Abstract
Janus kinase-2 (JAK2) supports breast cancer growth and clinical trials testing JAK2 inhibitors are underway. In addition to the tumor epithelium, JAK2 is also expressed in other tissues including immune cells; whether the JAK2 mRNA levels in breast tumors correlate with outcomes has not been evaluated. Using a case-control design, JAK2 mRNA was measured in 223 archived breast tumors and associations with distant recurrence were evaluated by logistic regression. The frequency of correct pairwise comparisons of patient rankings based on JAK2 levels versus survival outcomes, the concordance index (CI), was evaluated using data from 2,460 patients in 3 cohorts. In the case-control study, increased JAK2 was associated with a decreasing risk of recurrence (multivariate p=0.003, n=223). Similarly, JAK2 was associated with a protective CI (<0.5) in the public cohorts: NETHERLANDS CI=0.376, n=295; METABRIC CI=0.462, n=1,981; OSLOVAL CI=0.452, n=184. Furthermore, JAK2 strongly correlated with the favorable prognosis LYM metagene signature for infiltrating T cells (r=0.5, p<2×10−16, n=1,981) and with severe lymphocyte infiltration (p=0.00003, n=156). Moreover, the JAK1/2 inhibitor ruxolitinib potently inhibited the anti-CD3-dependent production of interferon-γ, a marker of the differentiation of T-helper cells along the tumor-inhibitory Th1 pathway. The potential for JAK2 inhibitors to interfere with the antitumor capacities of T cells should be evaluated.
Keywords: Breast cancer, JAK2, ruxolitinib, LYM metagene, T cells
INTRODUCTION
Janus kinase-2 (JAK2) is essential for the signaling of a variety of cytokine receptors including receptors for erythropoietin during erythropoiesis and for prolactin during mammary differentiation (1, 2). JAK2 has emerged as an important target in myeloproliferative disorders, and increasingly, in solid tumors such as breast cancer. Recent studies have implicated JAK2 in interleukin (IL)-6-dependent breast cancer stem cell self-renewal (3), and in both IL-6- and IL-8-dependent growth of triple-negative breast cancers (4). Furthermore, JAK2 signaling has been implicated as a mechanism of escape from other targeted breast cancer therapies (5). Thus, JAK2 inhibitors are being evaluated in patients with breast cancer (6). JAK2 is also expressed in diverse cell types, including immune cells, and whether the overall JAK2 mRNA levels in breast tumors are associated with clinical outcomes has not been evaluated.
Studies of mRNA levels in primary breast tumors have been useful for classifying breast cancers into subtypes that correlate with prognosis and drug responsiveness (7–9), for predicting recurrence (10, 11), and for delineating gene expression signatures that correlate with prognosis (12–15). Here, we evaluated the association between tumor mRNA levels of JAK2 and clinical outcomes in a novel case-control study and in 3 public cohorts. Outcomes included distant metastatic recurrence in a matched case-control study (n=223); recurrence-free survival in the Netherlands Cancer Institute cohort that was used to develop the MammaPrint recurrence risk test (n=295) (11); overall and disease-specific survival in METABRIC, currently the largest collection of gene expression and copy number data linked to long-term breast cancer outcomes (n=1,981) (14); and overall survival in OSLOVAL, a recent cohort that along with METABRIC formed the basis of the Sage Bionetworks DREAM breast cancer prognosis challenge (n=184) (12, 15).
MATERIALS AND METHODS
Selection of cases and controls
The protocol to use a breast cancer research database for case selection, to access institution-archived leftover tumor tissue, and to undertake molecular biology studies of the tissue was approved by the Institutional Review Boards of the Fred Hutchinson Cancer Research Center (File #6643) and the Swedish Medical Center (File #4924C-10). Patient consent was not required. The breast cancer research database at the Swedish Cancer Institute contains patient, tumor, treatment and outcomes data collected since 1989 for over 12,000 patients. The dataset was reduced to women followed for at least 2 years with invasive carcinoma with T1–3 primary tumors and treated by partial mastectomy plus breast irradiation or total mastectomy, sentinel node biopsy or axillary dissection, and adjuvant chemotherapy. Patients with multiple primaries, T4 primaries or distant metastases, and those receiving neoadjuvant chemotherapy were excluded. Matching variables included extra-nodal extension of metastasis, lymphovascular invasion, estrogen receptor (ER)/progesterone receptor (PR)/human epidermal growth factor receptor-2 (HER2) status, T-stage, and N-stage. A T-N interaction term allowed for the fact that tumor size is more important for women without positive nodes than for women with positive nodes. Within each matched pair, diagnosis dates of the recurring and non-recurring patients were no more than 2 years apart. Propensity scoring was used to match 112 cases of distant recurrence following surgery to 112 non-recurring controls using the “Optmatch” package (16) and R (17).
Quantitative RT-PCR
RNA was extracted from 4×10 μm sections using the Absolutely RNA FFPE system (Stratagene). The amount of tumor versus normal tissue in each section was greater than 50% for 84% of samples and greater than 90% for 47% of samples as determined by pathologists’ inspection of hematoxylin and eosin stained slides. cDNA was synthesized using random hexamers and Superscript III (Invitrogen) and was preamplified for 14 cycles using the Taqman preamplification system (Applied Biosystems). All probes bound exon junctions to prevent genomic DNA amplification (Supplementary Table S1). Diluted cDNA was used to seed triplicate real time PCR reactions for each Taqman assay using standard cycling conditions. Cycle threshold (Ct) values were determined using Sequence Detection Software (Applied Biosystems). Relative quantification was calculated as 2^−delta Ct, where delta Ct values were calculated by subtracting the indicated control gene mean Ct value from the target gene mean Ct value.
JAK2 mRNA levels and distant recurrence in the case-control study
Associations between JAK2 mRNA and the likelihood of recurrence were evaluated by logistic regression. In the continuous model, coefficients were calculated as estimates of the change in the log of the odds that an individual experienced a recurrence for every twofold increase in JAK2 levels, where a negative coefficient indicates that increasing JAK2 levels are associated with a decreasing likelihood of recurrence. Since participants had either one (n=184), two (n=26), or three tumor specimens (n=14), generalized estimating equations were used to account for varying numbers of tissues per individual (“Geepack” package) (18). Regressions were also performed using only primary tumors. For this, among the 26 individuals with both a primary and node specimen, only the primary tissue was included, and data from 31 individuals with only a node specimen were excluded. In the dichotomous model, coefficients were calculated with above-median versus below-median JAK2 levels as a predictor of recurrence, where multiple specimens were averaged to one value per individual. Multivariate analysis adjusted for clinical factors with which JAK2 expression was significantly correlated.
JAK2 mRNA levels and survival outcomes in the public cohorts
The inclusion criteria, clinical characteristics, and follow-up of the NETHERLANDS, METABRIC, and OSLOVAL cohorts were described (11, 12, 14). Data are available via Sage Bionetworks (www.synapse.org) under the following identifiers: doi:10.7303/syn4517.1, doi:10.7303/syn1688369; and doi:10.7303/syn1688370. Concordance index values were determined as described (12, 19).
RESULTS
Case-control study of JAK2 mRNA levels and distant recurrence
We previously optimized methods for measuring JAK2 mRNA in formalin-fixed, paraffin-embedded tumors by quantitative RT-PCR despite the degradation that characterizes RNA extracted from these samples (20). We applied this approach to tumor specimens from 112 women receiving surgery for breast cancer who subsequently experienced a distant metastatic recurrence and 112 women who did not. With the exception of a borderline significant increase in the number of ER-negative tumors among recurrences, there were no significant differences in clinical characteristics between cases and controls (Supplementary Table S2). Of note, while ER was a matching variable, it was not the only variable, which accounts for the residual effect of this strong prognostic factor even after propensity score matching. Sufficient RNA was available in 223 tumor specimens for JAK2 mRNA determinations. The validity of our mRNA measurements was confirmed by 1) the strong correlation in values obtained using probes for both JAK2 exon8/9 and exon23/24; 2) the reproducible mRNA levels across 3 separate tumor specimens for 14 tumors for which this comparison was possible; and 3) the strong concordance in mRNA levels of ESR1, PGR, ERBB2, and the corresponding clinical immunohistochemistry results (Supplementary Figure S1).
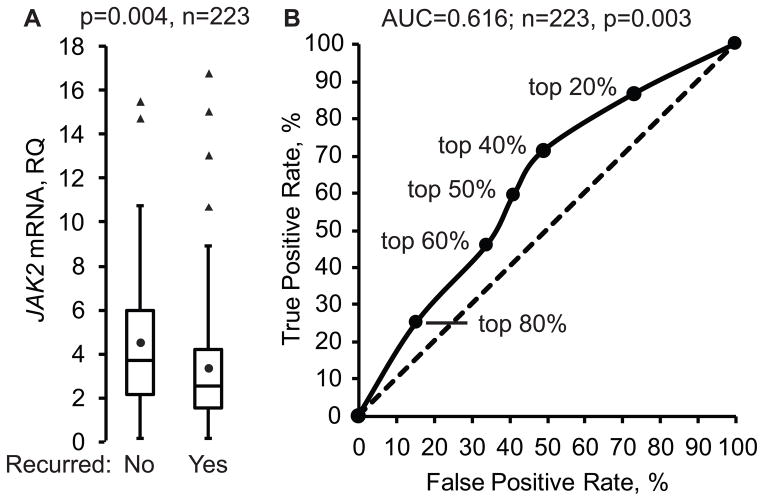
JAK2 mRNA levels were significantly higher in tumors from women who experienced no distant recurrence compared to those who experienced a distant recurrence (Figure 1A). This association was significant for both JAK2 exon8/9 and exon23/24 probes in logistic regression when JAK2 mRNA was treated as a continuous or dichotomous variable (Supplementary Table S3). Furthermore, with the exception of the JAK2 exon8/9 probe in the dichotomous model, significance was maintained in multivariate analysis. The association between increasing JAK2 mRNA and decreasing distant recurrence was also significant when analysis was restricted to primary tumors. A receiver operator curve revealed that the association between higher JAK2 mRNA and reduced recurrence was maximal when tumor samples with the top 40–50% of JAK2 expression level were defined as high JAK2 (Figure 1B).
Figure 1. JAK2 mRNA is associated with reduced distant breast cancer recurrence.
A) JAK2 mRNA was measured by quantitative RT-PCR using RNAs extracted from 223 breast tumor samples. Values for the JAK2 exon23/24 junction probe are shown normalized to the endogenous control gene HMBS. Box plots depict the distribution of normalized JAK2 mRNA values in quartiles. The circle in the box represents the mean value while the horizontal line represents the median value. Outliers are shown as triangles. The p-value was calculated using the T-test. RQ, relative quantification. B) JAK2 mRNA was modeled in a receiver operator curve as a predictor of reduced recurrence across different thresholds for defining high JAK2. The true positive rate versus false positive rate of distant recurrence for each model versus actual outcomes is plotted for each threshold of defining high JAK2. The percent of samples that are defined as high JAK2 are shown for each model (top %). The p-value for the area under the curve (AUC) was calculated using the Z-test.
JAK2 mRNA levels and survival outcomes in the public cohorts
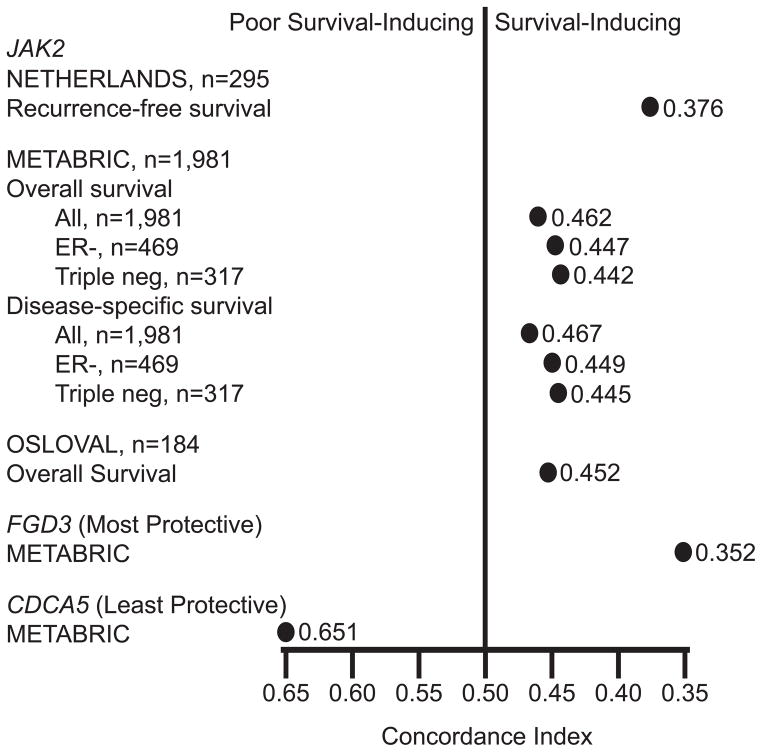
Next, we evaluated the association between JAK2 mRNA and outcomes in the NETHERLANDS, METABRIC, and OSLOVAL cohorts. We used the concordance index (19), which provides a convenient measure of the strength and direction of an association between a single gene and outcomes and was used to score submissions in the Sage Bionetworks DREAM breast cancer prognosis challenge (12). The concordance index is the relative frequency of correct pairwise comparisons of patient rankings based on gene expression levels versus survival outcomes. A concordance index >0.5 indicates that higher expression is associated with shorter survival while a value <0.5 indicates that higher expression is associated with longer survival. For example, using disease-specific survival data in METABRIC, the single-gene mRNA with the poorest prognosis was previously found to be CDCA5 with a concordance index of 0.651, indicating that if 2 patients were randomly selected, the patient with the higher CDCA5 level will have shorter survival 65.1% of the time (12). Conversely, the single most protective gene was FGD3 with a concordance index of 0.352, indicating that if 2 patients were randomly selected, the patient with the higher FGD3 level will have the longer survival 64.8% (100%-35.2%) of the time.
JAK2 mRNA exhibited a protective concordance index in all 3 datasets (Figure 2). The strongest effect was observed in the NETHERLANDS cohort, where the concordance index of 0.376 indicates that if 2 patients were randomly selected, the patient with the higher tumor JAK2 mRNA level will have the longer recurrence-free survival 62.4% (100%-37.6%) of the time. Similarly, JAK2 mRNA was consistently protective, albeit to a lesser extent, in the METABRIC and OSLOVAL cohorts. Since METABRIC provided sufficient sample size, we also evaluated the concordance index for JAK2 mRNA in ER- and ER-/PR-/HER2- (triple-negative) subtypes; JAK2 mRNA was even more protective for both overall and disease-specific survival in these subtypes.
Figure 2. JAK2 mRNA is associated with a protective concordance index in the NETHERLANDS, OSLOVAL, and METABRIC cohorts.
The concordance index in patient rankings based on JAK2 mRNA versus survival outcomes is shown for each indicated cohort. For comparison, the concordance indexes for the least and most protective single genes in METABRIC (CDCA5 and FDG3) are shown.
JAK2 mRNA and protein levels
To explore the mechanism by which JAK2 mRNA is associated with favorable prognosis, we investigated the relationship between JAK2 mRNA and protein levels. The specificity of a total JAK2 antibody was validated by the strong correlation between JAK2 mRNA and protein levels (as measured by western blotting) in a panel of 18 human breast cancer cell lines and by the ability to discriminate between JAK2-deficient γ2A cells (21) and JAK2-transfected γ2A cells in immunohistochemistry (Supplementary Figures S2 and S3). We then measured 10 tumors from the case-control study randomly selected from the highest quartile of JAK2 mRNA expression and 10 tumors from the lowest quartile. Immunohistochemical staining ranged from absent (0) to robust (3+) and was prominent in the tumor epithelial cells. However, JAK2 antibody staining levels in the tumor epithelial cells were not correlated with overall tumor JAK2 mRNA levels (r=−0.17, n=20) (Supplementary Table S4). This suggests that the association between higher JAK2 mRNA levels and favorable outcomes in breast cancer may not be a result of JAK2 protein function in breast tumor epithelial cells, and that other cell types in the primary tumors that are associated with prognosis contribute to overall JAK2 mRNA levels.
JAK2 mRNA levels and tumor-infiltrating T cells
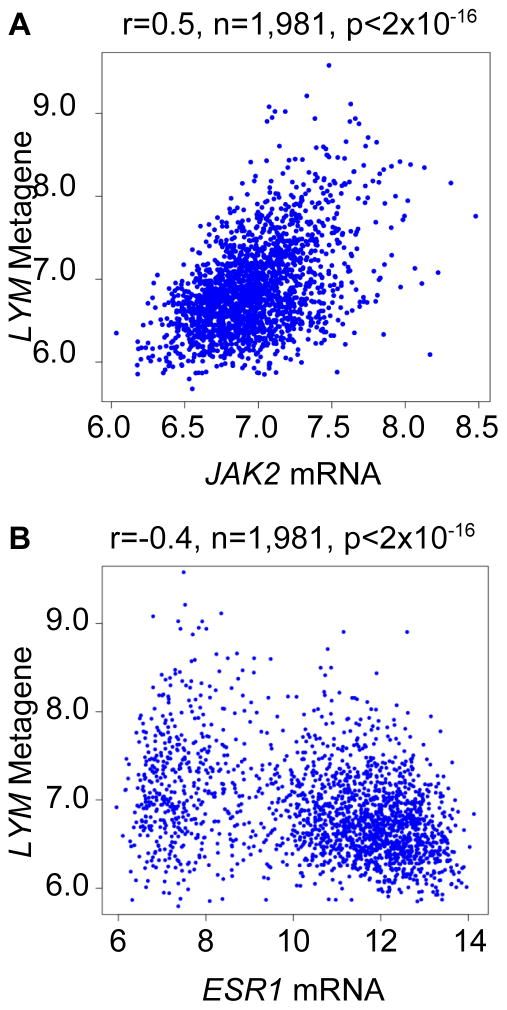
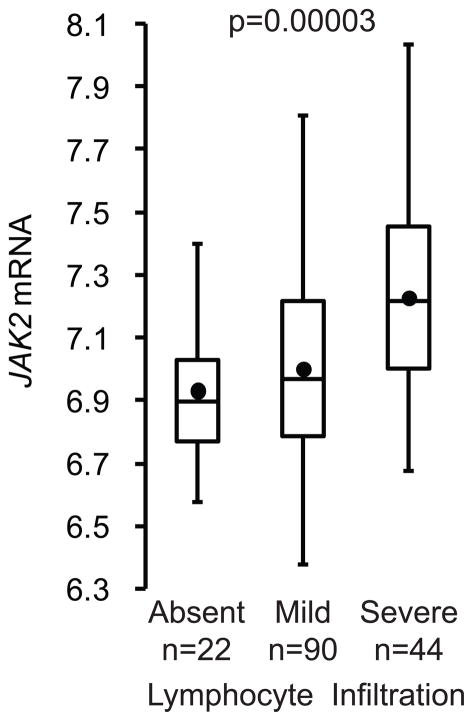
JAK2 is expressed in immune cells, and tumor-infiltrating lymphocytes, especially T cells, and have been associated with favorable breast cancer prognosis (14, 22). We therefore tested whether breast tumor JAK2 mRNA levels correlate with the T cell transcript-enriched LYM metagene signature. The LYM metagene is associated with favorable prognosis in breast cancer, in particular in ER-negative breast cancer and even more so in the absence of multiple positive lymph nodes, and recently formed part of the winning prognostic model in the Sage Bionetworks DREAM breast cancer prognosis challenge (12, 15). The LYM metagene was recently defined with increased accuracy following mining from data sets from multiple cancer types available from The Cancer Genome Atlas (23). Indeed, there was a highly significant correlation between JAK2 mRNA levels and the LYM metagene in tumor samples from METABRIC (Figure 3A). In contrast, the LYM metagene had an inverse correlation with the breast epithelial-associated transcript ESR1 (Figure 3B). Furthermore, JAK2 mRNA levels correlated strongly with levels of infiltrating lymphocytes as determined by pathologic assessment in a subset of 156 tumors for which these data were available (Figure 4). These tumor samples belonged to METABRIC integrative cluster 4, which was previously associated with favorable prognosis and a strong adaptive immune response signature (14). Finally, consistent with a functional role for JAK2 in supporting cytokine receptor signaling during T cell activation, we found that the JAK1/2 inhibitor ruxolitinib markedly inhibited the anti-CD3-dependent production of interferon-γ (IFNγ), a marker of the differentiation of T-helper cells along the tumor-inhibitory Th1 pathway (24) (Supplementary Figure S4). These results suggest that the consistently protective effect of JAK2 mRNA is related, at least in part, to the levels of infiltrating T cells.
Figure 3. JAK2 mRNA correlates with the LYM metagene signature.
A) The average expression of the top-ranked genes of the LYM metagene signature (SASH3, CD53, and NCKAP1L) in each tumor sample from METABRIC is shown relative to JAK2 mRNA. B) The scatter plot for ESR1, which is restricted to epithelial cells, is shown for comparison.
Figure 4. JAK2 mRNA correlates with levels of tumor-infiltrating lymphocytes.
JAK2 mRNA levels are shown relative to levels of tumor-infiltrating lymphocytes in tumor samples from the favorable prognosis METABRIC integrative cluster 4 that is enriched for an adaptive immune response signature. Box plots depict the distribution of JAK2 mRNA values in quartiles. The circle in the box represents the mean value while the horizontal line represents the median value. The p-value was calculated using ANOVA.
DISCUSSION
To our knowledge, this is the first time that a consistent association between increasing JAK2 mRNA levels and improved breast cancer outcomes has been demonstrated. This association was strongest in the case-control study that matched for variables associated with recurrence. Although the influence of JAK2 mRNA on survival outcomes in the unmatched public cohorts was predictably not as strong, the remarkably consistent association between higher JAK2 mRNA and favorable survival is unexpected because JAK2 proteins collaborate with a variety of cytokine receptors that were shown to promote breast cancer growth (3, 4). The association of JAK2 mRNA with favorable prognosis may reflect a lack of concordance between the levels of JAK2 mRNA and total JAK2 protein and the active phospho-JAK2 in breast tumor epithelial cells. The association between JAK2 mRNA and favorable prognosis also likely reflects the presence of additional JAK2-expressing cell types in the tumor specimens. Consistent with this, our previous laser capture microdissection studies demonstrated that JAK2 mRNA was expressed in both breast tumor epithelial and non-epithelial fractions, but was 8.3 (1.5–44.6) fold higher in non-epithelial fractions (25). Indeed, we presently observed a strong correlation between JAK2 mRNA and levels of tumor-infiltrating lymphocytes and the favorable prognosis LYM metagene signature. Our finding that a single gene correlates with a larger biomolecular metagene that is associated with prognosis is reminiscent of the frequent association between single genes and the PCNA and CIN metagene signatures for proliferation and chromosomal instability (12).
In addition to the present demonstration that the JAK1/2 inhibitor ruxolitinib inhibits CD3-dependent Th1 differentiation, support for a role of JAK2 in T cells is provided by studies demonstrating that JAK2 is involved in the signaling of IL-12 and IFNγ, key regulators of the tumor-inhibitory Th1 response (24, 26, 27). Furthermore, JAK inhibitors have been shown to impair production of these Th1 cytokines (28) and to inhibit IFNγ-dependent T cell trafficking in murine preclinical studies (29). Determining how specific inhibition of the individual JAK family members influences the repertoire and antitumor activities of tumor-infiltrating T cells represents an important area for future investigation. Such studies will provide insights into whether the benefits of inhibiting JAK2 in breast tumor cells outweigh the potential risks of inhibiting tumor-infiltrating T cells.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by grants from the National Cancer Institute (1R01CA135357 to CAB), Avon (P50CA083636-S2 to NDU), and the American Cancer Society (117682-MRSG-09-268-01-CCE to CPM).
We thank Brigham Mecham and Michèl Schummer for insightful discussions which improved the interpretation of the results, Mary Atwood for providing patient clinical characteristics, Desirée Iriarte and Kathy O’Briant for acquisition of pathology specimens, Ronald Tickman and Sean Thornton for tumor-block selection, Brian Johnson and Peggy Porter for immunohistochemistry and interpretation, Jeanne Lee for assistance in harvesting tumor tissue, Dan Herendeen, Ekram Gad, and Benjamin Curtis for providing murine splenocytes, Hsuan-Ni Lee, Nancy Zhang, and Galen Chen for assistance in analyzing data, and Erica Jonlin for assistance with human subjects.
Footnotes
Disclosure of Potential Conflicts of Interests: The authors have applied for a patent related to the content of this manuscript.
References
- 1.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–95. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 2.Wagner KU, Krempler A, Triplett AA, Qi Y, George NM, Zhu J, et al. Impaired alveologenesis and maintenance of secretory mammary epithelial cells in Jak2 conditional knockout mice. Mol Cell Biol. 2004;24:5510–20. doi: 10.1128/MCB.24.12.5510-5520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–35. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartman ZC, Poage GM, den Hollander P, Tsimelzon A, Hill J, Panupinthu N, et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 2013;73:3470–80. doi: 10.1158/0008-5472.CAN-12-4524-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britschgi A, Andraos R, Brinkhaus H, Klebba I, Romanet V, Muller U, et al. JAK2/STAT5 Inhibition Circumvents Resistance to PI3K/mTOR Blockade: A Rationale for Cotargeting These Pathways in Metastatic Breast Cancer. Cancer Cell. 2012;22:796–811. doi: 10.1016/j.ccr.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Lin N, Gelman R, Brock J, Bardia A, Mayer E, Overmoyer B, et al. Phase II study of ruxolitinib in patients with pStat3+ breast cancer. J Clin Oncol. 2013;31(suppl):abstr TPS1134. [Google Scholar]
- 7.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 9.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 11.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 12.Cheng WY, Ou Yang TH, Anastassiou D. Development of a prognostic model for breast cancer survival in an open challenge environment. Sci Transl Med. 2013;5:181ra50. doi: 10.1126/scitranslmed.3005974. [DOI] [PubMed] [Google Scholar]
- 13.Cheng WY, Ou Yang TH, Anastassiou D. Biomolecular events in cancer revealed by attractor metagenes. PLoS Comput Biol. 2013;9:e1002920. doi: 10.1371/journal.pcbi.1002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy N. Prognostic models: rising to the challenge. Nat Rev Cancer. 2013;13:378. doi: 10.1038/nrc3530. [DOI] [PubMed] [Google Scholar]
- 16.Hansen BB, Klopfer SO. Optimal full matching and related designs via network flows. Journal of Computational and Graphical Statistics. 2006;15:609–27. [Google Scholar]
- 17.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. URL http://www.R-project.org/ [Google Scholar]
- 18.Halekoh U, Højsgaard S, Yan J. The R package geepack for generalized estimating equations. Journal of Statistical Software. 2006;15:1–11. [Google Scholar]
- 19.Pencina MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23:2109–23. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 20.Miller CP, Lowe KA, Valliant-Saunders K, Kaiser JF, Mattern D, Urban N, et al. Evaluating erythropoietin-associated tumor progression using archival tissues from a phase III clinical trial. Stem Cells. 2009;27:2353–61. doi: 10.1002/stem.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohlhuber F, Rogers NC, Watling D, Feng J, Guschin D, Briscoe J, et al. A JAK1/JAK2 chimera can sustain alpha and gamma interferon responses. Mol Cell Biol. 1997;17:695–706. doi: 10.1128/mcb.17.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29:1949–55. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 23.Cheng W, Ou Yang TH, Shen H, Laird PW, Anastassiou D the Cancer Genome Atlas. Multi-cancer molecular signatures and their interrelationships. arXiv: 2013;1306.2584v2. [Google Scholar]
- 24.Pardoll D. Metastasis-promoting immunity: when T cells turn to the dark side. Cancer Cell. 2009;16:81–2. doi: 10.1016/j.ccr.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Miller CP, Urban N, Blau CA. Quantitative comparison of erythropoietin receptor levels in the epithelial versus endothelial fractions of primary breast tumors. Anticancer Res. 2011;31:1189–95. [PMC free article] [PubMed] [Google Scholar]
- 26.Bacon CM, McVicar DW, Ortaldo JR, Rees RC, O’Shea JJ, Johnston JA. Interleukin 12 (IL-12) induces tyrosine phosphorylation of JAK2 and TYK2: differential use of Janus family tyrosine kinases by IL-2 and IL-12. J Exp Med. 1995;181:399–404. doi: 10.1084/jem.181.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn BA, Quelle FW, et al. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-gamma signal transduction pathway. Nature. 1993;366:166–70. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 28.Fridman JS, Scherle PA, Collins R, Burn TC, Li Y, Li J, et al. Selective inhibition of JAK1 and JAK2 is efficacious in rodent models of arthritis: preclinical characterization of INCB028050. J Immunol. 2010;184:5298–307. doi: 10.4049/jimmunol.0902819. [DOI] [PubMed] [Google Scholar]
- 29.Choi J, Ziga ED, Ritchey J, Collins L, Prior JL, Cooper ML, et al. IFNgammaR signaling mediates alloreactive T-cell trafficking and GVHD. Blood. 2012;120:4093–103. doi: 10.1182/blood-2012-01-403196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.