Abstract
The growing number of studies suggested that inhibition of autophagy enhances the efficacy of Akt kinase inhibitors in cancer therapy. Here, we provide evidence that ML-9, a widely used inhibitor of Akt kinase, myosin light-chain kinase (MLCK) and stromal interaction molecule 1 (STIM1), represents the ‘two-in-one' compound that stimulates autophagosome formation (by downregulating Akt/mammalian target of rapamycin (mTOR) pathway) and inhibits their degradation (by acting like a lysosomotropic agent and increasing lysosomal pH). We show that ML-9 as a monotherapy effectively induces prostate cancer cell death associated with the accumulation of autophagic vacuoles. Further, ML-9 enhances the anticancer activity of docetaxel, suggesting its potential application as an adjuvant to existing anticancer chemotherapy. Altogether, our results revealed the complex effect of ML-9 on autophagy and indentified ML-9 as an attractive tool for targeting autophagy in cancer therapy through dual inhibition of both the Akt pathway and the autophagy.
Keywords: ML-9, autophagy, cell death, lysosomotropic agents, calcium
The phosphatidylinositol 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) signaling pathway is a vital pathway that is implicated in a variety of cellular processes, deregulation of which could contribute to the malignant transformation. Accordingly, this pathway has been linked to tumorigenesis and resistance to anticancer therapy in different cancers.1, 2 Considering this, a number of novel inhibitors targeting PI3K, Akt or mTOR are currently being investigated in clinical trials for cancer therapy. However, it was reported that inhibition of the different components of the PI3K/Akt/mTOR pathway often stimulates prosurvival processes, such as autophagy, limiting the anticancer efficacy of the aforementioned inhibitors.3, 4
Autophagy, a cellular process comprising degradation and recycling of protein aggregates, long-lived proteins and damaged organelles, is essential to maintain cellular homeostasis.5, 6 At basal and stress conditions, autophagy assures cytoprotective response to support cell survival. In contrast, a growing number of studies suggest that autophagy may contribute to cell death.7 Defects in autophagy are linked to a number of pathological states including cancer.8 Indeed, accumulated evidence suggests that autophagy inhibition facilitates apoptotic cell death, inhibits tumor cells growth and sensitizes tumor cells to chemotherapy treatments.9, 10, 11, 12, 13
Recently, it was proposed that inhibition of autophagy represents a prospective strategy to increase the therapeutic efficacy of Akt inhibitors.14, 15, 16
A naphthalene sulphonamide derivative, 1-(5-chloronaphthalene-1-sulphonyl)-1H-hexahydro-1,4-diazepine (ML-9), is a membrane permeable organic substance (Supplementary Figure S1), which is widely used as an inhibitor of Akt kinase,17, 18 myosin light-chain kinase (MLCK)19 and stromal interaction molecule 1 (STIM1).20 ML-9 has been previously shown to promote apoptotic cell death and inhibit invasion and adhesion in certain cancer cell lines.21, 22, 23 Recently, ML-9 was identified as a compound that increases the number of green fluorescent protein (GFP)-LC3 vesicles in human glioblastoma H4 cells.24
Given the increasing interest in development of novel autophagy modulators as well as inhibitors of the Akt/mTOR pathway for cancer treatment, we investigated the mechanisms of autophagy regulation by ML-9 and assessed its potential application as an anticancer drug.
Results
ML-9 stimulates autophagy by downregulating Akt/mTOR pathway in prostate cancer cells
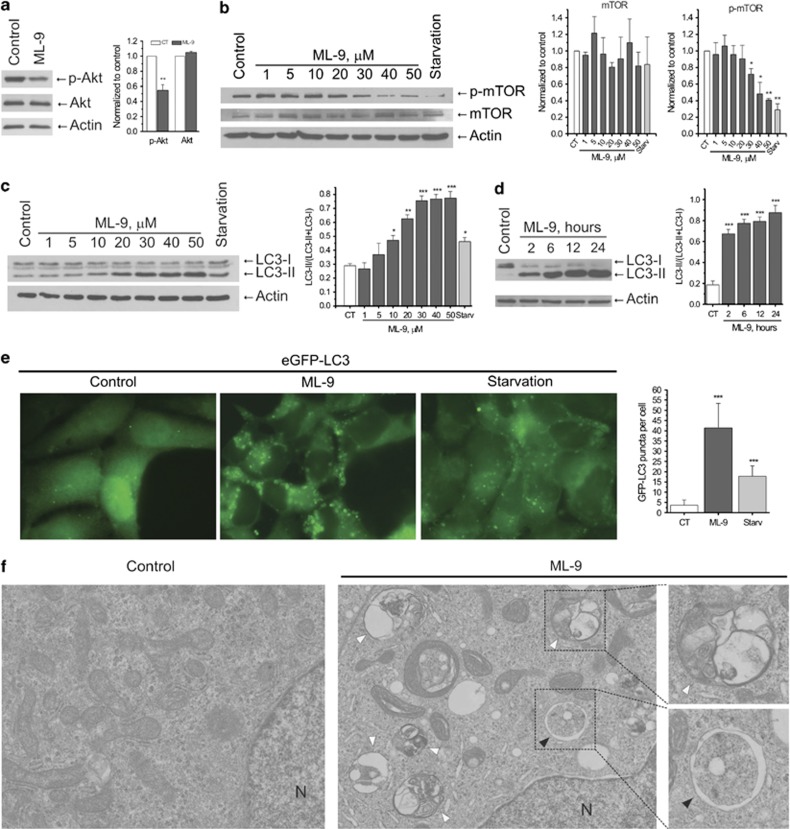
Akt phosphorylation is widely established to be one of the key-stone events involved in the regulation of prostate cancer cell survival.25, 26, 27, 28 Since ML-9 has been suggested to be the potent inhibitor of Akt phosphorylation in other cell models, we first checked for this function of ML-9 in prostate cancer cells. In lymph node carcinoma of the prostate (LNCaP) cells, ML-9 (30 μM) reduced the phosphorylation of Akt kinase suggesting its inhibition (Figure 1a). Since mTOR kinase acts downstream of Akt kinase, we further examined the effect of ML-9 on mTOR activity. We found that phosphorylation of mTOR was significantly reduced by ML-9 in a dose-dependent manner, which suggests the inhibition of mTOR kinase activity (Figure 1b).
Figure 1.
ML-9 stimulates autophagy by downregulating Akt/mTOR pathway in prostate cancer cells. (a) ML-9 inhibits Akt. To assess Akt phosphorylation, LNCaP cells were treated with full or 30 μM ML-9-containing medium for 12 h. Densitometric quantitation for normalized p-Akt and total Akt relative to Actin is shown. Values represent means±S.E.M. n=3. (b) ML-9 inhibits mTOR. LNCaP cells were incubated in full medium, serum-starved medium or full medium with indicated concentrations of ML-9 for 12 h. The expression levels of p-mTOR, mTOR and beta-Actin were analyzed. Densitometric quantitation for normalized p-mTOR and mTOR relative to Actin is shown. Values represent means±S.E.M. n=3. (c) ML-9 increases LC3-II levels in LNCaP cells in a dose-dependent manner. LNCaP cells were incubated in full medium, serum-starved medium or full medium with indicated concentrations of ML-9 for 6 h. Densitometric quantitation for LC3-II/(LC3-II+LC3-I) ratio is shown. Values represent means±S.E.M. n=3. (d) ML-9 increases LC3-II levels in LNCaP cells in a time-dependent manner. LNCaP cells were untreated or treated with 30 μM ML-9 for 2, 6, 12 and 24 h. Densitometric quantitation for LC3-II/(LC3-II+LC3-I) ratio is shown. Values represent means±S.E.M. n=4. (e) LNCaP cells stably expressing eGFP-LC3 (green) were incubated in full, serum-starved or 30 μM ML-9-containing media for 6 h. Quantitation shown on the right represents means±S.D. GFP-positive puncta per cell (n=50) from three independent experiments. (f) TEM images of LNCaP cells untreated and treated with 30 μM ML-9 for 12 h showing autophagosome (black arrow) and autolysosomes/amphisomes (white arrows). N: nucleus
As mTOR kinase represents a central player in autophagy regulation, we next tested the effect of ML-9 on autophagy by analyzing the levels of endogenous LC3-II protein, the most widely used marker for autophagosomes.29 ML-9 significantly increased LC3-II levels in LNCaP cells in a dose- and time-dependent manner (Figures 1c and d). In addition, in LNCaP cells stably expressing eGFP-LC3 (LNCaP-eGFP-LC3), treatment with ML-9 resulted in accumulation of eGFP-LC3 puncta (Figure 1e).
Further, transmission electron microscopy (TEM) analysis revealed the increased number of large autophagosome-like and autolysosome-like vacuoles in ML-9-treated cells compared with the control ones (Figure 1f). Moreover, the accumulation of multi-lamellar membrane structures, small vesicles, cytoplasmic material and organelles at different stages of degradation has been detected within the lumen of the numerous expanded vacuoles after ML-9 treatment (Figure 1f). Importantly, we have noted that apparent autolysosomes were significantly greater in number than autophagosomes (Figure 1f; Supplementary Figure S2). In addition, immuno-TEM analysis showed the presence of immunogold-labeled lysosomal protein lysosomal-associated membrane protein 2 (Lamp2) in these vacuoles (Supplementary Figure S3). These findings suggest autophagic/lysosomal origin of these vacuoles.
ML-9 also caused an increase in LC3-II levels in prostate cancer PC-3 cells, human embryonic kidney 293 (HEK-293) cells, pancreatic cancer ASPC1 and BxPC3 cells, but not in DU-145 cells lacking full-length autophagy-related gene 5 (ATG5)30 (Supplementary Figures S4a and b), suggesting the necessity of the functional autophagic pathway.
Overall, these data suggest that ML-9 stimulates autophagy and induces accumulation of autophagic vacuoles and functional autophagic machinery is required for this.
ML-9 blocks autophagic flux
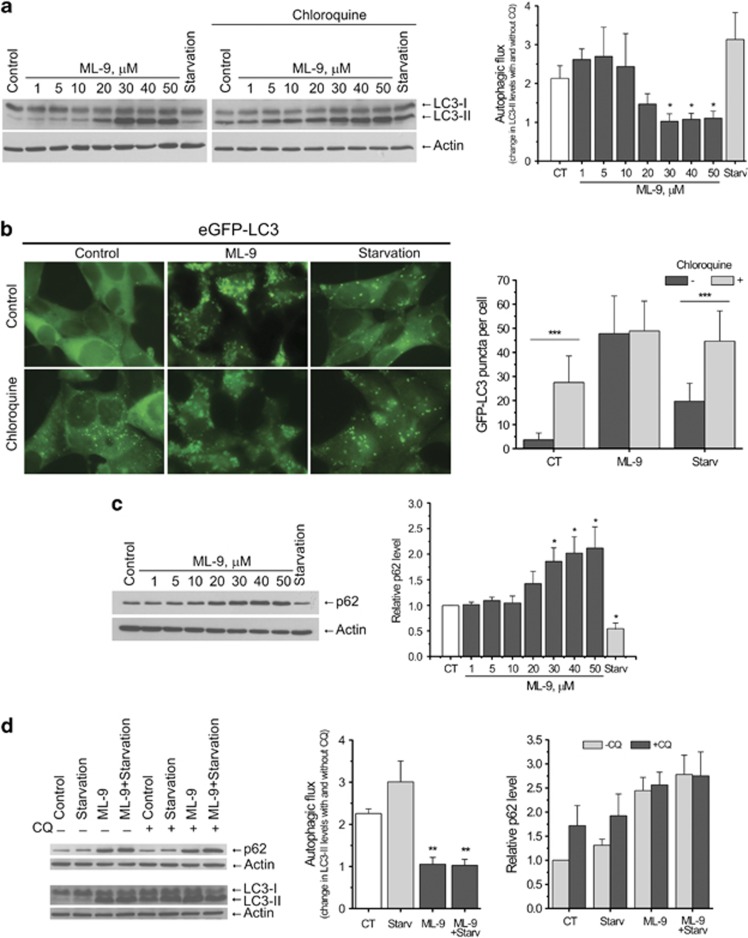
An accumulation of autophagic vacuoles can be the consequence of two different processes: increased autophagosome formation and/or block in autophagosome maturation/degradation. Therefore, we analyzed autophagic flux in LNCaP cells using the lysosome inhibitor chloroquine (CQ). We showed that at concentrations up to 20 μM ML-9 induced LC3-II accumulation without inhibiting autophagic flux, as clear increase in LC3-II levels was detected upon addition of CQ (50 μM for 1 h). In contrast, CQ failed to increase LC3-II levels induced by higher concentrated ML-9 (30–50 μM) (Figure 2a). In addition, combined treatment of LNCaP cells with ML-9 (30 μM) and bafilomycin A1 (another inhibitor of lysosomal acidification) did not elevate LC3-II levels compared with the treatment with ML-9 alone (Supplementary Figure S5). These results suggest that at concentrations >30 μM ML-9 effectively inhibits autophagic flux.
Figure 2.
ML-9 blocks autophagic flux. (a) Chloroquine does not increase LC3-II levels induced by elevated concentrations of ML-9. LNCaP cells were incubated in full, serum-starved or ML-9-containing media for 6 h in the absence or presence of 50 μM chloroquine for the last hour. Densitometric quantitation showing autophagic flux (change in LC-II levels induced by chloroquine) is represented. Values represent means±S.E.M. n=3. (b) Chloroquine fails to increase the number of eGFP-positive puncta following ML-9 treatment. eGFP-LC3 expressing LNCaP cells were treated with full, serum-starved or 30 μM ML-9-containing media for 6 h in the absence or presence of 50 μM chloroquine for the last hour. Quantitation shown on the right represents means±S.D. GFP-positive puncta per cell (n=50) from three independent experiments. (c) ML-9 induces p62 accumulation. LNCaP cells were treated with full, serum-starved or ML-9 containing media for 12 h. Densitometric quantitation for normalized p62 relative to Actin is shown. Values represent means±S.E.M. n=3. (d) ML-9 blocks starvation induced autophagic flux. LNCaP cells were treated with full, serum-starved, 30 μM ML-9-containing full medium or 30 μM ML-9-containing serum-starved medium for 6 h in the absence or presence of 50 μM chloroquine for the last hour. Densitometric quantitation showing autophagic flux (change in LC-II levels induced by chloroquine) as well as normalized p62 relative to Actin is shown. Values represent means±S.E.M. n=3
Consistent with immunoblotting experiments CQ failed to enhance ML-9-induced eGFP-LC3 puncta accumulation in LNCaP-eGFP-LC3 cells, while a clear increase in eGFP-LC3 puncta has been observed under basal and serum-starved conditions (Figure 2b).
We next confirmed these results by analyzing the levels of autophagy substrate protein p62—another widely used autophagy marker, which is degraded by autophagy.31, 32 ML-9 induced p62 accumulation in LNCaP cells in a dose-dependent manner, once again supporting the inhibitory role for ML-9 in autophagy (Figure 2c).
Interestingly, we found that ML-9 blocked serum starvation-induced autophagic flux. Addition of CQ to LNCaP cells treated with ML-9+ starvation caused no further increase in LC3-II and p62 levels (Figure 2d). Thus, these results demonstrate that ML-9 arrests basal and induced autophagic flux.
ML-9 affects maturation of autophagosomes and increases lysosomal pH
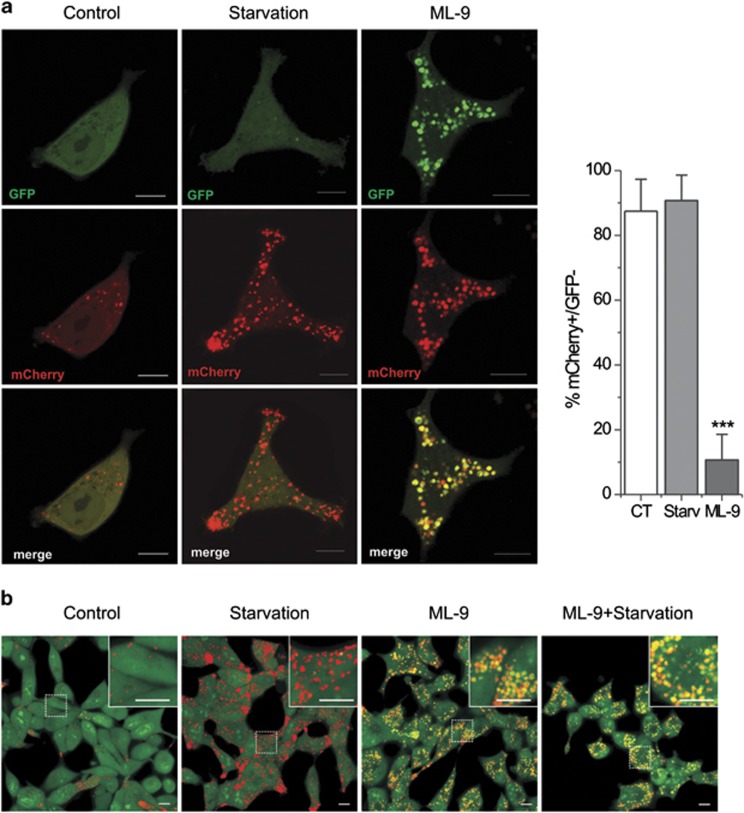
Next, to confirm these results we utilized a tandem mCherry-GFP reporter fluorescence assay designed to monitor autophagic flux (see Materials and methods).32 LNCaP cells were transiently transfected with mCherry-GFP-LC3B and treated with normal, serum-starved or ML-9 (30 μM) containing medium for 12 h. The colocalization of GFP and mCherry signals was analyzed. Under starvation conditions both the number of puncta and fraction of red-only puncta increased, indicating that fusion with acidic endosomes/lysosomes occurs rapidly after autophagosome formation. In contrast, ML-9 treatment caused a dramatic decrease in the proportion of red-only puncta, thus the majority of the puncta were double positive mCherry+/GFP+ (Figure 3a). Thus, the accumulation of enlarged double positive mCherry+/GFP+ LC3-puncta (as it follows from confocal microscopy) together with the accumulation of autolysosomes (as it goes from TEM) following ML-9 treatment suggest that ML-9 inhibits autophagic flux mainly through the increase in endosomal/lysosomal pH and impairment of proteolytic degradation of autophagy substrates inside the autolysosomes, but not through the prevention of fusion between autophagosomes and endosomes/lysosomes.
Figure 3.
ML-9 affects maturation of autophagosomes and increases lysosomal pH. (a) LNCaP cells were transiently transfected with mCherry-GFP-LC3B and treated with full, serum-starved, or ML-9 (30 μM) containing medium for 12 h. The colocalization of GFP and mCherry signals was analyzed. Quantitation shown on the right represents means±S.D. mCherry-positive/GFP-negative puncta per cell (n=20). (b) ML-9 decreases lysosomal acidification. LNCaP cells were treated with full, serum-starved, 30 μM ML-9-containing full medium or 30 μM ML-9-containing serum-starved medium for12 h, stained with acridine orange and analyzed using confocal microscopy
To test this assumption we utilized acridine orange, an acidotropic dye that is widely used as an indicator of acidification. LNCaP cells stained with acridine orange exhibited green fluorescence with a few red puncta in basal medium. Serum starvation induced accumulation of red puncta, indicating an increase in autolysosomes number. In contrast, ML-9-treated cells showed an accumulation of enlarged vesicles emitting in both green and red, indicating an increase in intraorganellar pH (Figure 3b). This effect of ML-9 could be explained by its weak base properties (pKa=8.04, calculated using the Marvin software by ChemAxon, Budapest, Hungary). Thus, it seems that ML-9 behaves like a lysosomotropic agent which is accumulated in the lysosomes and leads to an increase in lysosomal pH. We confirmed these results by assessing Lamp1 immunofluorescence in LNCaP cells transfected with mCherry-GFP-LC3B. ML-9 visibly increased the size of Lamp1-positive vesicles/puncta (Supplementary Figure S6a). In addition, ML-9 induced accumulation of mCherry-GFP-LC3B puncta, which were largely colocalized with Lamp1-positive vesicles/puncta (Supplementary Figure S6b).
Altogether these data suggest that ML-9 represents a ‘two-in-one' compound, which stimulates autophagosome formation (by downregulating Akt/mTOR pathway) and inhibits their degradation (by acting like a lysosomotropic agent and increasing lysosomal pH).
ML-9 stimulates autophagy via both Vps34-dependent and -independent mechanisms
Next, we assessed the levels of another important autophagy protein Beclin1 upon treatment with ML-9. Western blot analysis showed that ML-9 induced an increase in Beclin1 levels (Figure 4a). To test whether Vps34-Beclin1 complex is important in ML-9-induced accumulation of autophagic vacuoles, we treated LNCaP cells with ML-9 or serum-starved medium, followed by the addition of 100 nM Wortmannin (an inhibitor of Vps34) for 1.5 h. Wortmannin effectively reduced the LC3-II levels in LNCaP cells treated with low concentrations of ML-9 (up to 20 μM). In contrast, Wortmannin showed no significant effect when cells were treated with higher concentrations of ML-9 (Figure 4b). This could be due to the inability of Wortmannin to influence autophagosome degradation. To test this possibility, we used another Vps34 inhibitor 3-methyladenine (3-MA) and pretreated our cells with 5 mM 3-MA for 1 h before treatments with full, serum-starved or ML-9 containing medium for 6 h in the continuous presence of 3-MA. 3-MA effectively reduced LC3-II levels under basal and serum-starved conditions as well as in LNCaP cells treated with 20 μM ML-9. In contrast, 30 μM ML-9 induced LC3-II accumulation even following pretreatment with 3-MA (Figure 4c). In line with this, ML-9 (30 μM) was effective in increasing the number of eGFP-LC3-positive puncta in LNCaP-eGFP-LC3 cells pretreated with Wortmannin (Figure 4d).
Figure 4.
ML-9 stimulates autophagy via both Vps34-dependent and -independent mechanisms. (a) ML-9 increase Beclin1 levels in LNCaP cells. LNCaP cells were incubated in full medium, serum-starved medium or full medium with indicated concentrations of ML-9 for 12 h. Densitometric quantitation for normalized Beclin1 relative to Actin is shown. Values represent means±S.E.M. n=3. (b) Wortmannin effectively reduces LC3-II levels induced by ML-9 at low concentrations but is almost ineffective in reducing LC3-II levels induced by ML-9 at high concentrations. LNCaP cells were incubated in full medium, serum-starved medium or full medium with indicated concentrations of ML-9 for 6 h in the absence or presence of 100 nM Wortmannin for the last 1.5 h. Densitometric quantitation for normalized LC3-II relative to Actin is shown. Values represent means±S.E.M. n=3. (c) 3-MA does not preclude ML-9 induced LC3-II accumulation. LNCaP cells were incubated in full medium, serum-starved medium, 20 μM ML-9-containing medium or 30 μM ML-9-containing medium for 6 h. Alternatively, LNCaP cells were pretreated with 5 mM 3-MA for 1 h followed by the 6-h treatment with full medium, serum-starved medium, 20 μM ML-9-containing medium or 30 μM ML-9-containing medium in the continuous presence of 5 mM 3-MA. Densitometric quantitation for normalized LC3-II relative to Actin is shown. Values represent means±S.E.M. n=3. (d) eGFP-LC3 expressing LNCaP cells were treated with full, serum-starved or 30 μM ML-9-containing media for 6 h in the absence or presence of 100 nM Wortmannin. Quantitation shown on the right represents means±S.D. GFP-positive puncta per cell (n=30)
Overall, these results suggest that ML-9 stimulates autophagy via both Vps34-dependent and -independent mechanisms.
ML-9 induces prostate cancer cell death
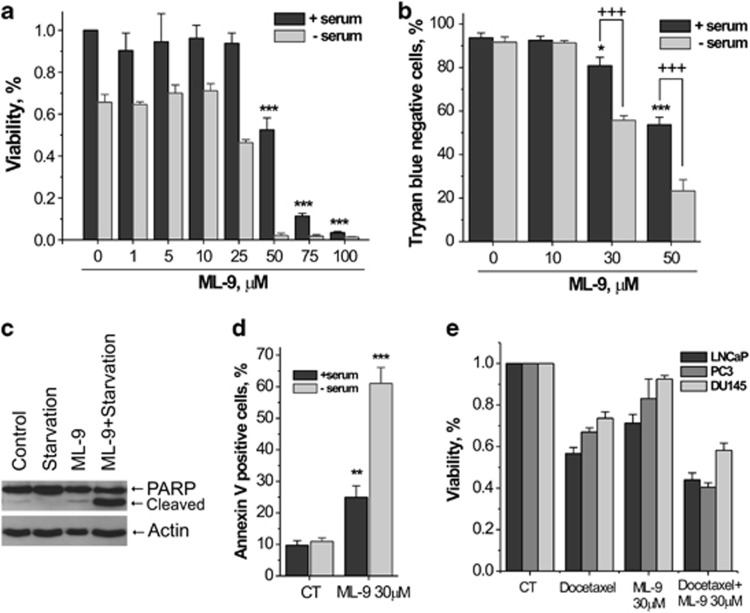
Considering the previously reported data, suggesting that combination of Akt inhibitors with late-stage autophagy inhibitors promotes cancer cell death,14, 15, 16 we next assessed the effect of ML-9 on prostate cancer cell viability. MTS and trypan blue staining assays revealed that ML-9 reduced the viability of LNCaP cells in a concentration-dependent manner. Moreover, this cell-killing effect of ML-9 was strikingly potentiated in a serum-free medium (Figures 5a and b). Next, we assessed whether ML-9 induces apoptosis in LNCaP cells. First, we checked whether ML-9 could induce cleavage of poly (ADP-ribose) polymerase (PARP). Treatment of LNCaP cells with ML-9 (30 μM) for 24 h resulted in the appearance of a cleaved PARP 89-kD fragment characteristic of apoptosis. PARP cleavage was significantly higher when cells were treated with ML-9 under serum-starved conditions (Figure 5c). These results indicate that ML-9 induces apoptosis in LNCaP cells. To confirm this, we determined the level of apoptosis by Annexin V/Propidium iodide double staining. Treatment with ML-9 (30 μM) for 24 h significantly increased the percentage of Annexin V-positive LNCaP cells and serum-free medium greatly potentiated this effect (Figure 5d).
Figure 5.
ML-9 induces prostate cancer cell death. (a) ML-9 reduced cell viability in a concentration-dependent manner. LNCaP cells, plated on 96-well plate, were treated with different doses of ML-9 in full or serum-starved media for 24 h. Cell viability was monitored using the CellTiter 96 Aqueous One Solution cell proliferation assay (MTS assay). Following treatment, the cells were incubated with reagent solution and absorbance was recorded at 490 nm wavelength using an ELISA plate reader. Values represent means±S.E.M. n=3. (b) Cell viability was assessed by trypan blue exclusion assay. LNCaP cells were treated with different concentrations of ML-9 in full or serum-starved media for 24 h, the cells were collected by trypsin-EDTA, incubated with trypan blue and viable cells as well as dead (blue) cells were counted using haemocytometer. Values represent means±S.E.M. +++P<0.001. n=4. (c) ML-9 induces PARP cleavage. LNCaP cells were treated with full, serum-starved, 30 μM ML-9-containing full medium or 30 μM ML-9-containing serum-starved medium for 24 h. (d) ML-9 induces apoptosis in LNCaP cells. LNCaP cells were treated as in (c). At the end of the treatment, cells were collected, stained with Annexin V/PI and subjected to fluorescence microscopy analysis. Data are represented as means±S.E.M. n=4. (e) LNCaP, PC3 and DU145 cells, plated on 96-well plate, were treated with docetaxel (5 nM), ML-9 (30 μM) or combination of docetaxel+ML-9 for 48 h. Cell viability was monitored using the CellTiter 96 Aqueous One Solution cell proliferation assay (MTS assay)
In addition to LNCaP cells ML-9 effectively reduced the viability of PC-3, DU-145 as well as HEK-293 cells, although these cell lines exhibited greater resistance to ML-9-induced cell death compared with LNCaP cells, with DU-145 being the most resistant (Supplementary Figure S7). At high concentrations (50–100 μM) ML-9 induced pronounced rounding up and detachment of LNCaP cells within the first 2 h of treatment. Again, this effect was intensified by the removal of serum from medium (data not shown).
Of note, after 4 h of treatment with ML-9 (30–50 μM) we detected vacuolization of the majority of LNCaP cells under the light microscope (Supplementary Figure S8). This effect correlates well with the weak base properties of ML-9 and the accumulation of autophagic vacuoles (as revealed by TEM and fluorescence microscopy).
Thus, we conclude that ML-9 as a monotherapy effectively induces prostate cancer cell death associated with the accumulation of autophagic vacuoles.
ML-9 accelerates cell death in combination with chemotherapy
Given that ML-9 effectively induces cell death in prostate cancer cells, our next step was to see whether ML-9 could be useful as an adjuvant to anticancer chemotherapy. LNCaP, PC3 and DU-145 cells were treated with docetaxel (5 nM), ML-9 (30 μM) or combination of docetaxel+ML-9 for 48 h. As shown in Figure 5e, cell viability was significantly reduced when docetaxel was combined with ML-9 compared with either drug alone. These results suggest that ML-9 could potentially be considered as an adjuvant to existing anticancer chemotherapy.
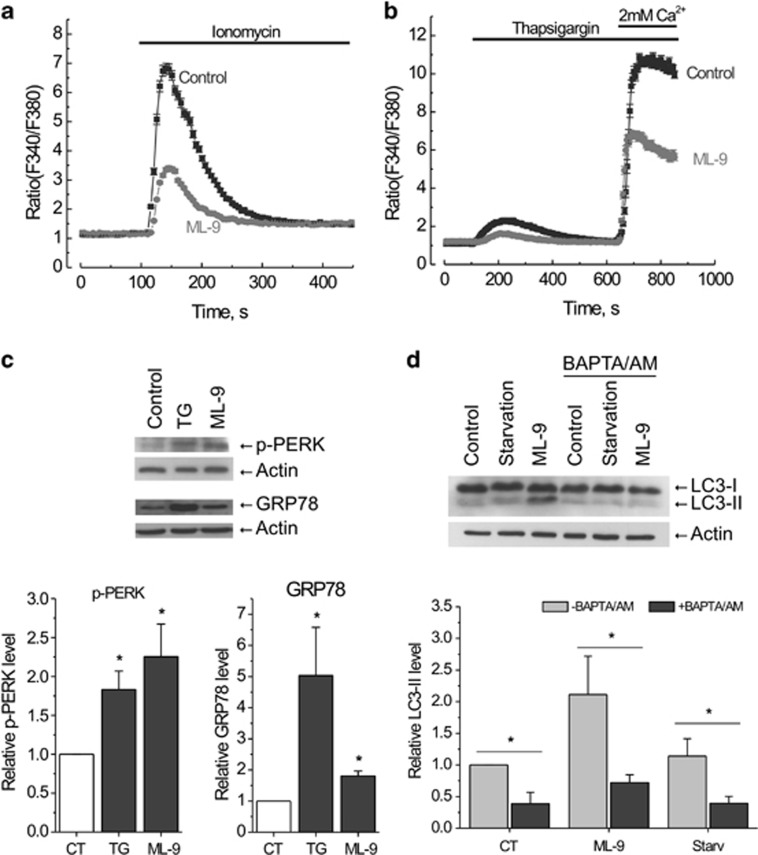
ML-9 modulates Ca2+ homeostasis and stimulates autophagy in a calcium-dependent manner
Previous reports demonstrated that ML-9 could influence cellular calcium homeostasis.20, 33, 34, 35 As calcium was shown to be an important autophagy regulator, we next investigated whether calcium-related mechanisms are involved in ML-9-induced autophagy. First, we checked whether ML-9 influences cytosolic calcium levels in LNCaP cells using a Ca2+ ionophore ionomycin and an inhibitor of sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) thapsigargin (TG). Application of 2 μM ionomycin in the absence of extracellular Ca2+ caused a transient increase in cytosolic calcium, which was significantly less when cells were pretreated with 30 μM ML-9 for 6 h (Figure 6a). Consistent with this, ML-9 treatment significantly reduced both TG-induced calcium release and store operated calcium entry (SOCE) (Figure 6b), suggesting that ML-9 reduces both calcium content of intracellular calcium stores and SOCE in LNCaP cells.
Figure 6.
ML-9 modulates Ca2+ homeostasis and stimulates autophagy in a calcium-dependent manner. (a) LNCaP cells were pretreated with full medium or 30 μM ML-9-containing medium for 6 h, loaded with Fura2/AM probe and subjected to calcium imaging experiment. 2 μM ionomycin-induced transients in calcium-free extracellular medium were analyzed. (b) ML-9 reduces both calcium content of intracellular calcium stores and SOCE in LNCaP cells. LNCaP cells were treated as in (a). 2 μM TG-induced transients in calcium-free extracellular medium as well as SOCE were analyzed. (c) ML-9 induces ER-stress. LNCaP cells were treated with full medium, 1 μM TG-containing medium or 50 μM ML-9-containing medium for 24 h. The levels of p-PERK and GRP78 were analyzed. Densitometric quantitation for normalized p-PERK and GRP78 relative to Actin is shown. Values represent means±S.E.M. n=3. (d) Ca2+ is required for ML-9-induced autophagy. LNCaP cells were treated with full, serum-starved, 30 μM ML-9-containing full media in the absence or presence of 20 μM BAPTA/AM for 3 h. Densitometric quantitation for normalized LC3-II relative to Actin is shown. Values represent means±S.E.M. n=3
It is well known that depletion of ER calcium content can trigger ER stress.36, 37 ER stress has been linked to both autophagy and cell death.38, 39 Given that ML-9 induces Ca2+ release from intracellular stores and slows down its refilling by the inhibition of SOCE, we hypothesized that in prolonged treatments ML-9 could induce ER stress. To test this hypothesis, we assessed the GRP78 (a widely used ER stress marker) level and phosphorylation of protein kinase RNA-like endoplasmic reticulum kinase (PERK), a protein kinase representing a major sensor of unfolded protein response. Treatment with ML-9 (50 μM) induced an increase in both the GRP78 level and the phosphorylation level of PERK, suggesting ER stress induction (Figure 6c), which could potentially contribute to ML-9-induced autophagy and cell death.
It is known that Ca2+ mobilizing agents, such as ionomycin and TG, regulate autophagy in a calcium-dependent manner.40 We therefore tested whether ML-9-induced autophagy is calcium dependent using an intracellular calcium chelator BAPTA/AM (1,2-Bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis/acetoxymethyl ester). Addition of 20 μM BAPTA/AM together with 30 μM ML-9 to LNCaP cells for 3 h completely abolished the ML-9-induced increase in LC3-II levels (Figure 6d), indicating that Ca2+ is required for ML-9-induced autophagosome formation.
Recent reports demonstrated that the inhibitory effect of ML-9 on SOCE is attributable to its inhibition of Stim1 redistribution.20 We therefore tested whether the effect of ML-9 on autophagy is mediated by the inhibition of Stim1. Silencing of Stim1 by small interfering RNA (siRNA) did not induce accumulation of LC3-II in LNCaP cells (Supplementary Figure S9a). In line with this result, treatment of LNCaP cells with BTP2, another inhibitor of SOCE, was without effect on LC3-II (Supplementary Figure S9b). These results indicate that ML-9 exerts its effects on autophagy independently of Stim1 and SOCE inhibition.
Discussion
In this study, we provide evidence that ML-9, a widely used inhibitor of Akt kinase, MLCK and STIM1, represents the ‘two-in-one' compound which stimulates autophagosome formation and inhibits their degradation. We also demonstrate that cytosolic calcium is essential for ML-9-induced autophagosome formation; however, neither STIM1 nor SOCE is necessary. Further, we show that ML-9 as a monotherapy effectively induces prostate cancer cell death associated with the accumulation of autophagic vacuoles. In addition, ML-9 enhances the anticancer activity of docetaxel, suggesting its potential application as an adjuvant to existing anticancer chemotherapy. We also demonstrate that ML-9 significantly reduces ER calcium content and induces ER stress, which could contribute to its cytotoxicity. Altogether our results revealed the complex effect of ML-9 on autophagy and indentified ML-9 as an attractive tool for targeting autophagy in cancer therapy through dual inhibition of both the Akt pathway and the autophagy.
Recently, Zhang et al.24 performed high-throughput image-based screening for small-molecule regulators of autophagy and among others identified ML-9 as a compound that increases GFP-LC3 vesicles in human glioblastoma H4 cells. In another study ML-7, a structurally related analog of ML-9, was shown to induce accumulation of vesicle-like structures in Schwann cells.41
The evidence presented here clearly demonstrates that the reported increase in GFP-LC3 vesicles by ML-9 is in fact the consequence of both the stimulation of autophagosome formation and the inhibition of their degradation. To reach this conclusion, we used a number of approaches including LC3 protein detection, electron microscopy for direct visualization of autophagic process, fluorescence and confocal microscopy for GFP-LC3+ and mCherry-GFP-LC3+ puncta analysis as well as endogenous autophagy substrate (p62) degradation analysis.
We demonstrated that ML-9 stimulates autophagy through inhibition of mTOR kinase. Several mechanisms may contribute to the inhibition of mTOR by ML-9. First, as mTOR acts downstream of Akt and Akt is known to positively regulate mTOR, inhibition of Akt by ML-9 could consequently induce mTOR inhibition and activation of autophagy. Second mechanism is based on the fact that during autophagy the nutrients generated by degradation of cargo in the autolysosomes stimulate mTOR, representing a feedback regulatory loop. Accordingly, ML-9-induced lysosomal dysfunction leads to a decrease in autophagic flux thereby decreasing nutrients availability and promoting mTOR inhibition.
Intriguingly, we discovered that ML-9 being an autophagy activator inhibits autophagy in the late stages. This effect of ML-9 could be attributed to its weak base properties. We propose that ML-9 as a lipophilic weak base enters the cell by simple diffusion. At neutral pH, ML-9 is in its non-protonated form. When ML-9 enters acidic vacuoles (such as lysosomes and amphisomes) it becomes positively charged through protonation and trapped in these vacuoles. Then, the increased osmotic pressure in these vacuoles stimulates water influx and vacuoles enlargement. All these events result in the increased intravacuolar pH, subsequent inhibition of lysosomal enzymes and block in degradation of the autolysosomal content. Based on the immuno-TEM and immunofluorescence analysis, we concluded that the majority of the vacuoles represent autolysosomes favoring the hypothesis that ML-9 does not prevent fusion between autophagosomes and lysosomes.
Surprisingly, the early-stage autophagy inhibitors 3-MA and Wortmannin did not prevent accumulation of autophagic vacuoles induced by high doses of ML-9 (>30 μM). This result could indicate that ML-9 could at once induce both canonical and non-canonical autophagy via different pathways.
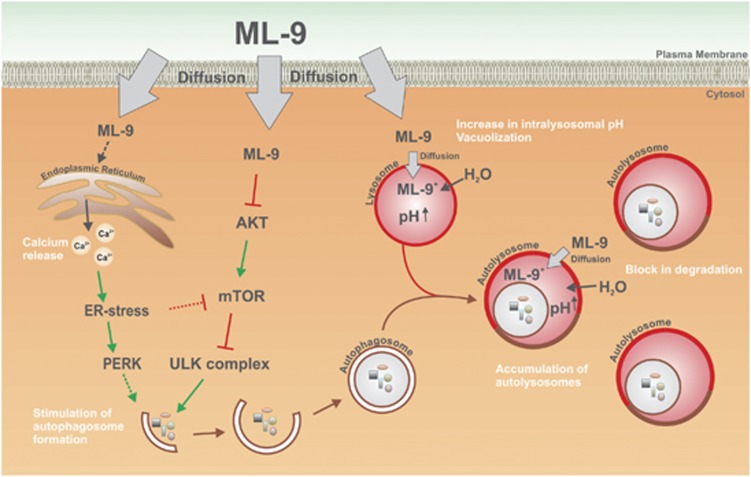
A number of studies reported the modulation of cellular Ca2+ homeostasis by ML-9.20, 34, 35 ML-9 has been shown to inhibit redistribution of STIM1 and it is widely used as an SOCE inhibitor.20 However, our data demonstrate that STIM1 and SOCE do not contribute to ML-9-induced autophagy. In contrast, ML-9 induces an increase in GRP78 level and PERK activation (suggesting ER-stress induction) presumably through the decrease in ER calcium content. Interestingly, ER stress has been shown to negatively regulate Akt/mTOR pathway and as such stimulate autophagy.42 Of note, it was proposed that ML-9 promotes calcium release through the IP3 receptor.33 The accumulated data suggested a complex role for IP3R in autophagy regulation.43, 44, 45 Thus, IP3R pathway as well as PERK pathway46, 47 could potentially contribute to both ML-9-induced autophagy and cell death. In addition, we demonstrated that cytosolic calcium is essential factor for ML-9-induced autophagy, as its removal by chelation blocks autophagy at an early stage before autophagosome formation. A graphic model for ML-9's mode of action is represented in Figure 7.
Figure 7.
A model for ML-9's mode of action. ML-9 as a lipophilic weak base enters the cell by simple diffusion. ML-9 downregulates Akt/mTOR pathway and as such stimulates autophagosome formation. ML-9 induces calcium release from ER and provokes ER stress. ER stress is a well-known autophagy inducer. At neutral pH, ML-9 is in its non-protonated form. When ML-9 enters acidic vacuoles (such as lysosomes, late endosomes, amphisomes and autolysosomes) it becomes positively charged through protonation and trapped in these vacuoles. Then, the increased osmotic pressure in these vacuoles stimulates water influx and vacuoles enlargement. All these events result in the increased intravacuolar pH, subsequent inhibition of lysosomal enzymes and block in degradation of the autolysosomal content as well as accumulation of autophagic vacuoles
Recently, it was proposed that dual inhibition of Akt kinase and autophagy represents a prospective strategy in anticancer therapy.14, 15, 16 Indeed, one of the most important functions of Akt kinase is cell survival. This kinase is often upregulated in cancer and promotes cell proliferation, cell growth and resistance to apoptosis. Thus, inhibiting Akt kinase appears to be an effective approach in cancer treatment. Similarly, autophagy is thought to be predominantly a cell-survival mechanism. Elevated autophagy is often detected in cancer cells in response to radiation and chemotherapy.13, 48, 49 Furthermore, autophagy seems to contribute to the therapeutic resistance of some cancers. Thus, autophagy inhibition also seems to represent a promising therapeutic strategy in the treatment of cancer. The combination of these two approaches shows additive efficacy in anticancer therapy.
Our results suggest that ML-9 represents a dual inhibitor of both Akt pathway and autophagy and thus is potentially interesting for cancer treatment. Indeed, ML-9 as a monotherapy effectively induces prostate cancer cell death as well as enhances the anticancer activity of docetaxel, indicating its potential application as an adjuvant to existing anticancer chemotherapy. These data are in line with the previously published studies showing that ML-9 and its structurally related analog ML-7 promote apoptotic cell death in a variety of cell lines, although these effects have been linked to MLCK inhibition.21, 22, 23 Moreover, ML-7 was proposed as a promising candidate for treating cancer.22 Interestingly, we found that cell-killing effect of ML-9 was strikingly potentiated in a serum-free medium. This result can be explained by the dual inhibition of androgen receptor (AR) and Akt/mTOR signaling pathways in these conditions. Indeed, serum-free medium mimics steroid-deprived conditions and thus inhibits AR signaling pathway. It should be noted that interaction between AR and Akt/mTOR pathways has been reported to be involved in tumorigenesis.25, 50 Moreover, the use of Akt/mTOR inhibitors in combination with AR antagonists has been demonstrated to improve anticancer efficacy.51, 52 Thus, combining inhibitors of Akt, AR and autophagy could potentially constitute a novel strategy in prostate cancer therapy.
Overall, our data suggest that ML-9 represents an attractive tool for targeting autophagy in cancer through dual inhibition of AKT pathway and autophagy. Further, the chemical structure of ML-9 could serve as a ‘template' for the synthesis of improved structurally related and more selective compounds, which could potentially be used for cancer treatment.
MATERIALS AND METHODS
Antibodies and reagents
Rabbit anti-LC3B (L7543) and mouse anti-betaActin (A5441) were from Sigma-Aldrich (Saint-Quentin-Fallavier, France). Guinea pig anti-p62 (GP62-C) was from PROGEN Biotechnik GmbH (Heidelberg, Germany). Rabbit anti-mTOR (2972S), rabbit anti-p-mTOR (2971S), rabbit anti-p-Akt (4058S), rabbit anti-Akt (9272S) and rabbit anti-PARP (9542) were from Cell Signaling (Danvers, MA, USA). Rabbit-anti-p-PERK (sc-32577), rabbit anti-GRP78 (H-129), mouse anti-Lamp1 (H4A3) and mouse-anti-Lamp2 (sc-18822) were from Santa Cruz Biotechnology (Heidelberg, Germany).
ML-9 (105637) was from Enzo Life Sciences (Villeurbanne, France). Acridine orange (A-3568) was from Life Technologies (Grand Island, NY, USA). Bafilomycin A1 (1334) was from Tocris (Bristol, UK). Chloroquine (C6628), Wortmannin (W1628) and trypan blue solution (T8154) were from Sigma-Aldrich.
Constructs
The pDest-mCherry-eGFP-LC3B plasmid was kindly provided by Prof. Terje Johansen (Institute of Medical Biology, University of Tromso, Tromso, Norway). The pcDNA3.1(-)-GFP-LC3 plasmid was a kind gift from Prof. Geert Bultynck (KU Leuven, Leuven, Belgium).
Cell culture and transfection
Prostate cancer cell lines LNCaP, PC3 and DU-145 were purchased from the American Type Culture Collection (ATCC) and cultured in RPMI-1640 medium (31870; Gibco-Life Technologies, Grand Island, NY, USA) supplemented with 5 mM L-glutamine (25030; Gibco-Life Technologies) and 10% fetal bovine serum (F7524; Sigma-Aldrich). HEK-293 cells were cultured in Dulbecco's minimal essential medium (DMEM)+GlutaMAX (31966, Invitrogen, Life Technologies) supplemented with 10% fetal bovine serum (F7524; Sigma-Aldrich). Pancreatic cancer cell lines ASPC1 and BxPC3 were purchased from the ATCC and cultured in RPMI-1640 medium (31870; Gibco-Life Technologies) supplemented with 5 mM L-glutamine (25030; Gibco-Life Technologies) and 10% FCS Gold (PAA Laboratories GmbH, Colbe, Germany).
LNCaP cells stably expressing eGFP-LC3 (LNCaP-eGFP-LC3) were generated by stable transfection with eGFP-LC3 plasmid using Xtremegene HP transfection reagent (Qiagen, Courtaboeuf, France) and selection with 500 μg/mL G418 (Sigma-Aldrich).
LNCaP cells were transiently transfected with mCherry-eGFP-LC3B construct using Nucleofector Technology (Lonza, Basel, Switzerland) as described by the manufacturer. Briefly, one million of cells were transfected with 2 μg of mCherry-eGFP-LC3B plasmid and seeded on tissue culture dishes with cover glass bottom (FluoroDish, FD35; World Presicion Instruments, Inc., Hertfordshire, UK). Two days after plating, cells were used for treatments and subsequent confocal imaging. Tandem mCherry-GFP reporter fluorescence assay is based on the use of the pH-sensitive fluorescent tag consisting of a tandem fusion of the red, acid-insensitive mCherry and the acid-sensitive GFP.32 Under neutral pH, both mCherry and GFP fluoresce and colocalize indicating an autophagosome which is not fused with acidic lysosome. Alternatively, colocalization of mCherry and GFP signals could point on an amphisome or autolysosome with decreased acidification and/or impaired proteolytic degradation. In contrast, mCherry signal without GFP corresponds to an amphisome or an autolysosome with physiologically acidic interior.32
LNCaP cells were transfected with 40 nM of siRNA against STIM1 (Eurogentec, Angers, France) using 6 μl Hyperfect transfection reagent (Qiagen), following the manufacturer's instructions.
Cell viability
Cells were seeded at 10 000 cells/well on 96-well plates in normal medium. The cells were treated either with DMSO or with the indicated concentrations of ML-9 for up to 72 h in full or serum-starved media. Cell viability was monitored using the CellTiter 96 Aqueous One Solution cell proliferation assay (Promega, Madison, WI, USA), on the basis of the cellular conversion of the colorimetric reagent MTS [3,4-(5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt] into soluble formazan by dehydrogenase enzymes found only in metabolically active cells. Following treatment, the cells were incubated with reagent solution and absorbance was recorded at 490 nm wavelength using an ELISA plate reader (Molecular Devices corp., Sunnyvale, CA, USA).
The cell viability was also assessed by trypan blue exclusion assay. After treatment with either DMSO or ML-9, the cells were collected by trypsin-EDTA, incubated with trypan blue and the viable cells as well as dead (blue) cells were counted.
Apoptosis assays
The level of apoptosis was determined by Alexa Fluor 488 Annexin V/Propidium iodide double staining. At the end of the treatments, both floating and attached cells were collected by trypsinization, centrifuged, washed with PBS and stained with Alexa Fluor 488 Annexin V and Propidium iodide according to the manufacturer instructions (Alexa Fluor 488 Annexin V/Dead Cell Apoptosis Kit; Life Technologies). Cells were examined by fluorescence microscopy on Zeiss Axio Imager A1 microscope (Carl Zeiss S.A.S., Marly-le-Roi, France). The percentage of Alexa Fluor 488 Annexin V-positive cells was determined by counting at least 500 cells in random fields.
Electron microscopy
Cell pellets were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4 for at least 30 min at 4°C. After fixation, the specimens were thoroughly washed in 0.1 M cacodylate buffer and then postfixed with 1% osmium tetroxide in the same buffer for 1 h at room temperature, stained with 2% uranyl acetate in distilled water for 15 min, dehydrated in graded acetonitrile, and embedded in Epon. Ultrathin sections (80–90 nm thick) were cut on a Leica UC7 (Leica Microsystems, Nanterre, France), transferred on 150-mesh grids and contrasted with 2% uranyl acetate solution and Reynolds lead citrate solution. The electron micrographs were taken with a Hitachi H600 (Hitachi, Krefeld, Germany) transmission electron microscope at 75 kV accelerating voltage.
For immuno-electron microscopy, the cells were fixed overnight at 4°C in 8% paraformaldehyde and 0.05% glutaraldehyde in PBS buffer, thoroughly washed in the same buffer and infused in sucrose 2.3 M containing 20% polyvinyl pyrrolidone 10000 in phosphate buffer 0.1 M. The pellets were mounted on microtome supports and rapidly frozen in melting nitrogen. Ultrathin sections of about 90–100 nm were obtained using a Leica UC7 cryoultramicrotome equipped with an FC7 device. Sections were first incubated in blocking medium (0.05 M glycine, 5% fish gelatine in 0.1 M PBS buffer) for 30 min. The grids were incubated with the antibody anti-LAMP2 for 1 h at 37°C or overnight at 4°C. After washing, sections were incubated at room temperature for 30 min in the secondary gold conjugates (18 nm; Jackson Immuno Research, Suffolk, UK) diluted in the same buffer. Following a final thorough wash in PBS alone, the grids were fixed in 2% glutaraldehyde for 10 min at room temperature and washed in water. After staining with 0.5% uranyl acetate in 1.5% methyl cellulose, sections were observed on a Hitachi H600 transmission electron microscope at 75 kV accelerating voltage.
Western blotting
Cells were washed with cold PBS and lysed in ice-cold buffer containing: 1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 1% Sodium deoxycholate, 10 mM PO4Na2/K buffer, a protease inhibitor cocktail (Sigma-Aldrich) and a phosphatase inhibitor cocktail PhosSTOP (Roche, Basel, Switzerland). The lysates were centrifuged at 15 000 × g at 4°C for 15 min to remove cell debris and supernatant protein concentration was determined by the BCA protein assay kit (Thermo Fisher Scientific, Courtaboeuf, France). In all, 30 μg of total protein was subjected to SDS-PAGE followed by transfer onto PVDF membranes using the Trans-Blot SD semi-dry transfer cell (Bio-Rad, Marnes-la-Coquette, France). The membranes were blocked in a 5% fat-free milk containing TNT buffer (Tris-HCl, pH 7.5, 140 mM NaCl and 0.05% Tween-20) for 1 h at room temperature. The membranes were next incubated overnight at 4°C with primary antibodies, and then for 1 h at room temperature with secondary antibodies conjugated to horseradish peroxidase. After washing, the membranes were processed for chemiluminescence detection using Luminata Western HRP substrate (Millipore, Billerica, MA, USA). Image J software (NIH, Bethesda, MD, USA) was employed for quantitative analysis.
Immunocytochemistry and fluorescence microscopy
LNCaP-GFP-LC3 cells were grown on glass coverslips. Following treatments cells were rinsed with PBS, fixed with 4% paraformaldehyde-1 × PBS for 15 min. After three washes with PBS the slides were mounted with Mowiol (81381, Sigma-Aldrich) on glass slides and subjected to subsequent fluorescence analysis using Zeiss Axiovert microscope (Carl Zeiss S.A.S.).
Acridine orange staining
LNCaP cells were seeded on tissue culture dishes with cover glass bottom (FluoroDish, FD35; World Presicion Instruments, Inc.). Two days after plating, cells were treated with normal, serum-starved or ML-9 (30 μM) containing medium for 12 h. At the end of treatments, acridine orange was added to the cells (1 μg/ml final concentration) for 15 min in 37°C. Then, the cells were washed two times with appropriate medium and subjected to confocal imaging. Upon excitation by blue light acridine orange emits at 525 nm (green). Due to its weak base properties acridine orange accumulates in acidic organelles, such as lysosomes and autolysosomes, where it precipitates and emits at around 650 nm (red). Thus, healthy acidic vesicles appear as red puncta in green cytoplasm. When the pH inside the acidic organelles increases, acridine orange fluorescence switches from red to green.
Confocal microscopy
Live-cell images were obtained using confocal laser scanning microscope (LSM 700, Carl Zeiss MicroImaging GmbH, Jena, Germany) with a Plan Apochromat 40 × /1.3 numerical aperture oil immersion objective and equipped with a CO2 and thermocontrolled chamber. The images were analyzed in Zeiss LSM Image Browser software (Carl Zeiss MicroImaging GmbH) and prepared for publication in Adobe Photoshop.
Calcium imaging
Ratiometric dye Fura-2/AM was used as a Ca2+ indicator. LNCaP cells were loaded with 2 μM Fura-2/AM for 45 min at 37°C and 5% CO2 in RPMI medium and subsequently washed three times with external solution containing (in mM): 140 NaCl, 5KCl, 1 MgCl2, 2 CaCl2, 5 Glucose, 10 Hepes (pH 7.4). The coverslip was then transferred in a perfusion chamber on the stage of Nikon Eclipse Ti microscope (Nikon, Champigny-sur-Marne, France). Fluorescence was alternatively excited at 340 and 380 nm with a monochromator (Polychrome IV, TILL Photonics GmbH, Gräfelfing, Germany) and captured at 510 nm by a QImaging CCD camera (QImaging, Surrey, BC, Canada). Acquisition and analysis were performed with the MetaFluor 7.7.5.0 software (Molecular Devices Corp.).
Statistical analysis
Data were analyzed using Origin 7.0 (Microcal Software Inc., Northampton, MA, USA). Statistical analysis was performed using Student's t-test, and P<0.05 was considered as significant. Asterisks denote *P<0.05, **P<0.01 and ***P<0.001.
Acknowledgments
We thank Professor Terje Johansen for the pDest-mCherry-eGFP-LC3B plasmid, Professor Geert Bultynck for the pcDNA3.1(-)-GFP-LC3 plasmid and Professor Cristophe Biot (University Lille 1) for the valuable discussions. We acknowledge financial support from the INSERM, la Ligue Nationale Contre le Cancer, le Ministere de l‘Education Nationale, the Region Nord/Pas-de-Calais. Artem Kondratskyi was supported by fellowship from FRM (Fondation de Recherche Medicale). Maya Yassine was a recipient of a PhD scholarship from Erasmus Mundus. Kateryna Kondratska was an IonTrac Project fellow.
Glossary
- STIM1
stromal interaction molecule 1
- PI3K
phosphatidylinositol 3-kinase
- mTOR
mammalian target of rapamycin
- MLCK
myosin light-chain kinase
- GFP
green fluorescent protein
- LNCaP
lymph node carcinoma of the prostate
- ATG
autophagy-related gene
- TEM
transmission electron microscopy
- Lamp2
lysosomal-associated membrane protein 2
- HEK-293
human embryonic kidney 293
- CQ
chloroquine
- 3-MA
3-Methyladenine
- SERCA
sarco/endoplasmic reticulum Ca2+ ATPase
- TG
thapsigargin
- SOCE
store operated calcium entry
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- BAPTA/AM
1,2-Bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis/acetoxymethyl ester
- AR
androgen receptor
- siRNA
small interfering RNA
- PARP
poly (ADP-ribose) polymerase
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by GM Fimia
Supplementary Material
References
- Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Kempf CR, Chappell WH, Abrams SL, Stivala F, et al. Therapeutic resistance resulting from mutations in Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR signaling pathways. J Cell Physiol. 2011;226:2762–2781. doi: 10.1002/jcp.22647. [DOI] [PubMed] [Google Scholar]
- Cheng CK, Fan QW, Weiss WA. PI3K signaling in glioma–animal models and therapeutic challenges. Brain Pathol. 2009;19:112–120. doi: 10.1111/j.1750-3639.2008.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, et al. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118 (Pt 1:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- Shen S, Kepp O, Kroemer G. The end of autophagic cell death. Autophagy. 2012;8:1–3. doi: 10.4161/auto.8.1.16618. [DOI] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XL, Li D, Hu F, Song JR, Zhang SS, Deng WJ, et al. Targeting autophagy potentiates chemotherapy-induced apoptosis and proliferation inhibition in hepatocarcinoma cells. Cancer Lett. 2012;320:171–179. doi: 10.1016/j.canlet.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Selvakumaran M, Amaravadi RK, Vasilevskaya IA, O'Dwyer PJ. Autophagy inhibition sensitizes colon cancer cells to antiangiogenic and cytotoxic therapy. Clin Cancer Res. 2013;19:2995–3007. doi: 10.1158/1078-0432.CCR-12-1542. [DOI] [PubMed] [Google Scholar]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101–116. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux F, Thomas C, Crafter C, Kumano M, Zhang F, Davies BR, et al. Blocked autophagy using lysosomotropic agents sensitizes resistant prostate tumor cells to the novel Akt inhibitor AZD5363. Clin Cancer Res. 2013;19:833–844. doi: 10.1158/1078-0432.CCR-12-3114. [DOI] [PubMed] [Google Scholar]
- Lamoureux F, Zoubeidi A. Dual inhibition of autophagy and the AKT pathway in prostate cancer. Autophagy. 2013;9:1119–1120. doi: 10.4161/auto.24921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BG, Wei Y, Moron JA, Lin RZ, Javitch JA, Galli A. Akt is essential for insulin modulation of amphetamine-induced human dopamine transporter cell-surface redistribution. Mol Pharmacol. 2005;68:102–109. doi: 10.1124/mol.104.009092. [DOI] [PubMed] [Google Scholar]
- Smith U, Carvalho E, Mosialou E, Beguinot F, Formisano P, Rondinone C. PKB inhibition prevents the stimulatory effect of insulin on glucose transport and protein translocation but not the antilipolytic effect in rat adipocytes. Biochem Biophys Res Commun. 2000;268:315–320. doi: 10.1006/bbrc.2000.2145. [DOI] [PubMed] [Google Scholar]
- Saitoh M, Ishikawa T, Matsushima S, Naka M, Hidaka H. Selective inhibition of catalytic activity of smooth muscle myosin light chain kinase. J Biol Chem. 1987;262:7796–7801. [PubMed] [Google Scholar]
- Smyth JT, Dehaven WI, Bird GS, Putney JW., Jr Ca2+-store-dependent and -independent reversal of Stim1 localization and function. J Cell Sci. 2008;121 (Pt 6:762–772. doi: 10.1242/jcs.023903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell LE, Helfman DM. Myosin light chain kinase plays a role in the regulation of epithelial cell survival. J Cell Sci. 2006;119 (Pt 11:2269–2281. doi: 10.1242/jcs.02926. [DOI] [PubMed] [Google Scholar]
- Gu LZ, Hu WY, Antic N, Mehta R, Turner JR, de Lanerolle P. Inhibiting myosin light chain kinase retards the growth of mammary and prostate cancer cells. Eur J Cancer. 2006;42:948–957. doi: 10.1016/j.ejca.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Kaneko K, Satoh K, Masamune A, Satoh A, Shimosegawa T. Myosin light chain kinase inhibitors can block invasion and adhesion of human pancreatic cancer cell lines. Pancreas. 2002;24:34–41. doi: 10.1097/00006676-200201000-00005. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci USA. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitting RL, Armstrong AJ. Targeting the PI3K/Akt/mTOR pathway in castration-resistant prostate cancer. Endocr Relat Cancer. 2013;20:R83–R99. doi: 10.1530/ERC-12-0394. [DOI] [PubMed] [Google Scholar]
- Majumder PK, Sellers WR. Akt-regulated pathways in prostate cancer. Oncogene. 2005;24:7465–7474. doi: 10.1038/sj.onc.1209096. [DOI] [PubMed] [Google Scholar]
- Morgan TM, Koreckij TD, Corey E. Targeted therapy for advanced prostate cancer: inhibition of the PI3K/Akt/mTOR pathway. Curr Cancer Drug Targets. 2009;9:237–249. doi: 10.2174/156800909787580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourmand G, Ziaee AA, Abedi AR, Mehrsai A, Alavi HA, Ahmadi A, et al. Role of PTEN gene in progression of prostate cancer. Urol J. 2007;4:95–100. [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang DY, Xu LH, He XH, Zhang YT, Zeng LH, Cai JY, et al. Autophagy is differentially induced in prostate cancer LNCaP, DU145 and PC-3 cells via distinct splicing profiles of ATG5. Autophagy. 2013;9:20–32. doi: 10.4161/auto.22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Norwood N, Moore TM, Dean DA, Bhattacharjee R, Li M, Stevens T. Store-operated calcium entry and increased endothelial cell permeability. Am J Physiol Lung Cell Mol Physiol. 2000;279:L815–L824. doi: 10.1152/ajplung.2000.279.5.L815. [DOI] [PubMed] [Google Scholar]
- Tran QK, Watanabe H, Le HY, Pan L, Seto M, Takeuchi K, et al. Myosin light chain kinase regulates capacitative ca(2+) entry in human monocytes/macrophages. Arterioscler Thromb Vasc Biol. 2001;21:509–515. doi: 10.1161/01.atv.21.4.509. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Takahashi R, Zhang XX, Kakizawa H, Hayashi H, Ohno R. Inhibition of agonist-induced Ca2+ entry in endothelial cells by myosin light-chain kinase inhibitor. Biochem Biophys Res Commun. 1996;225:777–784. doi: 10.1006/bbrc.1996.1250. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. The unfolding tale of the unfolded protein response. Cell. 2001;107:827–830. doi: 10.1016/s0092-8674(01)00623-7. [DOI] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Leitman EM, Tewari A, Horn M, Urbanski M, Damanakis E, Einheber S, et al. MLCK regulates Schwann cell cytoskeletal organization, differentiation and myelination. J Cell Sci. 2011;124 (Pt 22:3784–3796. doi: 10.1242/jcs.080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6:239–247. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- Decuypere JP, Welkenhuyzen K, Luyten T, Ponsaerts R, Dewaele M, Molgo J, et al. Ins(1,4,5)P3 receptor-mediated Ca2+ signaling and autophagy induction are interrelated. Autophagy. 2011;7:1472–1489. doi: 10.4161/auto.7.12.17909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parys JB, Decuypere JP, Bultynck G. Role of the inositol 1,4,5-trisphosphate receptor/Ca2+-release channel in autophagy. Cell Commun Signal. 2012;10:17. doi: 10.1186/1478-811X-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Shih YL, Ko WC, Wei YH, Shih CM. Cadmium-induced autophagy and apoptosis are mediated by a calcium signaling pathway. Cell Mol Life Sci. 2008;65:3640–3652. doi: 10.1007/s00018-008-8383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avivar-Valderas A, Salas E, Bobrovnikova-Marjon E, Diehl JA, Nagi C, Debnath J, et al. PERK integrates autophagy and oxidative stress responses to promote survival during extracellular matrix detachment. Mol Cell Biol. 2011;31:3616–3629. doi: 10.1128/MCB.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verfaillie T, Salazar M, Velasco G, Agostinis P, Linking ER. Stress to autophagy: potential implications for cancer therapy. Int J Cell Biol. 2010;2010:930509. doi: 10.1155/2010/930509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol. 2005;26:1401–1410. [PubMed] [Google Scholar]
- Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- Schayowitz A, Sabnis G, Goloubeva O, Njar VC, Brodie AM. Prolonging hormone sensitivity in prostate cancer xenografts through dual inhibition of AR and mTOR. Br J Cancer. 2010;103:1001–1007. doi: 10.1038/sj.bjc.6605882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia C, Gravina GL, Angelucci A, Millimaggi D, Muzi P, Vicentini C, et al. Additive antitumor effects of the epidermal growth factor receptor tyrosine kinase inhibitor, gefitinib (Iressa), and the nonsteroidal antiandrogen, bicalutamide (Casodex), in prostate cancer cells in vitro. Int J Cancer. 2005;115:630–640. doi: 10.1002/ijc.20917. [DOI] [PubMed] [Google Scholar]
- Schayowitz A, Sabnis G, Njar VC, Brodie AM. Synergistic effect of a novel antiandrogen, VN/124-1, and signal transduction inhibitors in prostate cancer progression to hormone independence in vitro. Mol Cancer Ther. 2008;7:121–132. doi: 10.1158/1535-7163.MCT-07-0581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.