Abstract
Although plastid sedimentation has long been recognized as important for a plant's perception of gravity, it was recently shown that plastids play an additional function in gravitropism. The Translocon at the Outer envelope membrane of Chloroplasts (TOC) complex transports nuclear-encoded proteins into plastids, and a receptor of this complex, Toc132, was previously hypothesized to contribute to gravitropism either by directly functioning as a gravity signal transducer or by indirectly mediating the plastid localization of a gravity signal transducer. Here we show that mutations in multiple genes encoding TOC complex components affect gravitropism in a genetically sensitized background and that the cytoplasmic acidic domain of Toc132 is not required for its involvement in this process. Furthermore, mutations in TOC132 enhance the gravitropic defect of a mutant whose amyloplasts lack starch. Finally, we show that the levels of several nuclear-encoded root proteins are altered in toc132 mutants. These data suggest that the TOC complex indirectly mediates gravity signal transduction in Arabidopsis and support the idea that plastids are involved in gravitropism not only through their ability to sediment but also as part of the signal transduction mechanism.
Keywords: gravitropism, roots, TOC complex, plastid, signal transduction, Arabidopsis
Introduction
Root gravitropism allows plants to anchor themselves while exploring their environments to gain access to water and nutrients and to avoid obstacles and toxins. In roots, gravity sensing occurs primarily in the columella region of the cap where the cells contain dense, starch-filled amyloplasts that sediment in response to reorientation within the gravity field. Amyloplast sedimentation triggers changes in the localization of plasma membrane-associated auxin efflux facilitators, leading to the accumulation of auxin on the lower side of the root. Upon transmission to the elongation zones, the resulting auxin gradient promotes differential cellular elongation between the upper and lower flanks, resulting in downward curvature. Possible second messengers in this process include Ca2+, inositol 1,4,5-triphosphate, and protons. It is still unknown how amyloplast sedimentation leads to an auxin gradient (reviewed in Strohm et al., 2012).
Previously, we showed that ALTERED RESPONSE TO GRAVITY 1 (ARG1) is a peripheral membrane protein that is necessary for a full gravitropic response (Sedbrook et al., 1999; Boonsirichai et al., 2003). arg1 mutants do not display the characteristic cytoplasmic alkalinization or generate an auxin gradient across the root cap upon reorientation. Expressing ARG1 in the gravity-sensing cells (statocytes) of only the root or the shoot restores gravitropism only in that organ. Together, these data suggest that ARG1 functions in the statocytes in the early phases of gravity signal transduction. ARG1 localizes to some of the same components of the vesicle trafficking pathway as the PIN auxin efflux carriers and therefore may affect their localization or activity. However, GFP-ARG1 signal is absent from plastids (Boonsirichai et al., 2003). Because arg1 single mutants display only a partial gravitropic defect and still respond slowly to reorientation, they were used in an enhancer screen. This approach identified MODIFIERS OF ARG1 1 and 2 (MAR1 and 2), which encode components of the Translocon at the Outer envelope membrane of Chloroplasts (TOC) complex (Stanga et al., 2009). Although mar single mutants grow normally, arg1 mar double mutants show no response to gravity (Stanga et al., 2009).
TOC complexes transport nuclear-encoded proteins into plastids. These complexes consist of a pore (Toc75/MAR1), a Toc159 family receptor (Toc159, Toc132/MAR2, Toc120, or Toc90), and a Toc34 family receptor (Toc33 or Toc34/PPI3). The Toc159 family members contain an N-terminal cytoplasmic acidic domain, a GTP-binding domain, and a C-terminal membrane domain. Most of the variation between these family members occurs within the acidic domain, which has been implicated in substrate selectivity (Inoue et al., 2010). The Toc34/PPI3 members contain only the GTP-binding and membrane domains. Toc132, Toc120, Toc34, and Toc75 are thought to assemble into complexes that tend to import plastid-associated proteins not directly involved in photosynthesis, whereas the import of photosynthesis-related proteins into chloroplasts seems to be mediated mainly by complexes that include Toc159, Toc33, and Toc75 (Ivanova et al., 2004; Kubis et al., 2004).
Because ARG1 and the TOC complex localize to different parts of the cell and have different functions, it is not obvious why the corresponding mutations show a strong genetic interaction within the gravitropism signaling pathway. Several hypotheses have been proposed to explain this result (Stanga et al., 2009). In the direct interaction hypothesis, Toc132 directly functions as an amyloplast-associated ligand that interacts with an endoplasmic reticulum (ER)- or plasma membrane-associated receptor upon sedimentation onto these structures (Figure 1A). The genetic interaction between TOC75 and ARG1 is consistent with this hypothesis if Toc75 is required for the proper plastid targeting of Toc132. In the targeted interaction hypothesis, the TOC complex mediates the plastid membrane localization of a molecule that interacts as a ligand with a receptor on the ER or plasma membrane (Figure 1B). In the indirect interaction model, the TOC complex facilitates the plastid import of a molecule that does not physically interact with a receptor but is needed for a gravitropic response in an arg1 background (Figure 1C). The work described here is aimed at testing the direct interaction hypothesis as a first step toward clarifying the role of plastids in the signal transduction cascade between amyloplast sedimentation and auxin redistribution.
Figure 1.
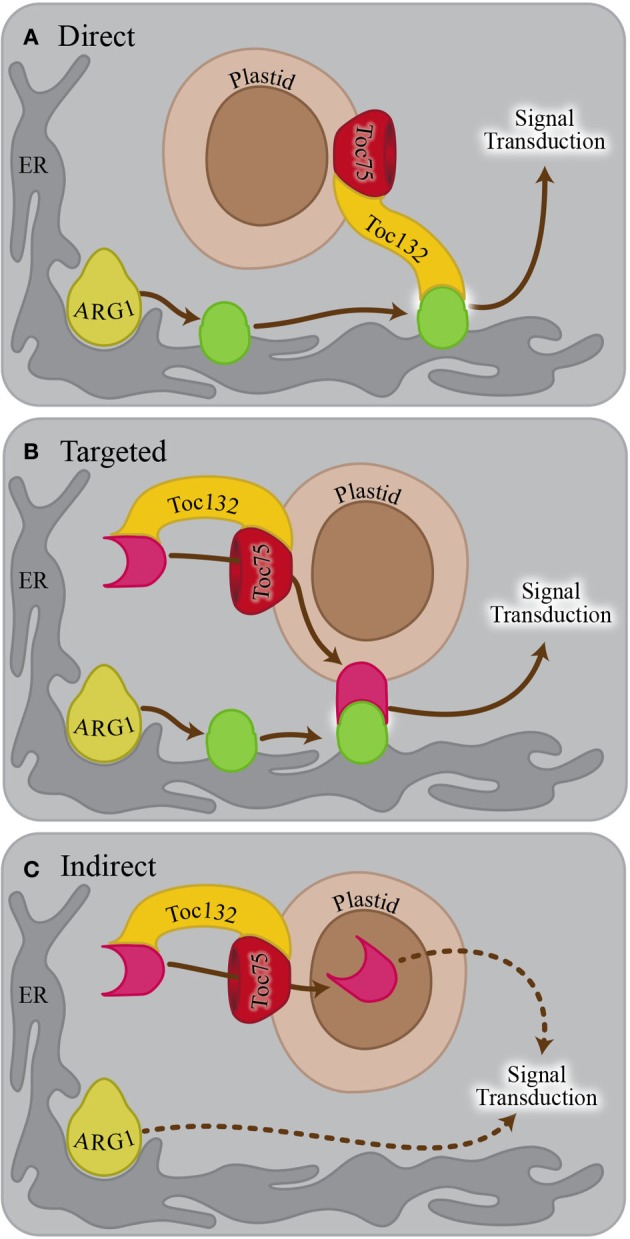
Possible models explaining the genetic interaction between ARG1 and TOC132. (A) In the direct interaction model, Toc132 acts as a ligand that interacts with a receptor (green oval) on the ER or plasma membrane. The localization or the activity of the receptor is mediated by ARG1. (B) In the targeted interaction model, Toc132 facilitates the plastid localization of a molecule acting as a ligand (pink shape) that interacts with a receptor (green oval) on the ER or plasma membrane upon amyloplast sedimentation. The localization or activity of the receptor is mediated by ARG1. (C) In the indirect interaction model, Toc132 facilitates the plastid localization of a molecule that does not act as a ligand but is still required for a gravitropic response in an arg1 background. In all three panels, the Toc33/34 receptor was omitted from the drawing for the sake of clarity and simplification of the model.
Materials and methods
Plant materials and growth conditions
toc120-3 (SALK_017374) and ppi3-1 were provided by Paul Jarvis (Constan et al., 2004; Kubis et al., 2004), and toc120-3 was backcrossed to WS wild type before use. mar2-1 was previously described as a mutation in the TOC132 gene (AT2G16640) (Stanga et al., 2009). To comply with the Arabidopsis nomenclature, we have renamed this allele toc132-4mar2−1 in this manuscript. The arg1-2 and arg1-2 toc132-4mar2−1 mutants have also been described previously (Sedbrook et al., 1999; Stanga et al., 2009).
The seeds were sterilized by washing with 95% ethanol. They were plated on half-strength buffered Linsmaier and Skoog medium containing macro- and micro-nutrients, vitamins, and 1.5% sucrose (Caisson Laboratories, North Logan, UT) supplemented with 1.5% agar type E (Sigma-Aldrich, St. Louis, MO) unless otherwise indicated. The seedlings were grown in a Conviron (Asheville, NC) TC16 growth chamber set at 22°C and a 16 h light/8 h dark cycle. The light intensity was 50–70 μmol m−2s−1 and was provided by cool white fluorescent bulbs (Grainger, Lake Forest, IL).
Transgenic constructs
The bases encoding the Toc132 GTP-binding and membrane domains (bases 1365–3618 from the start codon) were amplified from Col-0 DNA with the addition of a start codon. This region was cloned in between the AttL1 and AttL2 sites in the Gateway entry vector pENTR/D-TOPO (Life Technologies, Carlsbad, CA). An LR reaction was then performed to transfer this region into the binary vector pMDC32, which placed Toc132GM under the control of the CaMV 35S promoter and the NOS terminator (Curtis and Grossniklaus, 2003; Xu and Li, 2008).
This construct was sequenced and introduced into arg1-2 toc132-4mar2−1 plants using the Agrobacterium-mediated floral dip method (Clough and Bent, 1998). T1 transformants were selected and self-pollinated (Harrison et al., 2006), and T3 seeds likely to carry two copies of the transgene as determined by antibiotic resistance were used.
Root reorientation kinetics
The seeds were embedded within the medium described above supplemented with 0.7% agar. After at least 2 days of stratification, the seedlings were grown vertically as described above for 8 days. The plates were then turned horizontally, and photographs were taken at select time points. The root tip angle of each seedling at each time point was measured using Adobe Photoshop.
Protein extraction and mass spectrometry
WS and toc132-4mar2−1 seedlings were grown vertically for 2 weeks on medium supplemented with 0.6% agarose with one genotype grown on medium containing natural abundance ammonium nitrate and potassium nitrate (Sigma-Aldrich) and the other grown on 15N-enriched ammonium nitrate and potassium nitrate (Cambridge Isotope Laboratories, Tewksbury, MA) as previously described (Kline et al., 2010; Minkoff et al., 2012). The roots from approximately 300 seedlings per sample were dissected, combined, frozen in liquid nitrogen, and ground using a Mixer Mill 200 (Retsch, Haan, Germany). This process was repeated at non-overlapping times for a total of three trials. For each trial, each genotype was grown on both nitrogen sources for a total of six samples, each containing WS wild-type and toc132-4mar2−1 tissue. Therefore, for each trial, one sample contained WS seedlings grown on natural abundance nitrogen media and toc132-4mar2−1 seedlings grown on 15N-enriched media, while the second sample contained toc132-4mar2−1 seedlings grown on natural abundance nitrogen media and WS seedlings grown on 15N-enriched media. For each sample and in each trial, the proteins were extracted, trypsin digested, subjected to a solid-phase extraction, and analyzed on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher, Waltham, MA) as previously described (Minkoff et al., 2012). Mascot software v2.2.2 (Perkins et al., 1999) was used to compare the mass spectrometra to sequences present in The Arabidopsis Information Resource protein database (Lamesch et al., 2012). The settings for Mascot searches included permitting up to two missed cleavages. Deamidation of asparagine and glutamine residues and oxidation of methionine residues were set as variable modifications. Cysteines were searched in carbamidomethylated form as a fixed modification. The precursor mass tolerance was set to 20 ppm (allowing for the selection of precursors from the monoisotopic or first or second 13C isotopes), and fragment tolerance was set to 0.5 Da. The Mascot output was filtered to a 1.0% false discovery rate using a concatenated decoy database strategy with an in-house written script. Census software (Park et al., 2008) and additional in-house scripts (http://www.biotech.wisc.edu/sussmanlab/research/supporting_Minkoff_2013) were used to make quantitative ratio comparisons between the 14N- and 15N-labeled signals, and the ratios were normalized to 1 based on the median of each trial. 14N/15N (light/heavy) ratios were determined for each peptide and were then averaged for all the peptides in each protein. The 14N/15N ratios of the samples in which WS wild type was labeled with 14N were compared to the 14N/15N ratios of the samples in which WS wild type was labeled with 15N using a Student's T-Test and a significance level of p < 0.1. For the proteins in which one of these ratios was greater than 1.15 and the other was less than 0.85, the ratio between the genotypes was calculated by taking the inverse of the 14N/15N ratio for the samples in which toc132-4mar2−1 was labeled with 15N and averaging the six toc132-4mar2−1/WS values. The data are available in Data Sheet 1.
RNA isolation and qRT-PCR
Seedlings were grown as described for the mass spectrometry experiment using medium supplemented with natural abundance ammonium nitrate and potassium nitrate. The roots were dissected with a scalpel, and RNA was prepared using the QIAGEN RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The RNA samples were treated with RQ1 DNase (Promega, Madison, WI) according to the instructions. This procedure was conducted at three non-overlapping times for a total of three biological replicates. cDNA synthesis and quantitative real-time PCR (qRT-PCR) were conducted simultaneously using a qScript™ One-Step qRT-PCR Kit (Quanta Biosciences, Gaithersburg, MD) as recommended. Four technical replicates per sample were included. The samples were run on a Roche Light Cycler 480 and analyzed using LinRegPCR (Ramakers et al., 2003). Reference genes with similar expression levels to the candidate genes were chosen (Czechowski et al., 2005); At1g58050 was used for AT4G23690 and GDH3, At4g27960 was used for MAJOR LATEX PROTEIN LIKE 1, and At1g13320 was used for all other genes.
Results
Mutations in TOC120 and TOC34/PPI3 enhance the arg1 phenotype
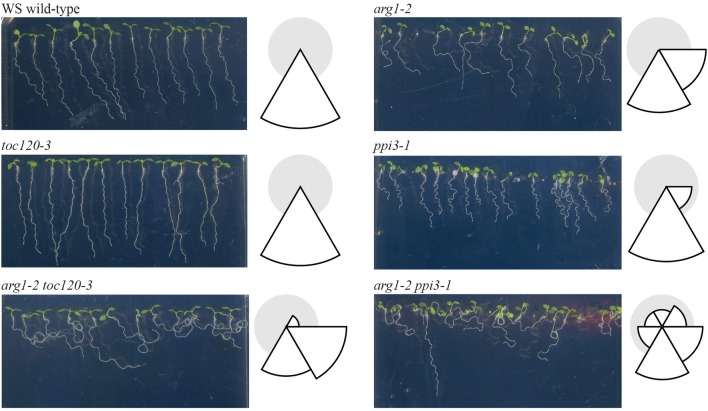
If Toc132 specifically acts as a ligand as proposed in the direct interaction model, mutations in other TOC complex receptors are unlikely to enhance the arg1 gravitropic defect. However, if Toc132 mediates the gravitropic response through its role in plastid protein import, then mutations in other TOC complex receptor genes such as TOC120 and TOC34/PPI3 may also enhance the arg1 mutant phenotype. Therefore, we created arg1-2 toc120-3 and arg1-2 ppi3-1 plants and analyzed their gravitropic responses. Although the roots of these double mutants did not grow in completely random directions as did arg1-2 toc132-4mar2−1 and arg1-2 mar1-1 roots (Stanga et al., 2009), they showed significantly more variable root growth on hard agar surfaces relative to the single mutants (Figure 2). This result suggests that mutations in multiple TOC complex receptors can enhance the arg1 gravitropic defect, and therefore, protein import via the TOC complex is important for gravitropism. These data indicate that the direct interaction model is unlikely.
Figure 2.
arg1-2 toc120-3 and arg1-2 ppi3-1 seedlings showed increased variability of root growth angles compared to WS wild type and arg1-2, toc120-3, and ppi3-1 single mutants (F-test, p < 0.05, and n = 32–44). The seedlings were grown vertically for 3 days. They were then tilted backward 30° and grown for an additional 4 days. The root angles were then measured from the root-hypocotyl junction to the root tip using Adobe Photoshop. These angles were placed into one of six 60° bins, and the corresponding histograms were drawn to the right of each panel. In these histograms, the size of each wedge is proportional to the fraction of plants whose roots grew in the direction of the corresponding bin. This experiment was repeated three independent times with similar results.
The acidic domain of Toc132 is not required for its role in gravitropism
If the direct interaction model is not supported and Toc132 does not act directly as a ligand, we expect a Toc132 construct that lacks the acidic region but still retains the GTP-binding and membrane domains (Toc132GM) to restore the gravitropic response in an arg1 toc132 mutant background to that of arg1 single mutants. Indeed, such a truncated protein is likely to retain its function in protein import into plastids (Inoue et al., 2010) even though it lacks the acidic domain. This domain is the region most likely to act as a ligand because it protrudes into the cytosol and is the most divergent among the TOC complex receptors. We expressed TOC132GM in arg1-2 toc132-4mar2−1 mutant plants under the constitutive CaMV 35S promoter and found that the transformed seedlings showed similar root reorientation kinetics to arg1-2 single mutants (Figure 3). This result demonstrates that the acidic domain of Toc132 is not required for a gravitropic response, and it is consistent with both the targeted and the indirect interaction models.
Figure 3.
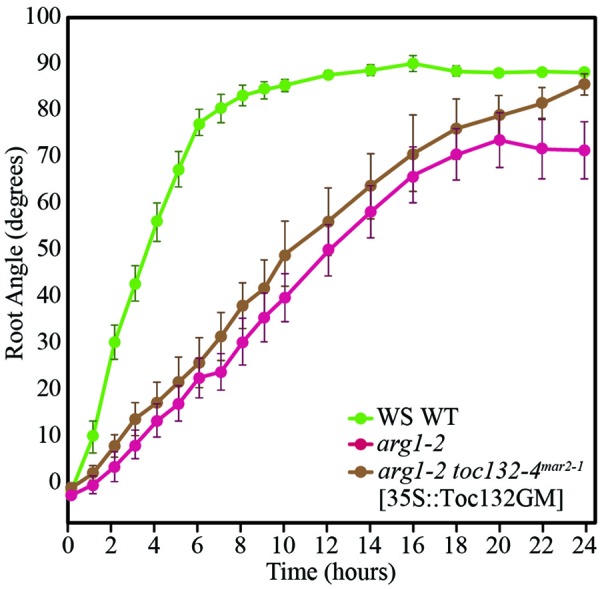
arg1-2 toc132-4mar2−1 [35S::Toc132GM] plants showed similar root tip reorientation kinetics compared to arg1-2 single mutants but slower kinetics than wild type. The values in degrees for arg1-2 toc132-4mar2−1 roots, which did not respond to reorientation, for the time points shown on the graph from left to right are −121 ± 29, −120 ± 28, −120 ± 29, −121 ± 29, −120 ± 29, −120 ± 29, −120 ± 29, −120 ± 29, −120 ± 28, −129 ± 29, −120 ± 29, −120 ± 28, −129 ± 28, −129 ± 28, −121 ± 28, −129 ± 29, −121 ± 29, and −120 ± 28. arg1-2 and arg1-2 toc132-4mar2−1 [35S::Toc132GM] root tip angles were not significantly different from each other at any time point (p > 0.05, Student's T-Test). The error bars represent the standard error, and n = 10. Similar results were obtained in two additional independent experiments.
Mutations in TOC132 enhance the pgm1 phenotype
We next sought to distinguish between the targeted and indirect interaction models. In the targeted interaction model, the signal transducer imported by the TOC complex triggers signal transduction upon amyloplast sedimentation, whereas the indirect interaction model postulates a role for this plastid-localized transducer that may not rely on amyloplast sedimentation. Therefore, to determine whether the TOC complex functions in conjunction with amyloplast sedimentation, we generated toc132-4mar2−1 pgm1-1 double mutants. pgm1-1 single mutants lack starch, and their amyloplasts do not sediment, although they still display some response to gravity (Caspar and Pickard, 1989; Kiss et al., 1989). We found that toc132-4mar2−1 pgm1-1 double mutants displayed stronger gravitropic defects than pgm1-1 single mutants (Figure 4). This result is not directly compatible with the targeted interaction model, in which we would expect no enhancement of the gravitropic defect, although it does not rule it out either as the double mutant still retains some gravitropic response.
Figure 4.
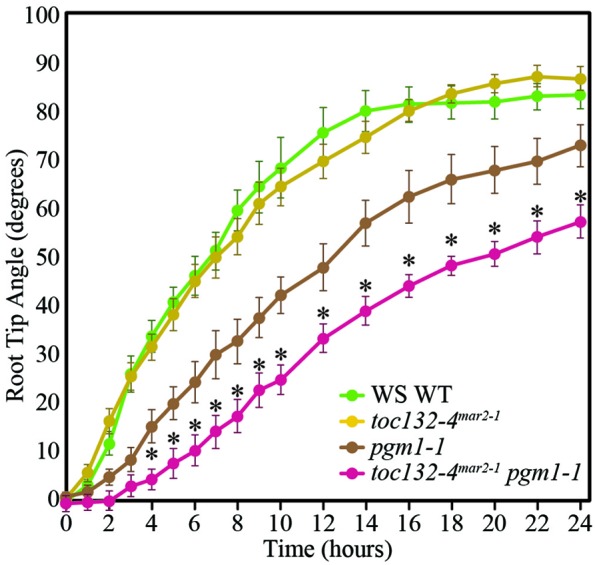
toc132-4mar2−1pgm1-1 plants showed slower root reorientation kinetics than toc132-4mar2−1 and pgm1-1 single mutants. The error bars represent the standard error, and n = 10. The asterisks represent significant differences between pgm1-1 toc132-4mar2−1 and the corresponding single mutants (p < 0.05, Student's T-Test). Similar results were obtained in two additional independent experiments.
The proteome and transcriptome are altered in toc132 mutants
To identify candidate proteins that might not be properly imported into plastids in toc132 mutants, we analyzed the wild-type and toc132-4mar2−1 proteomes of whole root tissue. A similar approach was recently used to investigate the levels of plastid-associated proteins using toc159 whole leaf tissue (Bischof et al., 2011). We expected proteins that were not properly imported into plastids in toc132 mutants to be degraded or to accumulate and therefore to show differences in expression between the two genotypes. We identified only one protein present at different levels between wild type and toc132-4mar2−1 mutants that was highly likely to localize to plastids (Baginsky and Gruissem, 2009), NUCLEOSIDE DIPHOSPHATE KINASE 3 (NDPK3) (Table 1). However, we found 25 nucleus-encoded proteins present at different levels between wild type and toc132-4mar2−1 that localize to other regions of the cell or whose localizations are unknown. Sixteen of these proteins were more abundant in toc132-4mar2−1, and nine were less abundant (Table 1). Nine of the proteins that were more abundant in toc132-4mar2−1 and seven of those that were less abundant are annotated as functioning in stress responses (Provart and Zhu, 2003).
Table 1.
Proteins present at different levels in wild-type and toc132-4mar2−1 roots.
| Locus | Annotation | toc132-4mar2-1/ WT ± SE | p |
|---|---|---|---|
| AT2G16005 | MD2-related lipid recognition domain-containing protein | 2.76 ± 0.49 | 0.081 |
| AT4G08770 | Peroxidase, putative | 2.40 ± 0.16 | 0.019 |
| AT1G54010 | Myrosinase-associated protein | 2.01 ± 0.07 | 0.005 |
| AT1G66200 | GLUTAMINE SYNTHETASE 1;2 (GLN1;2) | 1.76 ± 0.26 | 0.036 |
| AT1G78320 | GLUTATHIONE S-TRANSFERASE TAU 23 | 1.73 ± 0.24 | 0.004 |
| AT2G41840 | 40S Ribosomal protein S2 (RPS2C) | 1.60 ± 0.08 | 0.019 |
| AT4G11290 | Peroxidase, putative | 1.60 ± 0.26 | 0.027 |
| AT2G01520 | MAJOR LATEX PROTEIN LIKE 1 | 1.57 ± 0.12 | 0.034 |
| AT4G22380 | Ribosomal protein | 1.46 ± 0.10 | 0.043 |
| AT1G74020 | STRICTOSIDINE SYNTHASE 2 | 1.45 ± 0.09 | 0.094 |
| AT1G17170 | GLUTATHIONE S-TRANSFERASE TAU 24 | 1.45 ± 0.14 | 0.052 |
| AT1G66270 | BETA-GLUCOSIDASE 21 | 1.44 ± 0.06 | 0.010 |
| AT2G38390 | Peroxidase, putative | 1.43 ± 0.14 | 0.052 |
| AT3G13930 | Dihydrolipoamide S-acetyltransferase, putative | 1.43 ± 0.18 | 0.089 |
| AT1G45145 | THIOREDOXIN H-TYPE 5 | 1.40 ± 0.12 | 0.094 |
| AT1G19570 | DEHYDROASCORBATE REDUCTASE | 1.33 ± 0.08 | 0.030 |
| AT2G36580 | Pyruvate kinase, putative | 1.22 ± 0.07 | 0.055 |
| AT3G56070 | ROTAMASE CYCLOPHILIN 2 | 0.84 ± 0.02 | 0.002 |
| AT1G47128 | RESPONSIVE TO DEHYDRATION 21 | 0.80 ± 0.07 | 0.100 |
| AT2G38380 | PEROXIDASE 22 | 0.79 ± 0.04 | 0.058 |
| AT4G11010 | NUCLEOSIDE DIPHOSPHATE KINASE 3 | 0.76 ± 0.02 | 0.003 |
| AT5G07440 | GLUTAMATE DEHYDROGENASE 2 | 0.72 ± 0.03 | 0.0003 |
| AT4G23690 | Disease resistance-responsive family protein | 0.71 ± 0.11 | 0.063 |
| AT3G03910 | GLUTAMATE DEHYDROGENASE 3 | 0.67 ± 0.04 | 0.015 |
| AT4G23670 | MAJOR LATEX PROTEIN LIKE 6 | 0.52 ± 0.12 | 0.025 |
| AT1G20450 | EARLY RESPONSE TO DEHYDRATION 10 | 0.49 ± 0.08 | 0.079 |
The annotations were obtained from The Arabidopsis Information Resource. The ratios between the genotypes and the associated standard errors were determined as described in the Materials and Methods section, and a Student's T-test was used to generate the p-values.
We then used qRT-PCR to determine if some of the genes encoding these proteins are differentially expressed in toc132-4mar2−1 compared to wild type. The primers are shown in Table 2. Of the eight genes selected for analysis, four showed differences at the transcript level in the same directions predicted by the proteomic analysis (Figure 5).
Table 2.
Primers used to quantify gene expression.
| Locus | Forward primer | Reverse primer |
|---|---|---|
| AT1G54010 | atcccaacggcaaattctc | cgatcggaatcctcatgaat |
| AT1G66200 | cagctgctgatgaaatatggat | aaccacaccagcaatctctgt |
| AT4G11290 | caacatataaacaatgcaccctct | caagatggaaccatcacaacc |
| AT2G01520 | cgacggtgaatgggactc | ccttgaacacctccggttt |
| AT1G66270 | ccaactgttagccgtgtacttg | tttgtggataatctcccgaagt |
| AT2G38380 | tcgtcgatgaactgcagact | gatcgatgcatcacaaccac |
| AT4G23690 | ggcgacaatgtagcaaacg | acttgaagtttcctagtcctggag |
| AT3G03910 | ggaaccggtcctcagaca | aagatcaatgggttttccagtg |
| AT1G58050 | ccattctactttttggcggct | tcaatggtaactgatccactctgatg |
| AT1G13320 | taacgtggccaaaatgatgc | gttctccacaaccgcttggt |
| AT4G27960 | catcttgaaggagcagtggag | gggtttggatccgttaacaa |
The sequences are shown 5' to 3'.
Figure 5.
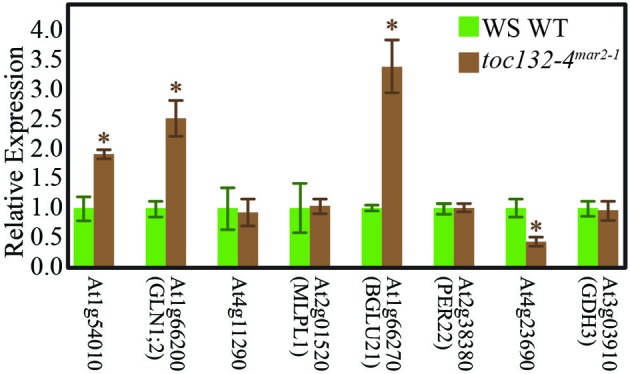
Many genes whose protein levels are altered in toc132-4mar2−1 also show different transcript levels in toc132-4mar2−1 relative to wild type. The first five genes from left to right encode proteins that are found at an increased abundance in toc132-4mar2−1, and the last three genes encode proteins that are found at decreased abundance in toc132-4mar2−1. The expression of each candidate gene was quantified relative to a reference gene as described in the Materials and Methods section, and the expression of each gene in wild type was set to 1. The error bars represent the standard error among three biological replicates. The asterisks indicate significant differences between wild type and toc132-4mar2−1 at p < 0.05 (Student's T-Test).
Discussion
We demonstrated that mutations in multiple TOC complex components caused enhanced gravitropic defects in an arg1 background (Figure 2). This result supports a model in which the TOC complex imports a molecule into plastids that is necessary for gravitropism (Figures 1B,C). However, we did not see completely random root growth when we mutated TOC120 or TOC34 in an arg1 background. This result may have occurred due to the abilities of other receptors such as Toc132 and Toc33 to partially compensate for the loss of these proteins (Kubis et al., 2003, 2004; Ivanova et al., 2004). In any case, these results indicate that Toc132 is unlikely to function as a ligand in gravity signal transduction. This result is in contrast to our previously published result that mutations in TOC120 do not enhance the arg1-2 phenotype (Stanga et al., 2009). A closer examination using more seedlings revealed this subtle but consistent phenotype.
Toc159 family members differ most significantly in the lengths and sequences of their N-terminal acidic domains. Although the acidic domain is thought to help regulate the specificity of imported proteins, it has little effect on overall import capacity (Inoue et al., 2010). We showed that a truncated version of Toc132 that lacks the cytoplasmic acidic domain is capable of restoring the gravitropism response of arg1-2 toc132-4mar2−1 seedlings back to that of arg1-2. Therefore, we conclude that the acidic domain is not required for Toc132's function in the gravitropic response (Figure 3). Because this construct still contained the GTP-binding and membrane domains, it likely retained most or all of the protein-import capability associated with full-length Toc132 (Inoue et al., 2010). We conclude that this construct likely rescued the arg1-2 toc132-4mar2−1 phenotype because it increased the overall protein import efficiency of the TOC complex. Therefore, Toc132 likely does not directly act as a ligand in gravity signal transduction because the large cytoplasmic acidic domain, which is the most likely region of the protein to interact with a receptor, is not necessary for its gravitropic function. This result reinforces the hypothesis that the TOC complex mediates gravitropism by modulating the targeting to plastids of another important molecule that contributes to gravity signal transduction (Figures 1B,C).
To further test the targeting and indirect models (Figures 1B,C), we examined the gravitropic response of toc132-4mar2−1 pgm1-1 plants and found that they displayed slightly enhanced gravitropic defects compared to the pgm1-1 single mutant (Figure 4). This result suggests that TOC132 and PGM1 function in different genetic pathways and that the TOC complex contributes to gravitropism in a manner at least partially independent of amyloplast sedimentation. Therefore, the indirect interaction model is plausible (Figure 1C); however, we cannot rule out the targeted interaction model (Figure 1B).
In our analysis of the toc132-4mar2−1 root proteome, we expected many plastid-localized proteins to be differentially expressed between toc132-4mar2−1 and wild type, as previously shown for toc159 leaf proteins (Bischof et al., 2011). However, we identified only one protein likely to localize to plastids, NDPK3, in a group of 26 differentially expressed proteins between toc132-4mar2−1 and wild type. This protein has also been shown to localize to mitochondria (Sweetlove et al., 2001). NDPKs have been implicated in stress and light signaling, and the related protein NDPK2 has been shown to be involved in auxin-related processes at least partly by affecting auxin transport (Choi et al., 2005). Furthermore, NDPK3 was previously found to be redox regulated, and redox signals have been implicated in plastid-to-nucleus retrograde signaling (Fey et al., 2005). Therefore, addressing a possible role for NDPK3 in gravitropism is an interesting area of future research.
The low number of plastid-localized differentially expressed proteins may be due to compensation by Toc120 or other receptors. However, we did identify many nucleus-encoded proteins present at different levels in toc132-4mar2−1 relative to wild type, and several of the corresponding genes also showed differences at the transcript level (Figure 5). Plastids can regulate nuclear gene expression through retrograde signaling, especially when they are stressed (Nott et al., 2006). Indeed, many of the proteins present at different levels in toc132-4mar2−1 are involved in stress responses. Alternatively, it is also possible that some of the expression differences we observed between mutant and wild-type roots are indirect consequences of altered cell metabolism in the mutant. Such effects could occur at the transcriptional, posttranscriptional, translational, and posttranslational levels, potentially explaining why several of the differentially expressed proteins are encoded by genes whose transcript levels are similar between mutant and wild-type roots (Table 1 and Figure 5). In any case, our results suggest the possibility that altered plastid protein import results in the altered abundance of (a) nuclear-encoded protein(s) that is (are) required for a partial gravitropic response in an arg1 background. Interestingly, the genes encoding two of the proteins in Table 1 (MD-2-related lipid recognition domain-containing protein and MAJOR LATEX PROTEIN LIKE 6) increase in expression in root tips upon gravistimulation (Kimbrough et al., 2004).
Considered together, these experiments suggest that the direct interaction model is highly unlikely to explain the role of the TOC complex in gravitropism. Future work will determine which proteins must be imported into plastids for a normal gravitropic response in an arg1 background and which, if any, non-plastid-associated proteins whose abundance is consequently altered are involved in this process.
Author contributions
Allison K. Strohm designed and performed the experiments and wrote the paper. Greg A. Barrett-Wilt performed the experiments and revised the paper. Patrick H. Masson designed the experiments and wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by grants from the National Science Foundation (IOS-0821884 and IOS-1121694) to Patrick H. Masson and a National Science Foundation Graduate Research Fellowship to Allison K. Strohm. We thank Paul Jarvis for providing us with ppi3-1 and toc120-3 and Katherine Baldwin for help with figures. We also thank Ben Minkoff for help with mass spectrometry and data analysis.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00148/abstract
References
- Baginsky S., Gruissem W. (2009). The chloroplast kinase network: new insights from large-scale phosphoproteome profiling. Mol. Plant 2, 1141–1153 10.1093/mp/ssp058 [DOI] [PubMed] [Google Scholar]
- Bischof S., Baerenfaller K., Wildhaber T., Troesch R., Vidi P.-A., Roschitzki B., et al. (2011). Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 23, 3911–3928 10.1105/tpc.111.092882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsirichai K., Sedbrook J. C., Chen R., Gilroy S., Masson P. H. (2003). ALTERED RESPONSE TO GRAVITY is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes. Plant Cell 15, 2612–2625 10.1105/tpc.015560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T., Pickard B. G. (1989). Gravitropism in a starchless mutant of Arabidopsis: implications for the starch-statolith theory of gravity sensing. Planta 177, 185–197 10.1007/BF00392807 [DOI] [PubMed] [Google Scholar]
- Choi G., Kim J. I., Hong S. W., Shin B., Choi G., Blakeslee J. J., et al. (2005). A possible role for NDPK2 in the regulation of auxin-mediated responses for plant growth and development. Plan Cell. Physiol. 46, 1246–1254 10.1093/pcp/pci133 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Constan D., Patel R., Keegstra K., Jarvis P. (2004). An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 38, 93–106 10.1111/j.1365-313X.2004.02024.x [DOI] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey V., Wagner R., Braütigam K., Wirtz M., Hell R., Dietzmann A., et al. (2005). Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J. Biol. Chem. 280, 5318–5328 10.1074/jbc.M406358200 [DOI] [PubMed] [Google Scholar]
- Harrison S. J., Mott E. K., Parsley K., Aspinall S., Gray J. C., Cottage A. (2006). A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2, 19 10.1186/1746-4811-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Rounds C., Schnell D. J. (2010). The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. Plant Cell 22, 1947–1960 10.1105/tpc.110.074328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova Y., Smith M. D., Chen K., Schnell D. J. (2004). Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 15, 3379–3392 10.1091/mbc.E03-12-0923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrough J. M., Salinas-Mondragon R., Boss W. F., Brown C. S., Sederoff H. W. (2004). The fast and transient transcriptional network of gravity and mechanical stimulation in the Arabidopsis root apex. Plant Physiol. 136, 2790–2805 10.1104/pp.104.044594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss J. Z., Hertel R., Sack F. D. (1989). Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta 177, 198–206 10.1007/BF00392808 [DOI] [PubMed] [Google Scholar]
- Kline K. G., Barrett-Wilt G. A., Sussman M. R. (2010). In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 15986–15991 10.1073/pnas.1007879107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S., Baldwin A., Patel R., Razzaq A., Dupree P., Lilley K., et al. (2003). The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15, 1859–1971 10.1105/tpc.012955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S., Patel R., Combe J., Bédard J., Kovacheva S., Lilley K., et al. (2004). Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16, 2059–2077 10.1105/tpc.104.023309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C., Sasidharan R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkoff B. B., Burch H. L., Sussman M. R. (2012). A pipeline for 15N metabolic labeling and phosphoproteome analysis in Arabidopsis thaliana. Methods Mol. Biol. 1062, 353–379 10.1007/978-1-62703-580-4_19 [DOI] [PubMed] [Google Scholar]
- Nott A., Jung H. S., Koussevitzky S., Chory J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759 10.1146/annurev.arplant.57.032905.105310 [DOI] [PubMed] [Google Scholar]
- Park S. K., Venable J. D., Xu T., Yates J. R. (2008). A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322 10.1038/nmeth.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- Provart N., Zhu T. (2003). A browser-based functional classification SuperViewer for Arabidopsis genomics. Curr. Comput. Mol. Biol. 2003, 271–272 [Google Scholar]
- Ramakers C., Ruijter J. M., Deprez R. H. L., Moorman A. F. M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- Sedbrook J. C., Chen R., Masson P. H. (1999). ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 96, 1140–1145 10.1073/pnas.96.3.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga J. P., Boonsirichai K., Sedbrook J. C., Otegui M. S., Masson P. H. (2009). A role for the TOC complex in Arabidopsis root gravitropism. Plant Physiol. 149, 1896–1905 10.1104/pp.109.135301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohm A. K., Baldwn K. L., Masson P. H. (2012). Molecular mechanisms of root gravity sensing and signal transduction. WIREs Dev. Biol. 1, 276–285 10.1002/wdev.14 [DOI] [PubMed] [Google Scholar]
- Sweetlove L. J., Mowday B., Hebestreit H. F., Leaver C. J., Millar A. H. (2001). Nucleoside diphosphate kinase III is localized to the inter-membrane space in plant mitochondria. FEBS Lett. 16, 272–276 10.1016/S0014-5793(01)03069-1 [DOI] [PubMed] [Google Scholar]
- Xu R., Li Q. Q. (2008). Protocol: Streamline cloning of genes into binary vectors in Agrobacterium via the Gateway® TOPO vector system. Plant Methods 4, 4 10.1186/1746-4811-4-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.