Abstract
The pathogenic mechanisms underlying the progression of non-alcoholic fatty liver disease (NAFLD) are not fully understood. In this study, we aimed to assess the relationship between endoplasmic reticulum (ER) stress and autophagy in human and mouse hepatocytes during NAFLD. ER stress and autophagy markers were analyzed in livers from patients with biopsy-proven non-alcoholic steatosis (NAS) or non-alcoholic steatohepatitis (NASH) compared with livers from subjects with histologically normal liver, in livers from mice fed with chow diet (CHD) compared with mice fed with high fat diet (HFD) or methionine-choline-deficient (MCD) diet and in primary and Huh7 human hepatocytes loaded with palmitic acid (PA). In NASH patients, significant increases in hepatic messenger RNA levels of markers of ER stress (activating transcription factor 4 (ATF4), glucose-regulated protein 78 (GRP78) and C/EBP homologous protein (CHOP)) and autophagy (BCN1) were found compared with NAS patients. Likewise, protein levels of GRP78, CHOP and p62/SQSTM1 (p62) autophagic substrate were significantly elevated in NASH compared with NAS patients. In livers from mice fed with HFD or MCD, ER stress-mediated signaling was parallel to the blockade of the autophagic flux assessed by increases in p62, microtubule-associated protein 2 light chain 3 (LC3-II)/LC3-I ratio and accumulation of autophagosomes compared with CHD fed mice. In Huh7 hepatic cells, treatment with PA for 8 h triggered activation of both unfolding protein response and the autophagic flux. Conversely, prolonged treatment with PA (24 h) induced ER stress and cell death together with a blockade of the autophagic flux. Under these conditions, cotreatment with rapamycin or CHOP silencing ameliorated these effects and decreased apoptosis. Our results demonstrated that the autophagic flux is impaired in the liver from both NAFLD patients and murine models of NAFLD, as well as in lipid-overloaded human hepatocytes, and it could be due to elevated ER stress leading to apoptosis. Consequently, therapies aimed to restore the autophagic flux might attenuate or prevent the progression of NAFLD.
Keywords: NAFLD, NASH, autophagy, ER stress, apoptosis, hepatocytes
Non-alcoholic fatty liver disease (NAFLD) is an increasingly common chronic liver disease worldwide encompassing from simple steatosis (non-alcoholic steatosis (NAS)) to steatohepatitis (non-alcoholic steatohepatitis (NASH)) with progressive fibrosis and, ultimately, cirrhosis.1, 2 Although oxidative stress and proinflammatory cytokines have relevant roles in NASH pathogenesis, our understanding of the molecular mechanisms involved in liver cell injury contributing to progression from NAS to NASH is still very limited.3, 4
The unfolded protein response (UPR) is an adaptive cellular process characterized by translational arrest of protein synthesis in the endoplasmic reticulum (ER), which when dysregulated can perpetuate ER stress and oxidative and inflammatory signaling, both processes known to be important in the pathogenesis of NASH.5 Cellular stressors initiate a signal-transduction cascade mediated by three transmembrane ER sensors: PKR-like eukaryotic initiation factor 2 kinase (PERK), activating transcription factor 6 (ATF6) and inositol requiring 1α (IRE1α). These cascades are kept inactive as long as sensors are bound to the intraluminal chaperone glucose-regulated protein 78 (GRP78). If the cell fails to adapt, oxidative and inflammatory signaling pathways are triggered, leading to apoptosis through PERK-mediated C/EBP homologous protein (CHOP) expression, IRE1α-mediated recruitment of tumor necrosis factor receptor-associated factor 2 and signal-regulated kinase 1/C-jun N-terminal kinase (JNK) (reviewed by Hotamisligil et al.6).
Autophagy, or cellular self-digestion, is a lysosomal pathway that degrades intracellular organelles to maintain energy homeostasis during periods of nutrient deprivation and to remove damaged cellular components.7, 8 There are three major pathways that regulate macroautophagy. First, in response to nutrients, phosphatidylinositol 3-kinase (PI3K) activates the mammalian target of rapamycin (mTOR), which, in turn, blocks autophagy through inhibition of Atg1 from recruiting its partners Atg13 and Atg17.9 A secondary pathway is mediated by Atg6/beclin-1, which forms a complex with Vps34, the class III PI3K. Beclin-1 is an important interface between autophagy and cell death pathways, because the antiapoptotic proteins Bcl-2 and Bcl-xL bind beclin-1 to inhibit autophagy.10 The third pathway involves Atg3- and Atg7-mediated conjugation of microtubule-associated protein 1 light chain 3 (LC3-I) to the membrane lipid phosphatidylethanolamine to form LC3-II.11 LC3-II is present on both the inner and outer isolation membranes, serving as a recognition site for LC3-binding chaperones such as p62/SQSTM1 (p62) that deliver their cargo to autophagosomes.12 Once formed, autophagosomes traffic along microtubules to reach lysosomes where they fuse to form autophagolysosomes allowing the degradation of their contents by lysosomal acid hydrolases.
The implication of autophagy in hepatocyte lipid metabolism has been recently demonstrated.13, 14 In humans, alterations in autophagy may have a role in the pathophysiology of disorders that result from excessive lipid accumulation, such as manifestations of metabolic syndrome and NAFLD. In NAFLD, impairment in hepatocyte autophagy may not only promote steatosis but also might lead to NASH owing to the loss of autophagy's protective function against cell death. The latter hypothesis prompted us to determine the extent of ER stress and impairment of autophagy and the pathways involved in NAFLD patients. We have also evaluated the relationship between lipid accumulation, ER stress and the autophagy pathways in human hepatocytes and murine models of hepatic steatosis and NASH.
Results
Patient characteristics
Features of NL (normal liver) individuals and NAFLD patients studied are detailed in Table 1. All groups were well matched in terms of age and sex distribution. In the NAFLD cohort, NAS patients had significantly higher body mass index (BMI) mean (P=0.017), insulin (P=0.007), homeostasis model assessment of insulin resistance (HOMA-IR) score (P=0.009), triglycerides (P=0.031) and γ-GT (P<0.001), as well as lower HDL-cholesterol mean (P=0.039) than subjects with NL. In NASH patients, the BMI mean, insulin, HOMA-IR score, triglycerides, ALT, AST and γ-GT were significantly higher than in NL individuals (P<0.001 for all). In addition, HDL-cholesterol mean was significantly lower in NASH patients than in NL subjects (P=0.013). As expected, NASH patients had significantly higher serum ALT than NAS patients (P<0.001).
Table 1. Demographic, metabolic, biochemical and histopathological characteristics of patients with normal liver and NAFLD.
| Feature | NL (n=34) | NAS (n=26) | NASH (n=23) |
|---|---|---|---|
| Age (years) | 45.1±11.3 | 47.9±14.7 | 48.5±14.3 |
| Female gender | 20 (58.8%) | 16 (61.5%) | 14 (60.9%) |
| Body mass index (kg/m2) | 25.8±2.4 | 29.2±3.1a | 30.1±2.8a |
| Glucose (mg/dl) | 92.8±4.6 | 96.9±7.1 | 96.5±7.3 |
| Insulin (μU/l) | 5.7±2.5 | 12.1±6.5a | 15.4±5.6a |
| HOMA-IR | 1.48±0.43 | 3.63±1.17a | 3.92±1.54a |
| Triglycerides (mg/dl) | 103.2±43.4 | 132.1±47.4a | 198.1±56.1a,b |
| HDL-cholesterol (mg/dl) | 49.1±8.2 | 42.2±8.7a | 40.4±9.1a |
| ALT (IU/l) | 13.9±6.6 | 18.8±14.6 | 64.2±29.2a,b |
| AST (IU/l) | 15.5±3.5 | 22.6±8.7 | 36.8±12.4a |
| γ-GT (IU/l) | 28.1±12.4 | 79.2±60.4a | 86.4±72.7a |
| Steatosis (%) | |||
| Grade 0 | 34 (100%) | ||
| Grade 1 | 11 (42.3%) | 7 (30.4%) | |
| Grade 2 | 9 (34.6%) | 10 (43.5%) | |
| Grade 3 | 6 (23.1%) | 6 (26.1%) | |
| Fibrosis (%) | |||
| Stage 0 | 34 (100%) | 26 (100%) | |
| Stage 1 | 12 (52.1%) | ||
| Stage 2 | 7 (30.5%) | ||
| Stage 3 | 4 (17.4%) | ||
| Ballooning and lobular inflammation (%) | |||
| Grade 0 | 34 (100%) | 26 (100%) | |
| Grade 1 | 7 (30.5%) | ||
| Grade 2 | 11 (47.8%) | ||
| Grade 3 | 5 (21.7%) | ||
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; γ-GT, gamma-glutamyltransferase; HDL, high-density lipoprotein; HOMA-IR, homeostatic model assessment-insulin resistant; NAS, nonalcoholic steatosis; NASH, nonalcoholic steatohepatitis; NL, normal liver
Data are shown as mean±S.D. or as number of cases (%)
P<0.05 with respect to NL group
P<0.05 with respect to NAS group
Hepatic autophagy is impaired in NAS and NASH patients
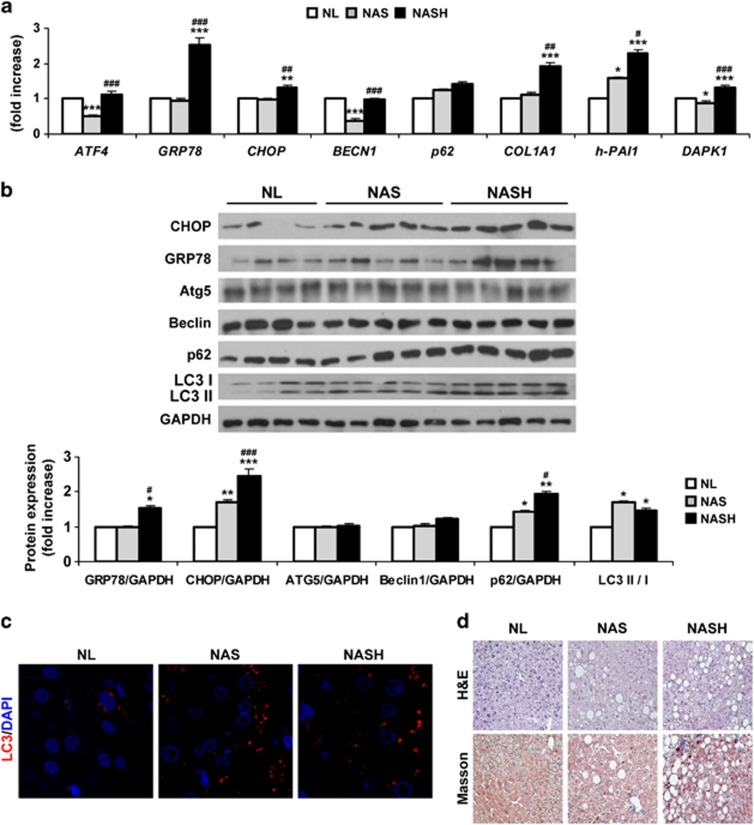
First, we analyzed ER stress markers in human liver biopsies from patients with NAFLD. Messenger RNA (mRNA) levels of ATF4 decreased in NAS patients compared with the other two groups (Figure 1a). Moreover, GRP78 and CHOP mRNA levels were increased in NASH patients compared with NAS patients or NL individuals. When protein analysis was carried out, an increase in CHOP was detected in NAS and NASH patients, whereas GRP78 only increased in the NASH group (Figure 1b).
Figure 1.
Hepatic autophagy flux is impaired in NAS and NASH patients. Analysis of ER stress and autophagy markers in human liver samples from NL (n=34) individuals and NAS (n=26) and NASH (n=23) patients. (a) qPCR analysis from all samples. The fold induction value for triplicate wells was averaged and data are presented as mean±S.E.M. relative to NL. (b) Representative western blots with the indicated antibodies. After densitometric analysis from blots corresponding to all patients, data are presented as mean±S.E.M. relative to NL. (c) Representative anti-LC3 immunostaining in liver sections. (d) Representative images from H&E and Masson's trichrome staining. *P<0.05, **P<0.01 and ***P<0.005, NAS or NASH, respectively, versus NL; #P<0.05, ##P<0.01 and ###P<0.005, NASH versus NAS
Next, we analyzed mRNA levels of autophagy mediators. Hepatic mRNA levels of BECN1 were significantly lower in patients with NAS as compared with individuals with NL (Figure 1a). However, hepatic mRNA levels of the autophagic substrate SQSTM1 (p62) increased in both NAS and NASH, although no statistical significance was found when these patients were compared with NL individuals.
As lipidation of LC3 and its association with autophagosome membranes has been established as useful sign for autophagy, we detected LC3 by immunoblotting and fluorescence microscopy. Importantly, a significant increase in LC3-II/LC3-I ratio was detected in both NAS and NASH patients compared with NL subjects (Figure 1b). Moreover, LC3-II punctuate was clearly visible by immunofluorescence in biopsies from both patient groups, whereas diffuse staining was visualized in NL samples (Figure 1c). However, these techniques should be analyzed carefully, as positive results clearly indicate increased numbers of autophagosomes, but do not always mean upregulation of the autophagic flux.8 True autophagic function can be measured by the change in LC3-II levels in the presence versus absence of lysosomal inhibitors, which measures the rate of LC3-II degradation. As these approaches were not possible in human liver samples, we measured protein levels of p62, a selective substrate of autophagy, because activation of the autophagic flux leads to a decline in p62 expression, and vice versa.15, 16 As Figure 1b shows, a significant increase in p62 was detected in both NAS and NASH patients compared with individuals with NL, indicating impairment in the autophagic flux. Furthermore, p62 was significantly elevated in NASH compared with NAS patients.
Fibrogenic markers (h-COL1A1 and h-PAI1) were significantly elevated in NASH patients compared with NL and NAS groups. Likewise, mRNA levels of DAPK1 were exclusively elevated in the NASH group (Figure 1a). The histological analysis by hematoxylin/eosin (H&E) and Masson's trichrome staining of these representative samples is shown in Figure 1d. The analysis of cell death by TUNEL previously reported17 revealed the presence of apoptosis only in the NASH group.
Increased ER stress correlates with the accumulation of p62, LC3-II and autophagosomes in livers of mice fed with HFD or MCD
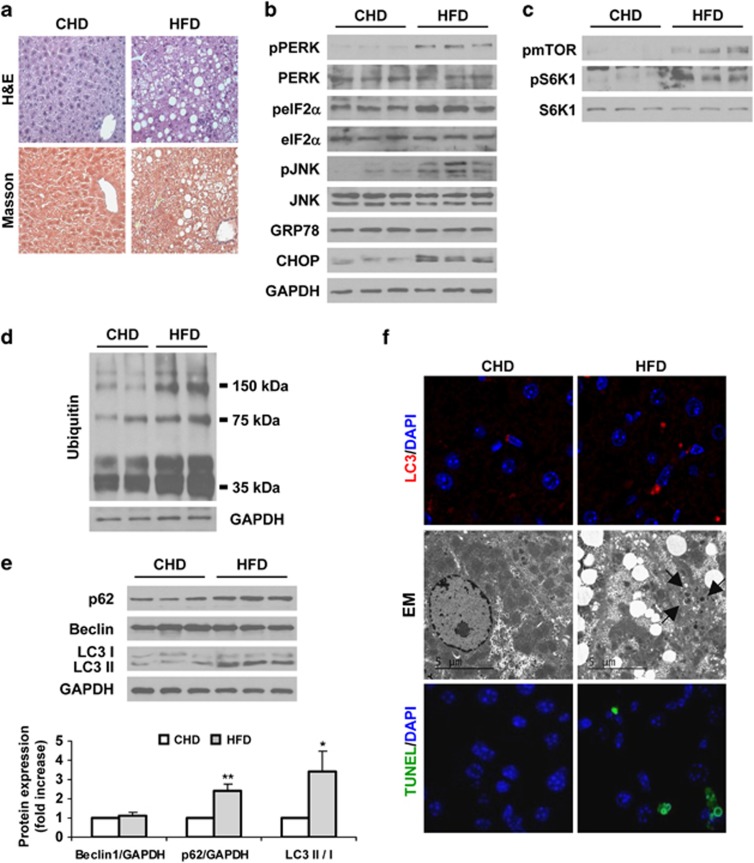
We next examined whether activation of the ER stress signaling was accompanied by changes in the autophagic flux during the progression of experimental NAFLD in mice. For this purpose, male C57BL6 mice were fed with high fat diet (HFD) for 30 weeks to generate NAS or with methionine-choline-deficient diet (MCD) for 4 weeks as a well-established model of NASH. Control mice were fed with chow diet (CHD). Upon histological examination, the livers of mice fed with HFD exhibited steatosis (Figure 2a). Phosphorylation of PERK, eIF2α and JNK, the latter activated via IRE,18 and CHOP protein levels were significantly increased in mice with hepatic steatosis compared with the control group (Figure 2b). However, protein levels of GRP78 were similar in both groups. These results agree with other studies.19, 20 As nutrient-mediated activation of mTOR suppresses autophagy,9 we evaluated the phosphorylation of mTOR and its downstream target S6K1. Figure 2c shows significant increases in both phosphorylated proteins in HFD-fed mice compared with mice receiving CHD. Consequently, the autophagic flux was blocked as revealed by significant increase in levels of polyubiquitinated proteins, accumulation of p62 and increased LC3-II/LC3-I ratio in mice fed with HFD that was also evidenced by the increased LC3-II punctuate in liver sections (Figures 2d–f). Electron microscopy (EM) revealed accumulation of double membrane structures (autophagosomes) in HFD-fed mice compared with control mice (CHD). Moreover, TUNEL analysis showed the presence of apoptotic hepatic cells only in HFD-fed mice (Figure 2f).
Figure 2.
Increased ER stress correlates with accumulation of p62 and LC3-II in livers of HFD-fed mice. Analysis of ER stress, autophagy and apoptosis in liver samples from C57BL6 mice fed with CHD or HFD for 30 weeks (n=8 animals per condition). (a) Representative images from H&E and Masson's trichrome staining. (b–e) Representative western blots with the indicated antibodies. After densitometric analysis from blots corresponding to all samples, data are presented as mean±S.E.M. relative to CHD liver. (f) Representative anti-LC3 immunostaining (upper panel), EM images (middle panel) and TUNEL assay staining images (lower panel). *P<0.05 and **P<0.01, HFD versus CHD
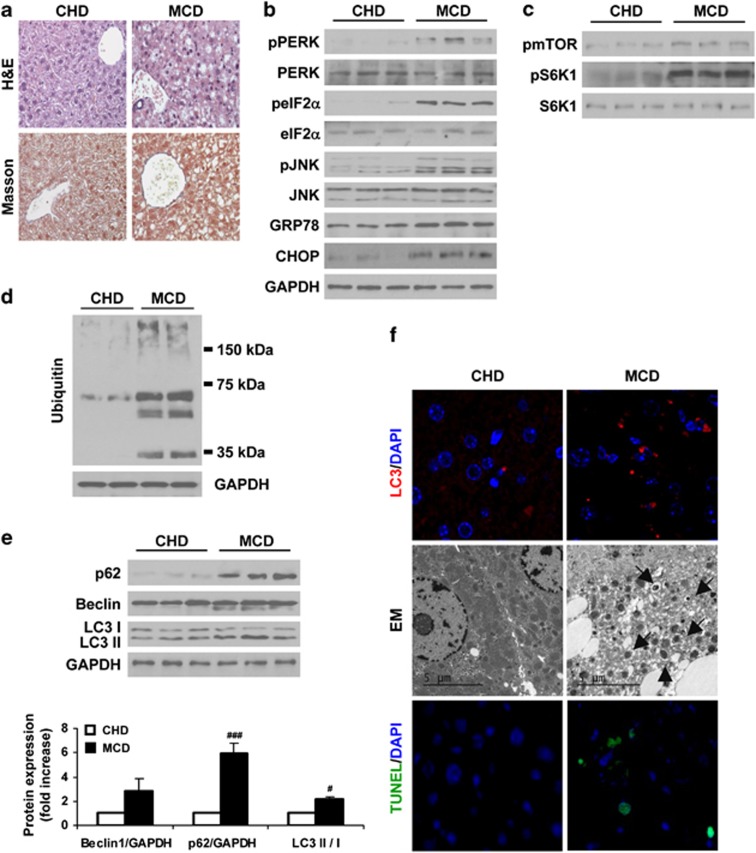
Mice fed with MCD diet developed NASH manifested by fat accumulation, ballooning and infiltration of inflammatory cells in the histological evaluation (Figure 3a) and also in the elevation of serum ALT (CHD, 30.1±2.6 U/l and MCD, 206.3±10.5 U/l). As observed in HFD-fed mice, mice fed with MCD showed elevated phosphorylation of ER stress markers, as well as increased GRP78 and CHOP protein expression compared with the control group (Figure 3b). Moreover, mTOR and S6K1 phosphorylations were elevated in livers of MCD-fed mice (Figure 3c) in parallel to an increase in the amount of polyubiquitinated proteins, an accumulation of p62, higher LC3-II/LC3-I ratio and visualization of punctuate LC3-II staining and autophagosomes compared with CHD controls (Figures 3d–f). These effects were coincident with increases in apoptosis in the MCD group as assessed by TUNEL (Figure 3f). Altogether, these results indicate that increased ER stress, blockade of the autophagic flux and apoptosis are hallmarks during NAFLD.
Figure 3.
Increased ER stress correlates with accumulation of p62 and LC3-II in livers of MCD diet-fed mice. Analysis of ER stress, autophagy and apoptosis in liver samples from C57BL6 mice fed with CHD or MCD for 4 weeks (n=10 animals per condition). (a) Representative images from H&E and Masson's trichrome staining. (b–e) Representative western blots with the indicated antibodies. After densitometric analysis from blots corresponding to all samples, data are presented as mean±S.E.M. relative to CHD liver. (f) Representative anti-LC3 immunostaining (upper panel), EM images (middle panel) and TUNEL assay staining images (lower panel). #P<0.05 and ###P<0.005, MCD versus CHD
Short-term treatment with PA induces autophagy in human hepatic cells
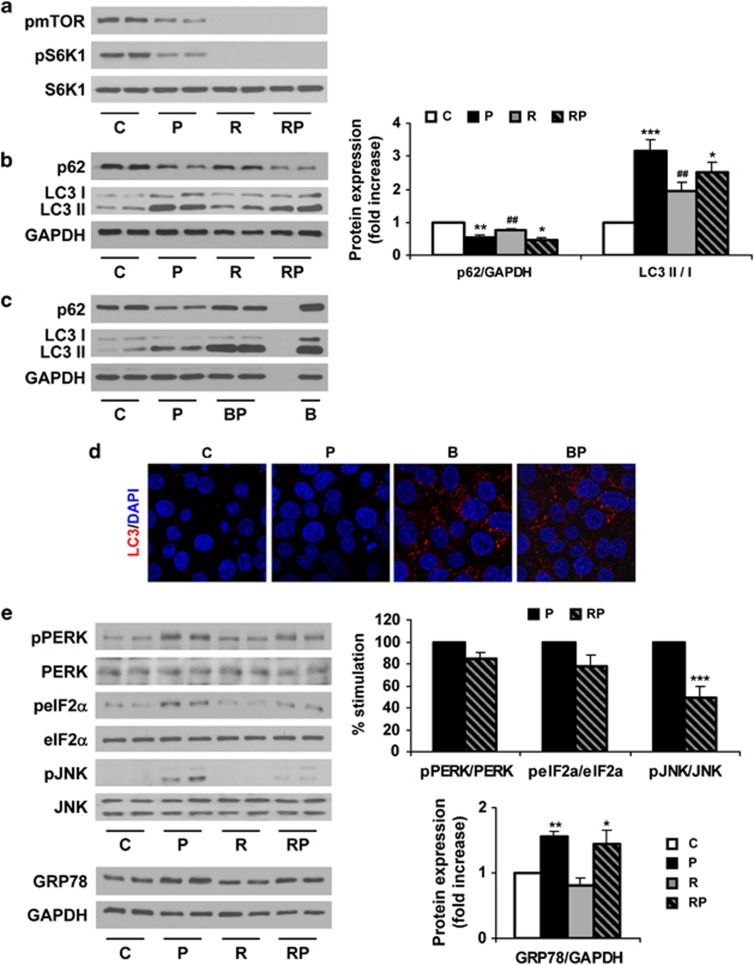
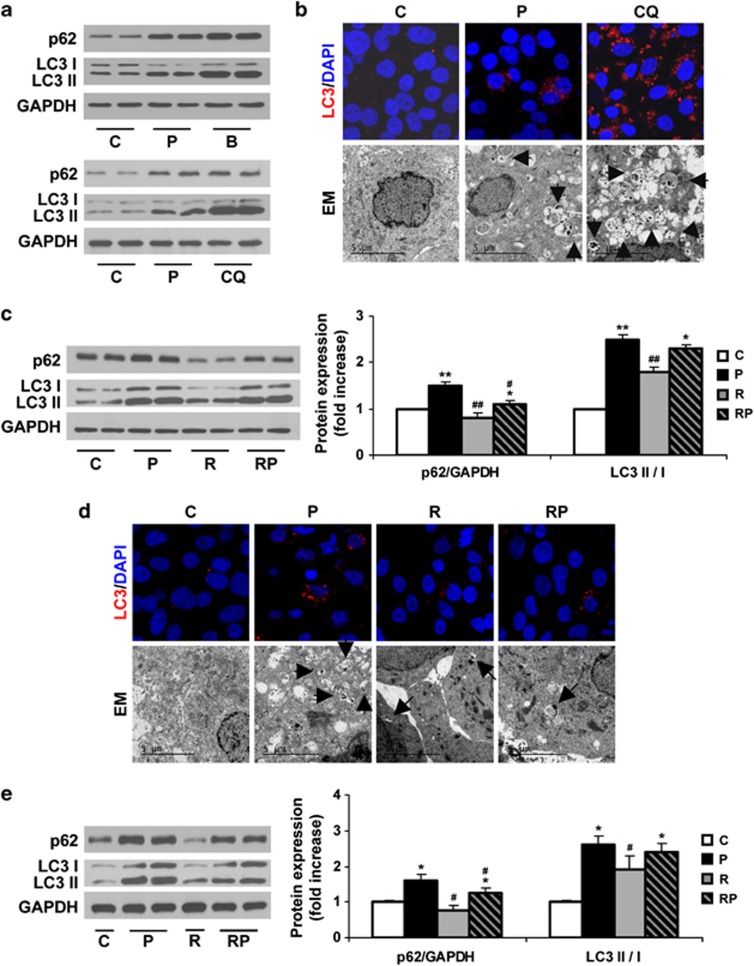
We next investigated the possibility to overcome the blockade of the autophagic flux by fatty acid overload by performing in vitro experiments in Huh7 human hepatic cells treated with palmitic acid (PA)21 combined with rapamycin, a well-known inducer of autophagy.22 Surprisingly, autophagic flux was not impaired after a short-term (8 h) treatment with PA, as assessed by reduced phosphorylation of mTOR and S6K1 in parallel to a decrease in p62 levels and increase in the LC3-II/LC3-I ratio. This effect was even more pronounced than that of rapamycin, which also activated the authophagic flux compared with untreated cells (Figures 4a and b). Additionally, we monitored the autophagic flux analyzing LC3 turnover assay using bafilomycin A1, which inhibits lysosomal function and the late degradation stage of autophagy, thereby triggering the accumulation of autophagosomes.16 The combination of PA and bafilomycin A1 increased p62 expression and also induced a higher accumulation of LC3-II compared with PA-treated cells (Figures 4c and d). These data demonstrated that LC3-II accumulation induced by PA results from the activation of autophagosome formation, but not from an inhibition of the autophagosome degradation steps.
Figure 4.
Short-term treatment with PA induces autophagy in human Huh7 cells. Analysis of ER stress and autophagy markers in samples from Huh7 cells treated with PA (P, 800 μM) for 8 h with or without rapamycin (R, 25 ng/ml) or bafilomycin A1 (B, 10 nM). (a–c and e). Representative western blots with the indicated antibodies. After densitometric analysis from blots corresponding to four independent experiments performed in triplicate, data are presented as mean±S.E.M. (d) Representative anti-LC3 immunostaining and representative EM images. *P<0.05, **P<0.01 and ***P<0.005, P versus C or RP versus R; ##P<0.01, R versus C or RP versus P
Rapamycin prevents ER stress treatment with PA in human hepatic cells
Regarding ER stress, PA treatment (8 h) induced PERK, eIF2α and JNK phosphorylations in Huh7 cells compared with the untreated controls (Figure 4e). This effect was accompanied by increased expression of GRP78. Cotreatment of PA and rapamycin reduced these events induced by PA and significantly blocked JNK phosphorylation. These results showed that activation of the UPR concurs with an active authophagic flux at early time periods of saturated fatty acid loading. Notably, cell death was not observed in Huh7 cells treated with PA for 8 h (results not shown).
Rapamycin recovers the autophagic flux and decreases cell death in Huh7 cells treated 24 h with PA
When we evaluated the effect of prolonged treatment (24 h) with PA in Huh7 cells, we found that accumulation of p62 was paralleled to an increase in the LC3-II/LC3-I ratio, reflecting a loss of the autophagic flux (Figure 5a). This effect was similar to the blockade of the autophagic flux induced by bafilomycin A1 or chloroquine (CQ).16 To verify these data, we performed LC3-II immunostaining and EM. Punctuate LC3-II was observed in Huh7 cells treated with PA for 24 h, but not in vehicle-treated cells. In addition, a marked accumulation of double-membrane structures containing cellular organelles was visualized by EM in PA-treated Huh7 cells (Figure 5b). This effect was higher in CQ-treated cells as compared with PA, indicating a more potent inhibition of the authophagic flux by this chemical compound.
Figure 5.
Rapamycin recovers the autophagic flux after prolonged treatment with PA in human Huh7 cells and human primary hepatocytes. Analysis of autophagy-related proteins expression in samples from Huh7 cells treated with PA (P, 800 μM) for 24 h or bafilomycin A1 (B, 10 nM) or CQ (50 nM) (a and b) or treated with PA (P, 800 μM) for 24 h in the absence or presence of rapamycin (R, 25 ng/ml) (c and d). (a and c) Representative western blots with the indicated antibodies. After densitometric analysis from blots corresponding to four independent experiments performed in triplicate, data are presented as mean±S.E.M. (b and d). Representative anti-LC3 immunostaining and EM images. (e) Analysis of autophagy-related protein expression in human primary hepatocytes treated with PA (P, 600 μM) for 24 h and/or rapamycin (R, 25 ng/ml). Representative western blots with the indicated antibodies. After densitometric analysis from blots corresponding to four independent experiments performed in triplicate, data are presented as mean±S.E.M. *P<0.05 and **P<0.01, P versus C or RP versus R; #P<0.05 and ##P<0.01, R versus C or RP versus P
Under these experimental conditions, rapamycin decreased p62 and LC3-II/LC3-I ratio in PA-treated cells for 24 h as compared with cells treated with PA alone, indicating a recovery of the authophagic flux (Figure 5c). This effect was parallel to decreased LC3-II punctuate and accumulation of autophagosomes as visualized in the EM images (Figure 5d). Similar effects of PA and rapamycin were found in p62 levels and LC3-II/LC-I ratio in primary cultures of human hepatocytes (Figure 5e).
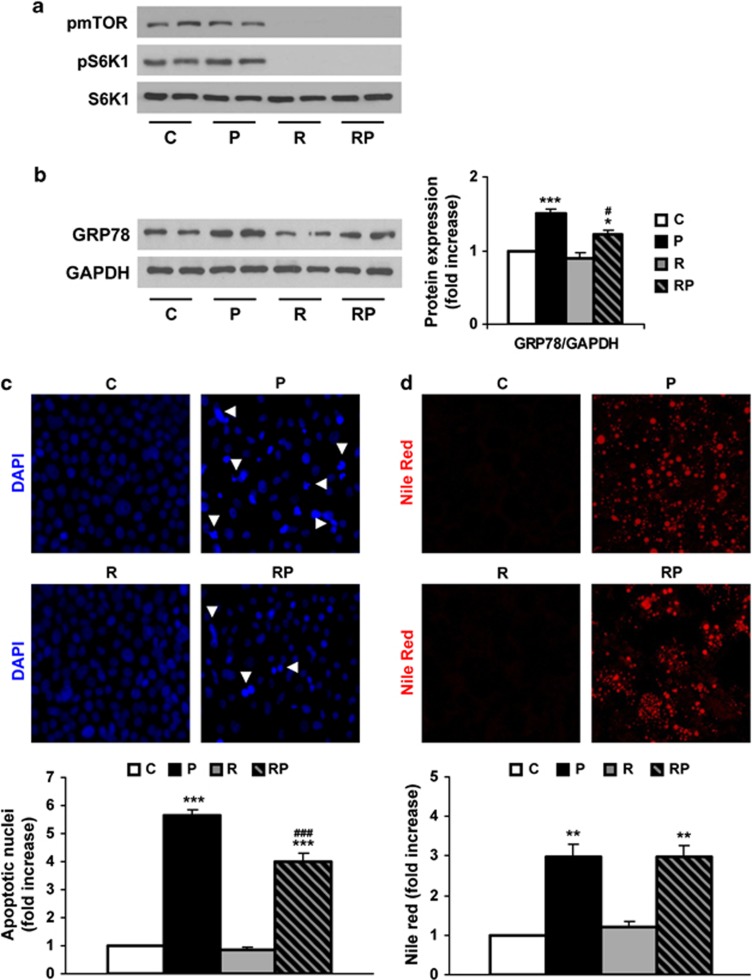
During the prolonged treatment of Huh7 cells with PA, the phosphorylation of mTOR and S6K1 remained similar to untreated cells in contrast to the effect observed at 8 h (Figure 6a). The ongoing ER stress activation was assessed by the expression of GRP78 and this effect was ameliorated in the presence of rapamycin (Figure 6b). In addition, we evaluated the impact of the restoration of the autophagic flux by rapamycin in apoptotic cell death and lipid accumulation. For this purpose, Huh7 cells were treated with PA, rapamycin or the combination of both for 24 h, and 4, 6-diamidino-2-phenylindole (DAPI) staining of nuclei and Nile Red staining of lipid droplets was analyzed. As depicted in Figure 6c, 24 h of PA treatment increased the number of apoptotic nuclei as described previously.23 Treatment with rapamycin alone did not alter the number of apoptotic nuclei; meanwhile, cotreatment with rapamycin and PA induced a significant decrease as compared with the effect of PA alone. Conversely, treatment with rapamycin did not modify PA-induced lipid droplets formation (Figure 6d).
Figure 6.
Rapamycin decreases cell death after prolonged treatment with PA in human Huh7 cells. Analysis of ER stress markers and apoptosis in samples from Huh7 cells treated with PA (P, 800 μM) for 24 h and/or rapamycin (R, 25 ng/ml). (a and b) Representative western blots with the indicated antibodies. After densitometric analysis from blots corresponding to four independent experiments performed in triplicate, data are presented as mean±S.E.M. (c). Representative DAPI staining images. Quantification of fragmented nuclei after DAPI staining. Results correspond to four independent experiments performed in triplicate and are presented as mean±S.E.M. (d) Representative Nile Red staining images. Quantification of Nile Red staining of the intracellular lipid bodies measured by flow cytometry. Results correspond to two independent experiments performed in triplicate and are presented as mean±S.E.M. *P<0.05, **P<0.01 and ***P<0.005, P versus C or RP versus R; #P<0.05 and ###P<0.005, R versus C or RP versus P
Reduction of CHOP levels decreases both blockade of the autophagic flux and cell death triggered by prolonged treatment with PA in Huh7 cells
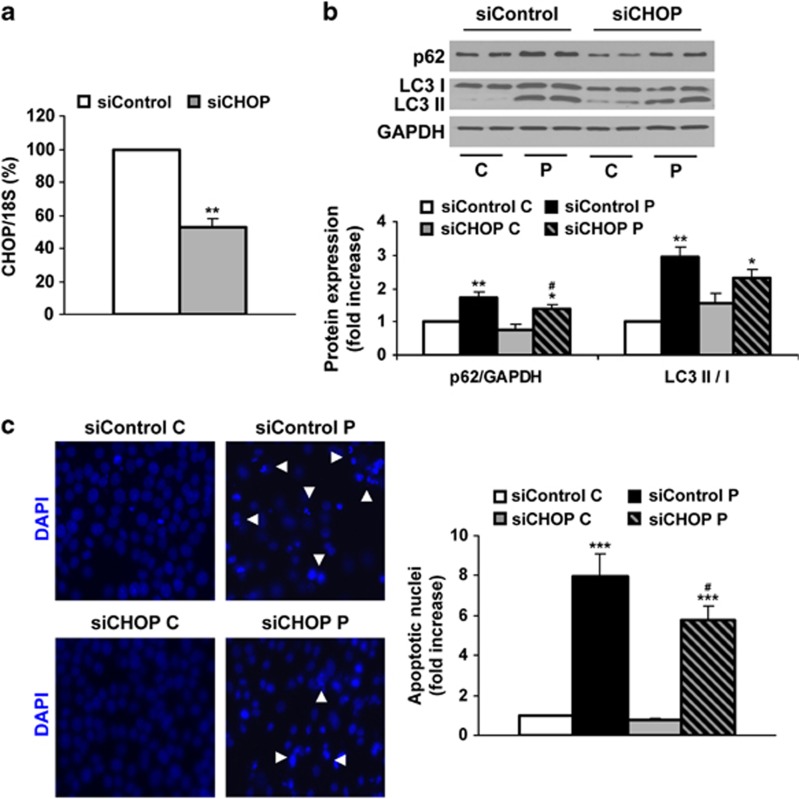
To clarify if elevated ER stress triggers disruption of the autophagy flux, we performed experiments inhibiting ER stress by reduction of CHOP by siRNA. As Figure 7a shows, CHOP mRNA levels were reduced by almost 60% in Huh7 cells treated with CHOP siRNA (siCHOP) compared with those cells treated with a control oligo (siControl). When we evaluated the effect of prolonged treatment with PA in Huh7 cells, we observed that silencing of CHOP reduced p62 together with LC3-II accumulation (Figure 7b). Interestingly, addition of siCHOP to PA-treated cells decreased apoptosis (Figure 7c).
Figure 7.
Efect of silencing of CHOP on the autophagic flux and cell death after prolonged treatment with PA in human Huh7 cells. Analysis of autophagy-related proteins expression and apoptosis in samples from Huh7 cells treated with control siRNA (siControl, 5 nM) or siCHOP (5 nM) for 36 h, followed by PA (P, 800 μM) for 24 h. (a) CHOP mRNA levels determined by real-time PCR. The fold induction value for triplicate wells was averaged and data are presented as mean±S.E.M. relative to siControl cells. (b) Representative western blots with the indicated antibodies. After densitometric analysis from blots corresponding to three independent experiments performed in triplicate, data are presented as mean±S.E.M. (c). Representative DAPI staining images. Quantification of fragmented nuclei after DAPI staining. Results correspond to three independent experiments performed in triplicate and are presented as mean±S.E.M. *P<0.05, **P<0.01 and ***P<0.005, P versus C or RP versus R; #P<0.05 siCHOP versus siControl
Discussion
During the past recent years, it has been established that defective autophagy promotes ER stress and insulin resistance in the liver,14, 24, 25 whereas degradation of damaged organelles by autophagy ameliorates liver diseases such as ethanol-induced steatosis26, 27 and HCC,28 suggesting that autophagy could have a relevant pathogenic role in liver diseases.
Previously, one study in human liver samples has found that the progression from NAS to NASH was accompanied by increased phosphorylation of JNK and increased ER stress reflected by elevated GRP78 mRNA.5 Lake et al.29 recently reported increased nuclear accumulation of total XBP-1 protein in NASH human livers. This paper also reported an enrichment of upregulated genes of autophagy by performing microarrays, although their validation was not shown. Thus, in the present study, we have analyzed for the first time the relationship between specific markers of ER stress and autophagy in the liver of NAS and NASH patients in comparison with individuals with normal liver. We found that protein amount of p62 was accumulated within the liver of patients with NAS or NASH in parallel with an increase in the LC3-II/LC3-I ratio. Of note, levels of p62 were significantly higher in NASH compared with NAS patients, suggesting that this effect is related with the disease progression in humans. Moreover, NASH patients displayed more elevated ER stress markers, such as GRP78 and CHOP mRNAs, as well as protein levels, reinforcing the notion that enhanced ER stress within liver cells may be relevant in the progression from NAS to NASH.
It has been shown that activation of the autophagic flux leads to a decline in p62 expression and, contrarily, an accumulation of p62 reflects a decrease in the autophagic flux.15, 16 When autophagy is off, LC3-II distributes diffusely throughout the cytoplasm. However, upon induction of autophagy, LC3-II is recruited via its lipid moiety to the inner and outer surfaces of autophagosome membranes forming LC3-autophagic punctuate.30 Therefore, accumulation of LC3-II and autophagosomes, event previously interpreted as induction of autophagy, is in fact a consequence of a blockade of the autophagic flux.31 Therefore, it is conceivable that the combination of elevated p62 and punctuate LC3-II, which was observed in liver samples from both NAS and NASH patients, could be reflecting an inhibition of autophagosome degradation. Considering findings mentioned so far, a potential mechanistic explanation for the pattern of p62 and LC3-II/LC3-I ratio during the progression of NAFLD would be that elevated ER stress due to lipid and nutrient overload might increase accumulation of unfolded proteins and/or damaged organelles, triggering liver injury through the disruption of the autophagic flux and leading to apoptotic cell death. In this regard, our previous results demonstrated that apoptosis was evident in NASH patients.17
In the liver, persistent UPR has been described in HFD-fed mice19 and in ob/ob mice that display hepatic steatosis,14 and also in MCD fed mice.32, 33, 34 In the present study, we found an accumulation of polyubiquitinated proteins in hepatocytes of HFD- and MCD-fed mice, which probably lead to chronic activation of the UPR, as demonstrated by elevated PERK and eIF2α phosphorylation and increased GRP78 and CHOP protein expression, overall reflecting ER overload. Whereas polyubiquitinated proteins and p62 levels are commonly increased in HFD-fed mice showing defective autophagic function, there are studies showing both decreased35 or increased LC3-II (Singh et al.36 and our study). Since, as stated above, the LC3-II/LC3-I ratio by itself does not assess the activation of the autophagic flux,16 it is possible that LC3-II levels might vary depending on the conditions of the HFD intervention. Nevertheless, our data clearly show that the autophagic flux is decreased in mice fed with HFD as well as in the MCD model, leading to the accumulation of autophagosomes, indicating for the first time that all the machinery that connects ER stress with the blockade of the autophagic flux is already activated in mice with hepatic steatosis. These results may have relevant clinical implications, suggesting that therapies aimed to restore the autophagic flux at early stages of NAFLD, such as NAS, might ameliorate progression to more advanced and irreversible forms of liver disease, such as NASH.
Free fatty acid (FFA)-induced lipotoxicity has a critical role in the pathogenesis of NAFLD (reviewed by Komatsu and Ichimura12 and Malhi and Gores37), being saturated FFAs the more toxic lipid species.38, 39 In the present study, we found that UPR was triggered in Huh7 human hepatic cells loaded with PA for 8 h concomitantly with the activation of the autophagic flux that was reflected by reduced phosphorylation of mTOR and S6K1, decreased p62, increased LC3-II/LC3-I ratio and by the effect of bafilomycin A1. These results suggest that this early response conferred protection against cell death as no apoptotic cells were detected at this time period. In contrast, a more prolonged exposure of Huh7 cells to ER stress by long PA treatment (24 h) induced cell death manifested by the abundance of apoptotic nuclei as compared with untreated cells. Notably, this later effect was coincident with the visualization of a high number of autophagosomes. These data clearly indicate that prolonged PA treatment induced a switch to the inhibition of the autophagic flux. To our knowledge, this is the first study that shows this transition monitored by the sequential accumulation of the autophagic substrate p62 between 8 and 24 h of PA exposure that was parallel to an increase in LC3-II punctuate and, importantly, to the induction of cell death. The fact that the reduction of the autophagic flux in Huh7 cells and primary human hepatocytes loaded with PA for 24 h was not accompanied by increased mTOR/S6K1 phosphorylation is intriguing and contrasts with our in vivo study in mice and data from Khamzina et al.40 One possible explanation for such difference is the more complex regulation of mTOR-mediated signaling pathway in liver tissue that integrates a number of signals emerging from different non-parenchymal cell types. In addition, an mTOR-independent regulation of autophagy and PA-mediated cell death through the activation of protein kinase C has also been described.41 Despite the lack of induction of mTOR phosphorylation by long-term treatment with PA in Huh7 cells, its inhibition by rapamycin reactivated the autophagic flux by decreasing p62 and the accumulation of autophagosomes (also observed in primary human hepatocytes) and ameliorated PA-induced increase in apoptotic cells. Reduction of ER stress by decreasing CHOP levels also prevented the blockade of autophagic flux by long-term treatment with PA and partly reduced cell death. The involvement of CHOP in ER stress-induced autophagy and cell death agrees with the previous data.42, 43 Thus, the prosurvival function of autophagy suggests a potential therapeutic strategy in the protection of hepatic cells against lipotoxicity.
In conclusion, we have demonstrated that autophagic flux is impaired in livers from both patients and murine models of NAFLD, as well as in long-term PA-loaded human hepatocytes, providing evidences that disruption of autophagy could be due to elevated ER stress leading to apoptosis. Our results strongly suggest that therapies aimed to restore the autophagic flux might prevent or attenuate the progression of NAFLD.
Materials and Methods
Reagents
Fetal bovine serum (FBS), culture media DMEM, TRIzol reagent, SuperScript III First-Strand Synthesis System and DAPI (D1306) were obtained from Invitrogen (Grand Island, NY, USA). PA (P0500), rapamycin, CQ, bafilomycin A1 and bovine serum albumin (BSA) were from Sigma-Aldrich (St. Louis, MO, USA). H&E and Masson's trichrome solutions were from Panreac Química (Barcelona, Spain). Bradford reagent, acrylamide and Immunoblot PVDF membrane were from Bio-Rad (München, Germany). Immobilon Western Chemiluminescent HRP Substrate was purchased from Millipore (Billerica, MA, USA).
Patients
This study comprised 49 non-diabetic patients with a clinical diagnosis of NAFLD who underwent a liver biopsy by a percutaneous route during programmed cholecystectomy. Inclusion criteria for NAFLD patients were based on the absence of alcohol intake (<20 g per day), the presence of biopsy-proven steatosis with/without necroinflammation and/or fibrosis, and no evidence of hepatitis B virus (HBV), hepatitis C virus (HCV) and human immunodeficiency virus (HIV) infections. Patients with other causes of chronic liver disease or those receiving potentially hepatotoxic drugs were excluded. We further studied 34 patients to whom a liver biopsy was taken during programmed laparoscopic cholecystectomy and were diagnosed of histologically normal liver (NL). All these NL patients had normal fasting glucose, cholesterol and triglycerides, normal serum ALT levels, and negative anti-HBV, anti-HCV and anti-HIV tests. In addition, all of them drank <20 g of alcohol per day and none used potentially hepatotoxic drugs. This study was performed in agreement with the Declaration of Helsinki, and with local and national laws. The Institution's Human Ethics Committee approved the study procedures, and written informed consent was obtained from all patients before inclusion in the study.
Clinical and analytical assessment
After a 12-h overnight fast, clinical and anthropometric data, as well as venous blood samples of each patient were obtained at the time of liver biopsy to test serum levels of liver enzymes and metabolic parameters. Serum insulin was determined by a chemiluminescent microparticle immunoassay (ARCHITECT insulin; Abbot Laboratories, Abbot Park, IL, USA). Insulin resistance was calculated by the HOMA method.44 BMI was calculated as weight (kg) divided by height (m) squared. Antibodies against HCV and HIV, as well as hepatitis B surface antigen were tested by immunoenzymatic assays (Murex, Dartford, UK).
Histopathology assessment
H&E and Masson's trichrome-stained paraffin-embedded liver biopsy sections were evaluated by a single experienced liver pathologist (JV-C) blinded to the clinical data. All liver biopsies measured >1.5 cm in length and showed more than 10 complete portal tracts. The percentage of hepatocytes containing lipid droplets was determined for each 10 × field. An average percentage of steatosis was then determined for the entire specimen.
Steatosis was assessed as outlined by Kleiner et al.45 grading percentage involvement by steatotic hepatocytes as follows: grade 0, <5% grade 1, 5–33% grade 2, >33–66% and grade 3, >66%.45 In addition, Brunt's histological scoring system was used to evaluate the degree of hepatocellular ballooning and lobular inflammation (grade of activity), as well as the stage of fibrosis.46 These patients were classified into two groups: those with simple steatosis (NAS) and those with definite steatohepatitis (NASH). Minimal criteria for the histological diagnosis of definite NASH included the combined presence of grade 1 steatosis, hepatocyte ballooning and lobular inflammation with or without fibrosis.
Animals and diets
Male 8-week-old C57BL/6 mice purchased to Charles River Laboratories (Charles River, Barcelona, Spain) were maintained in light/dark (12 h light/12 h dark), temperature (22 °C) and humidity-controlled rooms with free access to drinking water. Mice were fed with CHD (SAFE A04-10 Panlab, Barcelona, Spain), HFD (TD-88137; Harland-Teklad, Indianapolis, IN, USA) for 30 weeks or MCD (TD-90262; Harland-Tecklad) for 4 weeks. All animal experimentation was controlled following the recommendations of the Federation of European Laboratory Animal Science Associations (FELASA) on health monitoring, whereas use of animals in experimental procedures was approved by the Consejo Superior de Investigaciones Científicas (CSIC, Madrid, Spain) Animal Care and Use Committee.
Histopathology assessment
Paraffin-embedded liver biopsy sections (5 μm) were stained with H&E and Masson's trichrome solution and evaluated by a single-blinded hepatopathologist. Histopathology assessment was performed as described above.
Determination of serum transaminase levels
Serum ALT activity was determined using Reflotron strips (Roche Diagnostics, Barcelona, Spain), in accordance with the manual instructions.
EM and immunofluorescence in liver samples
For EM, liver biopsies were fixed in a glutaraldehyde (2%) plus p-formaldehyde (4%) for 60 min. Samples were postfixed in 1% osmium tetroxide for 60 min at 25 °C, stained with uranyl acetate (5 mg/ml) for 1 h at 25 °C, dehydrated in acetone and embedded in Epon 812 (EMbed 812; Electron Microscopy Science, Hatfield, PA, USA). Ultra-thin sections, unstained or poststained with uranyl acetate and lead hydroxide, were examined under a Morgagni 268D transmission electron microscope (FEI, Hillsboro, OR, USA) equipped with a Mega View II charge-coupled device camera (SIS, Soft Imaging System GmbH, Munster, Germany) and analyzed with AnalySIS software (SIS). For LC3 immunostaining, paraffin-embedded liver biopsy sections (5 μm) were stained with anti-LC3 antibody and anti-rabbit conjugate Alexa 546. For cell death detection staining, paraffin-embedded liver biopsy sections (5 μm) were stained using DeadEnd Fluorimetric TUNEL system (Promega, Madison, WI, USA), in accordance with the manual instructions. After corresponding inmunostaining, liver sections were mounted with fluorescent mounting medium and images were taken using a confocal microscope LSM710 (Zeiss, Barcelona, Spain).
Hepatocyte culture and PA treatment
Human hepatocytes were prepared from liver biopsies obtained from 12 patients (eight males, four females aged 58±4.0 years) submitted to a surgical resection for liver tumors after obtaining patients' written consent. Hepatocytes isolation was based on the two-step collagenase procedure.47 Huh7 cell line was kindly provided by Dr. Kern (Department of General Pathology, University Hospital Heidelberg, Heidelberg, Germany). Huh7 cell line authentication and intraspecies cell line cross-contamination were analyzed using Promega's StemElite ID System (Madison, WI, USA) for the following short tandem repeats (STRs): D21S11, TH01, TPOX, vWA, CSF1PO, D16S539, D7S820, D13S317 and D5S818; Amelogenin was also analyzed for gender identification together with a specific marker for mouse DNA. STRs were analyzed with the Applied Biosystems 3130xl Genetic Analyzer in the Genomics Core Facility of the Instituto de Investigaciones Biomédicas Alberto Sols CSIC-UAM. Cells were cultured in DMEM supplemented with 10% (v/v) heat-inactivated FBS at 37 °C with 5% CO2. PA was dissolved in isopropyl alcohol at a stock concentration of 80 mM and added to DMEM containing 1% BSA to assure a physiological ratio between bound and unbound PA in the medium.48 The concentration of PA used in the experiments (800 μM) was similar to the fasting PA plasma concentrations observed in human non-alcoholic steatohepatitis49, 50 and was used before for mimicking hepatic lipoapoptosis.21 Confluent cells were treated with PA (800 μM) for the indicated time periods in the absence or presence of rapamycin (25 ng/ml). Bafilomycin A1 (10 nM) and CQ (50 nM) were used as positive control of autophagosome accumulation.
Transient transfection with siRNA
siRNA oligonucleotides were synthesized by Dharmacon RNAi Technologies (Fisher Scientific-USA, Pittsburgh, PA, USA) for gene silencing of human CHOP. Huh7 cells were transfected with control or siCHOP (5 nM) following DharmaFECT General Transfection Protocol (Dharmacon RNAi Technologies). After 36 h, cells were used for experiments.
EM and immunofluorescence in Huh7 cells
For EM, Huh7 cells were grown in chamber slides and treated as described above. Then, cells were washed two times with PBS, fixed in a glutaraldehyde (2%) plus p-formaldehyde (4%) for 60 min and processed similar to liver biopsies. For LC3 immunostaining, Huh7 cells were grown in glass coverslips and treated with PA as described in Results and Figure legends. Then, cells were washed two times with PBS, fixed in p-formaldehyde (4%) for 10 min and processed for LC3 immunofluorescence as described above.
Quantitation of apoptosis
Huh7 cells were grown in glass coverslips and treated as described above. Then, cells were washed two times with PBS, fixed in p-formaldehyde (4%) for 10 min and characteristic morphological changes of apoptosis were assessed by staining nuclei with DAPI, followed by the analysis by fluorescence microscopy.
Nile Red staining
After treatment, Huh7 cells were fixed in p-formaldehyde (4%) and resuspended in Nile Red working solution (0.4 μg/ml). The fluorescence was determined using a FACScan flow cytometer (BD Biosciences, San Jose, CA, USA).
Quantitative real-time PCR analysis and primer sequence
Total RNA of liver biopsy samples was extracted by using TRIzol reagent and was reverse transcribed using a SuperScript III First-Strand Synthesis System for quantitative real-time polymerase chain reaction (qPCR) following manufacturer's indications. qPCR was performed with an ABI 7900 sequence detector (Life Technologies, Carlsbad, CA, USA) using the SYBR Green method and d(N)6 random hexamer with primers purchased from Invitrogen. PCR thermocycling parameters were 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Each sample was run in triplicate and was normalized to 18S RNA. Fold changes were determined using the ΔΔCt method. Primer sequences are available upon request.
Preparation of protein extracts and western blot
Liver biopsy samples were homogenized in a medium containing 10 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 0.5% 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate (CHAPS), 1 mM β-mercaptoethanol and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). To obtain total cell lysates, attached cells were scraped off and incubated for 10 min on ice with lysis buffer (25 mM HEPES, 2.5 nM EDTA, 0.1% Triton X-100, 1 mM PMSF and 5 μg/ml leupeptin). After protein content determination with Bradford reagent, total protein were boiled in Laemmli sample buffer and submitted to 8–15% SDS-PAGE. Proteins were transferred to Inmunoblot PVDF membrane, and after blocking with 3% BSA or 5% non-fat dry milk, incubated overnight with several antibodies as indicated. Immunoreactive bands were visualized using the ECL Western blotting protocol (Millipore). Densitometric analysis of the bands was performed using the Image J software (NIH, Bethesda, MD, USA). Anti-phospho-PERK (Thr 980) for mouse samples (no. 3179), anti-phospho-eIF2α (Ser 51) (no. 9721), anti-LC3 (no. 4108), anti-phospho-mTOR (Ser 2448) (no. 2971), anti-phospho-JNK (no. 4668), anti-S6K1 (no. 9202) and anti-ubiquitin (no. 3936) antibodies were from Cell Signaling Technology (Boston, MA, USA). Anti-p62 (SQSTM1) antibody was from MBL International (Woburn, MA, USA). The anti-phospho-S6K1 (Thr 389) (sc-11759), anti-JNK (sc-571), anti-IRβ (sc-711), Beclin (sc-11427), anti-phospho-PERK (Thr 980) for human samples (sc-32577), anti-PERK (sc-13073), anti-eIF2α (sc-11386), anti-GRP78 (sc-376768), anti-CHOP for human samples (sc-575) and anti-CHOP for mouse samples (sc-7351) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-GAPDH antibody (AM4300) was from Ambion (Grand Island, NY, USA). Anti-β-actin antibody (A-5441) was from Sigma Aldrich. Anti-rabbit-conjugated Alexa 546 (A11035) was from Life Technologies (Grand Island, NY, USA). Anti-Atg5 antibody was a gift from Dr. Patricia Boya (CSIC).
Statistical analysis
Categorical variables are presented as frequency and percentage. Continuous variables are shown as median and range. The baseline characteristics of the patients studied were compared by the Pearson χ2 test for categorical variables and the unpaired t-test or Mann–Whitney U-test for continuous variables. Data from qPCR and western blotting are presented as mean±S.E.M., and were compared by using the Bonferroni ANOVA test. All statistical analyses were performed using the IBM SPSS Statistics 21.0 (SPSS Inc., IBM, Armonk, NY, USA) software with two-sided tests, with a P-value of <0.05 considered as statistically significant.
Acknowledgments
We acknowledge the following grant support: SAF2012-33283 (MINECO, Spain), Comunidad de Madrid S2010/BMD-2423, EFSD and Amylin Paul Langerhans Grant and Centro de Investigación Biomédica en Red de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM, ISCIII, Barcelona, Spain) to AMV.; SAF2010-16037, SAF2013-43713-R (MINECO) and Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBEREHD, ISCIII) to PMS. RD12/0042/0019 (ISCIII) and S2010/BMD-2478 (Comunidad de Madrid) to LB, PI 13/01299 and Fundación Mutua Madrileña 2012 to C G-M and AIRC IG-2012 to GMF. We thank Dr. Patricia Boya and Dr. Ana I. Arroba for helpful discussions.
Glossary
- NAFLD
non-alcoholic fatty liver disease
- NAS
non-alcoholic steatosis
- NASH
non-alcoholic steatohepatitis
- HFD
high-fat diet
- MCD
methionine-choline-deficient diet
- HOMA
homeostasis model assessment of insulin resistance
- PCR
polymerase chain reaction
- mRNA
messenger RNA
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Younossi ZM, Stepanova M, Afendy M, Fang Y, Younossi Y, Mir H, et al. Changes in the prevalence of the most common causes of chronic liver diseases in the United States from 1988 to 2008 Clin Gastroenterol Hepatol 20119524–530.e521; quiz e560. [DOI] [PubMed] [Google Scholar]
- Day CP. Non-alcoholic fatty liver disease: a massive problem. Clin Med. 2011;11:176–178. doi: 10.7861/clinmedicine.11-2-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuschwander-Tetri BA. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology. 2010;52:774–788. doi: 10.1002/hep.23719. [DOI] [PubMed] [Google Scholar]
- Cheung O, Sanyal AJ. Recent advances in nonalcoholic fatty liver disease. Curr Opin Gastroenterol. 2010;26:202–208. doi: 10.1097/MOG.0b013e328337b0c4. [DOI] [PubMed] [Google Scholar]
- Puri P, Mirshahi F, Cheung O, Natarajan R, Maher JW, Kellum JM, et al. Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology. 2008;134:568–576. doi: 10.1053/j.gastro.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja MJ. Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology. 2011;140:1895–1908. doi: 10.1053/j.gastro.2011.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Czaja MJ. Autophagy in health and disease. 2. Regulation of lipid metabolism and storage by autophagy: pathophysiological implications. Am J Physiol Cell Physiol. 2010;298:C973–C978. doi: 10.1152/ajpcell.00527.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Monzon C, Lo Iacono O, Mayoral R, Gonzalez-Rodriguez A, Miquilena-Colina ME, Lozano-Rodriguez T, et al. Hepatic insulin resistance is associated with increased apoptosis and fibrogenesis in nonalcoholic steatohepatitis and chronic hepatitis C. J Hepatol. 2011;54:142–152. doi: 10.1016/j.jhep.2010.06.021. [DOI] [PubMed] [Google Scholar]
- Kato H, Nakajima S, Saito Y, Takahashi S, Katoh R, Kitamura M. mTORC1 serves ER stress-triggered apoptosis via selective activation of the IRE1-JNK pathway. Cell Death Differ. 2012;19:310–320. doi: 10.1038/cdd.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazanave SC, Mott JL, Elmi NA, Bronk SF, Werneburg NW, Akazawa Y, et al. JNK1-dependent PUMA expression contributes to hepatocyte lipoapoptosis. J Biol Chem. 2009;284:26591–26602. doi: 10.1074/jbc.M109.022491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni HM, Bockus A, Boggess N, Jaeschke H, Ding WX. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55:222–232. doi: 10.1002/hep.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H, Bronk SF, Werneburg NW, Gores GJ. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J Biol Chem. 2006;281:12093–12101. doi: 10.1074/jbc.M510660200. [DOI] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol Histopathol. 2002;17:897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Zhang H, Li M, Xiong X, Chen X, Dong XC, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Takehara T, Hikita H, Kodama T, Tsunematsu H, Miyagi T, et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer. 2012;131:548–557. doi: 10.1002/ijc.26374. [DOI] [PubMed] [Google Scholar]
- Lake AD, Novak P, Hardwick RN, Flores-Keown B, Zhao F, Klimecki WT, et al. The adaptive endoplasmic reticulum stress response to lipotoxicity in progressive human nonalcoholic fatty liver disease. Toxicol Sci. 2014;137:26–35. doi: 10.1093/toxsci/kft230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117 (Part 13:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Wong PM, Jiang X. Thapsigargin distinguishes membrane fusion in the late stages of endocytosis and autophagy. Autophagy. 2011;7:1397–1399. doi: 10.4161/auto.7.11.17651. [DOI] [PubMed] [Google Scholar]
- Greene MW, Burrington CM, Ruhoff MS, Johnson AK, Chongkrairatanakul T, Kangwanpornsiri A. PKC{delta} is activated in a dietary model of steatohepatitis and regulates endoplasmic reticulum stress and cell death. J Biol Chem. 2010;285:42115–42129. doi: 10.1074/jbc.M110.168575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinella ME, Siddiqui MS, Gardikiotes K, Gottstein J, Elias M, Green RM. Dysregulation of the unfolded protein response in db/db mice with diet-induced steatohepatitis. Hepatology. 2011;54:1600–1609. doi: 10.1002/hep.24553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon RK, Jr, Yan JS, Grenert JP, Maher JJ. Stress signaling in the methionine-choline-deficient model of murine fatty liver disease. Gastroenterology. 2010;139:1730–1739. doi: 10.1053/j.gastro.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H, Gores GJ. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28:360–369. doi: 10.1055/s-0028-1091980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Jr, Ory DS, et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S, Ni HM, Manley S, Bockus A, Kassel KM, Luyendyk JP, et al. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther. 2011;339:487–498. doi: 10.1124/jpet.111.184341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamzina L, Veilleux A, Bergeron S, Marette A. Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology. 2005;146:1473–1481. doi: 10.1210/en.2004-0921. [DOI] [PubMed] [Google Scholar]
- Tan SH, Shui G, Zhou J, Li JJ, Bay BH, Wenk MR, et al. Induction of autophagy by palmitic acid via protein kinase C-mediated signaling pathway independent of mTOR (mammalian target of rapamycin) J Biol Chem. 2012;287:14364–14376. doi: 10.1074/jbc.M111.294157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Han J, Zhai B, Ming X, Zhuang L, Liu Y, et al. Blocking autophagy enhances the apoptosis effect of bufalin on human hepatocellular carcinoma cells through endoplasmic reticulum stress and JNK activation. Apoptosis. 2013;19:210–223. doi: 10.1007/s10495-013-0914-7. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Miyazaki S, Matsuyama S, Takeda M, Kawano M, Nakagawa H, et al. Selection of autophagy or apoptosis in cells exposed to ER-stress depends on ATF4 expression pattern with or without CHOP expression. Biol Open. 2013;2:1084–1090. doi: 10.1242/bio.20135033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander-Tetri BA, Bacon BR. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- Pichard L, Raulet E, Fabre G, Ferrini JB, Ourlin JC, Maurel P. Human hepatocyte culture. Methods Mol Biol. 2006;320:283–293. doi: 10.1385/1-59259-998-2:283. [DOI] [PubMed] [Google Scholar]
- Richieri GV, Kleinfeld AM. Unbound free fatty acid levels in human serum. J Lipid Res. 1995;36:229–240. [PubMed] [Google Scholar]
- Belfort R, Harrison SA, Brown K, Darland C, Finch J, Hardies J, et al. A placebo-controlled trial of pioglitazone in subjects with nonalcoholic steatohepatitis. N Engl J Med. 2006;355:2297–2307. doi: 10.1056/NEJMoa060326. [DOI] [PubMed] [Google Scholar]
- Sanyal AJ, Campbell-Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]