Abstract
Loss of the nucleus is a critical step in keratinocyte terminal differentiation. To elucidate the mechanisms involved, we focused on two characteristic events: nuclear translocation of N-terminal fragment of profilaggrin and caspase-14-dependent degradation of the inhibitor of caspase-activated DNase (ICAD). First, we demonstrated that epidermal mesotrypsin liberated a 55-kDa N-terminal fragment of profilaggrin (FLG-N) and FLG-N was translocated into the nucleus. Interestingly, these cells became TUNEL positive. Mutation in the mesotrypsin-susceptible Arg-rich region between FLG-N and the first filaggrin domain abolished these changes. Furthermore, caspase-14 caused limited proteolysis of ICAD, followed by accumulation of caspase-activated DNase (CAD) in TUNEL-positive nuclei. Knockdown of both proteases resulted in a significant increase of remnant nuclei in a skin equivalent model. Immunohistochemical study revealed that both caspase-14 and mesotrypsin were markedly downregulated in parakeratotic areas of lesional skin from patients with atopic dermatitis and psoriasis. Collectively, our results indicate that at least two pathways are involved in the DNA degradation process during keratinocyte terminal differentiation.
Keywords: CAD, caspase-14, filaggrin, ICAD, mesotrypsin
Persistent presence of undigested nuclei in cornified cells (parakeratosis) is correlated with impaired barrier function, as shown by higher values of transepidermal water loss (TEWL), an indicator of barrier function.1 Although this is an important issue in connection with formation of the protective barrier of the skin, the mechanisms of DNA degradation during keratinocyte terminal differentiation remain unclear.
It has long been suggested that DNase 1L2 is responsible for denucleation in epidermis. However, DNase 1L2-deficient mice have a unique phenotype, with undigested nuclei being present only in hair shaft and nail plate.2 DNase 2, the predominant DNase on the mammalian skin surface, primarily targets exogenous DNA.3 Caspases are cysteine proteases that regulate apoptosis and inflammation.4, 5 Executioner caspases cause limited proteolysis of ICAD (inhibitor of caspase-activated DNase), releasing CAD (caspase-activated DNase) that is translocated into the nucleus, where it causes DNA degradation. However, proapoptotic caspases are not activated during terminal differentiation.6, 7, 8
Unlike other caspase family members, caspase-14 shows restricted expression, being found only in epidermis, skin appendages and thymic Hassall's bodies.9, 10 It has been suggested that caspase-14 is not involved in apoptosis or inflammation, but participates in keratinocyte terminal differentiation. Denecker et al.11 reported that although caspase-14 knockout mice showed defects in filaggrin (FLG) degradation, there was no effect on denucleation. However, the same group recently reported that caspase-14-deficient mice are prone to development of parakeratosis (persistent presence of nuclei in the cornified layer) in conditions triggering epidermal hyperproliferation.12 Another characteristic event related to the nuclear changes during terminal differentiation is translocation of the N-terminal fragment of profilaggrin into the nucleus. This translocation is associated with triggering of morphological changes of nuclei,13 although the mechanisms are not established.
In order to clarify the mechanism of denucleation of keratinocytes, we focused on two characteristic events that are associated with nuclear changes. One of our targets was the liberation and translocation of N-terminal fragment of profilaggrin, and the other was caspase-14, as its activation occurs at the onset of denucleation. Using liquid chromatography coupled with electrospray tandem mass spectrometry (LC/MS/MS), we found that the N-terminal fragment of profilaggrin interacts with epidermal mesotrypsin (trypsinogen 5)14 and caspase-14. Our results suggest that these proteases work cooperatively as well as independently via different pathways. Thus, at least two pathways appear to be involved in the DNA degradation process, and downregulation of both caspase-14 and mesotrypsin is associated with persistent presence of nuclei in the cornified layer.
Results
N-Terminal fragment of profilaggrin, consisting of A domain, B domain and truncated FLG repeat, is translocated into the nucleus
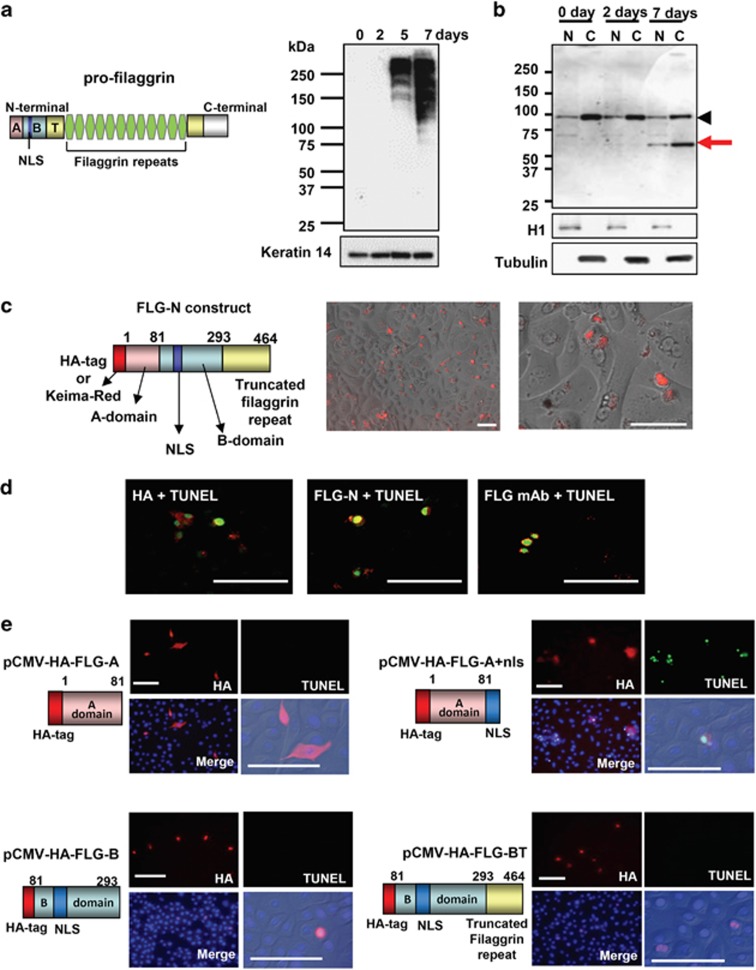
First, we examined the presence of N-terminal fragment of profilaggrin in nuclear fractions of cultured keratinocytes. Profilaggrin itself was detected at day 5 and strongly expressed at day 7 of prolonged culture after confluency (Figure 1a). Anti-A+B domain antibody (H-300) detected a fragment of 55 kDa in the nuclear extract as well as in the cytoplasmic extract obtained from the day 7 culture (Figure 1b), indicating that a large fragment consisting of the A–B domains together with a truncated domain is translocated into the nucleus after profilaggrin processing. Then, we prepared an expression vector with a 1392 bp insert encoding a 464-amino-acid N-terminal fragment of profilaggrin (FLG-N) (Figure 1c). This N-terminal region was selected based on the observed fragment size and the presence of possible cleavage sites for serine proteases around Arg464 of profilaggrin, as there is an Arg-rich region located at Arg455-Arg464. The molecular weight of FLG-N expressed in keratinocytes was similar to that of the fragment detected in nuclear extract (Supplementary Figure S1A). FLG-N was also recognized by anti-FLG monoclonal antibody (mAb). A time-lapse study using a construct of fluorescent protein Keima-Red (KR) fused to FLG-N (KR-FLG-N) showed that fluorescence emerged from 7 h after transfection and subsequently accumulated in the nucleus (Figure 1c and Supplementary Figure S1B). The transfected cells showed the increase of fluorescence and it gradually disappeared. Immunohistochemical study demonstrated that after FLG-N transfection in the growth phase, only HA-positive and FLG mAb-positive cells exhibited DNA degradation, as judged in terms of TUNEL positivity (Figure 1d). Translocation of FLG-N into the nucleus was confirmed with two different antibodies: anti-A+B domain antibody (H-300) and anti-FLG mAb that recognizes only the FLG repeat. These results show that FLG-N in the nucleus contains not only the A and B domains, but also a truncated FLG domain. Next we tried to identify which domain is responsible for DNA degradation. We constructed expression vectors for A domain (FLG-A), A domain with NLS (FLG-A+nls), B domain (FLG-B) and B domain+truncated FLG repeat (FLG-BT) (Figure 1e). After transfection of FLG-A, only cytoplasm was stained with anti-HA antibody (Ab), and TUNEL staining was completely negative. In contrast, cells expressing FLG-A with nuclear localization signal (NLS) showed accumulation of this fragment in the nucleus and became TUNEL positive. FLG-B- or FLG-BT-transfected cells did not develop TUNEL positivity, although the fragments were translocated into the nucleus. Thus, B domain by itself can enter the nucleus, in accordance with the observation of Pearton et al.15 However, our results indicate that the A domain is responsible for DNA degradation.
Figure 1.
Translocation of N-terminal fragment that includes truncated filaggrin repeat. (a) Profilaggrin structure and profilaggrin expression in cultured keratinocytes. Profilaggrin (∼500 kDa) consists of an N-terminal domain, 10 to 12 filaggrin repeats and a C-terminal domain. Keratinocytes were cultured until confluency (day 0). At 2, 5 and 7 days after confluency, keratinocyte extracts were examined with anti-filaggrin mAb. Keratin 14 was used as a loading control (bottom). (b) Presence of N-terminal fragment in the nuclear fraction. Cytoplasmic and nuclear extracts were obtained at confluence (day 0), and day 2 and day 7 after confluency. The presence of N-terminal fragment was detected using H-300. Note the appearance of a 55-kDa band at day 7 (red arrow). Histone H1 was used as a loading control of nuclear extracts. Tubulin was used as a loading control for cytoplasmic extracts. Arrowheads indicate nonspecific bands. (c) Schematic illustration of the profilaggrin N-terminal construct (FLG-N) and images of live FLG-N-transfected keratinocytes. Fluorescent protein, Keima-Red (KR), was fused to profilaggrin N-terminal construct of 464 amino acids, containing A domain, B domain and a truncated filaggrin repeat. After transfection into growing keratinocytes, time-lapse studies were carried out using PASCAL confocal microscopy. An image taken at 24 h after transfection is shown in the enlarged figure. Scale bars, 100 μm. (d) Presence of FLG-N in nuclei with TUNEL positivity. FLG-N was detected with anti-HA antibody (left), anti-A+B domain Ab (H300) (middle) and anti-filaggrin mAb (right). Images with TUNEL staining are shown as merged figures. FLG-N was detected with anti-filaggrin mAb (right). Scale bars, 100 μm. (e) The A domain of profilaggrin N-terminal is responsible for the induction of DNA degradation. A schematic illustration of each domain construct and results of expression studies are shown. After transfection with pCMV-HA-FLG-A, pCMV-HA-FLG-A+nls, pCMV-HA-FLG-B or pCMV-HA-FLG-BT vector, keratinocytes in the growth phase were stained for HA and TUNEL. Merged images with nuclear staining (DAPI) are shown. Enlarged images with differential interference contrast are also shown. Scale bars: 50 μm
N-terminal fragment of profilaggrin interacts with trypsinogen 5 and caspase-14
To identify molecules interacting with the N-terminal fragment of profilaggrin, we carried out affinity purification-mass spectrometry. An N-terminal fragment consisting of the A and B domains of profilaggrin was prepared as a glutathione-S-transferase (GST)-fusion protein (GST-N-FLG), following Pearton et al.15 GST-N-FLG was incubated with five kinds of extracts from keratinocytes and cornified cells, and interacting molecules were subjected to LC/MS/MS analyses. Molecules interacting with GST alone were also identified for exclusion as nonspecific binders. Structural proteins and ribosomal proteins were also excluded (Supplementary Table S3). We finally selected two proteases, trypsinogen 3 and caspase-14, as candidate interactors. These proteases were commonly detected in nuclear extracts of keratinocytes and in cornified cell extracts. Interestingly, the sequence identified by the analysis is identical with one that we have previously cloned, a specific isoform of the PRSS3 gene products that we named trypsinogen 5.14 After activation, identical mesotrypsin is produced from all three isoforms (trypsinogens 3–5). These proteases are of particular interest because their activation coincides spatiotemporally with denucleation.
N-terminal fragment of profilaggrin is released by mesotrypsin, and caspase-14 may contribute to this reaction
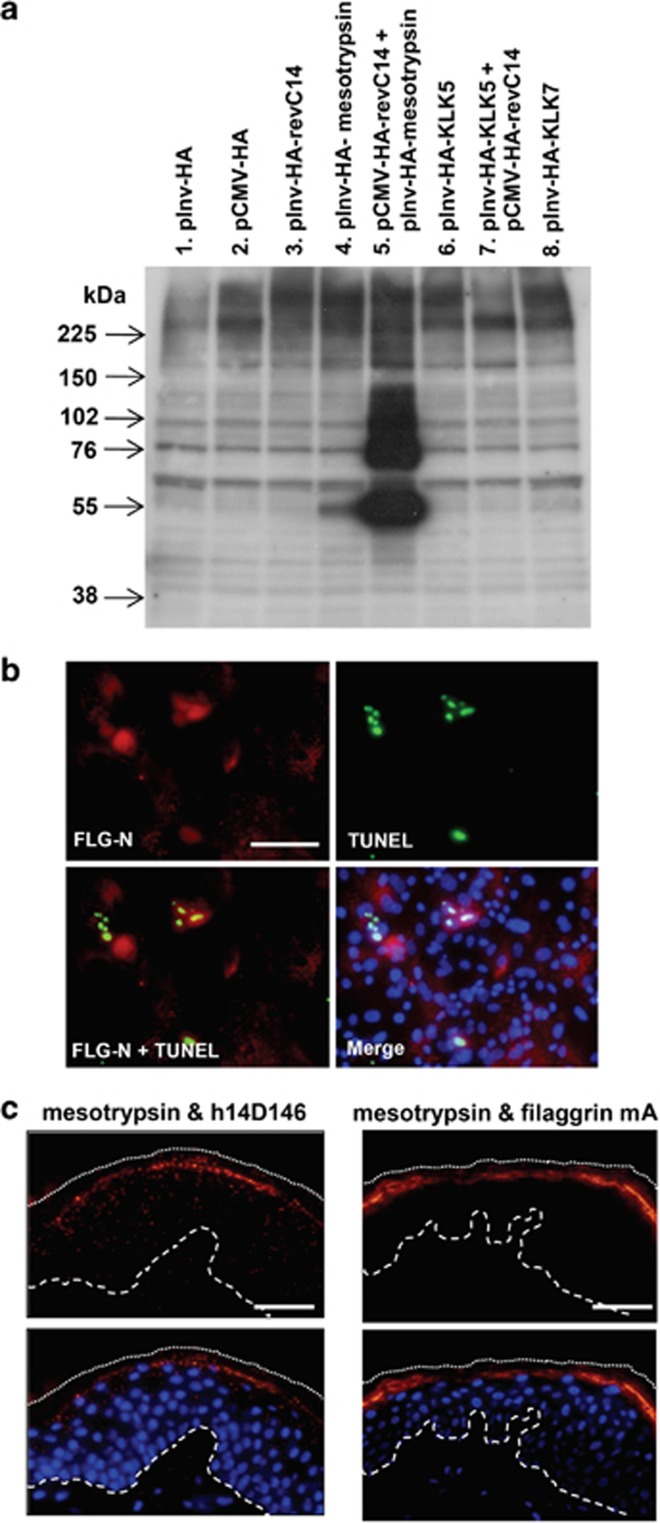
We constructed expression vectors for constitutively active mesotrypsin, caspase-14, kallikrein-related peptidase-5 (KLK5), and KLK7. KLK5 was chosen as it is suggested to be an initiator protease in desquamation protease cascades, and KLK7 was selected to examine the effect of a chymotrypsin-like enzyme. As caspase-14 is not activated in the monolayer culture system,6, 7, 8 we used constitutively active caspase-14 generated by reversing the large and small subunits (revC14), prepared in our laboratory.16 The revC14 showed equivalent enzyme activity to caspase-14 purified from the extract of human cornified cells17 (Supplementary Figure S2A). We used two kinds of promoters. The CMV promoter made it possible to express revC14, as well as other active proteases, in proliferating keratinocytes. On the other hand, the involucrin promoter enabled us to express these proteases in differentiated keratinocytes. Keratinocytes were transfected with these expression vectors and continuously cultured after confluency. Expression of the proteases was confirmed by means of western blotting with anti-HA antibody (Supplementary Figure S2B). Mesotrypsin alone liberated an N-terminal fragment of profilaggrin with a molecular weight of 55 kDa (Figure 2a), to a small extent. However, neither caspase-14 nor KLK5 and KLK7 alone produced the N-terminal fragment. Interestingly, we found that this fragment was efficiently generated when active caspase-14 and mesotrypsin were coexpressed. When constitutively active mesotrypsin was introduced into keratinocytes, the N-terminal fragment was accumulated in nuclei that became TUNEL positive (Figure 2b). Only mesotrypsin-expressing cells showed DNA degradation after confluence had been reached, as judged by colocalization of HA- and TUNEL-positive cells (Supplementary Figure S2C). We confirmed that expression of active mesotrypsin itself did not induce DNA degradation in the growth phase (Supplementary Figure S2D). As KLK5 is a trypsin-like protease, it may influence mesotrypsin activity. We tested this possibility, but found that KLK5 had no effect on mesotrypsinogen activation (Supplementary Figure S3A). Furthermore, we investigated the ratio of TUNEL-positive cells to HA-positive cells. Most of the revC14-expressing cells were TUNEL positive (95%). Active mesotrypsin-expressing cells showed rather lower TUNEL positivity (72%). Coexpression of these proteases resulted in 95% TUNEL positivity. Possible reasons why revC14-expressing cells showed a higher rate of TUNEL positivity would be a higher efficiency of DNA degradation by the ICAD–CAD system and/or a slower rate of degradation by the FLG-N–mesotrypsin system. Proximity ligation assay (PLA) suggested interaction between mesotrypsin and active caspase-14 as well as between mesotrypsin and filaggrin in normal human epidermis (Figure 2c). These results are consistent with a physiological role of epidermal mesotrypsin as an N-terminal-processing enzyme in differentiated keratinocytes.
Figure 2.
Epidermal mesotrypsin liberates an N-terminal fragment from profilaggrin. (a) Effect of epidermal proteases expressed in cultured human keratinocytes. Expression vectors with HA-tag driven by CMV promoter or involucrin promoter were prepared for constitutively active forms of caspase-14, mesotrypsin, KLK5 and KLK7. After transfection of vectors, keratinocyte extracts were obtained from day 5 confluent cultures and analyzed by western blotting with anti-A+B domain Ab (H-300). Lane 1, involucrin promoter-driven HA vector (pInv-HA); lane 2, pCMV-HA-vector; lane 3, pInv-HA-revC14 (constitutively active caspase-14); 4, pInv-HA-active mesotrypsin; 5, pCMV-HA-revC14+pInv-HA-active mesotrypsin; lane 6, pInv-HA-active KLK5; lane 7, pInv-HA-active KLK5+pCMV-HA-revC14; lane 8, pInv-HA-KLK7. (b) Immunohistochemical analysis of mesotrypsin-expressing keratinocytes from day 2 confluent culture. Constitutively active mesotrypsin was expressed using pInv-HA-mesotrypsin vector. Anti-N-terminal Ab (H-300) staining and TUNEL staining were carried out. DAPI was used for nuclear staining and a merged figure is also shown. Scale bars, 50 μm. (c) Association of mesotrypsin and active caspase-14 (left), mesotrypsin and filaggrin (right) in normal epidermis. PLA was carried out using anti-trypsin Ab/h14D146 and anti-trypsin Ab/anti-filaggrin mAb combinations. Lower panel shows merged figures with nuclear staining (DAPI). The broken line indicates the epidermal–dermal junction. The dotted line shows the border of the cornified layer. Blue staining (DAPI) indicates nuclei. Scale bars, 100 μm
Mutation in the Arg-rich region abolished mesotrypsin-induced nuclear translocation of FLG-N
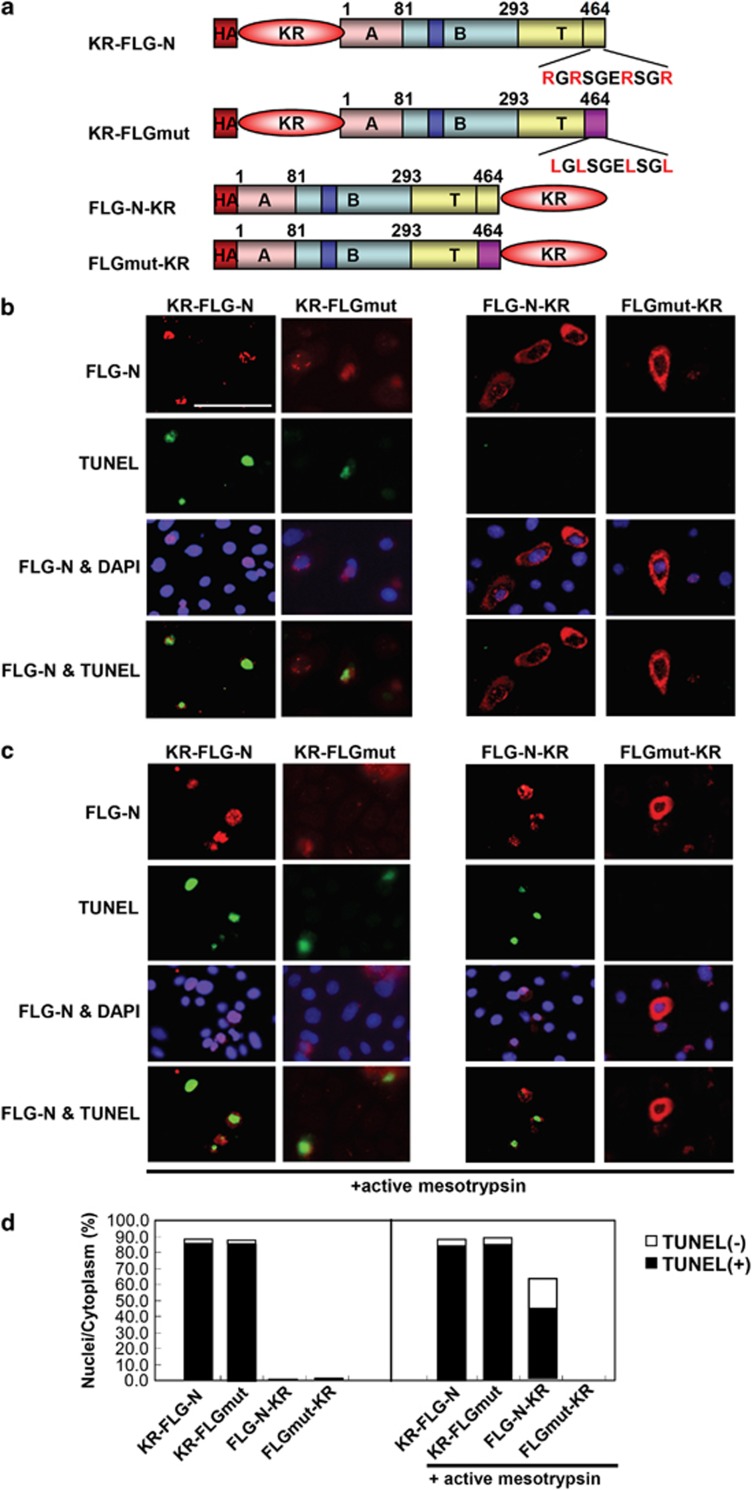
There are four arginines present in 10 residues (Arg455, Arg457, Arg461 and Arg464) near the C-terminal end of FLG-N. These were changed to Leu (FLGmut), and various expression vectors (FLG-N-KR, AG-FLGmut and FLGmut-KR) were constructed in addition to KR-FLG-N for elucidation of the physiological role of mesotrypsin (Figure 3a). When fluorescent proteins were fused to the N-terminal of FLG-N (KR-FLG-N) or FLGmut (AG-FLGmut), the fusion proteins were translocated into nuclei and the cells became TUNEL positive (Figure 3b). Accumulation of KR-FLG-N as well as AG-FLGmut in nuclei was evident in both cases. In contrast, fusion of the fluorescent proteins to the C-terminal of FLG-N (FLG-N-KR) or FLGmut (FLGmut-KR) completely suppressed the translocation and TUNEL staining was negative. Next, we co-transfected the active mesotrypsin construct into these cells. In the presence of active mesotrypsin, FLG-N-KR-expressing cells became TUNEL-positive, whereas coexpression of FLGmut-KR and active mesotrypsin had no effect (Figure 3c). The results of quantitative analyses are summarized in Figure 3d. Expression of KR-FLG-N and AG-FLGmut gave similar results, showing 88.3% and 91.1% nuclear localization, respectively. More than 95% of these cells were TUNEL positive. In contrast, FLG-N-KR or FLGmut-KR remained almost entirely in cytoplasm, and nuclei showed <1% TUNEL positivity. When FLG-N-KR was co-transfected with mesotrypsin, 63% of FLG-N-KR-expressing cells showed nuclear localization, and 69% of those cells became TUNEL positive. In the case of FLGmut-KR, coexpression caused no observable change. Furthermore, we performed biochemical analysis. Using recombinant FLG-N and FLGmut proteins as substrates, cleavage of the linker region by mesotrypsin was investigated. If the linker region of FLG-N (55 kDa) is cleaved, ∼3 kDa less peptide would be generated. Our results clearly showed that a 52-kDa peptide was generated from the recombinant FLG-N by the incubation with mesotrypsin. The recombinant FLGmut protein, where four Arg residues in the linker region were changed to Leu residues, did not show any changes in the incubation with mesotrypsin, showing insusceptible nature against mesotrypsin digestion (Supplementary Figure S3D). These results indicate that liberation of the FLG N-terminal is essential for translocation into the nucleus, and is sufficient to initiate DNA degradation. Epidermal mesotrypsin appears to be involved in the cleavage of the Arg-rich linker region.
Figure 3.
Mutation of Arg-rich region at the end of FLG-N abolished nuclear translocation. (a) Schematic illustration of various profilaggrin N-terminal constructs. pCMV-HA vector was used for expression of KR-FLG-N, KR-FLGmut, FLG-N-KR and FLGmut-KR. (b) These expression vectors were transfected into growing keratinocytes that were double stained using anti-FLG-N antibody (upper panel) and TUNEL stain (middle panel). Merged figures with nuclear staining are also shown (lower panel). When fluorescent proteins were fused to the C-terminal end, they remained in the cytoplasm and TUNEL staining was completely negative. (c) Keratinocytes were co-transfected with each FLG construct and active mesotrypsin and stained as described above. When mesotrypsin was coexpressed with FLG-N-KR, the cells became TUNEL positive. Mutation of the Arg-rich region before KR suppressed translocation into the nucleus. Scale bars: 20 μm. (d) Quantification of TUNEL-positive cells after transfection of various FLG-N constructs. Each N-terminal construct was coexpressed with mesotrypsin and nuclear and cytoplasmic localizations were evaluated. In each case, positive cells in 12 fields were counted and summed. The ratio of nucleus-positive cells to total N-terminal fragment-positive cells or that of TUNEL-positive cells among cells with nuclear localization was calculated
Caspase-14 alone induces DNA degradation in cultured keratinocytes
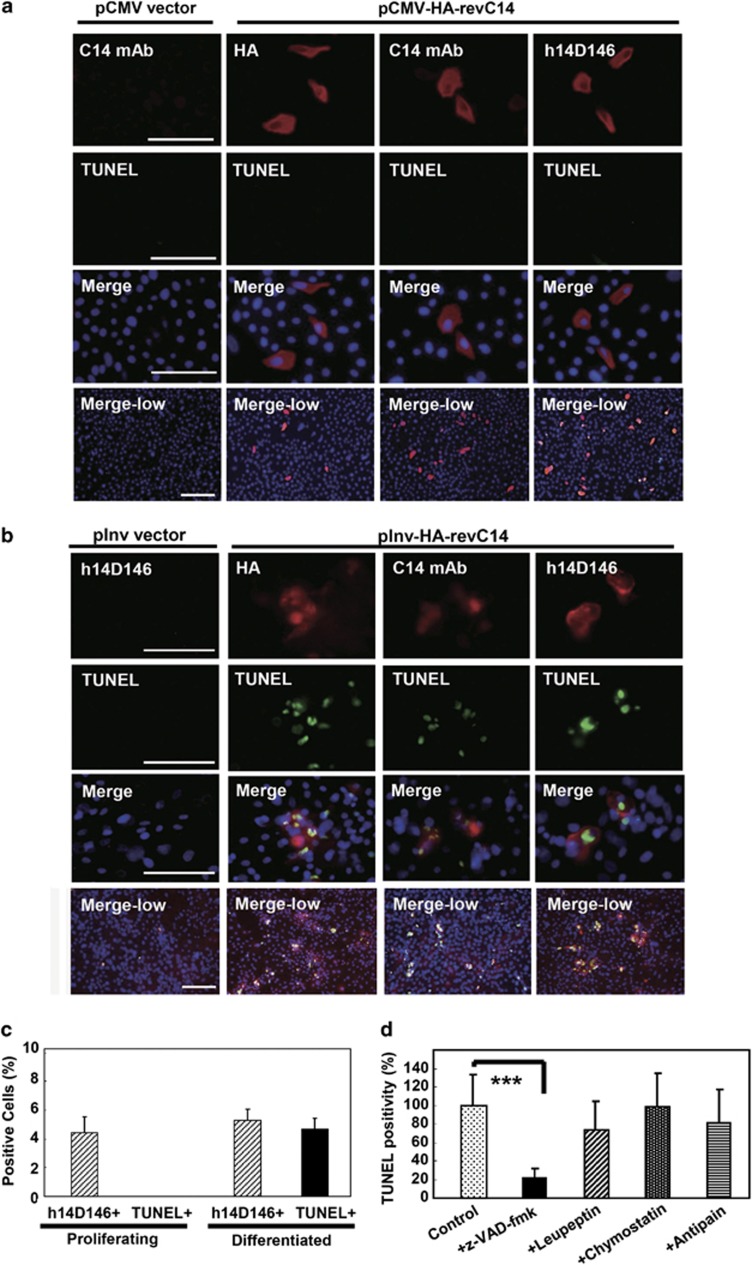
Keratinocytes were transfected with the CMV promoter-driven revC14 vector, and expression of revC14 was monitored by immunostaining with h14D146 (Figure 4). Although active caspase-14 was expressed in only ∼4.5% of keratinocytes, these cells in the growth phase did not show positive in TUNEL assay (Figure 4a). Control proliferating keratinocytes synthesized little or no caspase-14, as detected with anti-caspase-14 mAb. In another experiment, keratinocytes were transfected with the involucrin-promoter construct and cultured to confluency, then a further differentiation stimulus was applied (addition of 2 mM calcium). Figure 4b shows that involucrin promoter-driven revC14 emerged in the transfected cells after prolonged culture. In a typical experiment, revC14-positive cells amounted to ∼5.3% of the total, as judged by h14D146 antibody staining. Interestingly, most of the cells stained with these antibodies were found to be TUNEL positive (88% of the revC14-positive cells), indicating that DNA degradation occurred in the active caspase-14-expressing differentiated keratinocytes. The results of quantitative analysis are summarized in Figure 4c, confirming that active caspase-14 has the ability to induce DNA degradation in differentiated keratinocytes. Time-lapse study further confirmed that caspase-14-induced changes occurred only in differentiated keratinocytes (Supplementary Figure S4A). Among proliferating keratinocytes, caspase-14-expressing cells were actively moving, and some even showed cell division. In confluent culture, however, most of the keratinocytes transfected with the involucrin promoter-driven revC14 showed marked morphological changes after the induction of differentiation (Supplementary Figure S4B). As keratinocytes express many proteases,18 we further examined the effects of various protease inhibitors on the TUNEL positivity. DNA degradation was significantly suppressed only in the presence of z-VAD-fmk. Quantitative analysis demonstrated that in the presence of z-VAD-fmk, TUNEL-positive cells were markedly decreased to 21.5% of the number in the absence of the inhibitor (Figure 4d). Leupeptin, chymostatin and antipain, which are serine proteinase and lysosomal cysteine proteinase inhibitors, had no significant effect. Furthermore, we tested the effect of the enzyme-dead revC14 (revC14mut: 228Cys to 228Ala) in order to avoid nonspecific cell death. Expression of the inactive revC14 did not cause any significant effect and these expressing cells were TUNEL negative. Thus, DNA degradation was caspase-14 activity dependent.
Figure 4.
TUNEL-positive cells emerged among revC14-transfected cells. (a) Immunocytochemical analysis of revC14-transfected cells in the growth phase. Control (pCMV-HA vector only) wells were stained with anti-caspase-14 (C14) mAb. The pCMV-HA-revC14-transfected cells were stained with anti-HA antibody, anti-caspase-14 mAb and anti-h14D146 Ab. The second panel shows TUNEL staining. Merged figures with high magnification (third panel) and merged figures with low magnification (bottom) are shown. Scale bars, 100 μm. (b) Immunocytochemical analysis of revC14-transfected cells in the differentiated phase. Control (pInv-HA vector only) wells were stained with h14D146 Ab. pInv-HA-revC14-transfected cells were stained with the three Abs described above after keratinocyte differentiation. (c) Quantitative analysis of (a) and (b). Approximately 1500–2000 cells were counted for each assay and assays were repeated three times. Each bar indicates the mean±S.D. (n=7). (d) Effects of protease inhibitors on TUNEL positivity. The pInv-HA-revC14-transfected keratinocytes were cultured to confluence. Pan-caspase inhibitor, z-VAD-fmk, leupeptin, chymostatin or antipain was added and incubation was continued in a high calcium condition. After fixation with cold methanol, keratinocytes were stained for h14D146, TUNEL and nuclei (DAPI). TUNEL-positive cells were counted among 1500 cells in 7 fields and the results are summarized in this figure. Significant suppression of TUNEL reaction was found in z-VAD-fmk-treated cells. ***P<0.001
Caspase-14 cleaves ICAD and liberates CAD
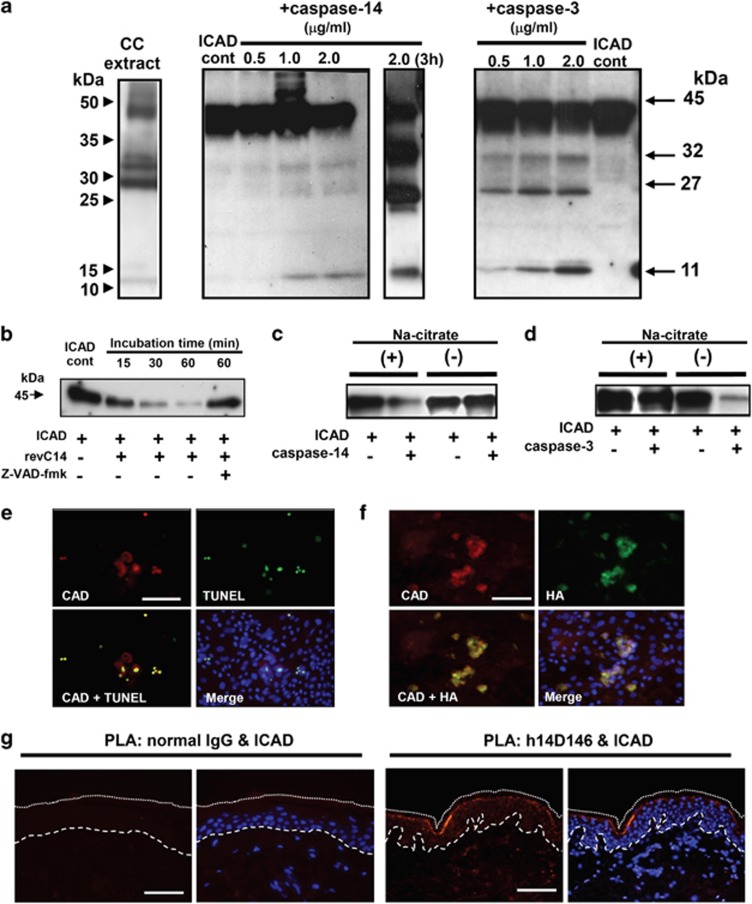
We further examined whether the ICAD–CAD system is involved in this pathway. Purified caspase-14 from human cornified cell extracts caused limited proteolysis of ICAD, releasing three major degradation products of 32, 27 and 11 kDa that appear to be similar to those generated by caspase-3 (Figure 5a). Prolonged incubation (3 h) increased the formation of the cleavage products. Interestingly, similar degradation bands were also observed in extracts of human cornified cells. We also tested the ICAD-degrading activity of revC14, and found that revC14 catalyzed time-dependent ICAD degradation (Figure 5b). Addition of zVAD-fmk strongly suppressed ICAD degradation by revC14. ICAD-degrading activity of caspase-14 was observed only in the presence of kosmotropic salt (Figure 5c) that is essential for enzymatic activity of caspase-14.19 However, in the presence of kosmotropic salt, ICAD degradation by caspase-3 was significantly suppressed compared with that by caspase-14 (Figure 5d). We next addressed the questions of whether CAD is liberated in this reaction and, if so, whether it is translocated into the nucleus. Keratinocytes transfected with revC14 expression vector were cultured for 2 days after confluency. Most CAD-expressing cells showed CAD accumulation in nuclei, and CAD-positive nuclei were also positive for TUNEL reaction (Figure 5e). Expression of revC14 was confirmed with anti-HA antibody (Figure 5f). HA-positive cells were also TUNEL positive. Close association between active caspase-14 and ICAD was demonstrated at the granular layer of normal human epidermis using PLA (Figure 5g). As profilaggrin is neither expressed nor processed in day 2 culture (Figure 1a), DNA degradation was caspase-14 activity dependent. Collectively, these results suggest that caspase-14 has the ability to degrade ICAD, liberating CAD and thereby leading to DNA degradation, although its action is restricted to differentiated keratinocytes.
Figure 5.
Caspase-14 induces DNA degradation via the ICAD–CAD system. (a) Western blot analysis of ICAD degradation products. Human caspase-14 was purified from corneocyte extract of healthy individuals using sequential chromatographic procedures.17 Purified active caspase-14 was incubated with ICAD, and ICAD degradation products were subjected to SDS-PAGE (15 μg each). Western blot analysis was carried out using anti-ICAD antibody (FL331). Western blot analyses of cornified cell extract (CC), ICAD+purified caspase-14 (30 min and 3 h incubation) and ICAD+caspase-3 (30 min incubation) are shown. (b) ICAD degradation by constitutively active caspase-14 (revC14). The effect of zVAD-fmk is also shown. (c) Effect of kosmotropic ion (sodium citrate) on ICAD degradation. ICAD degradation by purified caspase-14 was tested with or without kosmotropic ion. (d) Effect of kosmotropic ion on caspase-3 activity. ICAD-degrading activities of caspase-3 were examined with or without kosmotropic ion. (e) Accumulation of CAD after revC14 transfection. Keratinocytes were transfected with pInv-HA-revC14 and 2 days after confluency, examined with anti-CAD Ab and TUNEL staining. (f) Colocalization of CAD and revC14. The presence of revC14 was verified with anti-HA staining. Scale bars, 50 μm. (g) Association of active caspase-14 and ICAD in normal epidermis. PLA was carried out using antibody combinations with h14D146 and anti-ICAD Ab. Normal rabbit IgG was used as negative control. A merged figure with nuclear staining (DAPI) is also shown. The broken line indicates the epidermal–dermal junction. The dotted line shows the border of the cornified layer. Blue staining (DAPI) indicates nuclei. Scale bars, 50 μm
Knockdown of both caspase-14 and mesotrypsin increased DNA remnants in the cornified layer
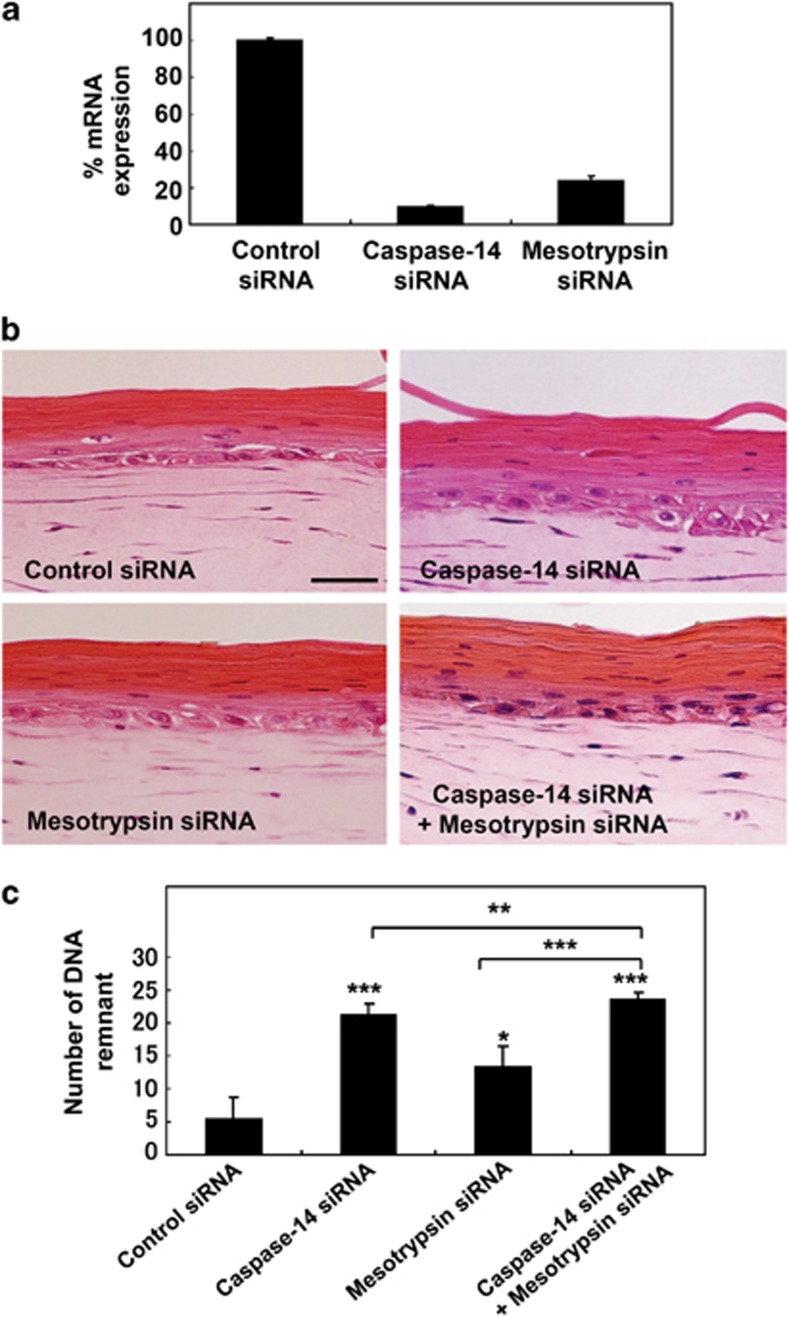
Our results suggested that both the caspase-14-dependent ICAD–CAD pathway and a FLG N-terminal fragment-dependent pathway may be involved in DNA degradation during terminal differentiation of keratinocytes. Thus, we examined the effect of siRNA knockdown of caspase-14 and mesotrypsin in a skin equivalent model. These siRNA treatments efficiently knocked down expression of the corresponding proteases to 9.9% and 24.0% of the control levels, respectively (Figure 6a). We detected considerable numbers of undegraded nuclei in the cornified layer after either caspase-14 knockdown or mesotrypsin knockdown. When both caspase-14 and mesotrypsin were knocked down together, many undigested nuclei were observed in the cornified layer (Figure 6b). Numbers of nuclear remnants were counted in each 450 μm field and a total length of 4.5 mm was examined in each model (Figure 6c). Interestingly, the number of remnants was increased significantly in the skin model with mesotrypsin knockdown and further increased in that with caspase-14 knockdown. The greatest effect was consistently observed in the model with double knockdown.
Figure 6.
Suppression of caspase-14 as well as mesotrypsin caused a marked increase of nuclear remnants in the cornified layer. (a) Effect of caspase-14 and mesotrypsin suppression. The siRNAs strongly suppressed the expression of these proteases, by suppressed by 90% and 76%, respectively. (b) Skin equivalent models were prepared using keratinocytes treated with nonspecific siRNA (control), siRNA to caspase-14, siRNA to mesotrypsin and combined siRNAs to caspase-14 and mesotrypsin. Images of H&E staining are shown. Scale bars: 50 μm. (c) Quantitative analysis of the presence of nuclear remnants in the cornified layer. Numbers of nuclear remnants in the cornified layer were counted in each 450 μm field for total of 4.5 mm length and expressed as mean±S.D. *P<0.05, **P<0.01, ***P<0.001
Caspase-14 and mesotrypsin were markedly downregulated in parakeratotic areas of diseased skin
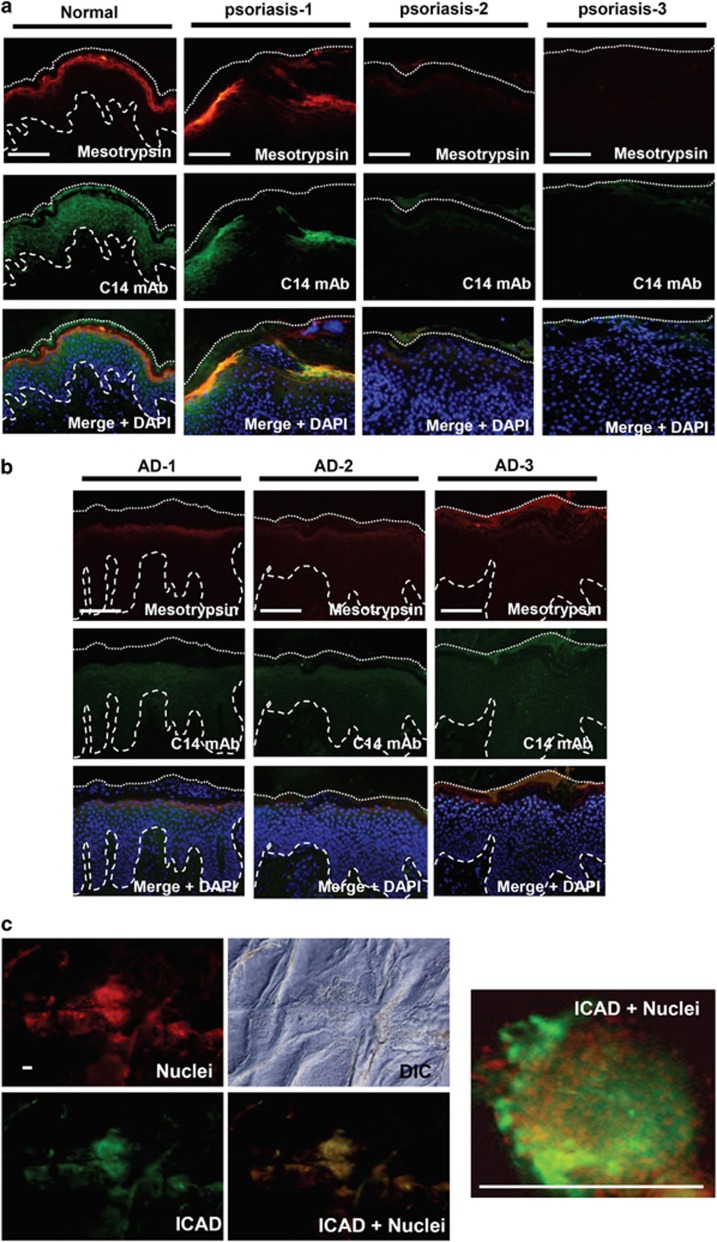
Finally, we examined the expression of mesotrypsin and caspase-14 in skin from patients with atopic dermatitis (AD) and psoriasis, as such skin frequently shows abnormal differentiation and parakeratosis.20, 21 Normal skin showed strong immunostaining for mesotrypsin in the granular layer and for caspase-14 in the spinous to granular layers. Immunostaining clearly demonstrated strong downregulation of procaspase-14 in the lesional skin with psoriasis (Figure 7a). Immunostaining of mesotrypsin was also significantly decreased in the same area. Nuclear staining revealed persistent presence of undigested nuclei in the areas where both enzymes were hardly detectable. Similar results were obtained for skin with AD (Figure 7b). Thus, the FLG-N pathway is likely to be suppressed in parakeratotic areas. We further investigated whether the ICAD–CAD pathway was concomitantly impaired in parakeratotic skin. Skin surface staining for ICAD showed isolated regions of various sizes that were positive for ICAD in AD skin (Figure 7c). Nuclear staining with propidium iodide (PI) showed that parakeratotic nuclei were always present in these patchy islands. Bright field microscopy of the superficial cornified layer showed a bumpy and rough surface compared with the smooth surface of normally keratinized areas. The merged images demonstrate that the parakeratotic areas coincided with ICAD-positive areas. These results indicate that suppression of the two pathways is associated with persistent presence of nuclei in the cornified layer.
Figure 7.
Caspase-14 and mesotrypsin were hardly detectable in parakeratotic skin areas. (a) Localizations of caspase-14 (C14) and mesotrypsin in normal (left) and psoriatic skin (right three panels). Representative images from three different patients are shown. (b) Localizations of caspase-14 and mesotrypsin in skin with AD. Top indicates localization of mesotrypsin, middle indicates localization of caspase-14 and bottom indicates merged images with nuclear staining (DAPI). (c) ICAD immunostaining and nuclear staining on the skin surface of a subject with AD. A high-magnification picture is also shown. Note the colocalization of ICAD and nuclei. Scale bar: 200 μm
Discussion
Loss of nuclei is a critical event in epidermal barrier formation. Our results indicate that at least two pathways are involved in the denucleation process during keratinocyte terminal differentiation, that is, profilaggrin N-terminal fragment-dependent and caspase-14-mediated CAD-dependent DNA degradation pathways.
First, we searched for molecules that interact with the N-terminal fragment of profilaggrin by means of LC/MS/MS analyses, and identified epidermal mesotrypsin and caspase-14 as interactors in extracts from keratinocytes and cornified cells (Supplementary Table S3). Identification of these interactions is consistent with the idea that profilaggrin is a natural substrate for these proteases.22
Mesotrypsin shows a distinct substrate specificity from the PRSS1 gene product, anionic trypsin, or PRSS2 gene product, cationic trypsin.23 It is not susceptible to intrinsic trypsin inhibitors, but instead has the ability to degrade them.24, 25 Furthermore, its proteolytic activity is quite limited, and it does not digest large protein substrates into small fragments, as other trypsins do. Mesotrypsin is required for the processing of FLG-N and liberated FLG-N is translocated into the nucleus because of the presence of NLS in the B domain. We also found that the translocated N-terminal of profilaggrin contained not only A and B domains but also a truncated FLG domain. This conclusion was also supported by the results of our mutation study. Our finding is contrary to that of Pearton et al.,15 who reported that only the A and B domains enter into the nucleus. The reason for this discrepancy is unclear, but it may be because of the difference in cell lines used for assays, that is, human keratinocytes and COS-7, as human keratinocytes express epidermal-specific mesotrypsin.14 Although several proteases have been reported to be involved in the profilaggrin processing cascade leading to filaggrin, our findings strongly suggest that epidermal mesotrypsin and caspase-14 are key profilaggrin-processing molecules.
On the other hand, we also established that caspase-14 has the ability to degrade ICAD, generating peptides similar to those formed by caspase-3. Indeed, expression of constitutively active caspase-14 (revC14) liberated CAD that was accumulated in nuclei. As expected, the resulting cells became TUNEL positive. In addition, we compared the TUNEL-positive rate in revC14-expressing cells and mesotrypsin-expressing cells. Most revC14-expressing cells were TUNEL positive (95%), whereas active mesotrypsin-expressing cells showed lower TUNEL positivity (72%). The reason why revC14-expressing cells showed a higher rate of TUNEL positivity may be higher efficiency of the ICAD–CAD system for DNA degradation and/or slower reactivity of the FLG-N–mesotrypsin system. We recently showed that mesotrypsin plays multiple roles in keratinocyte terminal differentiation.26 For example, it is secreted to interstitial areas of the cornified layer and is involved in proKLK activation. Thus, its availability may vary at different stages of differentiation. The PLA method demonstrated association of active caspase-14 and ICAD at the boundary area between the granular layer and the cornified layer, where nuclear loss takes place. Involvement of epidermal mesotrypsin and caspase-14 in the denucleation process is also supported by findings in skin equivalent models with siRNA-mediated knockdown of these proteases and in PLA interaction assays using skin samples. Downregulation of both enzymes resulted in a significant increase of undigested nuclei in the cornified layer. Furthermore, it was found that both caspase-14 and mesotrypsin were absent in areas where parakeratotic nuclei were evident. Therefore, these proteases appear to play major roles in denucleation during keratinocyte terminal differentiation.
Interestingly, we found that caspase-14 promotes the conversion of mesotrypsinogen to mesotrypsin. This is probably accomplished by cleavage near the enterokinase recognition sequence (DDDDKI),27 as caspase-14 cleaves only after Asp residues. Indeed, our unpublished data support the idea that active caspase-14 promotes mesotrypsinogen activation, even though enterokinase was several times more potent than caspase-14 as an activator. However, this does not exclude a role of caspase-14 as a possible activator, considering that the catalytic efficiency of enterokinase is 34 000-fold greater than that of trypsin.28
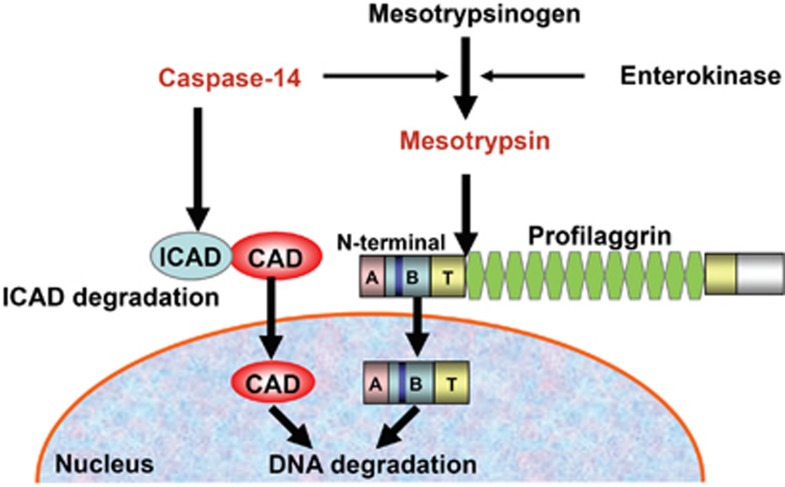
Collectively, our results indicate the presence of two distinct pathways of epidermal denucleation that appear to work both cooperatively and independently, depending upon the differentiation conditions (Figure 8). However, a defect of even a single pathway may affect the maintenance of epidermal homeostasis, because it has been shown that a defect of caspase-14 predisposes to the development of parakeratosis.12
Figure 8.
Involvement of multiple pathways in the denucleation process during keratinocyte terminal differentiation. Two distinct pathways are involved in epidermal denucleation. Activated caspase-14 degrades ICAD and liberated CAD is translocated into the nucleus, resulting in the degradation of chromosomal DNA. On the other hand, activated mesotrypsin cleaves profilaggrin, releasing its N-terminal domain. The free N-terminal domain enters the nucleus and induces DNA degradation. Mesotrypsinogen can be activated by both enterokinase and caspase-14. Thus, these systems appear to work cooperatively, as well as independently, depending on the differentiation conditions
Because the mechanisms of parakeratosis have been unknown, there has been no effective treatment directly targeting parakeratosis. Here, we have shown that at least two pathways are involved in denucleation during keratinocyte terminal differentiation, acting cooperatively for the formation of fully differentiated corneocytes. Simultaneous treatment directed toward both pathways might be effective to improve parakeratosis, as well as barrier disruption.
Materials and Methods
Materials
Antibodies used in this study are listed in Supplementary Table S1. Primer sequences used for human cDNA amplification are listed in Supplementary Table S2.
Cell culture
Human keratinocytes derived from normal foreskin (Kurabo, Osaka, Japan) were cultured in Humedia KG2 supplemented with epidermal growth factor (0.1 ng/ml), insulin (10 μg/ml), hydrocortisone (0.5 μg/ml), bovine pituitary extract (0.4%), gentamicin (50 μg/ml) and amphotericin B (50 ng/ml). All experiments were carried out on cells at the third or fourth passages.
Tissue specimens
This study was approved by the Ethical Committee of Tokyo Medical University and by the Shiseido Committee on Human Ethics. In total, 7 patients with AD and 14 patients with psoriasis vulgaris were recruited for this study after having given informed consent. Patients with AD (7 men, mean age 32.7 years) and psoriasis (11 men, mean age 44.6 years) had not received topical steroid treatment or UV therapy for at least 4 weeks before participation in the study. The severity of AD was evaluated by using the Eczema Area and Severity Index (EASI) score29 that ranged from 13.7 to 48 (mean score=29.9). The severity of psoriasis was evaluated in terms of the PASI score (2.0–52.8, mean score=21.1). For all patients, 4 mm punch biopsies were taken from acute or active lesions and nonlesional skin. Tissue samples were fixed in acetone and embedded in paraffin.
Preparation of cell extracts
Keratinocytes (KCs) were collected at 80% confluency (growth phase), 100% confluency (confluency) at and 2, 5 and 7 days after confluency in the presence of 1.2 mM calcium (differentiated phase). Cell pellets were homogenized using a sintered glass homogenizer (Wheaton Science Products, Millville, NJ, USA) with extraction buffer (10 mM Tris HCl (pH 7.5), 50 mM NaCl, 1% NP-40, 5 mM EDTA) and subjected to ultrasonication for 10 s, three times. Supernatant was obtained after centrifugation. Cornified cell extracts were also prepared from scraped heels of healthy individuals as reported previously.17 Each extract was reacted with GST-fusion N-terminal fragment (A+B domains) and revC14 coupled with GST-Agarose. After extensive washing, bound proteins were eluted with 100 mM glutathione, dialyzed against H2O and lyophilized for trypsin digestion. For western blot analysis, cell pellets were solubilized with SDS sample buffer and after centrifugation, supernatants were subjected to SDS-PAGE.
Purification of human caspase-14 and construction of revC14
Human caspase-14 was purified from cornified cells according to our previous report, using sequential chromatographies.17 Briefly, revC14 cDNA was inserted into the pET 15b vector at the BamHI/XhoI sites. Recombinant proteins were purified by means of Ni-NTA-Agarose (Qiagen, Venlo, Netherlands) and Mono Q chromatography.
Construction of expression vectors
Two kinds of expression vectors for constitutively active proteases were constructed using pCMV-HA vector (Clontech, Mountain View, CA, USA) and pH3700-pL2 vector (kindly given by Dr. LB Taichman (State University of New York, Stony Brook, NY, USA)) that was driven by the involucrin promoter. cDNA of revC14 was cloned into pCMV-HA vector (pCMV-HA-revC14). In addition, a PCR product (HA+revC14) was amplified using the common forward primer with NotI recognition sequence+HA sequence and the specific reverse primer with NotI site. It was inserted into pH3700-pL2 vector at the NotI site (pInv-HA-revC14). Two kinds of expression vectors (pCMV-HA and pInv-HA) for constitutively active KLK5 and active KLK7 were similarly prepared from full-length cDNA clones (OriGene Technologies, Inc., Rockville, MD, USA). Expression vectors for FLG N-terminal domains were also prepared using pCMV-HA. These were A domain construct (FLG-A), A domain with NLS (FLG-A-nls), B domain construct (FLG-B), B domain+truncated FLG repeat (FLG-BT) and N-terminal fragment with A domain+B domain+truncated FLG repeat (FLG-ABT). For time-lapse studies, fluorescent protein, Azami-Green (AG) or KR (Amalgam-MBL, Nagoya, Japan), was inserted after the HA-tag of these vectors (pCMV-HA-AG-revC14, pInv-HA-AG-revC14 and pCMV-HA-KR-FLG-ABT, respectively). These constructs were introduced into keratinocytes during the growth phase using FuGENE (Roche Diagnostics, Tokyo, Japan). As controls, pCMV-HA vector and pH3700-pL2 were similarly transfected. Primers used are listed in Supplementary Table S2.
Construction of mutated expression vectors
Expression vectors with Arg mutations in the linker region between the FLG-N and the first FLG domain were constructed as described below. This region contains 4 arginines within 10 residues. All four Arg residues were changed to Leu using PCR primers, 5′-GGCAAGCTTCATCTGCAGTCAGCGATCGTGGAC-3′ and 5′-TGCTCACCTGGTAGAGGGAAGACCCTGAAAGTCCAGACAGTTCCCCTGACAGGCCAAGTGTGGACTCTTG-3′. The PCR product was inserted into the HindIII/EcoNI site of pCMV-HA-FLG-ABT and named pCMV-HA-FLGmut. pCMV-HA-FLGmut-KR was prepared by inserting KR cDNA into the XhoI/NotI site of pCMV-HA-FLG-ABT. pCMV-HA-KR-FLGmut was also constructed by inserting KR cDNA into the SfiI/EcoRI site of pCMV-HA-FLGmut.
Measurement of caspase-14 activity
Caspase-14 activity was measured using Ac-WEHD-MCA as a substrate. The assay buffer consisted of 0.1 M HEPES buffer (pH 7.5), 0.06 M NaCl, 0.01% CHAPS ( 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonic acid), 5 mM DTT and 1.2 M sodium citrate (final concentration). revC14 (5 μg/ml, 10 μl) was preincubated in the assay buffer (80 μl) for 15 min at room temperature. After addition of 20 μl of 100 μM Ac-WEHD-MCA, the mixture was incubated for 30 min. The activity of revC14 was measured using a Fluoroskan Ascent FL (Thermo Electron Co., Wolsam, MA, USA) with excitation at 355 nm and emission at 460 nm.
ICAD degradation by caspase-14
The assay mixture containing 5 μl of revC14 or purified caspase-14 from cornified cells in 40 μl of the assay buffer was preincubated for 15 min at room temperature, and 5 μl of ICAD (0.1 mg/ml) (inhibitor of caspase-activated DNase) was added. At each time point, 3 μl of the incubation mixture was removed and diluted with 27 μl of H2O. SDS-PAGE samples were prepared by adding 10 μl of 4 × sample buffer and analyzed by western blotting with the anti-ICAD antibody (FL331; Santa Cruz Biotechnology, Dallas, TX, USA).
Western blot analysis
SDS-PAGE was carried out using a 5–20% gradient gel. After electrophoresis, proteins were transferred onto a polyvinylidine difluoride membrane (Immobilon-P, Millipore, Bedford, MA, USA) and incubated with appropriate primary antibodies. Peroxidase-labeled anti-rabbit IgG (Sigma, St. Louis, MO, USA) or anti-mouse IgG was used as a secondary antibody, and immunoreactive proteins were visualized by chemiluminescence using ECL-plus (Amersham, Little Chalfont, UK). Primary antibodies used are listed in Supplementary Table S1.
Real-time quantitative PCR
The mRNA was reverse transcribed with SuperScript II (Invitrogen Corporation, Carlsbad, CA, USA). Real-time quantitative PCR was performed on a LightCycler rapid thermal cycler system using LightCycler FastStart DNA master SYBR green I kit (Roche Diagnostics Ltd) according to the manufacturer's instructions. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a reference gene.
Immunostaining and TUNEL assays
Expression of revC14 was detected with anti-HA antibody, anti-caspase-14 mAb16 and h14D146 antibody.17 For the detection of epidermal mesotrypsin, we used anti-trypsin Ab (Athens Research & Technology, Athens, GA, USA). As active mesotrypsin and trypsin are ∼90% identical at the amino acid level, we confirmed that this antibody crossreacted with epidermal mesotrypsin without showing any difference in immunostaining or western blot analysis.14 Alexa Fluor 555 or 488 (Molecular Probes Inc., Eugene, OR, USA) was used as the secondary antibody. DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes Inc.) was used for visualization of nuclei. The TUNEL reaction was performed using a fluorescein in situ cell death detection kit (Roche Diagnostics Ltd) according to the manufacturer's instructions. After immunostaining, cells with DAPI-stained nuclei were counted. The transfection ratio was calculated from the number of h14D146-positive or anti-HA antibody-positive cells versus the total cell number. The ratio of TUNEL-positive cells to active caspase-14-expressing cells was calculated similarly. In a typical experiment, over 1500 cells in 7 fields were counted and experiments were repeated three times. Tissue sections from human skin and the skin equivalent models were stained in the same manner. Primary antibodies used are listed in Supplementary Table S1.
Proximity ligation assay
PLA reaction was performed using two antibody combinations (monoclonal anti-trypsin antibody/h14D146 antibody, rabbit anti-trypsin antibody/monoclonal anti-FLG antibody and h14D146 antibody/anti-ICAD antibody) according to the manufacturer's instructions. For the negative control, normal rabbit IgG was used as the primary antibody. Dilution was 0.5 μg/ml for each antibody.
Skin surface staining
For skin surface staining, medical adhesive Aron Alpha A (Sankyo, Tokyo, Japan) was applied on a glass slide and the glass was firmly pressed on the skin for 5 min. It was then carefully removed, and the attached cornified cells were stained for ICAD using anti-ICAD antibody (FL331). PI (1 μg/ml) was used for nuclear staining. Observation was done by means of confocal microscopy (LSM5 PASCAL, Carl Zeiss, Oberkochen, Germany). The lens was a Plan APOCHROMAT × 20 magnification.
Preparation of skin equivalent models
Keratinocytes were treated with control siRNA-A (sc-37007), caspase-14 siRNA (sc-37364) or PRSS3 Stealth RNAi (Invitrogen) and plated on dermal equivalent, Matrex (Toyobo, Osaka, Japan). At 2 days after plating, the cell surface was exposed to air and culture was continued for 10 days.
Statistical analysis
Data are expressed as the mean±S.D. We employed simple pair-wise comparison with Student's t-test (two-tailed distribution with two-sample equal variance). P<0.05 was considered significant.
Acknowledgments
We thank Drs. Masuyoshi Saito, Mami Saito and Yukari Okubo (Tokyo Medical University) for providing biopsy skin samples from patients with atopic dermatitis and psoriasis vulgaris. This work was supported in part by JSPS KAKENHI Grant Number 24781180 and Tokyo Medical University Research Grant to MY-T.
Glossary
- FLG
filaggrin
- FLG-N
a 464-amino acid N-terminal fragment of profilaggrin
- NLS
nuclear localization signal
- AG
Azami Green
- KR
Keima Red
- KLK
kallikrein-related peptidase
- ICAD
inhibitor of caspase-activated DNase
- CAD
caspase-activated DNase
- LC/MS/MS
liquid chromatography coupled with electrospray tandem mass spectrometry
- fmk
fluoromethyl ketone
- MCA
4-methylcoumaryl-7-amide
- Ab
antibody
- mAb
monoclonal antibody
- CHAPS
3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonic acid
- GST
glutathione-S-transferase
- PLA
proximity ligation assay
- PI
propidium iodide
- DAPI
4′,6-diamidino-2-phenylindole
- TEWL
transepidermal water loss
- AD
atopic dermatitis
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by E Candi
Supplementary Material
References
- Katagiri C, Iida T, Nakanishi J, Ozawa M, Aiba S, Hibino T. Up-regulation of serpin SCCA1 is associated with epidermal barrier disruption. J Dermatol Sci. 2010;57:95–101. doi: 10.1016/j.jdermsci.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Fischer H, Szabo S, Scherz J, Jaeger K, Rossiter H, Buchberger M, et al. Essential role of the keratinocyte-specific endonuclease DNase1L2 in the removal of nuclear DNA from hair and nails. J Invest Dermatol. 2011;131:1208–1215. doi: 10.1038/jid.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, Scherz J, Szabo S, Mildner M, Benarafa C, Torriglia A, et al. DNase 2 is the main DNA-degrading enzyme of the stratum corneum. PLoS One. 2011;6:e17581. doi: 10.1371/journal.pone.0017581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DK. The role of the Caspase family of cysteine proteases in apoptosis. Sem Immunol. 1997;9:35–49. doi: 10.1006/smim.1996.0058. [DOI] [PubMed] [Google Scholar]
- Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- Fischer H, Stichenwirth M, Dockal M, Ghannadan M, Buchberger M, Bach J, et al. Stratum corneum-derived caspase-14 is catalytically active. FEBS Lett. 2004;577:446–450. doi: 10.1016/j.febslet.2004.10.046. [DOI] [PubMed] [Google Scholar]
- Eckhart L, Declercq W, Ban J, Rendl M, Lengauer B, Mayer C, et al. Terminal differentiation of human keratinocytes and stratum corneum formation is associated with caspase-14 activation. J Invest Dermatol. 2000;115:1148–1151. doi: 10.1046/j.1523-1747.2000.00205.x. [DOI] [PubMed] [Google Scholar]
- Lippens S, Kockx M, Knaapen M, Mortier L, Polakowska R, Verheyen A, et al. Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing. Cell Death Differ. 2000;12:1218–1224. doi: 10.1038/sj.cdd.4400785. [DOI] [PubMed] [Google Scholar]
- Eckhart L, Ban J, Fischer H, Tschachler E. Caspase-14: analysis of gene structure and mRNA expression during keratinocyte differentiation. Biochem Biophys Res Commun. 2000;277:655–659. doi: 10.1006/bbrc.2000.3698. [DOI] [PubMed] [Google Scholar]
- Lippens S, VandenBroecke C, Van Damme E, Tschachler E, Vandenabeele P, Declercq W, et al. Caspase-14 is expressed in the epidermis, the choroid plexus, the retinal pigment epithelium and thymic Hassall's bodies. Cell Death Differ. 2003;10:257–259. doi: 10.1038/sj.cdd.4401141. [DOI] [PubMed] [Google Scholar]
- Denecker G, Hoste E, Gilbert B, Hochepied T, Ovaere P, Lippens S, et al. Caspase-14 protects against epidermal UVB photodamage and water loss. Nat Cell Biol. 2007;9:666–674. doi: 10.1038/ncb1597. [DOI] [PubMed] [Google Scholar]
- Hoste E, Denecker G, Gilbert B, Van Nieuwerburgh F, van der Fits L, Asselbergh B, et al. Caspase-14-deficient mice are more prone to the development of parakeratosis. J Invest Dermatol. 2013;133:742–750. doi: 10.1038/jid.2012.350. [DOI] [PubMed] [Google Scholar]
- Ishida-Yamamoto A, Takahashi H, Presland RB, Dale BA, Iizuka H. Translocation of profilaggrin N-terminal domain into keratinocyte nuclei with fragmented DNA in normal human skin and loricrin keratoderma. Lab Invest. 1998;78:1245–1253. [PubMed] [Google Scholar]
- Nakanishi J, Yamamoto M, Koyama J, Sato J, Hibino T. Keratinocytes synthesize enteropeptidase and multiple forms of trypsinogen during terminal differentiation. J Invest Dermatol. 2010;130:944–952. doi: 10.1038/jid.2009.364. [DOI] [PubMed] [Google Scholar]
- Pearton DJ, Dale BA, Presland RB. Functional analysis of the profilaggrin N-terminal peptide: identification of domains that regulate nuclear and cytoplasmic distribution. J Invest Dermatol. 2002;119:661–669. doi: 10.1046/j.1523-1747.2002.01831.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Kamata Y, Iida T, Fukushima H, Nomura J, Saito M, et al. Quantification of activated and total caspase-14 with newly developed ELISA systems in normal and atopic skin. J Dermatol Sci. 2011;61:110–117. doi: 10.1016/j.jdermsci.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Hibino T, Fujita E, Tsuji Y, Nakanishi J, Iwaki H, Katagiri C, et al. Purification and characterization of active caspase-14 from human epidermis and development of the cleavage site-directed antibody. J Cellular Biochem. 2010;109:487–497. doi: 10.1002/jcb.22425. [DOI] [PubMed] [Google Scholar]
- Raymond AA, Mechin MC, Nachat R, Toulza E, Tazi-Ahnini R, Serre G, et al. Nine procaspases are expressed in normal human epidermis, but only caspase-14 is fully processed. Br J Dermatol. 2007;156:420–427. doi: 10.1111/j.1365-2133.2006.07656.x. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk J, Scott FL, Krajewski S, Sutherlin DP, Salvesen GS. Activation and substrate specificity of caspase-14. Biochem. 2004;43:10560–10569. doi: 10.1021/bi0498048. [DOI] [PubMed] [Google Scholar]
- Sakurai K, Sugiura H, Matsumoto M, Uehara M. Occurrence of patchy parakeratosis in normal-appearing skin in patients with active atopic dermatitis and in patients with healed atopic dermatitis: a cause of impaired barrier function of the atopic skin. J Dermatol Sci. 2002;30:37–42. doi: 10.1016/s0923-1811(02)00047-6. [DOI] [PubMed] [Google Scholar]
- Weiss VC, van den Broek H, Barrett S, West DP. Immunopathology of psoriasis: a comparison with other parakeratotic lesions. J Invest Dermatol. 1982;78:256–260. doi: 10.1111/1523-1747.ep12506623. [DOI] [PubMed] [Google Scholar]
- Olexa SA, Budzynski AZ. Binding phenomena of isolated unique plasmic degradation products of human cross-linked fibrin. J Biol Chem. 1979;254:4925–4932. [PubMed] [Google Scholar]
- Sahin-Toth M. Human mesotrypsin defies natural trypsin inhibitors: from passive resistance to active destruction. Prot Pep Lett. 2005;12:457–464. doi: 10.2174/0929866054395356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinderknecht H, Renner IG, Abramson SB, Carmack C. Mesotrypsin: a new inhibitor-resistant protease from a zymogen in human pancreatic tissue and fluid. Gastroenterology. 1984;86:681–692. [PubMed] [Google Scholar]
- Salameh MA, Soares AS, Hockla A, Radisky ES. Structural basis for accelerated cleavage of bovine pancreatic trypsin inhibitor (BPTI) by human mesotrypsin. J Biol Chem. 2008;283:4115–4123. doi: 10.1074/jbc.M708268200. [DOI] [PubMed] [Google Scholar]
- Miyai M, Matsumoto Y, Yamanishi H, Yamamoto-Tanaka M, Tsuboi R, Hibino T.Keratinocyte-specific mesotrypsin contributes to the desquamation process via kallikrein activation and LEKTI degradation J Invest Dermatol; e-pub ahead of print 3 January 2014; doi: 10.1038/jid.2014.3 [DOI] [PubMed]
- Light A, Janska H. Enterokinase (enteropeptidase): comparative aspects. Trends Biochem Sci. 1989;14:110–112. doi: 10.1016/0968-0004(89)90133-3. [DOI] [PubMed] [Google Scholar]
- Anderson RG, Hein CE. Distribution of anionic sites on the oviduct ciliary membrane. J Cell Biol. 1977;72:482–492. doi: 10.1083/jcb.72.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci G, Dondi A, Patrizi A. Useful tools for the management of atopic dermatitis. Am J Clin Dermatol. 2009;10:287–300. doi: 10.2165/11310760-000000000-00000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.