Abstract
Aim
We investigate patterns of phylogenetic diversity in relation to species diversity for European birds, mammals and amphibians, to evaluate their congruence and highlight areas of particular evolutionary history. We estimate the extent to which the European network of protected areas (PAs) network retains interesting evolutionary history areas for the three groups separately and simultaneously.
Location
Europe
Methods
Phylogenetic (QEPD) and species diversity (SD) were estimated using the Rao’s quadratic entropy at 10′ resolution. We determined the regional relationship between QEPD and SD for each taxa with a spatial regression model and used the tails of the residuals (QERES) distribution to identify areas of higher and lower QEPD than predicted. Spatial congruence of biodiversity between groups was assessed with Pearson’s correlation. A simple classification scheme allowed building a convergence map where a convergent pixel equalled to a QERES value of the same sign for the 3 groups. This convergence map was overlaid to the current PAs network to estimate the level of protection in convergent pixels and compared it to a null expectation built on 1000 randomization of PAs over the landscape.
Results
QERES patterns across vertebrates show a strong spatial mismatch highlighting different evolutionary histories. Convergent areas represent only 2.7% of the Western Palearctic, with only 8.4% of these areas being covered by the current PAs network while a random distribution would retain 10.4% of them. QERES are unequally represented within PAs: areas with higher QEPD than predicted are better covered than expected, while low QEPD areas are undersampled.
Main conclusions
Patterns of diversity strongly diverge between groups of vertebrates in Europe. Although Europe has the world’s most extensive PAs network, evolutionary history of terrestrial vertebrates is unequally protected. The challenge is now to reconcile effective conservation planning with a contemporary view of biodiversity integrating multiple facets.
Keywords: Phylogenetic diversity, protected areas, spatial biodiversity congruence, species diversity, terrestrial vertebrates, Europe
INTRODUCTION
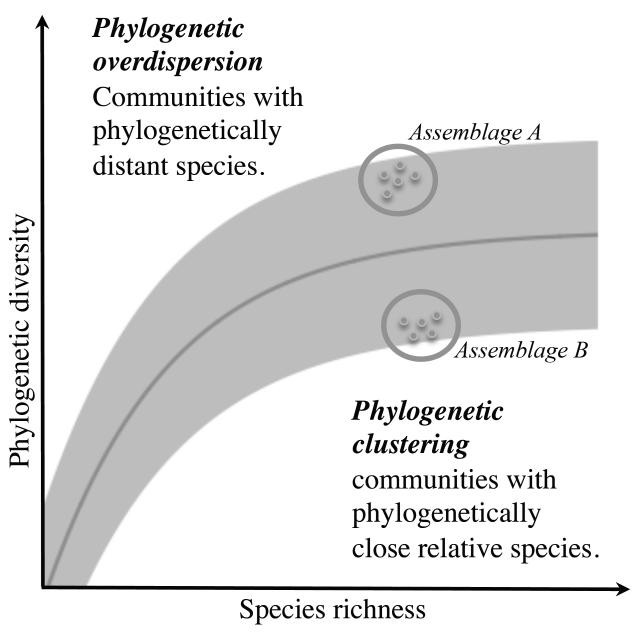
Species distributions, and ultimately biodiversity patterns, are shaped by the interplay of evolutionary, biological and anthropogenic processes (Ricklefs, 1987). With the rise of available distributional data, the last decades have seen an upsurge of studies exploring biodiversity patterns from local to broad geographical scales (Gaston, 2000), most of them focused on species richness (Currie & Paquin, 1987; Davies & Buckley, 2011) or species evenness (i.e. abundance distribution among species) (Hillebrand et al., 2008). Species richness has been the main focus of macro-ecological studies and is still widely used, mainly because of the easiness to quantify and interpret the data (Cadotte & Davies, 2010). In particular, conservation planning has traditionally used richness information combined to different irreplaceability measures (e.g. endemism or rarity) to prioritize some regions over others (e.g. “Biodiversity Hotspots”, Myers, 1988). However, focusing on species richness ignores the differences among species in terms of functional or evolutionary characteristics (Vane-Wright et al., 1991; Faith, 1992; Petchey & Gaston, 2002). To account for these other aspects of diversity, measures of phylogenetic and functional diversity have recently been developed (Pavoine & Bonsall, 2011 for a review). Both the increasing availability of molecular data in public databases (e.g. GenBank) and the advances in phylogenetic methods (Roquet et al., 2013) have enhanced the use of phylogenetic diversity measure (i.e. the amount of evolutionary history) as a powerful tool for featuring biodiversity. For instance, phylogenetic diversity measures are now widely used to understanding the diversity of current species distributions (e.g. Davies & Buckley, 2011) or the potential functioning of ecosystems (Lavergne et al., 2010; Mouquet et al., 2012). Although most phylogenetic diversity measures show a positive and monotonic link with species richness (Fig. 1) (Faith, 1992; Rodrigues et al., 2011; Morlon et al., 2011), this relationship can vary spatially (e.g. Forest et al., 2007) and this deviation can inform about the processes (speciation, extinction, lineage filtering, competition and migration) partly responsible for the current biodiversity patterns at large spatial scale (Davies & Buckley, 2011; Fritz & Rahbek, 2012). For instance, a region with high species richness and endemism but a low phylogenetic diversity (Fig. 1, bottom-right corner) might indicate areas where recent adaptive radiations have occurred (e.g. Cape floristic region of South Africa, Slingsby & Verboom, 2006).
Figure 1. Hypothetical relationship between phylogenetic diversity and species richness (SR) of species assemblages.
The grey region corresponds to the possible interval of phylogenetic diversity values for a given number of species while the darker line indicates the theoretical expected values of phylogenetic diversity. For an assemblage of few species, we would expect that the addition of one species will lead to a sharp increase in phylogenetic diversity value, this new species being likely to add new phylogenetic information, whereas at high level of SR all the combinations of phylogenetic diversity have already been sampled and the addition of a new species does not influence the value of phylogenetic diversity for the region. As an example, region A shows an assemblage where phylogenetic diversity is higher than expected by its common relationship with SR. This type of assemblage would probably include phylogenetically distant species, reflecting thus a low level of diversification. On the contrary, region B presents lower phylogenetic diversity than expected, and thus it will mostly contain phylogenetically close species, e.g. resulting from events of massive diversification in the recent history.
Assuming that closely related species have more chances to share common features (e.g. ecological niches, functional traits, Faith 1992; 1994) than randomly chosen species in the phylogeny, phylogenetic diversity could also serve as a proxy for functional diversity if traits related to these functions were highly conserved along the phylogeny (Webb et al., 2002). Under this assumption, prioritizing phylogenetic diversity in protected areas (PAs) networks would lead at the same time to the maximization of evolutionary history of Earth’s biota (Cadotte & Davies, 2010; Forest et al., 2007) and functional diversity.
Beyond the recent call to adopt a multifaceted approach to better understand and protect biodiversity as a whole (Devictor et al., 2010), there are still few large-scale studies analyzing patterns of phylogenetic diversity in relation to species richness and often limited to single taxonomic groups (e.g. plants, Forest et al., 2007; mammals, Davies & Buckley, 2011; Safi et al., 2011; birds, Devictor et al., 2010; fishes, Mouillot et al., 2011; and amphibians, Fritz & Rahbek, 2012). In this perspective, understanding how phylogenetic diversity and species richness relate across multiple taxa is of interest, not only to further infer the processes generating biodiversity patterns but also to be able to maximize the efficient use of limited conservation resources (Margules & Pressey, 2000) to preserve all biodiversity facets. Although the real impact of considering phylogenetic diversity in current conservation planning is still debated (Winter et al., 2013a) we miss large-scale studies on the congruence or mismatch between diversity facets of potential conservation interest across groups.
A limiting factor in conservation assessments is the lack of relevant data on spatial information (e.g. biodiversity distribution) upon which the effectiveness of conservation planning depends (Margules & Pressey, 2000). Consequently, conservationists often focus on a given group and use surrogates for which data can be obtained and assume that biodiversity features explicitly targeted in conservation efforts will also be effective in capturing unmapped biodiversity (Rodrigues & Brooks, 2007). Taxonomic surrogacy (whether one taxon is a good surrogate for another taxon when targeting species representation) has received substantial attention (Rodrigues & Brooks, 2007). Rodrigues et al. (2011) also explored whether taxonomic diversity is a good surrogate for phylogenetic diversity as measured with Faith’s phylogenetic diversity metric (Faith, 1992). However, the question of whether targeting a phylogenetic diversity measure for a group of organisms would also cover the one for another group has not been explored so far. Here, we propose a comparative approach to investigate spatial patterns of a phylogenetic diversity and species diversity (SD, combined measure of richness and evenness) for mammals, birds and amphibians over Europe while accounting for species habitat preferences within pixels. Using updated phylogenies and the Rao’s quadratic entropy to measure phylogenetic diversity (Rao, 1982) (hereafter referred as QEPD) and SD, we study their spatial distribution for each group separately and determine which regions show higher or lower phylogenetic diversity than expected. Finally, we undertake an assessment of the biodiversity coverage of the European network of PAs, and estimate whether and to which extent the current PAs network covers areas of higher/lower phylogenetic diversity than expected for these three groups of species simultaneously.
METHODS
Extent of the study area and spatial dataset
The study area includes the entire European sub-continent including Turkey (part of Asian continent) in order to have a complete picture of the Mediterranean coast. We used data on the spatial distribution of 275 mammals, 429 birds and 102 amphibians. These datasets were compiled from Maiorano et al., 2013 (see Appendix S1 in supporting information). For mammals and amphibians, the primary data were extent of occurrences (EOO) collected from the IUCN Global Mammal Assessment and Global Amphibian Assessment (IUCN, 2013). For bird species, EOO were obtained in combining data available from Hagemeijer & Blair (1997) with those available from the BWPi2.0.1 DVD-ROM (Birds of the Western Palearctic interactive 2006, version 2.0.1). For all species, habitat requirements were collected from expert opinion and published literature (Maiorano et al., 2013, Appendix S1). The collected data were used to assign a suitability score (0, unsuitable; 1, secondary habitat; and 2, primary habitat) to each of the 46 GlobCover land-use/land-cover classes (300m resolution). Scores were used to remove unsuitable cells (scored 0) and refine EOOs (no presence data were added, only false presence data were removed). Species distribution data were scaled up to a 10′ resolution. For each 10′ grid cell and for each species considered we kept the percentage of suitable habitat by summing the 300m pixels corresponding to primary or secondary habitat, we refer to this percentage as “potential suitable area” hereafter.
Phylogenetic data
Phylogenetic data for mammals were based on the updated super-tree of Fritz et al. (2009). We used 100 fully resolved phylogenetic trees, where polytomies were randomly resolved applying a birth-death model to simulate branch lengths (Kuhn et al., 2011). For birds, we extracted the 100 dated and fully dichotomous phylogenetic trees from Thuiller et al. (2011) and retained the 10 best ones as the variation between the trees was very low.
For amphibians, we conducted phylogenetic inference analyses based on DNA sequences extracted from GenBank (Appendix S2; Roquet et al., 2013). The phylogenetic analysis, conducted with RAxML (Stamatakis, 2006), included a search for 100 suboptimal trees, which yield identical topologies and similar branch lengths. The 100 phylogenies were transformed into cophenetic distance matrices and compared with Mantel tests. There were all highly correlated (correlation > 0.99). Because of that, we run all subsequent amphibian analyses using the best maximum likelihood tree (available on TreeBASE, accession number: S13561). This tree was dated with penalized-likelihood as implemented in r8s (Sanderson, 2003), using several fossil data to constrain certain nodes (Appendix S2). This is to our knowledge the most up-to date phylogenetic tree for European amphibian species.
Diversity measures
To measure both species and phylogenetic diversity, we used the Rao’s quadratic entropy (QE; Rao, 1982), a within-assemblage diversity measure (so-called alpha diversity) defined as the extent of dissimilarity between species in an assemblage (de Bello et al., 2010). For a given site (a 10′ cell), QE is defined as:
where dij is the dissimilarity between each pair of species i and j. pi and pj are the respective proportion of the species i and j, and can be expressed as any measure of relative species abundances (de Bello et al., 2010). In our study, pi and pj are taken from the “potential suitable area” estimated for each species. For measuring phylogenetic diversity (QEPD hereafter), dij was calculated as the patristic distance between species i and j derived from the phylogenetic trees. For species diversity (SD), dij was set to either 1 (when i ≠ j) or 0 (when i = j), in this particular case, QE equates to the Gini-Simpson index (de Bello et al., 2010). To make sure our indices were directly comparable, we transformed QEPD and SD values into equivalent number (Jost, 2007, Chao et al., 2010). The analyses were performed on 100 trees for mammals and 10 trees for birds to account for phylogenetic uncertainty. The results shown are median QEPD over the trees.
Phylogenetic diversity was originally estimated using the sum of the branch length of the species present in the assemblage (Faith, 1992), but since then several alternatives have been proposed (Pavoine & Bonsall 2011). Here, we used QEPD because it allows incorporating our measure of “potential suitable area”. In particular, it makes sure that pixels with equal number of species but very different proportion of suitable habitat for the respective species are distinguished. Practically, it allows a fine mapping and this is also particularly interesting for a conservation perspective, because it allows distinguishing sites to prioritize based on the potential population size of species (i.e. assuming that area is linked to population size).
Species diversity against phylogenetic diversity
Instead of using a null model to remove the effect-size of QEPD as usually done in community ecology to detect under or over-dispersion (e.g. Cavender-Bares et al., 2004), we used a model-based approach. The reason was two-fold: first, standardized effect size estimations requires a Gaussian distribution of phylogenetic distances, which was not the case here, and second, most of large-scale analyses have used a model-based approach, which facilitates comparisons (Fritz & Rahbek, 2012; Davies & Buckley, 2011). To analyse the spatial pattern of discrepancy between QEPD and SD in Europe, we built a spatial regression model between QEPD and SD for each vertebrate group. As the relationships between QEPD and SD were visually between linear and quadratic, (Fig. S1. Appendix S3), we tested both linear and quadratic terms. To account for spatial autocorrelation, we included geographic coordinates as a smooth factor (Wood, 2006). We chose this simplistic approach because models that account for a geographic correlation structure (e.g. generalised least squared regression) or more complex autocovariate (e.g. Eigen vector mapping, Peres-Neto & Legendre, 2010) were too data and time demanding to run at such resolution.
Pixels that deviated from the expected QEPD/SD relationship were thought to be the signature of particular evolutionary histories (Fig. 1, Fritz & Rahbek, 2012). To identify them, we used extremes positive and negative residuals depicting respectively areas with higher and lower QEPD than expected from the European QEPD/SD relationship. These residuals are called QERES hereafter. All models have been calibrated using the ‘mgcv’ package within R.2.12.1 (R Development Core Team, 2010).
Spatial co-variation of phylogenetic diversity across vertebrate groups
To examine how QEPD co-varied in space for the three taxonomic groups, we regressed QERES of each group against the other two. To evaluate congruency between the spatial distribution patterns of the different taxonomic groups, we classified QERES for each taxa within each cell as follow: values larger than 75% quantile were classified as 1, values lower than 25% quantile were classified as -1, and values falling in between were assigned a 0 value (we used this classification because we wanted to have the distribution tails of the residuals values). We then combined the values for the three taxonomic groups obtaining 27 codes (e.g. 1 for mammals, 0 for birds, 1 for amphibians results in the code 101). We referred the combinations “-1-1-1” and “111” as negative and positive convergence respectively, whereas “000” was called neutral convergence. The combinations differing in all three digits were referred to as divergent, whereas the remaining codes were noted as others. This classification allowed calculating the proportion of areas that show congruency (i.e. convergent sites) or mismatch (i.e. divergent and “others” sites) between the three taxonomic groups and was further used in the PAs assessment analysis (see next section). This classification might be seen as subjective but is close to hotspot definition based on species-area relationships (Guilhaumon et al., 2008). Here, it allows us to highlight the pixels where the three vertebrate groups have strikingly lower or higher than expected phylogenetic diversity.
Spatial congruence between protected areas and phylogenetic diversity patterns
We evaluated the current representation of each convergence-divergence category within three nested protected areas (PAs) networks. We first conducted the analyses on the complete list of PAs available from the World Database on Protected Area (WDPA, http://protectedplanet.net/) for our study area. To account for the broad range of PAs in WDPA that vary in terms of conservation action, we conducted analyses on a second network including only PAs with the most stringent conservation legislation (i.e. PAs belonging to IUCN category I and II). Finally, the third network concerned only Natura 2000 sites (http://www.eea.europa.eu/) and was reduced to European Union countries only. We first estimated the percentage of protection of each 10′ grid cell (NPROT). To assess the representation (R) of each convergence-divergence categories within the PAs we calculated the overlap between NPROT and the cells of each category (NcatPROT), we then divided NcatPROT by the total number of cells of each categories (NcatTOT).
To test the effectiveness of PAs network, we spatially randomized the distribution of NPROT (1000 times) and recalculated R of each category for each run, obtaining with this procedure a null distribution to be compared with the observed R for each category. This randomisation scheme explicitly tested whether the sites of QEPD convergence vs. divergence between species groups were more or less protected than under a random distribution of PAs.
RESULTS
The relationship between species diversity (SD) and phylogenetic diversity (QEPD) was nonlinear (Fig. S2, Appendix S4). For mammals and amphibians, a quadratic model had a better fit (R2 = 0.93, p < 0.001 and R2 = 0.83, p < 0.001 respectively, Table S1, Appendix S3) than any linear alternatives (mammals, R2 = 0.87, p < 0.001 and amphibians R2 = 0.82, p < 0.001, Table S2, Appendix S3). For birds, the difference between a linear and a quadratic fit was null (equal R2 = 0.59, p < 0.001, Table S1-S2, Appendix S3). To have consistent relationships for the three groups, the results presented hereafter refer to the quadratic models. The QEPD/SD relationships were linear for low and moderate levels of SD (i.e. the addition of a given species increased QEPD) and then became saturated for high SD values. In other words, when reaching a certain level of SD, QEPD cannot increase anymore, the overall tree of life for a given group being already entirely sampled.
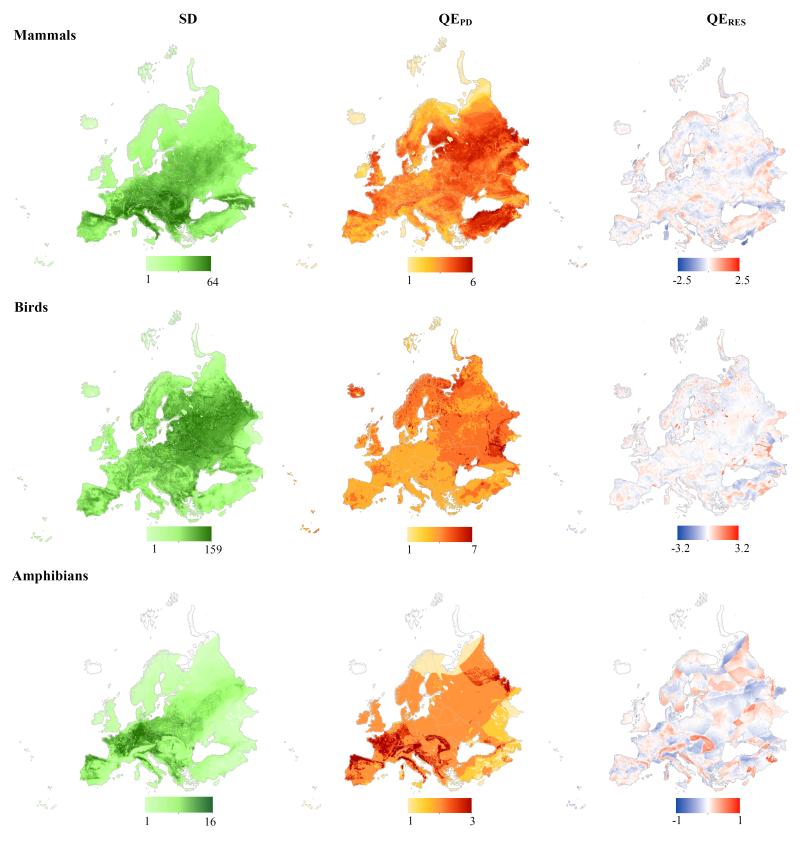
Comparing spatial patterns of SD, QEPD and QERES provided complementary results within and among the 3 groups of vertebrates. In particular, the distribution of QEPD for both mammals and birds showed a north-eastward increase with highest values in the Russian plains and Turkey for mammals, whereas this pattern was not found for amphibians (Fig. 2), which concentrate high QEPD values in southwest of Europe, in particular in the Po valley (Italy) and in Galicia (Spain). However, while the values of QERES for birds were negative in the major European mountain ranges (Alps, Carpathians, Apennins, Turkey mountains and Pyrenees), the opposite pattern was shown for mammals and amphibians (Fig. 2). In other words, the visible high QEPD for mammals and amphibians in European mountains was not only an effect of SD. Birds also showed areas of QEPD higher than expected from SD in regions associated to rivers (e.g. Volga Delta in Russia, Dniester and Dnieper estuary in Ukraine, Danube Delta in Romania) and lakes (e.g. Lacha lake in Russia, Värnen in Sweden, lake Van and Tuz in Turquey) (Fig. 2). There were also very diverging patterns in Cyprus and Corsica and in Mediterranean basin across the different groups: whereas QEPD of birds was generally high in those areas, there was correspondingly lower QEPD than expected with respect to SD for mammals (Fig. 2).
Figure 2. Spatial distribution patterns of species diversity (SD, left column), phylogenetic diversity (QEPD, middle column) and the residuals (QERES, right column) from the spatial regression between QEPD and SD for mammals (upper line), birds (middle line) and amphibians (lower line).
For SD, low to high values are represented by a green colour gradient from soft to dark green, the QEPD follows a yellow to red gradient for increasing values of QEPD and for QERES values, the blue colours depict negative values of residuals (lower diversity than expected by the relationship between QEPD and SD) while the red colours depict positive residuals (higher QEPD than expected).
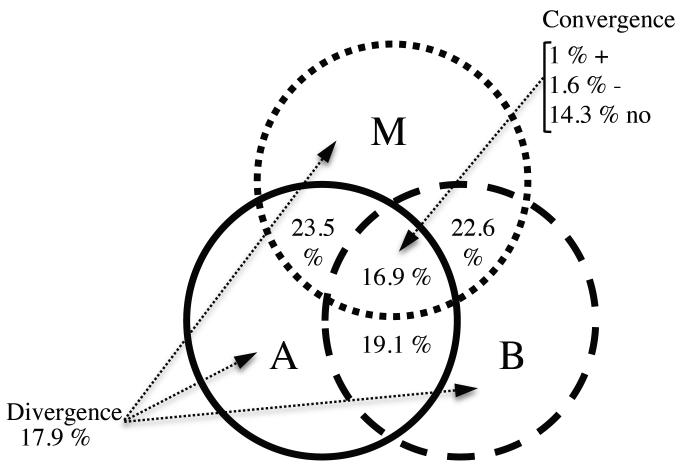
The co-variation of SD between taxonomic groups was positive with a high correlation between mammals and amphibians (Table 1a, Fig. S3, Appendix S4). Species-rich areas for one taxonomic group tended also, to some extent, to be rich areas for the other two groups. However, this apparent congruency did not hold for QEPD: as expected from the apparent mismatch of QEPD spatial distributions (Fig. 2), the strength of co-variation between the three groups did not show any kind of relationship for both QEPD and QERES (Table 1bc, Figs. S4-S5, Appendix S4). Moreover, strong spatial patterns emerged when comparing extreme values of QERES (Fig. 3). Only 1% of the Western Palearctic region (Fig. S6, Appendix S5) shows areas of positive convergence for the 3 taxonomic groups (i.e. areas with higher QEPD than expected for each group) and 1.6% of negative convergence (i.e. areas with lower QEPD than expected for each group), whereas 17% of the territory diverges completely between mammals, birds and amphibians (i.e. areas where QERES is positive for one taxa, negative for the second and null for the last one).
Table 1. Cross-taxon correlations (Pearson’s moment product) across birds, mammals and amphibians for a) species diversity (SD), b) phylogenetic diversity (QEPD) and c) residuals (QERES).
| (a) | ||||||
|---|---|---|---|---|---|---|
| Mammals | Birds | |||||
|
|
||||||
| Correlation | t | p | Correlation | t | P | |
| Birds | 0.44 | 116.09 | < 0.001 | / | / | / |
| Amphibians | 0.75 | 265.12 | < 0.001 | 0.46 | 120.08 | < 0.001 |
| (b) | ||||||
|---|---|---|---|---|---|---|
| Mammals | Birds | |||||
|
|
||||||
| Correlation | tvalue | pvalue | Correlation | tvalue | pvalue | |
| Birds | 0.0024 | 0.56 | 0.576 | / | / | / |
| Amphibians | 0.33 | 81.08 | < 0.001 | − 0.33 | − 80.87 | < 0.001 |
| (c) | ||||||
|---|---|---|---|---|---|---|
| Mammals | Birds | |||||
|
|
||||||
| Correlation | tvalue | pvalue | Correlation | tvalue | pvalue | |
| Birds | − 0.06 | − 11.82 | < 0.001 | / | / | / |
| Amphibians | 0.09 | 21.51 | < 0.001 | − 0.12 | − 28.96 | < 0.001 |
Figure 3. Venn diagram showing the congruence (in number of sites out of the total study area) in phylogenetic diversity (QEPD) patterns between mammals (M), birds (B) and amphibians (A).
Divergence represents areas where the residuals (QERES) for the 3 groups of vertebrates mismatch completely in space. Convergence encompasses areas where the 3 groups show higher values of QEPD than expected (+), lower values than expected (−) and finally areas where QERES was equal to 0 (no) for the 3 groups.
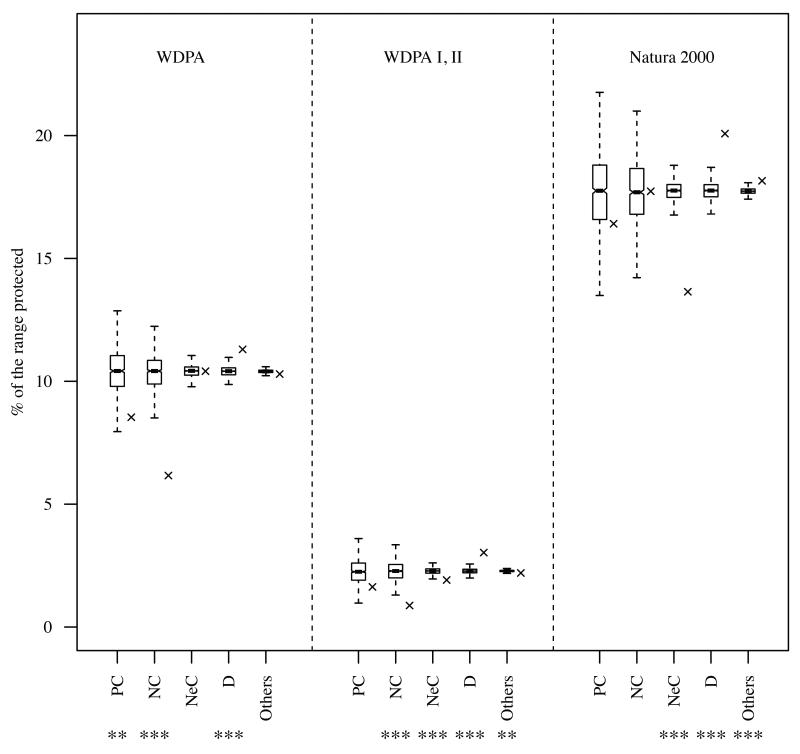
The percentage of QEPD representation in European PAs was not equal between the different PAs network (Fig. 4) with a higher representation of the QEPD congruency categories in Natura 2000 compared to the global World Protected Area network (WDPA) and the World Protected Area network with only IUCN categories I and II considered (WDPA I, II). This is not surprising as Natura 2000 covers more surface (17.7%) of Europe than the others do (10.4% for WDPA and 2.3% for WDPA I, II). In average, any PAs network tended to retain less QEPD than expected for birds and mammals while for amphibians PAs retained more QEPD than random (Table S4, Appendix S5). Regarding the areas of higher/lower QEPD relative to SD, results show an uneven protection: areas of high QEPD relative to SD tend to be well represented in PAs compared to random for any taxa and any PA network analyzed, but areas of low QEPD relative to SD tend to be underrepresented (Table S5, Appendix S4, significant for all taxa except for mammals). The representation of each category is consistent among PAs networks meaning that when one category is well represented by one network it is also the case in the other network. Sites with positive, negative and neutral convergence (PC, NC and NeC) are always less represented in PAs than random (Fig. 4, only significant for PC in WDPA, p < 0.01, for NC in WDPA I, II, p < 0.001, and for NeC for Natura 2000, p < 0.001). For instance, only 8.54%, 1.63% and 16.41% of the total PC cells are covered by WDPA, WDPA I,II and Natura 2000 respectively (Fig. 4 and Table S3, Appendix S5) when a random distribution of those PAs networks will cover these cells category better (10.43% ± 0.94 for WDPA, 2.26% ± 0.51 for WDPA I,II and 17.70% ± 1.68 for Natura 2000). On the contrary, divergent sites (D, Fig. 4 and Table S3, Appendix S5) are better covered by any PAs network than a random distribution of PAs would. Indeed, 11.3%, 3.03% and 20.8% of D cells are covered by WDPA, WDPA I, II and Natura 2000 respectively, whereas only 10.41% (± 0.21), 2.28% (± 0.11) and 17.75% (± 0.37) of D cells would be captured if WDPA, WDPA I, II and Natura 2000 respectively were randomly distributed.
Figure 4. Percentage of representation of congruency categories within protected areas.
PC, positive convergence, NC, negative convergence, NeC, neutral convergence and D, divergence. The black crosses are the observed percentage of protection while the box is the mean percentage of protected cells (relative to the total number of cell within the given category) over 1000 randomizations. The stars are the two-sided pvalues of the test comparing the observed and expected value. ***, p<0.001, **, p<0.005, *, p< 0.01
DISCUSSION
Patterns of spatial mismatch between the phylogenetic diversity of European vertebrates
Surrogate taxa are often used in conservation exercises due to the urgency in decision-making and the lack of comprehensive data for the majority of taxa (Rodrigues & Brooks, 2007). Such approaches assume that maximizing the diversity of one clade could lead to the maximization of overall biodiversity (e.g. other taxa). In our study, we showed positive co-variation of SD across vertebrates in Europe with highest correlation observed between mammals and amphibians compared to birds. Similar patterns have also been found at global (Lamoreux et al., 2006; Grenyer et al., 2006; Fritz & Rahbek, 2012), continental (Araújo et al., 2004) and national levels (Xu et al., 2008). This supports the idea that a species-rich region for one taxonomic group might be also expected, to some extent, to be rich for other taxonomic groups. However, these correlations are usually weak and sometimes simply explained by latitudinal gradients in diversity (Flather et al., 1997). Comparatively, the co-variation of QEPD patterns are weak between mammals and amphibians and almost null between birds and the two others taxonomic groups, meaning that high QEPD areas for one group is not at all representative of the QEPD level of the other groups. This suggests that in Europe and while accounting for species potential suitable area in the estimation of diversity, the surrogate’s principle cannot hold for other biodiversity facets than species richness, here phylogenetic diversity.
Potential mechanisms explaining biodiversity patterns
Disentangling the processes governing biodiversity patterns is not trivial (Gaston, 2000). Behavioural and ecological variation between the different groups of species might partly explain the observed patterns (Mittelbach et al., 2007). We showed that mammals have high SD in mountains, while this pattern is not found for amphibians and birds. Mammals are endotherm species and can stand in harsh climates (Mittelbach et al., 2007) while amphibians have difficulties to cope with values below zero (Araújo et al., 2006) and will tend to avoid extreme environments. Birds are also endotherm but might be more capable to avoid stressful environment due to their high dispersal ability or migration strategies (Mittelbach et al., 2007). But behavioural and ecological characters are probably not the only drivers of biodiversity.
Our approach to depict areas of higher and lower QEPD than expected for a given SD highlights regions with particularly rich or poor phylogenetic assemblages. Areas of positive residuals might reflect areas where the speciation rate has been low through time and lineages present in such region are likely to be old and suspend only few evolutionary distinct species (Isaac et al., 2007). Such sites might also be the mirror of ancient diversification or migration events but could also reflect high extinction rates (Davies & Buckley, 2011). Comparatively, assemblages with high SD but low QEPD can reflect a massive and recent diversification event only for some clades with a low extinction rate. We showed for mammals that islands (e.g. Corsica and Cyprus) present lower QEPD than expected; this could be explained partly by isolation from the main continent, with a species pools generated mostly by in-situ radiation through sympatric speciation resulting in assemblages composed of closely related species (Losos & Ricklefs, 2009).
Besides the ecological and historical drivers of species distribution, we cannot disregard the effects of anthropogenic influence (Mittelbach et al., 2007) and past climate change events (Araújo et al., 2006). Mammals, birds and amphibians are highly sensitive to human disturbance (Schipper et al., 2008; Stuart et al., 2004; Visconti et al., 2011). Anthropogenic forces are likely to have impacted species range and distributions by forcing species to migrate from their original habitat to new places. Such events (migration, introduction, extinction or range contraction) are likely to have modified the composition of assemblages and ultimately influenced phylogenetic diversity patterns differently for each groups, for instance we may lose large body size species first (Fritz et al., 2009).
Accounting for phylogenetically rich assemblages in conservation planning
We showed that areas characterized by either high or low QEPD for the three vertebrate groups simultaneously (i.e. convergent sites) are few in Europe and not better captured by PAs network than random. However, when taxonomic groups are analysed separately, areas of higher QEPD than predicted are better represented than random for any taxa, and for any PAs network. Such areas can be considered important to preserve because they are likely to contain profound nodes (great evolutionary history). Additionally, if we assume that assemblages with phylogenetically distinct species reflect assemblages of functionally different species, the protection of such areas would potentially maximize the preservation of ecosystem functioning (Cardinale et al., 2012; Cadotte et al., 2008). However, whether phylogenetic relatedness is a good proxy for functional similarity is controversial and recent analyses have shown that the assumption does not always hold (Mouquet et al., 2012; Lavergne et al., 2010). To verify such assumption, functional diversity, as measured directly from functional trait data, should be compared to phylogenetic diversity. Areas of lower phylogenetic diversity than expected could also be of conservation interest because they could potentially contribute to future evolutionary radiations under the hypothesis that they will continue to evolve at similar rates as in the past (Forest et al., 2007). In Europe these sites tend to be underrepresented in the PAs network.
However, we do not recommend targeting only the areas mentioned above as conservation priorities, because such a prioritization scheme would overlook species complementarity and cost-efficiency (Margules & Pressey, 2000). Indeed, two sites or regions having the same values of diversity (SD or QEPD) can reflect either similar or completely different species to the regional diversity and pools, meaning that in the maps presented here, there is no information on the redundancy between sites. A way to avoid redundancy between sites would be to not only maximize a set of high diversity sites (α-diversity) but also take into account the β-diversity (spatial turnover). This would tell us how much a site contributes to the regional diversity (γ-diversity) and the degree of compositional difference between sites. In any case, we believe that mapping the residuals as done here provides conservationists with a simple tool to contrast regions of high/medium/low congruencies between groups.
Underlying uncertainties
Although we used the best information available at European scale (Maiorano et al., 2013), it is evident that the resolution used in this study is too rough for practical management. We have partially addressed this problem by accounting for the amount of potential suitable area within pixel in the calculation of the phylogenetic diversity measure. However, the size of PAs in Europe still far exceed the resolution of the distribution data and our estimated percentage of protection should not be taken as exact quantitative estimates. Regional assessments with higher quality data should then follow such large-scale studies to accurately test the efficiency of PAs at protecting feature diversity.
Phylogenetic diversity in conservation: perspectives
Recent literature has questioned the rationale behind conserving phylogenetic diversity as well as the likelihood of adding this component in real conservation plans (Winter et al., 2013a,b; Rosauer & Mooers, 2013). Several reasons can justify the difficulty to use phylogenetic diversity as a relevant component for conservation. Obviously, one reason is ethical and does not need any biological justification: maximizing evolutionary history would preserve the “immense history of Earth” as a valuable dimension of biodiversity per se (Cadotte et al., 2010). The ecological reasons (i.e phylogenetic diversity as a proxy for ecological processes, evolutionary potential and ecosystem services) are less clear because many of the hypothesis behind cannot be taken for granted but need to be proved for each case considered. Moreover, adding a biodiversity component such as phylogenetic diversity to the one already used and accepted by conservation practitioners and policy makers is not an easy task. In this respect, species will probably still be considered as a simple and amenable currency for setting conservation action. However, when it comes that species are not representative of biodiversity as a whole, phylogenetic diversity offers an interesting alternative and is more or less already used in existing programs (e.g., CITES or EDGE). The growing availability of phylogenies for several groups and the development of handy softwares to estimate different indices of phylogenetic diversity (e.g. package picante in R, Kembel et al., 2010; Phylocom, Webb et al., 2008) help to produce maps, which are interesting tools for increasing the scope of conservation biogeography (Margules & Pressey 2000). Beyond these technical aspects, conservationists might communicate efficiently on the importance and meaning of phylogenetic diversity. A possible way of doing so could be to alert people on the natural heritage that phylogenetic diversity brings.
CONCLUSION
While global pattern of richness, threat and endemism have been widely investigated, still little is known on the distribution of other diversity facets among multiple taxa. In our study we offer a simple approach to identify areas of convergence of phylogenetic diversity for the 3 main groups of European terrestrial vertebrates. We show that phylogenetic diversity patterns strongly mismatch in space between groups and highlight that the diversity of one taxonomic group is not representative of the diversity of other groups. Moreover, we show that the current protected area network largely misses the few convergent regions and that protecting simultaneously several taxa and facets of diversity is challenging. Finally, we suggest that further research should be conducted on surrogate analyses, both to investigate other groups of taxa and to explore other facets of biodiversity (e.g. functional diversity) at different scales.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Münkemüller for her programming support and M. Winter together with two anonymous reviewers for their helpful comments. This research has received funding from the European Research Council under the European Community’s Seven Framework Programme FP7/2007-2013 Grant Agreement no. 281422 (TEEMBIO) and ANR-BiodivERsA project CONNECT (ANR-11-EBID-002), as part of the ERA-Net BiodivERsA 2010 call. Computations presented here were performed using the CIMENT infrastructure (https://ciment.ujf-grenoble.fr), supported by the Rhône-Alpes region (GRANT CPER07_13 CIRA: http://www.ci-ra.org).
BIOSKETCH
Laure Zupan interests lie at the interface between macroecology and biological conservation. Her focus has been on understanding global patterns of biodiversity for different groups of species to develop novel spatial conservation planning approaches to account for evolutionary history and ecosystem functioning.
REFERENCES
- Araújo MB, Densham PJ, Williams PH. Representing species in reserves from patterns of assemblage diversity. Journal of Biogeography. 2004;31:1037–1050. [Google Scholar]
- Araújo MB, Thuiller W, Pearson RG. Climate warming and the decline of amphibians and reptiles in Europe. Journal of Biogeography. 2006;33:1712–1728. [Google Scholar]
- de Bello F, Lavergne S, Meynard CN, Lepš J, Thuiller W. The partitioning of diversity: showing Theseus a way out of the labyrinth. Journal of Vegetation Science. 2010;21:992–1000. [Google Scholar]
- Cadotte MW, Cardinale BJ, Oakley TH. Evolutionary history and the effect of biodiversity on plant productivity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17012–17017. doi: 10.1073/pnas.0805962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadotte MW, Davies TJ, Regetz J, Kembel SW, Cleland E, Oakley TH. Phylogenetic diversity metrics for ecological communities: integrating species richness, abundance and evolutionary history. Ecology Letters. 2010;13:96–105. doi: 10.1111/j.1461-0248.2009.01405.x. [DOI] [PubMed] [Google Scholar]
- Cadotte MW, Davies TJ. Rarest of the rare: advances in combining evolutionary distinctiveness and scarcity to inform conservation at biogeographical scales. Diversity and Distributions. 2010;16:376–385. [Google Scholar]
- Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, Venail P, Narwani A, Mace GM, Tilman D, Wardle DA, Kinzig AP, Daily GC, Loreau M, Grace JB, Larigauderie A, Srivastava DS, Naeem S. Biodiversity loss and its impact on humanity. Nature. 2012;486:59–67. doi: 10.1038/nature11148. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. Phylogenetic overdispersion in Floridian oak communities. The American Naturalist. 2004;163:823–843. doi: 10.1086/386375. [DOI] [PubMed] [Google Scholar]
- Chao A, Chiu C-H, Jost L. Phylogenetic diversity measures based on Hill numbers. Philosophical Transactions of the Royal Society B, Biological Sciences. 2010;365:3599–609. doi: 10.1098/rstb.2010.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie DJ, Paquin V. Large-scale biogeographical patterns of species richness of trees. Nature. 1987;329:326–327. [Google Scholar]
- Davies TJ, Buckley LB. Phylogenetic diversity as a window into the evolutionary and biogeographic histories of present-day richness gradients for mammals. Philosophical Transactions of the Royal Society B, Biological Sciences. 2011;366:2414–2425. doi: 10.1098/rstb.2011.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devictor V, Mouillot D, Meynard C, Jiguet F, Thuiller W, Mouquet N. Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: the need for integrative conservation strategies in a changing world. Ecology Letters. 2010;13:1030–1040. doi: 10.1111/j.1461-0248.2010.01493.x. [DOI] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation. 1992;61:1–10. [Google Scholar]
- Faith DP. Phylogenetic pattern and the quantification of organismal biodiversity. Philosophical Transactions of the Royal Society B, Biological Sciences. 1994;345:45–58. doi: 10.1098/rstb.1994.0085. [DOI] [PubMed] [Google Scholar]
- Flather CH, Wilson KR, Dean DJ, McComb WC. Identifying gaps in conservation networks: Of indicators and uncertainty in geographic based analyses. Ecological Applications. 1997;7:531–542. [Google Scholar]
- Forest F, Grenyer R, Rouget M, Davies TJ, Cowling RM, Faith DP, Balmford A, Manning JC, Procheş S, Van der Bank M, Reeves G, Hedderson TAJ, Savolainen V. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature. 2007;445:757–760. doi: 10.1038/nature05587. [DOI] [PubMed] [Google Scholar]
- Fritz SA, Bininda-Emonds ORP, Purvis A. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecology Letters. 2009;12:538–549. doi: 10.1111/j.1461-0248.2009.01307.x. [DOI] [PubMed] [Google Scholar]
- Fritz SA, Rahbek C. Global patterns of amphibian phylogenetic diversity. Journal of Biogeography. 2012;39:1373–1382. [Google Scholar]
- Gaston KJ. Global patterns in biodiversity. Nature. 2000;405:220–227. doi: 10.1038/35012228. [DOI] [PubMed] [Google Scholar]
- Grenyer R, Orme CDL, Jackson SF, Thomas GH, Davies RG, Davies TJ, Jones KE, Olson V. a, Ridgely RS, Rasmussen PC, Ding T-S, Bennett PM, Blackburn TM, Gaston KJ, Gittleman JL, Owens IPF. Global distribution and conservation of rare and threatened vertebrates. Nature. 2006;444:93–96. doi: 10.1038/nature05237. [DOI] [PubMed] [Google Scholar]
- Guilhaumon F, Gimenez O, Gaston KJ, Mouillot D. Taxonomic and regional uncertainty in species-area relationships and the identification of richness hotspots. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15458–63. doi: 10.1073/pnas.0803610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer EJM, Blair MJ, editors. The EBCC Atlas of European Breeding Birds: their distribution and abundance. T & A.D. Poyser; London: 1997. [Google Scholar]
- Hillebrand H, Bennett DM, Cadotte MW. Consequences of dominance: a review of the evenness effects on local and regional ecosystem processes. Ecology. 2008;89:1510–1520. doi: 10.1890/07-1053.1. [DOI] [PubMed] [Google Scholar]
- Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PloS ONE. 2007;2:e296. doi: 10.1371/journal.pone.0000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN [Downloaded on 15 August 2013];The IUCN Red List of Threatened Species. (Version 2013.1). 2013 http://www.iucnredlist.org
- Jost L. Partitionning diversity into independent alpha and beta components. Ecology. 2007;88:2427–2439. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Kuhn TS, Mooers AØ, Thomas GH. A simple polytomy resolver for dated phylogenies. Methods in Ecology and Evolution. 2011;2:427–436. [Google Scholar]
- Lamoreux JF, Morrison JC, Ricketts TH, Olson DM, Dinerstein E, McKnight MW, Shugart HH. Global tests of biodiversity concordance and the importance of endemism. Nature. 2006;440:212–214. doi: 10.1038/nature04291. [DOI] [PubMed] [Google Scholar]
- Lavergne S, Mouquet N, Thuiller W, Ronce O. Biodiversity and Climate Change: Integrating Evolutionary and Ecological Responses of Species and Communities. Annual Review of Ecology, Evolution, and Systematics. 2010;41:321–350. [Google Scholar]
- Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457:830–836. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- Maiorano L, Amori G, Capula M, Falcucci A, Masi M, Montemaggiori A, Pottier J, Psomas A, Rondinini C, Russo D, Zimmermann NE, Boitani L, Guisan A. Threats from Climate Change to Terrestrial Vertebrate Hotspots in Europe. PLoS ONE. 2013;8:e74989. doi: 10.1371/journal.pone.0074989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margules CR, Pressey RL. Systematic conservation planning. Nature. 2000;405:243–253. doi: 10.1038/35012251. [DOI] [PubMed] [Google Scholar]
- Mittelbach GG, Schemske DW, Cornell HV, Allen AP, Brown JM, Bush MB, Harrison SP, Hurlbert AH, Knowlton N, Lessios HA, McCain CM, McCune AR, McDade L. a, McPeek M. a, Near TJ, Price TD, Ricklefs RE, Roy K, Sax DF, Schluter D, Sobel JM, Turelli M. Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecology Letters. 2007;10:315–331. doi: 10.1111/j.1461-0248.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- Morlon H, Schwilk DW, Bryant JA, Marquet PA, Rebelo AG, Tauss C, Bohannan BJM, Green JL. Spatial patterns of phylogenetic diversity. Ecology Letters. 2011;14:141–149. doi: 10.1111/j.1461-0248.2010.01563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillot D, Albouy C, Guilhaumon F, Ben Rais Lasram F, Coll M, Devictor V, Meynard CN, Pauly D, Tomasini JA, Troussellier M, Velez L, Watson R, Douzery EJP, Mouquet N. Protected and threatened components of fish biodiversity in the Mediterranean sea. Current Biology. 2011;21:1044–1050. doi: 10.1016/j.cub.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Mouquet N, Devictor V, Meynard CN, Munoz F, Bersier L-F, Chave J, Couteron P, Dalecky A, Fontaine C, Gravel D, Hardy OJ, Jabot F, Lavergne S, Leibold M, Mouillot D, Münkemüller T, Pavoine S, Prinzing A, Rodrigues ASL, Rohr RP, Thébault E, Thuiller W. Ecophylogenetics: advances and perspectives. Biological Reviews. 2012;87:769–785. doi: 10.1111/j.1469-185X.2012.00224.x. [DOI] [PubMed] [Google Scholar]
- Myers N. Threatened biotas: “hot spots” in tropical forests. The Environmentalist. 1988;8:187–208. doi: 10.1007/BF02240252. [DOI] [PubMed] [Google Scholar]
- Pavoine S, Bonsall MB. Measuring biodiversity to explain community assembly: a unified approach. Biological Reviews. 2011;86:792–812. doi: 10.1111/j.1469-185X.2010.00171.x. [DOI] [PubMed] [Google Scholar]
- Peres-Neto PR, Legendre P. Estimating and controlling for spatial structure in the study of ecological communities. Global Ecology and Biogeography. 2010;19:174–184. [Google Scholar]
- Petchey OL, Gaston KJ. Functional diversity (FD), species richness and community composition. Ecology Letters. 2002;5:402–411. [Google Scholar]
- Rao CR. Diversity and dissimilarity coefficients: A unified approach. Theoretical Population Biology. 1982;21:24–43. [Google Scholar]
- Ricklefs RE. Community diversity: relative roles of local and regional processes. Science. 1987;235:167–171. doi: 10.1126/science.235.4785.167. [DOI] [PubMed] [Google Scholar]
- Rodrigues ASL, Grenyer R, Baillie JEM, Bininda-Emonds ORP, Gittlemann JL, Hoffmann M, Safi K, Schipper J, Stuart SN, Brooks T. Complete, accurate, mammalian phylogenies aid conservation planning, but not much. Philosophical Transactions of the Royal Society B, Biological Sciences. 2011;366:2652–2660. doi: 10.1098/rstb.2011.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ASL, Brooks TM. Shortcuts for Biodiversity Conservation Planning: The Effectiveness of Surrogates. Annual Review of Ecology, Evolution, and Systematics. 2007;38:713–737. [Google Scholar]
- Roquet C, Thuiller W, Lavergne S. Building megaphylogenies for macroecology: taking up the challenge. Ecography. 2013;36:13–26. doi: 10.1111/j.1600-0587.2012.07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safi K, Cianciaruso MV, Loyola RD, Brito D, Armour-Marshall K, Diniz-Filho JAF. Understanding global patterns of mammalian functional and phylogenetic diversity. Philosophical transactions of the Royal Society B, Biological Sciences. 2011;366:2536–2544. doi: 10.1098/rstb.2011.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Schipper J, Chanson JS, Chiozza F, Cox NA, Hoffmann M, et al. The status of the world’s land and marine mammals: diversity, threat, and knowledge. Science. 2008;322:225–230. doi: 10.1126/science.1165115. [DOI] [PubMed] [Google Scholar]
- Slingsby JA, Verboom GA. Phylogenetic Relatedness Limits Co-occurrence at Fine Spatial Scales: Evidence from the Schoenoid Sedges (Cyperaceae: Schoeneae) of the Cape Floristic Region, South Africa. The American Naturalist. 2006;168:14–27. doi: 10.1086/505158. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stuart SN, Chanson JS, Cox NA, Young BE, Rodrigues ASL, Fischman DL, Waller RW. Status and trends of amphibian declines and extinctions worldwide. Science. 2004;306:1783–1786. doi: 10.1126/science.1103538. [DOI] [PubMed] [Google Scholar]
- Thuiller W, Lavergne S, Roquet C, Boulangeat I, Lafourcade B, Araújo MB. Consequences of climate change on the tree of life in Europe. Nature. 2011;470:531–534. doi: 10.1038/nature09705. [DOI] [PubMed] [Google Scholar]
- Vane-Wright RI, Humphries CJ, Williams PH. What to protect?—Systematics and the agony of choice. Biological Conservation. 1991;55:235–254. [Google Scholar]
- Visconti P, Pressey Robert L, Giorgini, D., Maiorano L, Bakkenes M, Boitani L, Alkemade R, Falcucci A, Chiozza F, Rondinini C. Future hotspots of terrestrial mammal loss. Philosophical Transactions of the Royal Society B, Biological Sciences. 2011;366:2693–2702. doi: 10.1098/rstb.2011.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M, Devictor V, Schweiger O. Phylogenetic diversity and nature conservation: where are we? Trends in Ecology & Evolution. 2013a;28:199–204. doi: 10.1016/j.tree.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Winter M, Devictor V, Schweiger O. Conquering current obstacles for avoiding the misuse of evolutionary diversity in nature conservation: a reply to Rosauer and Mooers. Trends in Ecology & Evolution. 2013b;28:323–324. doi: 10.1016/j.tree.2013.02.011. [DOI] [PubMed] [Google Scholar]
- Webb CO, Ackerly DD, McPeek M. a., Donoghue MJ. Phylogenies and Community Ecology. Annual Review of Ecology and Systematics. 2002;33:475–505. [Google Scholar]
- Webb CO, Ackerly DD, Kembel SW. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- Wood SN. Generalized Additive Models: an Introduction with R. Chapman & Hall/CRC Boca Raton; Florida, USA: 2006. [Google Scholar]
- Xu H, Wu J, Liu Y, Ding H, Zhang M, Wu Y, Xi Q, Wang L. Biodiversity Congruence and Conservation Strategies: A National test. BioScience. 2008;58:632–639. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.