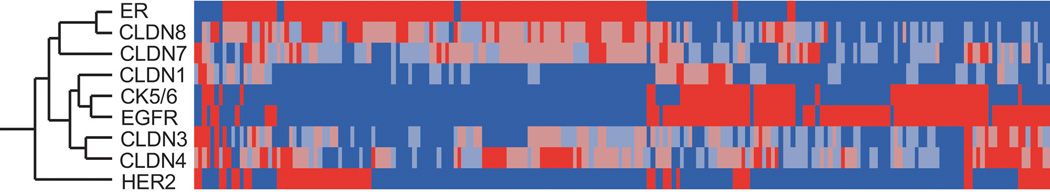
Hierarchical cluster analysis (Centroid method with data standardization) of claudin 1, 3, 4, 7, and 8 expressions and ER, HER2, CK5/6, and EGFR. Each column represents a different tumor, and each row represents a marker. Red: highest expression; blue: lowest expression. The analysis shows that Claudins 1, 3, and 4 clustered predominantly with cases positive for basal markers CK5/6 and EGFR, as indicated by short dendrogram branches linking these markers. Claudins 7 and 8 clustered with ER positive tumors.
While cluster analysis provided a comprehensive overview, correlation analysis revealed significant association between these markers (Table 2). When tumors were categorized into low (negative and weak expression) and high (moderate and strong expression) for each claudin, there were strong positive associations between claudin 1 and CK5/6 and EGFR (P=0.01 and 0.004, respectively) and significant negative association between claudin 1 and ER (P=0.0001). In addition, claudin 4 expression was positively associated with the expression of EGFR (P = 0.0306) and negatively associated with ER (P = 0.0125). In contrast to claudin 1 and 4, claudin 7 and 8 showed a strong direct association with ER expression (P=0.0028 and <0.0001, respectively) and negative association with CK5/6 (P=0.0084 and P=0.0072, respectively) and EGFR expression (P=0.0001 and P<0.0001, respectively).
Table 2.
Contingency analysis of claudin expression and other markers based on immunohistochemical expression.
| P-value | ER | HER2 | CK5/6 | EGFR |
|---|---|---|---|---|
| CLDN1 | 0.0001 | N.S | 0.01 | 0.004 |
| CLDN3 | N.S. | N.S | N.S. | N.S. |
| CLDN4 | 0.0125 | N.S | N.S. | 0.0306 |
| CLDN7 | 0.0028 | N.S. | 0.0084 | 0.0001 |
| CLDN8 | <0.0001 | N.S. | 0.0072 | <0.0001 |
Italic: Positive correlation; Bold: Negative correlation; N.S.: No significance (P>0.05).
Expression of claudins in different molecular subtypes of breast cancer
The relationship between claudins and ER, EGFR, and CK5/6 expression prompted us to assess claudin expression in the different molecular subtypes of breast cancer. A Chi-square analysis revealed that the luminal subtype contains significantly more tumors that were claudin 7 positive (45%, P < 0.0001) and claudin 8 positive (49%, P <0.0001, Table 3). In contrast, high levels of claudin 1 were observed in the basal-like subtype (63%, P=0.0132). Claudin 4 positive tumors demonstrated a tendency of association with basal-like tumors (40%).
Table 3.
Expression of Claudins in Different Molecular Subtypes
| Molecular Subtype | P value | |||||
|---|---|---|---|---|---|---|
| Total | Luminal | HER2 | Basal-like | Negative | ||
| Claudin 1 | 0.0132 | |||||
| Low | 186 (84%) | 54 (30%) | 53 (30%) | 62 (35%) | 9 (5%) | |
| High | 36 (16%) | 3 (9%) | 8 (25%) | 20 (63%) | 1 (3%) | |
| Claudin 3 | 0.9103 | |||||
| Low | 148 (67%) | 38 (27%) | 41 (29%) | 58 (41%) | 5 (4%) | |
| High | 73 (33%) | 19 (28%) | 21 (31%) | 24 (36%) | 3 (5%) | |
| Claudin 4 | 0.2944 | |||||
| Low | 128 (59%) | 38 (31%) | 32 (26%) | 47 (39%) | 4 (3%) | |
| High | 90 (41%) | 18 (21%) | 29 (34%) | 34 (40%) | 5 (6%) | |
| Claudin 7 | <0.0001 | |||||
| Low | 115 (57%) | 18 (17%) | 30 (28%) | 58 (54%) | 1 (1%) | |
| High | 87 (43%) | 37 (45%) | 21 (25%) | 19 (23%) | 6 (7%) | |
| Claudin 8 | <0.0001 | |||||
| Low | 129 (61%) | 20 (17%) | 27 (23%) | 66 (56%) | 6 (5%) | |
| High | 82 (39%) | 39 (49%) | 25 (31%) | 15 (19%) | 1 (1%) | |
| Claudins | <0.0001 | |||||
| All low | 30 (16%) | 1(3%) | 6(20%) | 23 (77%) | 0 | |
| Non-low | 190 (84%) | 64 (34%) | 59 (31%) | 63 (33%) | 10 (5%) | |
Identification of a claudin-low group
Low levels of expression (0–1+) were detected in 84%, 67%, 59%, 57%, and 61% of tumors stained for claudins 1, 3, 4, 7, and 8, respectively. Low expression of all five claudins was observed in 30 of 226 cases (14%) and this group was designated as claudin-low. The majority of the claudin-low group were basal-like cancers (23 of 30, 77%). In contrast, only 1 of 30 (3.3%) tumors was of the luminal phenotype and 6 of 30 (20%) cases were HER2+ (P<0.0001). Within the basal-like subgroup, 9 of 14 (64%) of the metaplastic and 14 of 72 (19%) of the non-metaplastic tumors were claudin-low.
Histologic features of claudin-low breast cancers
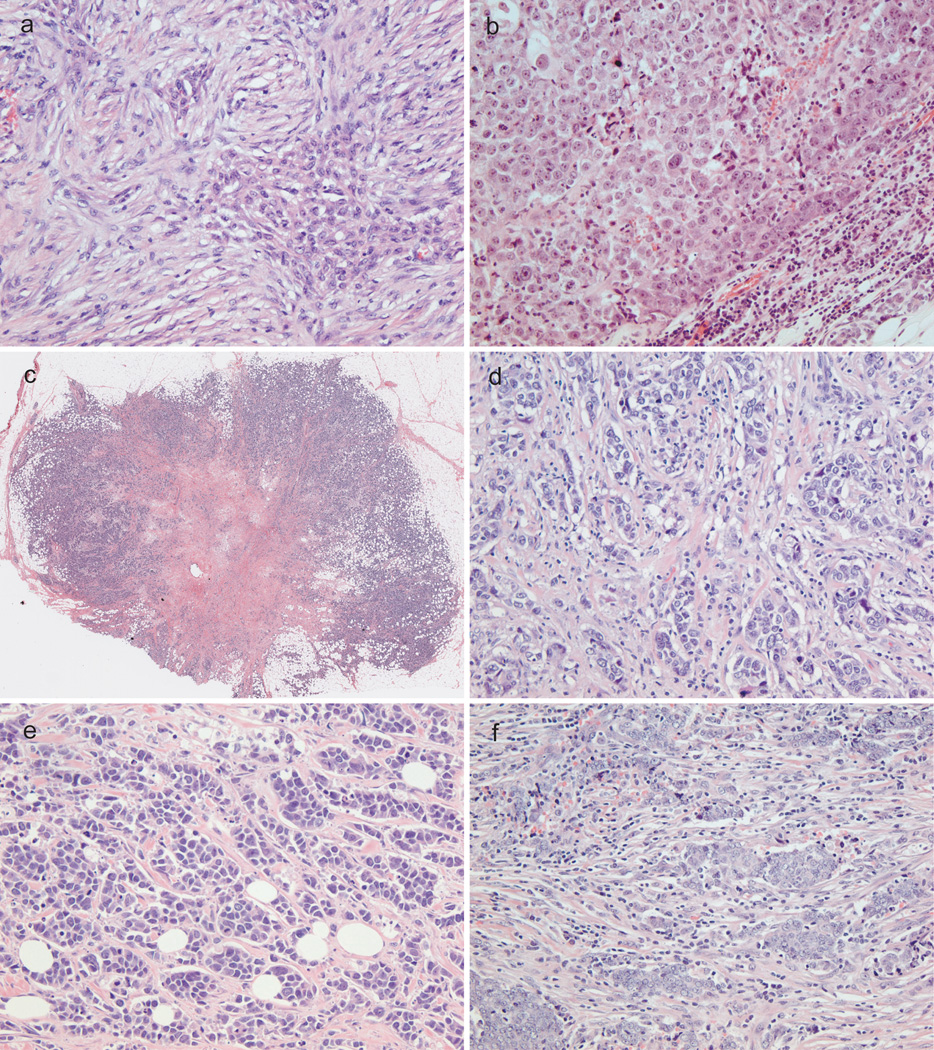
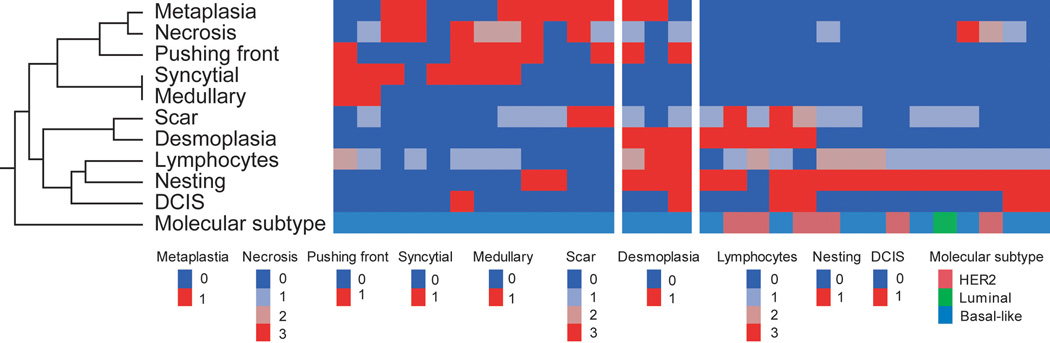
Given the findings that claudin-low tumors are related to the basal-like subtype carcinoma, we more closely examined the histology of these tumors (Figure 3). In order to quantify our observation, morphologic features were rated and mathematically clustered (Figure 4). Two main histologic subtypes of claudin-low tumors were identified. The first subtype consisted of 12 cases clustered exclusively with the basal-like group. These tumors showed metaplasia, necrosis, pushing border, as well as syncytial and medullary features. The second type contained 15 cases including basal-like, luminal, and HER2 subtypes, and was characterized by nesting growth pattern with desmoplasia, scar, lymphocytic infiltrate, and associated DCIS. Three cases shared histologic features between these two groups.
Figure 3.
Histologic features of claudin-low breast cancer. (a) Metaplastic cancer. (b) Medullary growth pattern. (c) Central scar. (d) Desmoplastic stromal reaction. (e) Nesting growth pattern. (f) Lymphocytic infiltrate.
Figure 4.
Hierarchical cluster analysis (Complete method with data standardization) of histologic features of claudin-low cancer. Each column represents a different tumor, and each row represents a histologic feature. Red: highest expression; blue: lowest expression. The analysis shows two main groups.
Of note, the vast majority (6 of 7, 86%) of metaplastic tumors with chondroid or osseous heterologous elements were claudin-low. In contrast, squamous differentiation was not a feature of claudin-low tumors. Only two of 9 claudin-low metaplastic tumors contained squamous components as opposed to 4 of 5 claudin non-low metaplastic tumors.
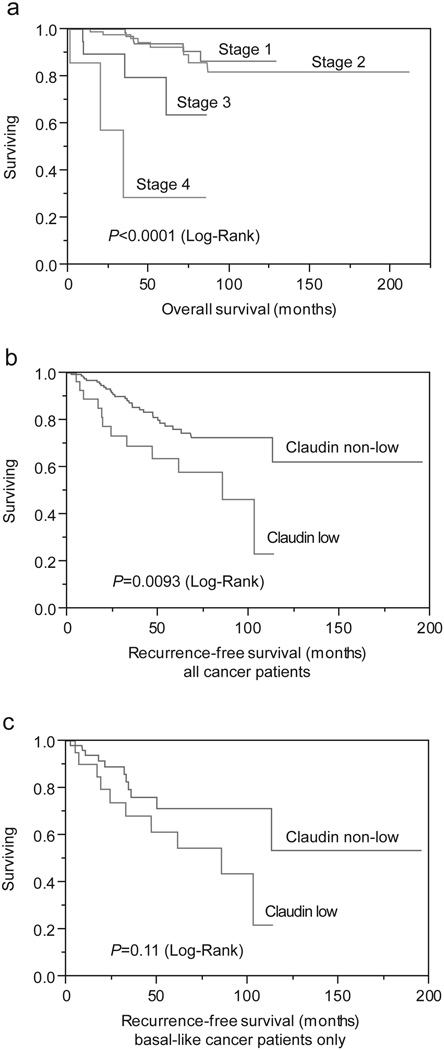
Claudin expression and survival
Univariate analysis of survival was performed in order to evaluate the impact of conventional prognostic predictors and claudins on patient survival. Kaplan-Meier survival curves were constructed, followed by the log-rank test. Univariate analysis revealed that tumor stage significantly influenced overall patient survival (P<0.0001, Figure 5a). Patients with claudin-low tumors had significantly worse recurrent free survival than the other patients (P=0.0093, Figure 5b). Within the basal-like group patients with claudin-low tumors also showed worse recurrent free survival as opposed to claudin-high tumors (P=0.11, Figure 5c). Multivariate analysis of survival revealed that the tumor stage (P=0.0025), and molecular subtype were the only independent predictors of survival (P=0.0025 and P=0.0345, respectively). Claudin expression was not an independent predictor of survival by multivariate analysis.
Figure 5.
Analysis of overall (a) and recurrence-free survival (b,c) in grade 3 invasive ductal carcinoma by stage (a), claudin-low expression in the entire cohort (b), and claudin-low expression in basal-like subtype only (c).
Discussion
Global gene expression studies identified multiple molecular subtypes of breast cancer including luminal, HER2-enriched, basal-like and normal breast-like. The subtypes of these tumors have significant differences in incidence, survival, and response to therapy. Recent genomic studies have identified a novel claudin-low molecular subtype of breast cancer (6). In this study we evaluated the expression patterns of claudins 1, 3, 4, 7, and 8 in different molecular subtypes of grade 3 breast cancer. Selection of claudins evaluated in this study was based on molecular evidence describing low gene expression of claudins 3, 4, and 7 in the claudin-low subtype (9). Claudin 1 and 8 were selected based on our previous observations of decreased expression of these tight junction proteins in colonic, gastric, and renal carcinomas (15, 16, 18).
Claudins are pivotal proteins in the formation and function of tight junctions. In normal epithelia the primary functions of the tight junctions are to seal the apical intercellular spaces of glandular epithelia creating a barrier against the paracellular passage of macromolecules and to separate the plasma membrane into the apical and basolateral domains (32, 33). There are currently at least 24 known members of the claudin family, which are expressed in a tissue-specific pattern in normal epithelia (10, 11). Neoplastic cells frequently exhibit structural and functional deficiencies in the tight junctions. Disruption of the tight junctions is believed to be one of processes that occur in carcinogenesis allowing for loss of cellular cohesion, aggressive growth, and differentiation of cancer cells (12, 34). This hypothesis is supported by loss of claudin expression in several cancers, including claudin 1 in colorectal carcinoma (15), claudins 1 and 7 in prostatic carcinoma (19), and claudins 3 and 4 in esophageal carcinoma (35). However, some malignant neoplasms do not follow this rule and demonstrate paradoxical overexpression of claudins. Our group has previously reported significantly stronger expression of claudin 4 in colonic and gastric adenocarcinoma as opposed to normal mucosa (15, 16). Upregulation of claudins 3 and 4 has been observed in ovarian cancer, but not in benign ovarian cystadenomas (36). Increased claudins 3 and 4 expression has been recently shown to contribute to the aggressive behavior of clear cell and serous papillary endometrial carcinoma (37).
Our findings demonstrate that expression of claudins in invasive ductal carcinoma of the breast is not only tissue specific, but is also unique in particular molecular subtypes of breast cancer. Luminal cancers appear to exhibit ER+/claudins 7, 8 high/ claudins 1, 4 low signature and therefore claudins 7 and 8 may be designated as “luminal”. In contrast, basal-like cancers demonstrate ER−/HER2−/ claudins 1, 4 high/ claudins 7, 8 low phenotype, and claudins 1 and 4 may be considered as “basal” claudins. To date, only a few studies have addressed the role of claudins in breast carcinoma. Studies of claudin 3 and 4 expression in invasive ductal carcinoma remain controversial. In one of the early studies Soini observed strong expression of claudins 3 and 4 in invasive ductal carcinoma; however, he did not find an association between claudin 3 and 4 expression and tumor grade or estrogen receptor status (38). Tokes et al reported significant loss of claudin 4 mRNA and protein expression in the majority of grade 1 ductal carcinomas, but increased expression in grade 2 and 3 tumors (39). Our findings of an association of claudins 1 and 4 expression with estrogen receptor negative tumors are in complete agreement with several recent reports (26, 27). Lanigan et al described the association of increased claudin 4 expression with high tumor grade, ER negative tumor status, and poor prognosis (26). Blanchard et al demonstrated correlation of claudins 1 and 4 with the basal-like subtype (27). Similar findings on claudin 4 overexpression in basal-like subtype of breast cancer have been reported by Kulka et al (28).
Only a few studies have examined the expression of claudin 7 in breast carcinomas. Kominsky et al reported loss of claudin 7 in high-grade invasive ductal carcinoma (22). Upon analyzing data of claudin 7 mRNA expression quantitated by real-time PCR from this study, in which breast cancer cell lines were used, we found that all ER− cell lines showed no detectable claudin 7 mRNA, while all ER+ cell lines showed detectable claudin 7 expression. In parallel with our data, reduced immunohistochemical expression of claudin 7 was associated with ER− breast carcinomas (23), and was less frequent in the triple negative group (24). Here we present, for the first time, immunohistochemical evidence of claudin 8 expression in invasive ductal carcinoma and demonstrated its association with the ER+ luminal subtype of breast cancer.
In this study we defined a group of tumors negative for all 5 claudins tested and demonstrated that this claudin-low group is strongly associated with basal-like breast cancer and metaplastic cancer. Claudin-low subtype of breast cancer was identified in 2007 by gene expression analysis of breast tumors (6). This group is characterized by low expression of genes involved in tight junctions and cell-cell adhesion, including claudin 3,4, and 7 (9). The claudin-low group is characterized by enrichment of epithelial to mesenchymal transition markers, immune response genes, and cancer stem cell markers. In contrast to other molecular subtypes of breast cancer, claudin-low tumors have been less studied. Recently, Prat et al comprehensively characterized this subtype at the molecular level by hierarchical clustering of approximately 1900 intrinsic genes and found that this group comprises approximately 12 % of breast cancers (9). Our data show similar frequencies of claudin-low group (13%). One would expect higher frequencies in this study group containing only high-grade tumors; however, our criteria required all 5 claudins to be low, thereby decreasing the number of claudin-low cases. As the expression of more claudin subtypes are evaluated in breast cancer, the definition of the claudin low variant will likely need to be modified.
Our results provide evidence that the basal-like subtype of breast cancer is a heterogeneous group and may be further subdivided into subgroup with loss of claudin expression (claudin-low) and a subgroup with gain of claudin expression (claudin-high). Recently, Myal et al discussed the significance of a subset of estrogen receptor negative breast cancers which express high levels of claudin 1 and proposed a possible “claudin high” subset of breast cancer (40). Our results demonstrate that claudin 1 high subgroup is a relatively frequent phenomenon in basal-like breast cancer. In both scenarios of claudin loss or gain it appears that abnormal claudin expression is associated with more aggressive tumor behavior.
In summary, we demonstrated a differential claudin expression in the four molecular subtypes of invasive ductal carcinoma and proposed “luminal” and “basal” claudins. Molecular subtypes of breast cancer are a heterogeneous group and can be further subdivided into a claudin-low group. Claudin-low subtype is a frequent phenomenon in metaplastic and basal-like breast cancer and appears to be a strong predictor of disease recurrence.
Acknowledgments
This study was supported by the Brown University Pathology Pilot Project Funds for Translational Research and the Molecular Pathology Core of the COBRE Center for Cancer Research Development, funded by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM103421. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
This study was presented in part at the annual meeting of the United States and Canadian Academy of Pathology in San Antonio, TX in 2011.
The authors declare no conflict of interest.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 5.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 6.Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taube JH, Herschkowitz JI, Komurov K, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morita K, Furuse M, Fujimoto K, et al. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci U S A. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 12.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 13.Soini Y. Expression of claudins 1, 2, 3, 4, 5 and 7 in various types of tumours. Histopathology. 2005;46:551–560. doi: 10.1111/j.1365-2559.2005.02127.x. [DOI] [PubMed] [Google Scholar]
- 14.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Resnick MB, Konkin T, Routhier J, et al. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol. 2005;18:511–518. doi: 10.1038/modpathol.3800301. [DOI] [PubMed] [Google Scholar]
- 16.Resnick MB, Gavilanez M, Newton E, et al. Claudin expression in gastric adenocarcinomas: a tissue microarray study with prognostic correlation. Hum Pathol. 2005;36:886–892. doi: 10.1016/j.humpath.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 17.Michl P, Barth C, Buchholz M, et al. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 2003;63:6265–6271. [PubMed] [Google Scholar]
- 18.Lechpammer M, Resnick MB, Sabo E, et al. The diagnostic and prognostic utility of claudin expression in renal cell neoplasms. Mod Pathol. 2008;21:1320–1329. doi: 10.1038/modpathol.2008.116. [DOI] [PubMed] [Google Scholar]
- 19.Sheehan GM, Kallakury BV, Sheehan CE, et al. Loss of claudins-1 and-7 and expression of claudins-3 and-4 correlate with prognostic variables in prostatic adenocarcinomas. Hum Pathol. 2007;38:564–569. doi: 10.1016/j.humpath.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Choi YL, Kim J, Kwon MJ, et al. Expression profile of tight junction protein claudin 3 and claudin 4 in ovarian serous adenocarcinoma with prognostic correlation. Histol Histopathol. 2007;22:1185–1195. doi: 10.14670/HH-22.1185. [DOI] [PubMed] [Google Scholar]
- 21.Jung JH, Jung CK, Choi HJ, et al. Diagnostic utility of expression of claudins in non-small cell lung cancer: different expression profiles in squamous cell carcinomas and adenocarcinomas. Pathol Res Pract. 2009;205:409–416. doi: 10.1016/j.prp.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Kominsky SL, Argani P, Korz D, et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–2033. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 23.Park D, Kåresen R, Axcrona U, et al. Expression pattern of adhesion molecules (E-cadherin, alpha-, beta-, gamma-catenin and claudin-7), their influence on survival in primary breast carcinoma, and their corresponding axillary lymph node metastasis. APMIS. 2007;115:52–65. doi: 10.1111/j.1600-0463.2007.apm_524.x. [DOI] [PubMed] [Google Scholar]
- 24.Bernardi MA, Logullo AF, Pasini FS, et al. Prognostic significance of CD24 and claudin-7 immunoexpression in ductal invasive breast cancer. Oncol Rep. 2012;27:28–38. doi: 10.3892/or.2011.1477. [DOI] [PubMed] [Google Scholar]
- 25.Kominsky SL, Vali M, Korz D, et al. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol. 2004;164:1627–1633. doi: 10.1016/S0002-9440(10)63721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanigan F, McKiernan E, Brennan DJ, et al. Increased claudin-4 expression is associated with poor prognosis and high tumour grade in breast cancer. Int J Cancer. 2009;124:2088–2097. doi: 10.1002/ijc.24159. [DOI] [PubMed] [Google Scholar]
- 27.Blanchard AA, Skliris GP, Watson PH, et al. Claudins 1, 3, and 4 protein expression in ER negative breast cancer correlates with markers of the basal phenotype. Virchows Arch. 2009;454:647–656. doi: 10.1007/s00428-009-0770-6. [DOI] [PubMed] [Google Scholar]
- 28.Kulka J, Szász AM, Németh Z, et al. Expression of tight junction protein claudin-4 in basal-like breast carcinomas. Pathol Oncol Res. 2009;15:59–64. doi: 10.1007/s12253-008-9089-x. [DOI] [PubMed] [Google Scholar]
- 29.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 30.Edge SB, Byrd DR, Compton CC. AJCC cancer staging manual. 2010. pp. 347–376. [Google Scholar]
- 31.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 32.Madara JL. Regulation of the movement of solutes across tight junctions. Annu Rev Physiol. 1998;60:143–159. doi: 10.1146/annurev.physiol.60.1.143. [DOI] [PubMed] [Google Scholar]
- 33.Tsukita S, Furuse M. The structure and function of claudins, cell adhesion molecules at tight junctions. Ann N Y Acad Sci. 2000;915:129–135. doi: 10.1111/j.1749-6632.2000.tb05235.x. [DOI] [PubMed] [Google Scholar]
- 34.Escudero-Esparza A, Jiang WG, Martin TA. The Claudin family and its role in cancer and metastasis. Front Biosci. 2011;16:1069–1083. doi: 10.2741/3736. [DOI] [PubMed] [Google Scholar]
- 35.Takala H, Saarnio J, Wiik H, Soini Y. Claudins 1, 3, 4, 5 and 7 in esophageal cancer: loss of claudin 3 and 4 expression is associated with metastatic behavior. APMIS. 2007;115:838–847. doi: 10.1111/j.1600-0463.2007.apm_656.x. [DOI] [PubMed] [Google Scholar]
- 36.Rangel LB, Agarwal R, D'Souza T, et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003;9:2567–2575. [PubMed] [Google Scholar]
- 37.Konecny GE, Agarwal R, Keeney GA, et al. Claudin-3 and claudin-4 expression in serous papillary, clear-cell, and endometrioid endometrial cancer. Gynecol Oncol. 2008;109:263–269. doi: 10.1016/j.ygyno.2008.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soini Y. Claudins 2, 3, 4, and 5 in Paget's disease and breast carcinoma. Hum Pathol. 2004;35:1531–1536. doi: 10.1016/j.humpath.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Tokés AM, Kulka J, Paku S, et al. Claudin-1-3 and-4 proteins and mRNA expression in benign and malignant breast lesions: a research study. Breast Cancer Res. 2005;7:R296–R305. doi: 10.1186/bcr983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myal Y, Leygue E, Blanchard AA. Claudin 1 in breast tumorigenesis: revelation of a possible novel"claudin high" subset of breast cancers. J Biomed Biotechnol. 2010;2010:956897. doi: 10.1155/2010/956897. [DOI] [PMC free article] [PubMed] [Google Scholar]