Abstract
Bacteria develop a broad range of phage resistance mechanisms, such as prevention of phage adsorption and CRISPR/Cas system, to survive phage predation. In this study, Pseudomonas aeruginosa PA1 strain was infected with lytic phage PaP1, and phage-resistant mutants were selected. A high percentage (~30%) of these mutants displayed red pigmentation phenotype (Red mutant). Through comparative genomic analysis, one Red mutant PA1r was found to have a 219.6 kb genomic fragment deletion, which contains two key genes hmgA and galU related to the observed phenotypes. Deletion of hmgA resulted in the accumulation of a red compound homogentisic acid; while A galU mutant is devoid of O-antigen, which is required for phage adsorption. Intriguingly, while the loss of galU conferred phage resistance, it significantly attenuated PA1r in a mouse infection experiment. Our study revealed a novel phage resistance mechanism via chromosomal DNA deletion in P. aeruginosa.
Bacteriophages and their bacterial hosts are constantly engaged in evolutionary competition1,2,3. On the one hand, bacteria employ a wide range of resistance mechanisms, including prevention of phage adsorption and phage DNA entry, digestion of phage nucleic acids, and development of abortive infection and CRISPR/Cas systems to evade phage infection or survive phage killing1,4,5,6,7. On the other hand, bacteriophages are capable of rapid adaptive responses to evolutionary changes in their hosts2. Adsorption of phages into bacterial hosts by specific interactions between phage proteins and receptors on the bacterial cell surface is the initial step of infection8,9. Several receptors, including lipopolysaccharides (LPS), capsular polysaccharides, flagella, and outer membrane proteins, have been reported5,8,9. As a defensive strategy, bacteria can modify these cell surface receptors and produce extracellular matrix or competitive inhibitors to prevent phage absorption1. As a counter-defensive measure, phages are able to modify their receptor-binding proteins, such as tail fibers, to achieve infection and kill the resistant bacterium10,11. For example, Pseudomonas fluorescens SBW25 has coevolved with its lytic phage phi2 for over 300 bacterial generations. This co-evolution is probably due to the continuous modification of bacterial receptors and phage receptor-binding proteins3,12.
P. aeruginosa is an important Gram-negative opportunistic pathogen that causes serious infections in cystic fibrosis patients, cancer patients, and other immunocompromised individuals13,14. The bacterium remains one of the leading pathogens found in most medical centers, partly because of its ability to develop resistance to conventional antimicrobials. Bacteriophages are a group of viruses that are able to infect and kill bacteria15,16,17; they have long been proposed as promising alternatives to conventional antibiotics for treating bacterial infections. Given increasing levels of drug resistance in a variety of pathogens and the shortage of new antimicrobials, phage research, particularly phage therapy, has recently gained intense research interest18,19,20. Thus, efforts to exploit the application of phage therapy in the treatment of bacterial infections have steadily grown. Phage resistance, an essential survival strategy of bacteria, is an important aspect of bacterial-phage interactions and poses serious obstacles in the application of phage therapy21. Therefore, a better understanding of the mechanism of phage-resistance is critical in helping design successful phage-therapy strategies to treat/prevent bacterial diseases, including P. aeruginosa-related infections4,22.
We have recently isolated and sequenced three P. aeruginosa phages to explore their biological properties and interaction with host bacteria11,23,24. In this study, using one of the lytic phages (PaP1) and a clinical isolate of P. aeruginosa strain (PA123), we identified a novel phage resistance mechanism by chromosomal DNA deletion.
Results
PaP1 infection of P. aeruginosa resulted in red-pigment producing, phage-resistant mutants (Red mutants)
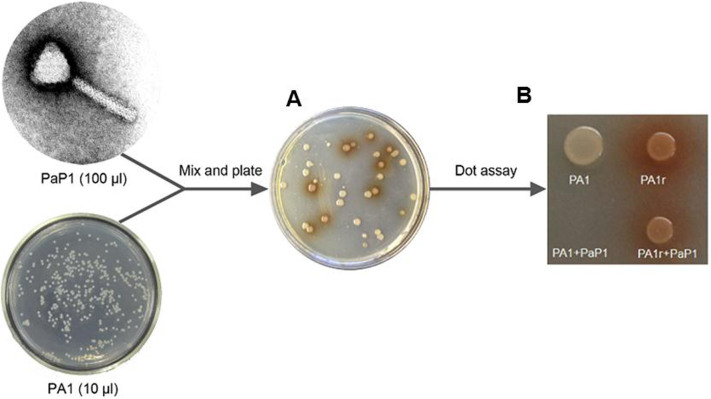
To investigate phage resistance mechanism in P. aeruginosa, wild-type strain PA1 was infected with phage PaP1 (MOI = 1000) and screened for phage-resistant mutants, which could be consistently recovered in high frequency (3.02 ± 0.85 *10−5) (Fig. 1A). Interestingly, a high percentage (31% ± 13.2%) of the mutants produced red pigment, and two coupled phenotypes can be stably maintained, indicating genetic mutations. One Red mutant, PA1r, was randomly selected for further analysis, and its PaP1 resistance, as well as red-pigment producing phenotypes, was confirmed by dot assay on an agar plate (Fig. 1B).
Figure 1. A subpopulation of phage resistant P. aeruginosa mutants produces red pigment.
(A): Red mutants were detected among other white phage resistant mutants. (B): Dot assay on agar plate shows that PA1r is phage resistant and produces red pigment.
Comparative genomic analysis revealed a 219.6 kb genomic fragment deletion in PA1r
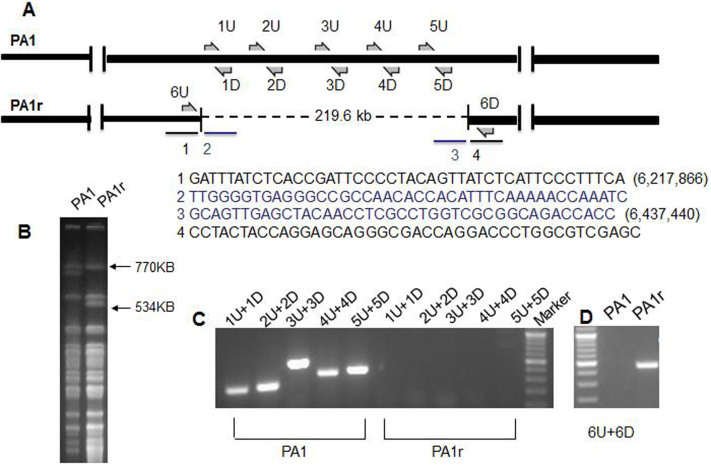
To identify the mutation(s) in PA1r that might confer PaP1 resistance, as well as red-pigment producing phenotypes, the genomes of both PA1 and PA1r were sequenced (GenBank: PA1: CP004054, PA1r: CP004055) and subjected to comparative genomic analysis. A 219,572 bp genomic fragment deletion (6,217,866 to 6,437,440) in PA1r compared with PA1 (Fig. 2A) was observed. Sequences flanking the end points are shown in Fig. 2A.
Figure 2. Genomic fragment deletion in phage-resistant strain PA1r.
(A): Diagram of P. aeruginosa genomes. 219.6 kb deleted fragment in PA1r is shown by dotted line. (B): PFGE of SpeI-digested genomic DNA from PA1 and PA1r. (C): PCR analysis of PA1 and PA1r. Positions of primers used for PCR analysis are indicated in Fig. A. (D): PCR analysis of the deletion site in PA1r.
The deletion in PA1r was confirmed by pulsed-field gel electrophoresis (PFGE) (Fig. 2B) and polymerase chain reaction (PCR) (Fig. 2C, Primer sets in Materials and Methods). The precise deletion site in PA1r was verified by PCR (Fig. 2D, Primer sets 6U/6D). Results showed that the deleted fragment in PA1r was flanked by gene cupA3 and PALES_33891 compared with another type of strain LESB5825.
Loss of hmgA gene as a result of genomic fragment deletion is responsible for red pigment production in PA1r
Bioinformatic analysis revealed that the deleted 219.6 kb genomic fragment contains 192 genes (Fig. S1), including hmgA, a gene encoding the enzyme homogentisate-1, 2-dioxygenase that converts homogentisic acid into 4-maleylacetoacetate. Inactivation of this gene results in the accumulation of homogentisic acid, which gives the cell a red pigmentation phenotype26,27.
An hmgA insertional deletion mutant (PA1ΔhmgA) was generated. PA1ΔhmgA produced red pigment but still retained sensitivity to phage PaP1 (Fig. 3), and complementation of hmgA gene in PA1r background did not restore the phage-resistant phenotype (Fig. 3). These results suggest that hmgA gene deletion does not confer PaP1 phage resistance.
Figure 3. Dot assay on agar plate.
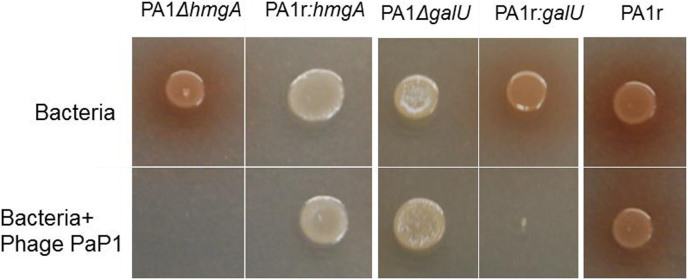
1 μl PA1ΔhmgA, PA1r: hmgA, PA1ΔgalU, PA1r: galU and PA1r were spotted on a LB plate alone or mixed with 2 μl 108 pfu/ml PaP1, and incubated at 37°C for 24 h.
Transposon mutagenesis revealed the involvement of PA1 lipopolysaccharides (LPS) in phage PaP1 infection
A random transposon mutant library of P. aeruginosa PA1 was infected with phage PaP1 to identify genes in P. aeruginosa that are essential for phage PaP1 infection. Two phage-resistant mutants (B10 and T2) (Table 1) were isolated and transposon insertion sites were determined by a two-step semi-degenerate PCR28. The mutation in B10 and T2 was mapped to genes PA0938 (wzz2) and PA5001, respectively, both of which have been shown to be involved in LPS biosynthesis29. Given that LPS often serve as phage-receptors in a variety of bacterial species8,30, our data suggested that PaP1 could potentially recognize PA1 LPS as receptors and initiate the infection process.
Table 1. Transposon mutants resistant to phage PaP1 infection.
PA1r carries defective LPS and can prevent phage adsorption
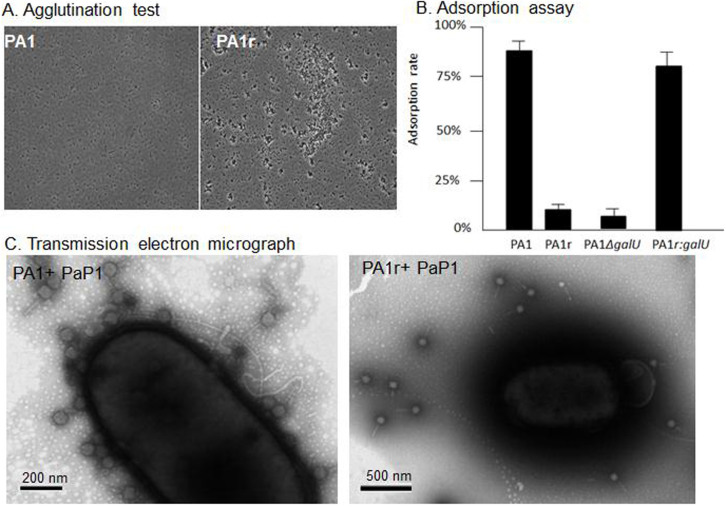
Agglutination test31 was performed with wild-type PA1 and PA1r strains to determine whether the resistance of PA1r to PaP1 infection was due to its defect in LPS biosynthesis. Results showed that PA1 remained as single cells, whereas PA1r tended to form cell clumps (Fig. 4A), suggesting that PA1r carries defective LPS.
Figure 4. PA1r is LPS negative and prevents phage adsorption.
(A): Agglutination test: PA1 remains single cells in 4% NaCl, but LPS negative bacteria PA1r are agglutinated. (B): Adsorption Assay of Bacteriophage PaP1 to P. aeruginosa PA1, PA1r, PA1ΔgalU and PA1r:galU. Phage adsorption to PA1 and PA1r were significantly different (Tukey's Multiple Comparison Test: P < 0.05). (C): Transmission electron micrograph shows that phage PaP1 binds to PA1 but not to PA1r.
Adsorption assays also revealed a significant reduction in phage adsorption to PA1r compared with wild-type PA1 strain (Fig. 4B). The result of the binding assay was further confirmed by transmission electron microscope data. After 3 min co-incubation, a significantly higher number of phage PaP1 particles can be seen attached to the surface of PA1 cell than those binding to PA1r (Fig. 4C).
Loss of galU in PA1r is responsible for phage resistance
Agglutination assay and phage adsorption data suggested that a possible defect in LPS biosynthesis might confer PaP1 phage resistance in PA1r. A detailed bioinformatic analysis revealed within the deleted fragment the presence of galU, a gene involved in LPS biosynthesis. This gene encodes UDP-D-glucose pyrophosphorylase which converts Glc-1-phosphate to UDP-D-Glc, which is the predominant sugar in the outer core of oligosaccharides. A galU mutant is devoid of O-antigen and synthesizes a defective LPS core32,33,34. Thus, a galU insertional deletion mutant (PA1 ΔgalU) was constructed. PA1ΔgalU showed a defect in LPS and was resistant to phage PaP1 infection. Moreover, galU was complemented into PA1r resulting in PA1r:galU, which was sensitive again toward this phage (Fig. 3).
High-frequency genomic fragment deletion is not site-specific
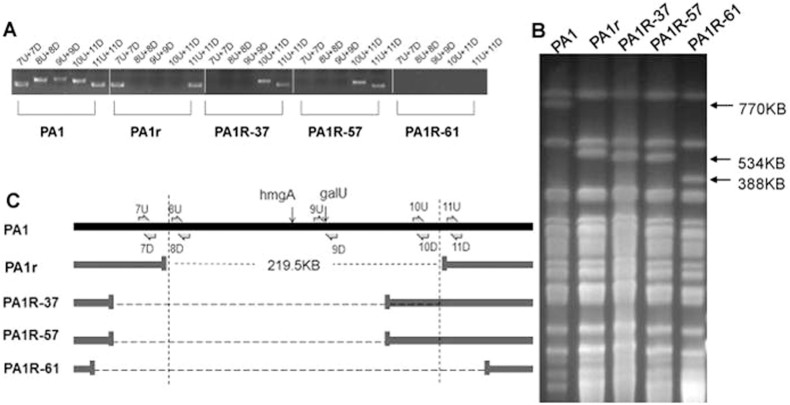
Three more Red mutants were randomly chosen and subjected to PCR and PFGE for further analysis to determine if Red mutants carry the same deletion. All three Red mutants (PA1R-37, PA1R-57, and PA1R-61) revealed genomic deletions with sizes similar to PA1r (Fig. 5). The deleted fragments ranged from approximately 200 kb to 300 kb, encompassing the region containing both galU and hmgA genes. However, the exact deletion sites among these mutants were different.
Figure 5. Genetic analysis of different phage-resistant Red mutants.
(A): PCR analysis of different resistant mutants. Primers used are indicated in Fig. 6C. (B): PFGE of SpeI-digested P. aeruginosa genomic DNA from different mutants. (C): Genomic diagram of PA1 and different mutant strains.
PA1r is significantly attenuated in a mouse infection model
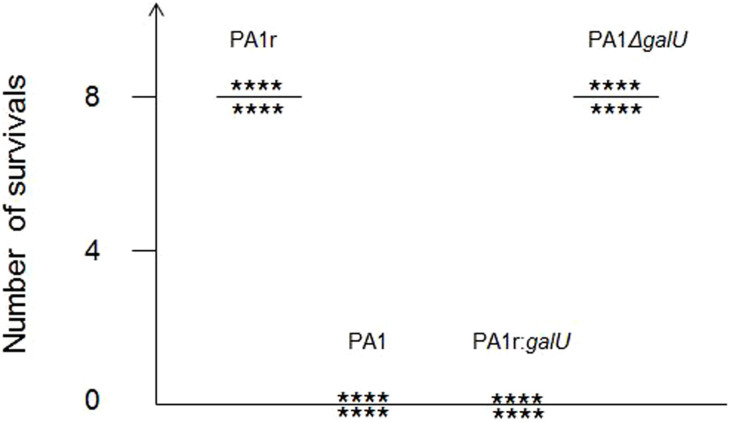
Our data suggested that PA1r acquired phage resistance by deleting a large genomic fragment that contains galU gene essential for LPS biosynthesis. Although the defect in LPS confers resistance to phage infection, this defect might reduce the mutant's ability to infect a host, because LPS is an important virulent factor in Gram-negative bacterial pathogens, including P. aeruginosa. To test this hypothesis, PA1r was used to infect mice intraperitoneally, and their survival was monitored. All eight mice died within 24 h after infection with either PA1 or PA1r:galU, whereas all eight mice survived the infection with PA1r or PA1ΔgalU (Fig. 6).
Figure 6. Phage resistant mutant PA1r is attenuated due to the loss of galU.
Survival was monitored in BALB/c mice at 24 h after challenged with 3*107 CFU wild type PA1, phage resistant mutant PA1r, galU mutant, PA1 ΔgalU or complementary strain PA1r:galU. Three independent replicates were performed, results were the same.
Red mutants are likely to arise spontaneously before phage infection
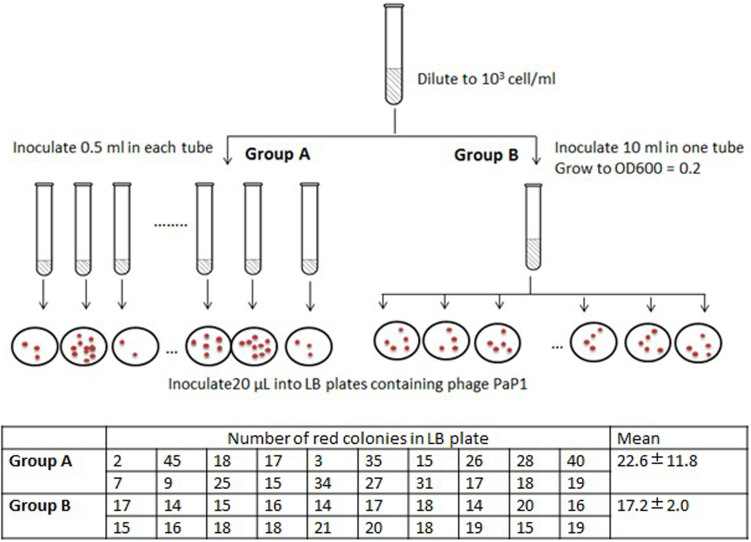
Fluctuation test was performed to demonstrate whether the genomic fragment deletion is induced after phage infection. As is shown in Figure 7, the number of red colonies on each plate varied drastically, from 2 to 45 colonies, in Group A; while the variation from group B is very small. This implies that deletion mutation arises before phage infection.
Figure 7. Red mutants arise before phage infection.
Diagram of fluctuation test is shown in the figure, and the numbers of red colonies are listed in the table. The numbers in Group A varied drastically, and the difference between group A and B is significant. (Student's t test, P < 0.05). Results from three independent replicates were similar, and this table presented the data from one replication.
Discussion
In this study, a series of phage-resistant P. aeruginosa mutants were isolated, and almost 30% of these mutants produced red pigment. However, by reviewing the phage resistance mechanisms1,2,35, no gene has been related to pigment production. Comparative genomic analysis revealed a 219.6 kb genomic fragment deletion in resistant mutant PA1r, including hmgA, which has been demonstrated to be associated with red pigment production (Fig. 3). However, the loss of hmgA could not confer phage resistance to PA1 (Fig. 3).
We further demonstrated that the loss of galU, a key gene involved in LPS biosynthesis, which is 19 kb in front of hmgA, confers phage resistance to PA1r by preventing phage adsorption because LPS is the likely receptor of PaP1 (Fig. 3). Adsorption to the receptor is the initial step for phage infection. Several phage receptors in Gram-negative bacteria, such as LPS, capsular polysaccharides, flagella, and outer membrane proteins5, have been identified. Mutation of the receptor is a common strategy for bacteria to avoid phage predation5,35. We used an agglutination assay to show that PA1r carried defective LPS (Fig. 4A). Given that the core oligosaccharide is abundant with negatively charged groups of 2-keto-3-deoxyoctulosonic acid and phosphate, strains that lack O polysaccharide will expose core oligosaccharide to the cell surface and cover up the negative charges. Therefore, LPS negative strains could be agglutinated by high salt concentrations31.
One of the major concerns regarding phage therapy is that phage-resistant mutants might result in the failure of phage therapy21. According to our experiment, phage-resistant bacteria are swift to emerge, and the frequency is much higher than those of drug-resistant mutants. However, phage-resistant mutant PA1r has defective LPS, and LPS is a major virulence factor in P. aeruginosa. Thus, LPS minus mutant PA1r is significantly attenuated in the mouse model (Fig. 6). The trade-off between phage resistance and fitness, such as growth rate or virulence, has also been reported in other phage-host experiments36, which suggests an advantage of phage therapy.
From a genetic perspective, the genomic fragment deletion as a response to phage predation and the underlying mechanism of phage resistance have not been reported. Phage resistance is often acquired by mutations1, DNA fragment insertion (CRISPR)37, or genomic rearrangements6. Our study demonstrated that bacteria could defend phage infection by deleting their own genomic fragment containing galU gene essential for phage infection. The two main causes of genomic fragment deletion are spontaneous and site-specific. Spontaneous deletion is caused by the dysfunction of DNA replication and repair system38. In this case, the deletion site is random and the deletion rate is quite low. For example, Tanji et al. isolated a phage-resistant Escherichia coli strain D198, which revealed a 209.4 kbp spontaneous deletion39. However, this deletion was spontaneous and not repeatable. In an evolution experiment with E. coli after 20,000 generations, only seven deletions were found, ranging from 1 bp to 23,293 bp40. In Salmonella enterica, the DNA loss rate was estimated to be 0.05 bp per chromosome per generation41. Sanna et al. estimated the apparent deletion rate in S. enterica with chromosomal variation ranging from 0.5 × 10−9 to 2.2 × 10−8/cell/generation42. Another experiment with P. aeruginosa also revealed random fragment deletion, but the deletion sites were unspecific and the frequency was quite low43.
By contrast, site-specific deletion is mediated by prophage or other mobile genetic elements44. These large DNA fragments are often genomic islands acquired by horizontal gene transfer and could be excised and integrated at a specific site mediated by integrase and other enzymes. This kind of deletion may usually occur at a high frequency. For example, an 89 kbp pathogenicity island in Streptococcus suis, which is flanked by a 15 bp direct repeat, could be excised from the genome with a certain frequency (~3.2 × 104)45.
In our experiments, the deletion events showed three clear novel features: high frequency (10−6), large genomic fragment deletions (200 kb to 300 kb), and no site-specificity. Thus, this deletion is neither site-specific nor spontaneous. Interestingly, smaller deletions are certainly favored in bacteria because the cost would be smaller, although the deletions in our experiment ranged from 200 kb to 300 kb. However, by reviewing the genes within the deleted region, none of the genes, including toxin genes, are obviously deleterious (Fig. S1). Therefore, this might be a novel deletion mechanism, and it is currently investigated in our laboratory.
Another interesting issue is whether the observed deletion was induced by phage infection or occurred before phage predation. The fluctuation test46 is a classical experiment to prove that phage resistant mutants arise before phage infection. If genomic fragment deletion was caused by an induced activation in bacteria, then each plate should contain roughly the same number of red colonies, because the deletion frequency should be consistent in the bacteria population. However, we found that the number of red colonies on each plate varied drastically, from 2 to 45 colonies in Group A; while the variation from group B is very small (Figure 7). This implies that deletion mutation arises before phage infection. What's more, the replication time of bacterium PA1 is 40 to 60 min at log phase (Figure S2), and the infection cycle of phage pap1 is about 40 min. Thus, within 40 min, the bacterium is unlikely to be able to delete its genomic fragment and losses LPS, because phage already killed bacteria before they replicate. Thus, it seems that the deletion mutants might have already existed in the bacterial population before phage infection.
The more intriguing question is why bacteria delete such a big fragment of their genome in the absence of phage. It is likely that phage resistant mutants were selected before phage treatment due to as-yet-unknown fitness benefits. In an effort to determine whether the selection could be due to mutants' growth advantage in culture medium, we compared the growth rate of PA1 and the deletion mutant PA1r. Compared to PA1 wildtype, PA1r displayed growth defect. (Figure S2). Furthermore, in a head-to-head competition experiment, PA1r was outcompeted by PA1(Figure S3). The fact that deletion mutants did not rise to a high frequency in the absence of phage could be due to the growth cost incurred by this deletion. This pleiotropic effect has also been reported in other phage resistant bacteria47,48. For example, phage resistant mutant of Pseudomonas fluorescens SBW25 was found to be less competitive when competing with wide type bacteria48. Thus, the unknown fitness of the deletion mutants before phage infection still needs further investigation in this system. In summary, we reported a novel phage resistance mechanism in which P. aeruginosa evades phage infection by genomic fragment deletion, a finding that could provide valuable insight into phage therapy.
Methods
Bacteria and bacteriophage growth conditions
Phages and hosts used in this study are listed in Table 2. P. aeruginosa and lytic phage PaP1 were stocked in our laboratory. P. aeruginosa strains were grown on Luria-Bertani (LB) agar plates or in LB broth with aeration at 37°C. When required, ampicillin, gentamicin, or tetracycline were used. Their concentrations in different media are described below.
Table 2. Bacterial strains, plasmids, and phage used in this study.
| Bacterial strain or plasmid | Relevant feature(s)a | Reference or source |
|---|---|---|
| P. aeruginosa Strains | ||
| PA1 | Wide type strain for phage PaP1 | [23] |
| PA1r | Phage resistant mutant | This study |
| PA1R-37 | Phage resistant mutant | This study |
| PA1R-57 | Phage resistant mutant | This study |
| PA1R-61 | Phage resistant mutant | This study |
| PA1ΔhmgA | Insertional deletion of hmgA | This study |
| PA1ΔgalU | Insertional deletion of galU | This study |
| PA1r: hmgA | Complementation of hmgA | This study |
| PA1r: galU | Complementation of galU | This study |
| E.coli strains | ||
| S17-1 (λ pir) | Conjugative donor strain | [51] |
| SM10pir | Conjugative donor strain | [28] |
| Bacteriophage | ||
| PaP1 | Lytic phage | [23] |
| Plasmid | ||
| pUCP24 | 4.036-kp pUC18-based broad-host-range vector, Gmr | [52] |
| pEX18Tc | Gene replacement vector, Tcr | [51] |
| pCM639 | ISphoA|hah-Tc delivery plasmid | [28] |
| pucp-galU | Broad-host-range vector containing galU, Gmr | This study |
| pucp-hmgA | Broad-host-range vector containing hmgA, Gmr | This study |
| pEX –galU | galU deletion vector, Tcr | This study |
| pEX -hmgA | hmgA deletion vector, Tcr | This study |
Abbreviations: Gmr, gentamicin resistance. Tcr, tetracycline resistance.
Selection for phage-resistant mutants
P. aeruginosa PA1 was grown at 37°C in LB broth with aeration. When the bacterial culture reached OD600 of 0.2, 10 μl of culture (approximately 1 × 106 cfu) was mixed with 100 μl and 1010 pfu/ml phage PaP1. The phage-infected culture was incubated at 4°C for 5 min and spread to one LB agar plate. Plates were then grown at 37°C for 24 h. Phage sensitivity of the isolated colonies were tested as previously described11. Briefly, 3 ml of 0.8% soft agar was mixed with 50 μl overnight culture of bacteria and was layered onto bottom agar. Finally, 10 μl of phages were deposited on the agar and incubated overnight at 37°C.
DNA extraction
Genomic DNA from P. aeruginosa wild-type strain PA1 and phage-resistant mutant PA1r were extracted using UNlQ-10 Column Bacterial Genomic DNA Isolation Kit (SK1202; Sangong Biotech, Shanghai) according to the manufacturer's instructions. The quality and quantity of extracted DNA were measured using spectrophotometry. Extracted DNA with a concentration of up to 500 ng/μl was used for the whole genome sequencing and PCR verification.
High-throughput genome sequencing
The complete genomes of the wild-type strain PA1 and the PaP1-resistant mutant PA1r were sequenced using Solexa to a depth of 200× (median coverage 161×), according to the manufacturer's protocols. The reference genome of P. aeruginosa strain LESB58 (GenBank accessions: NC_011770) was downloaded from NCBI and used for mapping the short sequencing reads produced by the Illumina Genome Analyzer. The sequencing reads of each isolate were mapped separately to their corresponding reference genome.
Verification of genomic fragment deletion by PCR
Primer sets (1U/1D, 2U/2D, 3U/3D, 4U/4D, and 5U/5D; Table 3) were designed to amplify five different regions within the 219.6 kb fragment. Primer set 6U/6D (Table 3) was used to detect the precise deletion site in PA1r. Primer sets (7U/7D, 8U/8D, 9U/9D, 10U/10D, and 11U/11D; Table 3) were designed to detect different deletion sites in different Red mutants. Their locations are indicated in Figs. 2A and 5A.
Table 3. Primers for detecting the deletion sites in phage resistant mutants.
| Primer name | Sequence |
|---|---|
| 1U | GCGGAGGCGAAGAAATGAAAGC |
| 1D | TTGACAGCGAACGGGTGAAACG |
| 2U | AGGGTCAGGTTCATAGCGGTCTCC |
| 2D | CGTCCAGTCTTCCTTGCGTTCC |
| 3U | GCCTGACAGCCAAGACGAGGAACA |
| 3D | GCCGAGCACGCAGGAAATGTAGC |
| 4U | CGCGTACAGGCTCATCAGCAACCC |
| 4D | TTCACCTTCGCCATCGCCATCA |
| 5U | GCCCACCGAATCTTCATAACACCA |
| 5D | AGGCGAACCAGAGCACCGTCAG |
| 6U | CTGGGAATGGATGACGAGACCTAC |
| 6D | GGATCGAGTGAGATCGGGTTGTA |
| 7U | GCAACAGGCGACCGATGAGGA |
| 7D | GGCGATGCCCGACAGCCAAGA |
| 8U | GCCACCAGTCCAACTACACCTA |
| 8D | ACGGCACGCTGAAGCTCGATAC |
| 9U | ATGATCAAGAAATGTCTTTTCCCG |
| 9D | TCAGTGAGCCTTGCCGGTCTTGT |
| 10U | AATTTAGAGCTGTCGCCTGTTTA |
| 10D | GATTGCCTATGCCAGTTAGTTTA |
| 11U | ACTATCTGAGCACGCAGAAGC |
| 11D | CGGTAGGTCTCGTCATCCATT |
PFGE
PFGE was performed as previously described49 with the following modifications. After digestion of the genomic DNA with SpeI, PFGE was performed at 14°C with CHEF MAPPER XA (Bio-Rad) at 200 V with switch times that ranged from 5 s to 52 s for 18 h.
Transposon mutagenesis
Transposon mutagenesis was performed as previously described28 with the following modifications. After mixing P. aeruginosa PA1 with E. coli strain SM10pir/pCM639 for 2 h, the cells were collected and resuspended in 1 ml of LB. Bacterial cells were then incubated with a 30-fold excess of the phage PaP1 for 20 min at 37°C and plated on LB medium containing tetracycline (65 μg/ml) and chloramphenicol (10 μg/ml) for the inhibition of the donor strain. The insertion of the transposon was identified by a two-stage semi-degenerate PCR and sequencing as described previously28.
Electron microscopy
Phage PaP1 and bacterial strain PA1 or PA1r were co-incubated for 3 min and placed on copper screen for 10 min before being negatively stained with 2% phosphotungstic acid (pH 4.8) for 1 min50. The samples were then scanned under TECNAI 10 electron microscope (Philips, The Netherlands) at 80 kV.
Agglutination test
LPS minus bacteria were agglutinated by high salt concentrations31. This procedure offers a simple way to distinguish LPS defective bacteria. Bacteria pellets from centrifuged cultures were resuspended in 4% NaCl for 5 min. Cells were then examined by microscopy. The cells of LPS defective strains were clumped together, whereas the LPS positive strains remained as single cells.
Bacteriophage adsorption assay
Bacteriophage adsorption assays were performed to determine whether insensitivity to bacteriophage was caused by receptor mutation11. Overnight culture of bacteria was collected by centrifugation at 13,000 × g for 1 min, and the cell pellet was washed with LB and resuspended to a final concentration of 109 cfu/ml in LB. Bacteriophage was added at a concentration of 105 pfu/ml to the bacterial suspension and incubated at 37°C with 150 rpm shaking under aerobic condition for 3 min. The samples were then filtered through a 0.2 μm filter, and phage titer was immediately determined by plaque assay. Percentage adsorption of the phage was calculated using the following equation: Percentage adsorption = [(initial titer − residual titer in the supernatant)/initial titer] × 100. Error bars represent the standard deviation of the three independent experiments.
Construction of galU and hmgA of P. aeruginosa mutants
P. aeruginosa galU insertional mutant was constructed by a described previously procedure51. With the use of PA1 genomic DNA as a template, a 598 bp fragment of galU was PCR-amplified with primer set q-GalU-U2 (forward: 5′-AAGCTT AAGGAAATGCTGCCGGTGGTGA-3′) and q-GalU-D2 (reverse 5′-CTATTCTAGA TTGCCCGGTTCAGTCTGCTCGA-3′). PCR products were gel-purified and ligated into T vector (pMD®19-T Simple vector, TaKaRa Code:D104A), resulting in pT-Q-galU. Fragment containing galU gene on the plasmid pT-Q-galU was cut out with XbalI/Hind III, and ligated into the XbaI/Hind III digested pEX18Tc, resulting in pEX -galU. Then, pEX -galU was transformed into E. coli strain S17-1 and conjugated into PA1 to generate galU insertional mutant via single crossover. The hmgA gene was knocked out as described above with the primers (forward 5′-tcatAAGCTTCGAGCAGCCCACCGACTTCCT-3′) and (reverse 5′-ctatTCTAGA GACCGTGCCGATGGTGTTGAA-3′).
Complementation of PA1r with galU or hmgA gene
The galU gene was PCR-amplified using forward (5′-GGATCCTATGATCAAGAAATGTCTTTTCCCG-3′) and reverse primer (5′-CTGCAGTCAGTGAGCCTTGCCGGTCTTGT-3′), and cloned into T vector (TaKaRa) resulting in pT-galU. The galU gene was then released from plasmid pT-galU via digestion with BamHI and PstI, and cloned into the same sites on pUCP2452 to generate pucp-galU. Then, pucp-galU was electroporated into strain PA1r to create PA1r:galU strains. The hmgA was complemented into PA1r using the same method with the primers forward hmgA-u (5′-ggatcctATGAACCTCGACTCCACTGCCC-3′), and reverse hmgA-d (5′-CTGCAGTTATCTCCGTTGCGGGTTGAAG-3′).
Mouse infection experiment
The Animal Research Ethics Committee of Third Military Medical University approved this mouse protocol. The animal experiment was performed in accordance with the Guidelines for the Care and Use of Laboratory Animals at Third Military Medical University. Bacteria were grown in 10 ml LB medium at 37°C until the early stationary phase. Cells were collected by centrifugation at 10,000 × g for 1 min. The cell pellet was washed and resuspended in saline to a final concentration of 3 × 107 cfu/ml. Each strain (1 ml) was injected intraperitoneally into 6- to 8-week-old BALB/c female mice, and animals were observed for 24 h. Three independent experiments were performed.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA, USA).
Fluctuation test
Log phage bacteria PA1 was diluted to 103 cell/ml. Then inoculate 0.5 ml into each of the 20 tubes (Group A), and inoculate 5 ml into one tube (Group B). When the culture reach OD600 = 0.2, 20 μL of cell culture from each tube in Group A was plated onto agar containing the PaP1 phage. For group B, all the 20 replicates were performed used the bacteria from the same tube. Then the red colonies were calculated after 24 h incubation.
Author Contributions
S.L., F.H. and W.S. conceived the study. S.L., X.Y., S.G.L., J.W., Y.Z. performed the experiments. Y.T., X.R., M.L., X.J., N.W. analyzed the sequence data. R.L. provided intellectual support. S.L. and H.X. wrote the paper.
Supplementary Material
Supplementary Informatio
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC,31171205) and State Scholarship Fund from China Scholarship Council. We thank H.P. Schweizer for providing plasmid pUCP24 and pEX18Tc. We thank Dr Jacobs for providing transposon plasmid pCM639.
References
- Labrie S. J., Samson J. E. & Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010). [DOI] [PubMed] [Google Scholar]
- Samson J. E., Magadan A. H., Sabri M. & Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 10.1038 (2013). [DOI] [PubMed] [Google Scholar]
- Pal C., Macia M. D., Oliver A., Schachar I. & Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450, 1079–1081 (2007). [DOI] [PubMed] [Google Scholar]
- Chibeu A. et al. The adsorption of Pseudomonas aeruginosa bacteriophage phi KMV is dependent on expression regulation of type IV pili genes. FEMS Microbiol. Lett. 296, 210–218 (2009). [DOI] [PubMed] [Google Scholar]
- Sorensen M. C. H. et al. Bacteriophage F336 Recognizes the Capsular Phosphoramidate Modification of Campylobacter jejuni NCTC11168. J. Bacteriol. 193, 6742–6749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A. E. et al. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 3, 1142–1151 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson T. R., Saroj S. D., Llewellyn A. C., Tzeng Y. L. & Weiss D. S. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497, 254–257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhuba D. V., Kolomiets E. I., Dey E. S. & Novik G. I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol 59, 145–155 (2010). [PubMed] [Google Scholar]
- Chatterjee S. & Rothenberg E. Interaction of bacteriophage l with its E. coli receptor, LamB. Viruses. 4, 3162–3178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A. & Brockhurst M. Bacteria-virus coevolution. Adv Exp Med Biol 751, 347–370 (2012). [DOI] [PubMed] [Google Scholar]
- Le S. et al. Mapping the Tail Fiber as the Receptor Binding Protein Responsible for Differential Host Specificity of Pseudomonas aeruginosa Bacteriophages PaP1 and JG004. Plos One 8, e68562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez P. & Buckling A. Bacteria-Phage Antagonistic Coevolution in Soil. Science 332, 106–109 (2011). [DOI] [PubMed] [Google Scholar]
- Wolfgang M. C. et al. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100, 8484–8489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Jelsbak L. & Molin S. Microbial ecology and adaptation in cystic fibrosis airways. Environ. Microbiol. 13, 1682–1689 (2011). [DOI] [PubMed] [Google Scholar]
- Krylov V. N. Bacteriophages of Pseudomonas aeruginosa: long-term prospects for use in phage therapy. Adv Virus Res 88, 227–278 (2014). [DOI] [PubMed] [Google Scholar]
- Khairnar K., Raut M. P., Chandekar R. H., Sanmukh S. G. & Paunikar W. N. Novel bacteriophage therapy for controlling metallo-beta-lactamase producing Pseudomonas aeruginosa infection in Catfish. BMC Vet Res 9, 264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endersen L. et al. Phage Therapy in the Food Industry. Annu Rev Food Sci Technol 092415 (2014). [DOI] [PubMed] [Google Scholar]
- Harper D. R. & Enright M. C. Bacteriophages for the treatment of Pseudomonas aeruginosa infections. J. Appl. Microbiol. 111, 1–7 (2011). [DOI] [PubMed] [Google Scholar]
- McVay C. S., Velasquez M. & Fralick J. A. Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob. Agents Chemother. 51, 1934–1938 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello E. et al. Pulmonary Bacteriophage Therapy on Pseudomonas aeruginosa Cystic Fibrosis Strains: First Steps Towards Treatment and Prevention. Plos One 6, e16963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai T. & Park S. C. Bacteriophage therapy of infectious diseases in aquaculture. Res. Microbiol. 153, 13–18 (2002). [DOI] [PubMed] [Google Scholar]
- Cady K. C., Bondy-Denomy J., Heussler G. E., Davidson A. R. & O'Toole G. A. The CRISPR/Cas Adaptive Immune System of Pseudomonas aeruginosa Mediates Resistance to Naturally Occurring and Engineered Phages. J. Bacteriol. 194, 5728–5738 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. et al. Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: establishment of genus PaP1-like phages. Plos One 8, e62933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y. et al. Whole genome sequencing of a novel temperate bacteriophage of P. aeruginosa: evidence of tRNA gene mediating integration of the phage genome into the host bacterial chromosome. Cell Microbiol. 9, 479–491 (2007). [DOI] [PubMed] [Google Scholar]
- Winstanley C. et al. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res 19, 12–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Duan J., Sibley C. D., Surette M. G. & Duan K. Identification of mutants with altered phenazine production in Pseudomonas aeruginosa. J Med Microbiol 60, 22–34 (2011). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rojas A. et al. Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology-(UK) 155, 1050–1057 (2009). [DOI] [PubMed] [Google Scholar]
- Jacobs M. A. et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100, 14339–14344 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. D., Kocincova D., Westman E. L. & Lam J. S. Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15, 261–312 (2009). [DOI] [PubMed] [Google Scholar]
- Garbe J., Bunk B., Rohde M. & Schobert M. Sequencing and Characterization of Pseudomonas aeruginosa phage JG004. BMC Microbiol. 11, 12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenyuan S., Stocker B. A. D. & JULIUS A. Effect of the surface composition of motile Escherichia coli and motile Salmonella species on the direction of galvanotaxis. J Bacteriol. 178, 1113–1119 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury B., Carlson R. W. & Goldberg J. B. The structure of the lipopolysaccharide from a galU mutant of Pseudomonas aeruginosa serogroup-O11. Carbohydr Res 340, 2761–2772 (2005). [DOI] [PubMed] [Google Scholar]
- Priebe G. P. et al. The galU Gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect Immun 72, 4224–4232 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C. R. & Goldberg J. B. Pseudomonas aeruginosa galU is required for a complete lipopolysaccharide core and repairs a secondary mutation in a PA103 (serogroup O11) wbpM mutant. FEMS Microbiol Lett 210, 277–283 (2002). [DOI] [PubMed] [Google Scholar]
- Avrani S., Wurtzel O., Sharon I., Sorek R. & Lindell D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474, 604–608 (2011). [DOI] [PubMed] [Google Scholar]
- Friman V. P. et al. High Temperature and Bacteriophages Can Indirectly Select for Bacterial Pathogenicity in Environmental Reservoirs. Plos One 6, e17651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- Lee M. C. M. C. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet 8, e1002651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji Y., Hattori K., Suzuki K. & Miyanaga K. Spontaneous deletion of a 209-kilobase-pair fragment from the Escherichia coli genome occurs with acquisition of resistance to an assortment of infectious phages. Appl. Environ. Microbiol. 74, 4256–4263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E. et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–U1274 (2009). [DOI] [PubMed] [Google Scholar]
- Nilsson A. I. et al. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci U S A. 102, 12112–12116 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi S., Sun S., Berg O. G. & Andersson D. I. Selection-Driven Gene Loss in Bacteria. PLoS Genet. 8, e100278710.1371/journal.pgen.1002787. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. E. et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103, 8487–8492 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D. F., Whiteson K. L. & Rice P. A. Mechanisms of Site-Specific Recombination. Annu. Rev. Biochem. Vol. 75 Annual Review of Biochemistry 567–605 (2006). [DOI] [PubMed] [Google Scholar]
- Li M. et al. GI-type T4SS-mediated horizontal transfer of the 89 K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol Microbiol 79, 1670–1683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E. & Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics 28(6), 491–511 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295, 2091–2094 (2002). [DOI] [PubMed] [Google Scholar]
- Hall A. R., Scanlan P. D., Morgan A. D. & Buckling A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett 14, 635–642 (2011). [DOI] [PubMed] [Google Scholar]
- Speijer H., Savelkoul P. H., Bonten M. J., Stobberingh E. E. & Tjhie J. H. Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J Clin Microbiol 37, 3654–3661 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. et al. Characterization and genome sequencing of phage Abp1, a new phiKMV-like virus infecting multidrug-resistant Acinetobacter baumannii. Curr Microbiol 66, 535–543 (2013). [DOI] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J. & Schweizer H. P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 (1998). [DOI] [PubMed] [Google Scholar]
- West S. E., Schweizer H. P., Dall C., Sample A. K. & Runyen-Janecky L. J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148, 81–86 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Informatio