This work uses a genetic screen based on overexpression to identify a gene encoding a polygalacturonase enzyme that degrades pectins, both as a purified protein and in intact plant cell walls, and is required for cellular growth in dark-grown seedlings and the normal developmental control of flower petal number in adult Arabidopsis thaliana plants.
Abstract
Pectins are acidic carbohydrates that comprise a significant fraction of the primary walls of eudicotyledonous plant cells. They influence wall porosity and extensibility, thus controlling cell and organ growth during plant development. The regulated degradation of pectins is required for many cell separation events in plants, but the role of pectin degradation in cell expansion is poorly defined. Using an activation tag screen designed to isolate genes involved in wall expansion, we identified a gene encoding a putative polygalacturonase that, when overexpressed, resulted in enhanced hypocotyl elongation in etiolated Arabidopsis thaliana seedlings. We named this gene POLYGALACTURONASE INVOLVED IN EXPANSION1 (PGX1). Plants lacking PGX1 display reduced hypocotyl elongation that is complemented by transgenic PGX1 expression. PGX1 is expressed in expanding tissues throughout development, including seedlings, roots, leaves, and flowers. PGX1-GFP (green fluorescent protein) localizes to the apoplast, and heterologously expressed PGX1 displays in vitro polygalacturonase activity, supporting a function for this protein in apoplastic pectin degradation. Plants either overexpressing or lacking PGX1 display alterations in total polygalacturonase activity, pectin molecular mass, and wall composition and also display higher proportions of flowers with extra petals, suggesting PGX1’s involvement in floral organ patterning. These results reveal new roles for polygalacturonases in plant development.
INTRODUCTION
The cell walls of plants are composed of several interacting networks of carbohydrate polymers (Somerville et al., 2004). These polymers, which include cellulose, hemicelluloses, and pectins, are synthesized by the cell and delivered to the apoplast (Keegstra, 2010), where they provide mechanical strength and protection to plant tissues. In growing tissues, the cell wall must resist the high turgor pressure that drives growth while simultaneously remaining flexible enough to selectively yield and expand in response to that pressure (Cosgrove, 2005). Wall loosening can be achieved by several molecular mechanisms, including disruption of intermolecular adhesion by proteins such as expansins (McQueen-Mason and Cosgrove, 1995), polymer rearrangements (Anderson et al., 2010), polymer lysis and religation by enzymes such as xyloglucan endotransglycosylases/hydrolases (Van Sandt et al., 2007), and/or enzymatic cleavage of polymer glycosyl linkages (Park and Cosgrove, 2012). However, the precise contributions of the cleavage of different carbohydrate polymers in wall loosening are not fully understood, and the fact that many different classes of glycosyl hydrolases and lyases are encoded by plant genomes (Cantarel et al., 2009) has made it challenging to define the relative contributions of each class of enzymes to wall expansion under physiological conditions.
Our current understanding of pectins (Atmodjo et al., 2013) adds complexity to conceptual models of cell wall modification during growth in two ways. First, the large number of linkages and structural motifs encompassed by different domains of pectins, which include homogalacturonan (HG), rhamnogalacturonan-I, rhamnogalacturonan-II, and xylogalacturonan, allows them to interact indirectly (Dick-Pérez et al., 2011) and/or via covalent bonds (Tan et al., 2013) with a wide range of apoplastic polymers. Second, pectins can generate mechanically tunable networks based on reversible calcium-mediated cross-linking between stretches of demethylated HG (also called pectate) (Vincken et al., 2003). HG methylation, which is high upon its initial synthesis, is reduced by the activity of apoplastic pectin methyl-esterases (Micheli, 2001), which are themselves regulated by pectin methyl-esterase inhibitor proteins (Jolie et al., 2010). Pectin demethylation can result in at least two alternative mechanical consequences for the wall, either by enabling the formation of calcium-mediated cross-links if the HG is demethylated in a blockwise fashion or by increasing the susceptibility of randomly demethylated HG to cleavage by two classes of pectin-degrading enzymes, pectin/pectate lyases, and polygalacturonases (PGs) (Peaucelle et al., 2012). Rigidification of the cell wall by HG demethylation and subsequent calcium-mediated cross-linking has been hypothesized to constrain the expansion of hypocotyl cells (Derbyshire et al., 2007a; Zhao et al., 2008; Abasolo et al., 2009), whereas pectin demethylation has been linked to the loosening of the cell wall that accompanies organ initiation at the Arabidopsis thaliana shoot apical meristem (Peaucelle et al., 2008, 2011).
Multiple functions have been identified for endogenous pectin-degrading enzymes in plants. Pectate lyases, which cleave HG backbones via β-elimination, have been implicated in pollen tube growth, fruit softening, and susceptibility to plant pathogens (Marín-Rodríguez et al., 2002) and are expressed in a wide range of tissues (Palusa et al., 2007; Sun and van Nocker, 2010), whereas PGs, which cleave HG backbones via hydrolysis, have mainly been shown to function in cell separation events (González-Carranza et al., 2007). Two PGs, QUARTET2 (QRT2) and QRT3, function in pollen tetrad separation (Rhee and Somerville, 1998; Rhee et al., 2003), and two additional PGs, ADPG1 and ADPG2, plus QRT3, are required for cell separation events during reproductive development (Ogawa et al., 2009). Interestingly, overexpression of a PG in apple (Malus domestica) results in abnormal leaf morphology and organ shedding (Atkinson et al., 2002), whereas downregulation of Md-PG1 in the same species results in putatively enhanced cell adhesion and reduced cell expansion in the fruit (Atkinson et al., 2012). Additionally, overexpression of a stress-inducible PG in rice (Oryza sativa) has recently been shown to reduce pectin content and cell adhesion while increasing sensitivity to abiotic stress (Liu et al., 2013).
Many plant pathogens and biomass-degrading organisms also possess pectinolytic enzymes (Tian et al., 2009), and pectins can inhibit the recognition of other wall polymers by antibodies (Marcus et al., 2008, 2010), implying that they might mask other wall polymers and that their degradation might be a key step in the deconstruction of plant cell walls (Jordan et al., 2012; Biswal et al., 2014). Many plant species also express inhibitor proteins that bind to pectin/pectate lyases (Bugbee, 1993) or PGs (Ferrari et al., 2003, 2012; Protsenko et al., 2008; Maulik et al., 2012), although whether these inhibitors exclusively target enzymes produced by pathogens or might also interact with endogenous enzymes is currently unclear.
One challenge in the study of glycosyl hydrolases/lyases in plants is the large number of genes that encode many of these enzyme families. For example, there are 69 annotated putative PG (GH28) genes in the Arabidopsis genome (Kim et al., 2006; González-Carranza et al., 2007; Cantarel et al., 2009); the likelihood of genetic redundancy within these families that could potentially mask the effects of loss-of-function mutations is thus high. In addition, the centrality of wall biosynthesis and expansion in plant growth means that loss-of-function mutations in nonredundant genes involved in this process can result in gametophytic or embryonic lethality (Persson et al., 2007). Thus, although loss-of-function genetic screens have revealed the role of many genes in cell wall biosynthesis and modification (Reiter et al., 1997; Somerville et al., 2004), there are likely to be other genes whose function in wall expansion has not yet been uncovered. An alternative genetic screening method called activation tagging, in which enhancers inserted randomly in the genome drive the overexpression of nearby genes, has been used to identify genes involved in a wide array of biological processes (Weigel et al., 2000; Tani et al., 2004; Ogawa et al., 2009), and we hypothesized that this approach could prove useful for identifying wall expansion-related genes in etiolated Arabidopsis seedlings, the hypocotyls of which elongate mainly via cell expansion (Derbyshire et al., 2007b) without requiring new wall synthesis during later stages of expansion (Refrégier et al., 2004).
In this work, we describe an activation tag screen designed to identify novel genes involved in cell expansion and the characterization of one such gene, which encodes a PG. This PG is required for normal etiolated hypocotyl elongation, enhances hypocotyl elongation when overexpressed, localizes to the apoplast as a fusion with green fluorescent protein (GFP), and displays in vitro PG activity when heterologously expressed. Plants overexpressing or lacking this PG display alterations in total PG activity, pectin molecular mass, and wall composition. In addition, we observed higher numbers of flowers with extra petals in loss-of-function and overexpression plants. These results indicate that PGs can function in cell expansion and developmental patterning in addition to their roles in cell separation events.
RESULTS
Identification of PGX1 by Activation Tag Screening for Enhanced Hypocotyl Elongation
To identify novel genes involved in cell wall expansion, we performed activation tag screening using a collection of Arabidopsis Columbia (Col) ecotype lines containing randomly inserted tetrameric 35S enhancer elements (Weigel et al., 2000). Seedlings from pools of lines were grown in the dark on Murashige and Skoog (MS) media for 6 d to allow for the growth of a lawn of etiolated seedlings. Tall seedlings were identified visually, transferred to vertical MS plates containing 1% Suc, grown in 24 h light for 10 to 14 d to allow for photomorphogenesis, and transferred to soil for maturation and seed harvesting. These taller seedlings represented the initial hits from the screen, and 637 hits were isolated from ∼1 million seedlings. The initial hits were secondarily screened (Supplemental Figure 1) for heritability of the long hypocotyl phenotype and Mendelian segregation of a marker encoded by the activation tag T-DNA (Weigel et al., 2000). Lines that displayed heritable long-hypocotyl phenotypes (P < 0.01, t test, two replicates) and were likely to contain single insertions (P > 0.1, χ2, two replicates) were identified, and the genomic insertion point for each line was identified using adapter ligation PCR (O’Malley et al., 2007) followed by sequencing and alignment to the Arabidopsis genome sequence. From the first 296 initial hits, we identified 27 independent candidate lines, two of which appeared twice in the total group of 29 candidate lines. The double identification of two lines validated our screening approach, since each activation tag line was represented by multiple seeds in the pooled collection. Within the set of 27 independent candidate lines, two insertion points were upstream of genes encoding putative PGs, and we chose one of these candidates, in which the activation tag insertion point was 1687 bp upstream of the predicted start codon of At3g26610, for further analysis. Within the PG gene family of Arabidopsis, At3g26610 is not closely related to previously characterized PGs (Supplemental Figure 2 and Supplemental Data Set 1). We named this gene POLYGALACTURONASE INVOLVED IN EXPANSION1 (PGX1) and performed detailed characterization of its expression, localization, biochemical activity, and effects on cell walls and plant growth and development as described below.
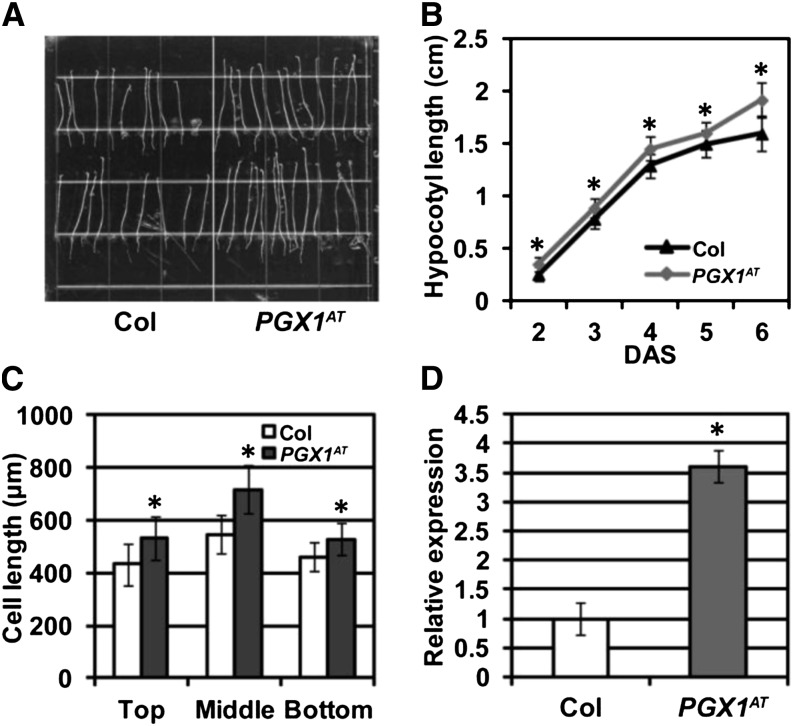
PGX1AT Activation Tag Plants Exhibit Longer Hypocotyls and Cells and Higher PGX1 Expression Levels Than Wild-Type Plants
We first confirmed the enhanced hypocotyl elongation phenotype of the PGX1 activation tag line (PGX1AT), finding that on average, PGX1AT seedling hypocotyls were 1.91 ± 0.16 (sd) cm long after 6 d of dark growth, whereas Col controls were 1.60 ± 0.13 (sd) cm long on average (Figures 1A and 1B). To determine whether this difference in hypocotyl length was due to increased cell length, we measured cell lengths in the top, middle, and bottom regions of hypocotyls of Col and PGX1AT seedlings and found that PGX1AT cells in all three regions were significantly longer than control cells (Figure 1C); the total number of cells along the lengths of PGX1AT hypocotyls did not differ significantly from Col controls (Supplemental Figure 3). To test whether PGX1 was overexpressed in the PGX1AT line, PGX1 mRNA levels were quantified in 6-d-old etiolated PGX1AT seedlings using quantitative real-time RT-PCR (qPCR) and were found to be more than 3.5 times those of Col controls (Figure 1D). PGX1 mRNA levels were also higher in stems, leaves, flowers, and siliques of PGX1AT plants compared with wild-type plants (Supplemental Figure 4). Together, these results indicate that enhanced PGX1 expression results in longer cells, and thus longer hypocotyls, in etiolated seedlings of the PGX1AT activation tag line identified in our screen.
Figure 1.
Activation Tag Line PGX1AT Displays Enhanced Hypocotyl Length, Cell Length, and PGX1 Expression.
(A) Hypocotyls of 6-d-old etiolated Arabidopsis Col (wild type, left) and activation tag line PGX1AT (right) grown on MS media without Suc.
(B) Quantification of hypocotyl length for 2- to 6-d-old etiolated Col and PGX1AT seedlings (n ≥ 63 seedlings per time point from three independent experiments). DAS, days after sowing.
(C) Cell length at the top, middle, and bottom of hypocotyls of 6-d-old etiolated Col and PGX1AT seedlings (n ≥ 60 seedlings from three independent experiments).
(D) qPCR quantification of PGX1 mRNA expression levels in 6-d-old etiolated Col and PGX1AT seedlings. ACT2 was used as an internal control. The expression level of PGX1 in Col was normalized to 1, and the relative expression level of PGX1 in PGX1AT line was calculated relative to this value. Error bars indicate sd (n = 3 replicates). Asterisks indicate significant differences (P < 0.001, t test).
Global Overexpression of PGX1 Results in Enhanced Hypocotyl and Cell Elongation
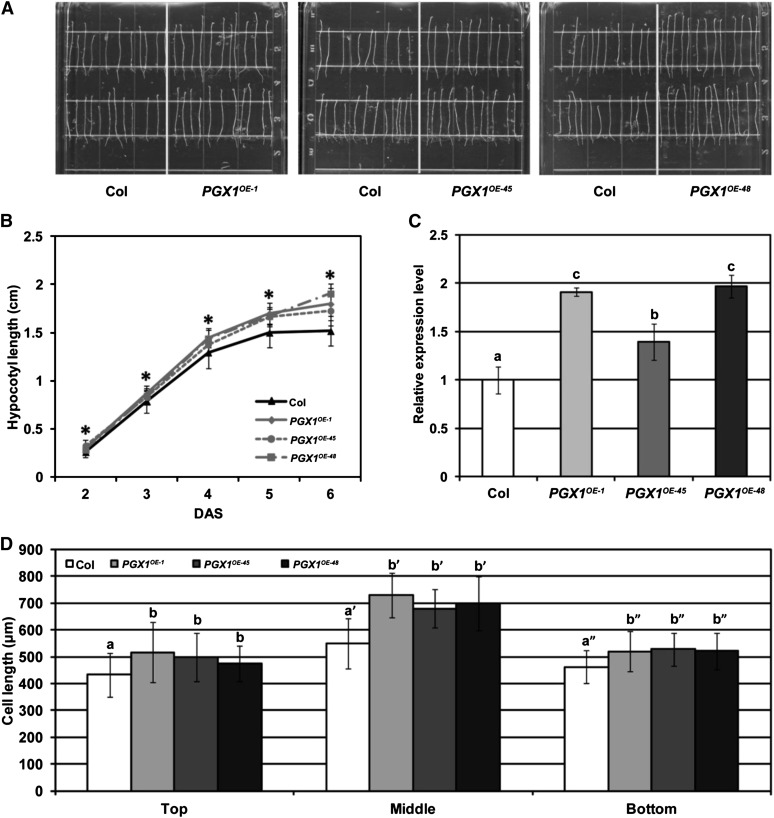
The activation tag upstream of PGX1 in the PGX1AT line is expected to amplify its endogenous expression pattern rather than drive constitutive expression across all tissues. In addition, activation tag enhancer elements can potentially enhance the expression of multiple genes in their vicinity (Weigel et al., 2000). To test whether the enhanced hypocotyl elongation observed in the PGX1AT line was attributable to the specific overexpression of PGX1, we generated transgenic lines in which the PGX1 coding sequence is expressed under the control of the constitutive strong cauliflower mosaic virus 35S promoter and tested them for etiolated hypocotyl and cell lengths, as well as PGX1 expression levels. For several independent p35S:PGX1 (PGX1OE) transformants, both the overall hypocotyl lengths (Figures 2A and 2B) and top, middle, and bottom hypocotyl cell lengths (Figure 2D) of 6-d-old etiolated seedlings were significantly longer than those of controls, whereas the numbers of cells along the lengths of hypocotyls from the overexpression lines did not differ significantly from controls (Supplemental Figure 3). PGX1 mRNA levels were significantly higher in the transformed lines than in wild-type controls as measured by qPCR (Figure 2C). These results further confirmed that high PGX1 expression results in enhanced hypocotyl cell elongation.
Figure 2.
Global Overexpression of PGX1 Promotes Hypocotyl Elongation.
(A) Hypocotyls of 6-d-old etiolated Arabidopsis Col and three independent transgenic lines expressing PGX1 driven by the cauliflower mosaic virus 35S promoter (PGX1OE-1, PGX1OE-45, and PGX1OE-48) grown on MS media without Suc.
(B) Quantification of hypocotyl length for 2- to 6-d-old etiolated Col and PGX1OE seedlings (n ≥ 66 seedlings from three independent experiments).
(C) mRNA expression levels of PGX1 in Col and three PGX1OE transgenic plants quantified by qPCR. ACT2 was amplified as a control.
(D) Cell length at the top, middle, and bottom of hypocotyls of 6-d-old etiolated seedlings of Col and three PGX1OE transgenic plants (n ≥ 30 seedlings from at least two independent experiments). Error bars indicate sd.
Asterisks in (B) indicate significant differences between Col and PGX1OE lines (P < 0.05, t test). Lowercase letters in (C) and (D) indicate significantly different groups (P < 0.05, one-way ANOVA and Tukey test).
PGX1 Is Required for Normal Etiolated Hypocotyl Elongation
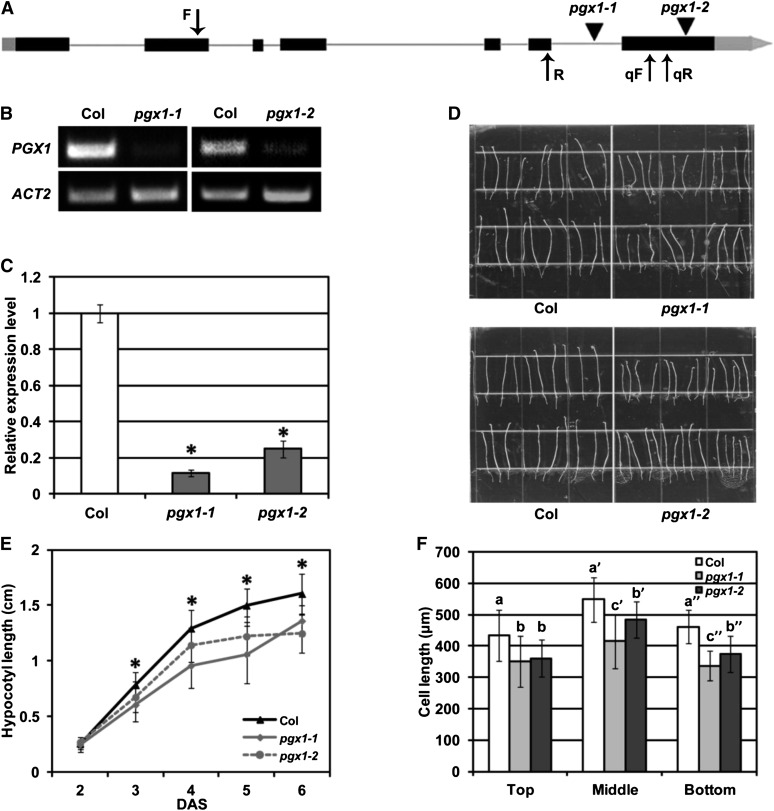
Given that PGX1 is one of 69 annotated Arabidopsis PG-encoding genes and that multiple PGs are expressed in most Arabidopsis tissues (Kim et al., 2006; González-Carranza et al., 2007), we hypothesized that disrupting PGX1 function would not result in visible growth phenotypes due to redundancy. Thus, we were surprised that upon isolating two homozygous T-DNA insertional mutants containing disruptions in the sixth intron and seventh exon of the gene (pgx1-1 and pgx1-2; Figure 3A) and confirming that these lines expressed lower PGX1 mRNA levels than wild-type controls (Figures 3B and 3C), 6-d-old etiolated seedlings of both mutant lines displayed reduced hypocotyl lengths (Figures 3D and 3E) and cell lengths (Figure 3F) compared with controls. However, cell number along the lengths of hypocotyls was not significantly lower in the mutants (Supplemental Figure 3). These data indicate that PGX1 functions nonredundantly to promote cell expansion in etiolated hypocotyls and that final cell size in this tissue is likely determined at least in part by the activity of this protein. In pooled data for Col, mutant, and overexpression plants, we observed positive correlations between overall hypocotyl length and individual cell length in the top, middle, and bottom regions of hypocotyls (Pearson correlation coefficients of 0.582, 0.644, and 0.638, respectively, P < 0.001, two-tailed t test), suggesting that cell lengths in all three regions contribute to overall hypocotyl length.
Figure 3.
T-DNA Insertion Mutants of PGX1 Exhibit Reduced Hypocotyl Length.
(A) Schematic gene structure of PGX1 (exons shown as solid boxes, introns as lines, 5′ and 3′ regions as gray boxes, T-DNA insertions as triangles, and primer binding sites as arrows).
(B) and (C) Detection of PGX1 transcript in T-DNA insertion mutants pgx1-1 (WiscDsLox262B06; CS850042) and pgx1-2 (Salk_026818) by RT-PCR (B) and qPCR (C). Error bars indicate sd (n = 6 technical replicates from two independent experiments); ACT2 was amplified as an internal control.
(D) Hypocotyls of 6-d-old etiolated seedlings of Col, pgx1-1, and pgx1-2 mutants grown on MS media without Suc.
(E) Quantification of hypocotyl length for 2- to 6-d-old etiolated seedlings of Col, pgx1-1, and pgx1-2 seedlings (n ≥ 60 seedlings from three independent experiments).
(F) Cell length at the top, middle, and bottom hypocotyls of 6-d-old etiolated Col, pgx1-1, and pgx1-2 seedlings (n ≥ 48 seedlings from two independent experiments). Error bars indicate sd.
Asterisks in (E) indicate significant differences between Col and pgx1 mutant lines (P < 0.001, t test). Lowercase letters in (F) indicate significantly different groups (P < 0.05, one-way ANOVA and Tukey test).
We also observed 25-d-old wild-type, pgx1-1, pgx1-2, and PGX1 overexpression plants and found that rosette diameter was not significantly smaller than wild-type controls in pgx1-1 or pgx1-2 plants, but was significantly larger than wild-type controls in PGX1AT, PGX10E-1, and PGX1OE-48 plants (Supplemental Figure 5). Adult plant stem height was slightly reduced in 45-d-old pgx1-1 and pgx1-2 plants, but was not significantly different from Col controls in PGX1 mutant or overexpression lines (Supplemental Figure 6). These data suggest that additional PGs might function redundantly with PGX1 at later developmental stages, but that overexpression of PGX1 is sufficient to enhance the expansion of rosette leaves.
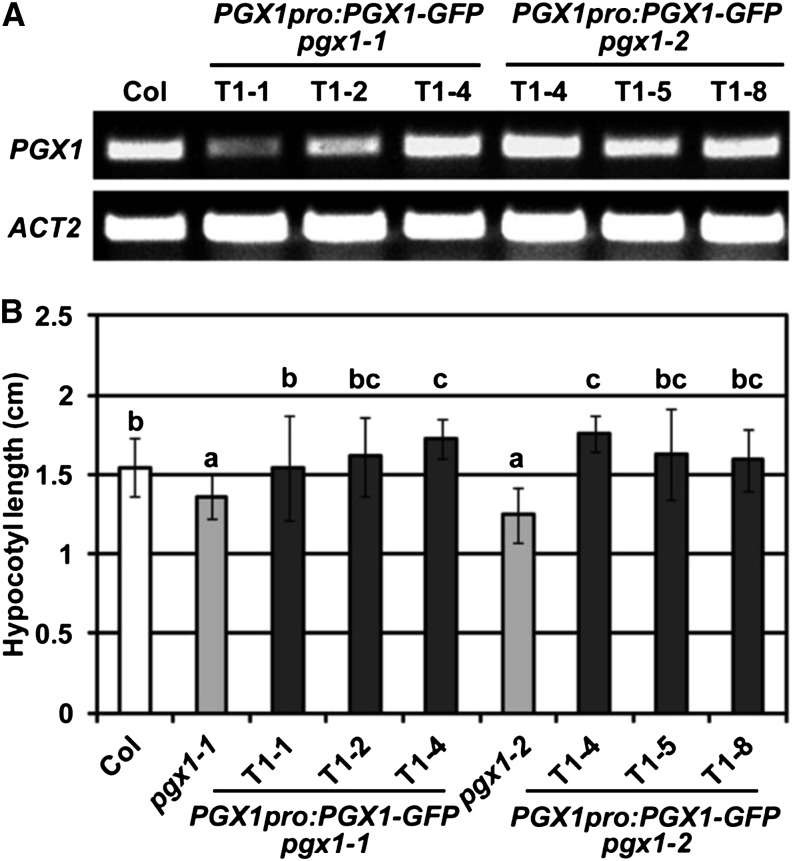
To further confirm the function of PGX1 in cell expansion in etiolated seedlings, we generated an expression construct containing a C-terminally tagged PGX1-GFP fusion driven by a promoter fragment that corresponded to the 2.5 kb upstream of the PGX1 start codon and transformed this construct into the pgx1-1 and pgx1-2 lines. Multiple transformant lines for each background were isolated and assayed for PGX1 expression and complementation of the short hypocotyl phenotype we observed in pgx1-1 and pgx1-2 mutants. As shown in Figure 4A, PGX1pro:PGX1-GFP pgx1-1 and PGX1pro:PGX1-GFP pgx1-2 lines expressed PGX1 transcript levels that were often comparable to wild-type levels. These lines also displayed hypocotyl lengths that were longer than the pgx1-1 and pgx1-2 lines and were not significantly different from, or in two lines significantly longer than, Col controls (Figure 4B; Supplemental Figure 7), indicating that the transgene rescued the growth defects observed in the mutant alleles. These data further support a function for PGX1 in hypocotyl cell expansion.
Figure 4.
PGX1 Complements the Hypocotyl Length Phenotype of pgx1-1 and pgx1-2 Mutants.
(A) RT-PCR analysis of PGX1 expression in Col and lines expressing PGX1-GFP driven by the PGX1 promoter in the pgx1-1 or pgx1-2 background. ACT2 was amplified as an internal control. Total RNA was isolated from 10-d-old light-grown seedlings of transgenic plants grown on MS media with 25 µg/mL hygromycin to select against segregants lacking the PGX1pro:PGX1-GFP transgene.
(B) Quantification of hypocotyl length in 6-d-old etiolated seedlings of Col and complementation lines grown on MS media without Suc. Bars indicate sd (n ≥ 40 seedlings from two independent experiments). Lowercase letters in (B) indicate significantly different groups (P < 0.05, one-way ANOVA and Tukey test).
PGX1 Is Expressed in Tissues Undergoing Cell Expansion or Separation and Localizes to the Apoplast
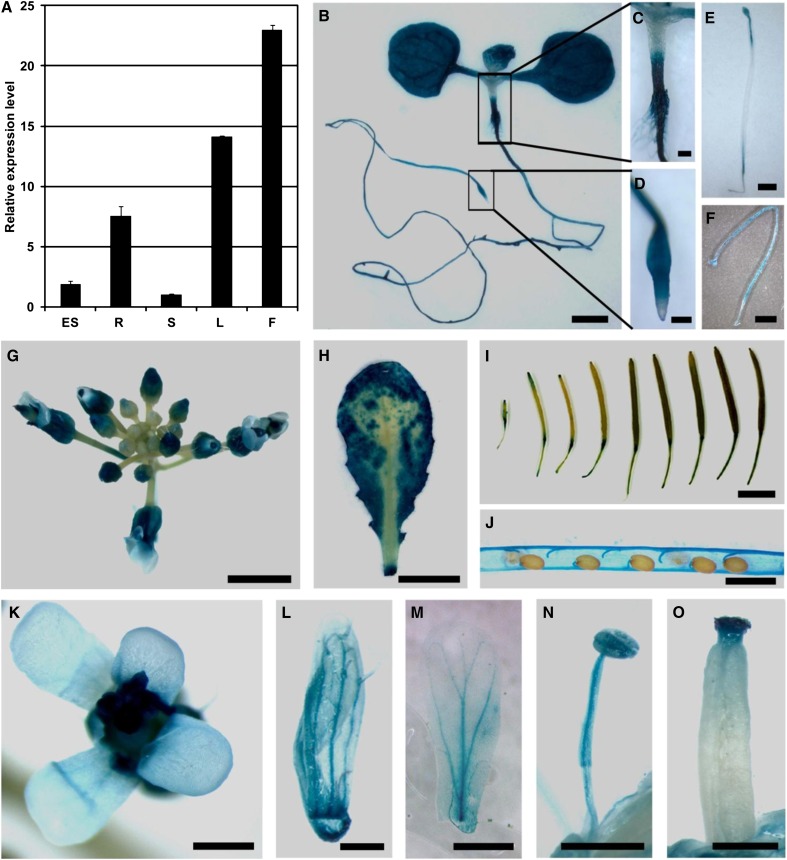
Based on existing microarray data, PGX1 is putatively expressed in multiple tissues and is most highly expressed in root tissues (Winter et al., 2007). We performed qPCR measurements of PGX1 mRNA levels to detect its relative expression levels in etiolated seedlings, roots, stems, leaves, and flowers and found higher expression in roots, leaves, and flowers (Figure 5A), with the highest expression level in flowers. To further analyze the expression pattern of PGX1, we generated a PGX1 promoter-GUS (β-glucuronidase) fusion construct, transformed it into Col plants, and performed GUS staining for multiple transformant lines. In young light-grown seedlings, strong GUS expression was evident in cotyledons, basal hypocotyls, and roots (Figures 5B to 5D), whereas in etiolated seedlings, GUS expression was detectable in the hypocotyl and cotyledons (Figures 5E and 5F). PGX1pro:GUS expression was also detected in flowers (Figures 5G and 5K to 5O), expanding rosette leaves (Figure 5H), and the bases of developing siliques (Figures 5I and 5J). Taken together, these data indicate that PGX1 is highly expressed in some, but not all, actively growing tissues (i.e., roots, hypocotyls, cotyledons, and leaves, but not stems), as well as tissues where cell separation occurs (i.e., flowers and siliques).
Figure 5.
PGX1 Expression in Different Tissues.
(A) Relative expression levels of PGX1 in different tissues including 6-d-old etiolated seedlings (ES) and 6-week-old Col roots (R), stems (S), leaves (L), and flowers (F) as measured by qPCR. ACT2 was used as an internal control, and stem PGX1 expression was normalized to 1. Bars show averages ± sd (n = 3 replicates).
(B) to (O) GUS staining of PGX1pro:GUS transgenic plants.
(B) Five-day-old light-grown PGX1pro:GUS seedling.
(C) and (D) Close-up images of hypocotyl and root tip of PGX1pro:GUS seedling in B.
(E) and (F) Two-hour and 12-h staining images of 6-d-old etiolated PGX1pro:GUS seedlings.
(G) to (J) Inflorescence, leaf, and silique of 6-week-old PGX1pro:GUS plants.
(K) to (O) Flower, sepal, petal, pistil, and stamen of PGX1pro:GUS plants.
Bars = 500 μm in (B), (E), and (J) to (O), 100 μm in (C) and (D), 2 mm in (F) and (G), and 5 mm in (H) and (I).
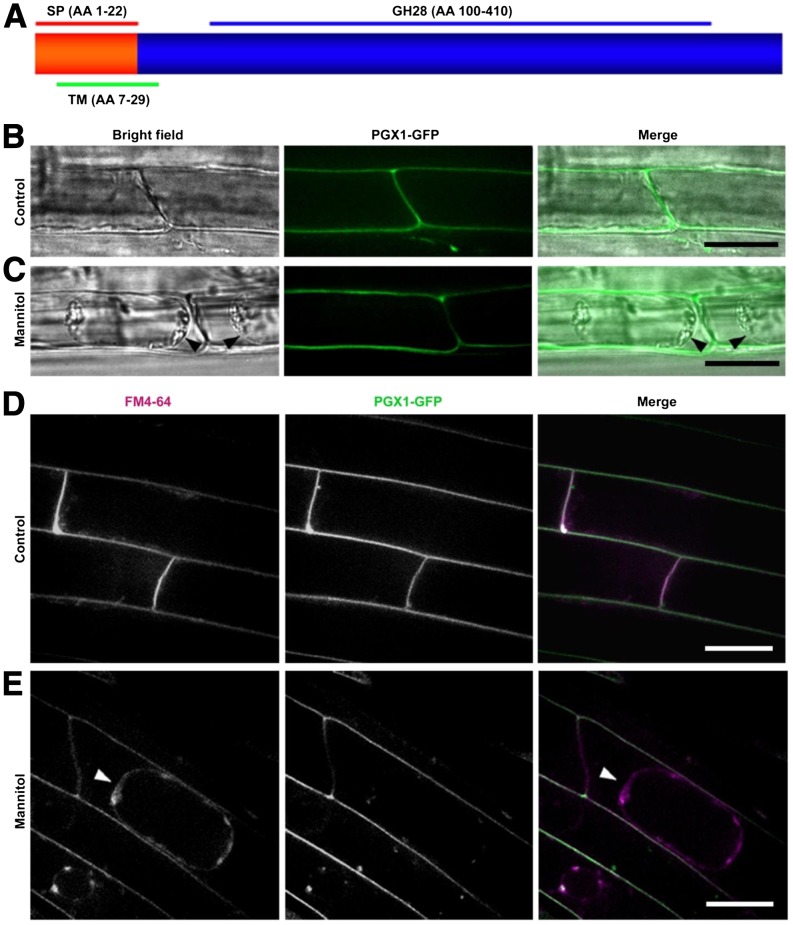
PGX1 is predicted to be 470 amino acids long and to contain a signal peptide that ends with a cleavage site as predicted by SignalP 4.1 (Figure 6A). It also contains a GH28 domain from amino acids 100 to 410 (Figure 6A) and is predicted to localize to the endoplasmic reticulum, vacuole, or cell wall/apoplast (Heazlewood et al., 2007). To determine its subcellular localization experimentally, we cloned PGX1 downstream of the endogenous PGX1 promoter and upstream of a GFP tag, generated transformants expressing this construct, and observed PGX1pro:PGX1-GFP seedlings using spinning disk confocal microscopy. We found that PGX1-GFP localized mainly at the edges of root epidermal cells (Figure 6B). To test whether this localization pattern represents apoplastic or plasma membrane localization, seedlings were incubated with 1 M mannitol to induce plasmolysis and examined. The majority of the PGX1-GFP signal remained at the cell edges rather than within the protoplast (Figure 6C), indicating that the fusion protein localizes to the cell wall. To confirm this localization pattern, we colabeled PGX1-GFP–expressing cells with the membrane dye FM4-64 (Bolte et al., 2004) and found that both markers localized similarly to cell edges in control cells (Figure 6D), but that this colocalization was disrupted after mannitol-induced plasmolysis (Figure 6E).
Figure 6.
PGX1-GFP Localizes to the Apoplast.
(A) Schematic protein structure of PGX1, including predicted signal peptide (SP; red line), transmembrane domain (TM; green line), and GH28 (blue line) sequences. The cleavage site of the signal peptide is between amino acids 22 and 23.
(B) and (C) Confocal images show the presence of PGX1-GFP fluorescence at the edges of root cells of 5-d-old PGX1pro:PGX1-GFP seedlings without (B) or with (C) plasmolysis induced by 1 M mannitol treatment for 5 min. Left panels, bright-field images; middle panels, fluorescence images of PGX1-GFP; right panels, merged bright-field and fluorescence images.
(D) and (E) Confocal images of FM4-64 staining without (D) or with (E) 1 M mannitol treatment. Five-day-old light-grown seedlings were stained in 5 μM FM4-64 for 10 min at room temperature, then mounted in 1 M mannitol for 5 min before observing fluorescence with 561- and 488-nm excitation for FM4-64 staining and GFP, respectively. Left panels, fluorescence images of FM4-64 staining; middle panels, fluorescence images of PGX1-GFP; right panels, merged fluorescence images of FM4-64 staining (magenta) and PGX1-GFP (green). Arrowheads in (C) and (E) indicate the plasma membrane where the protoplast has separated from the cell wall after plasmolysis. Bars = 50 μm.
Heterologously Expressed PGX1 Exhibits in Vitro PG Activity
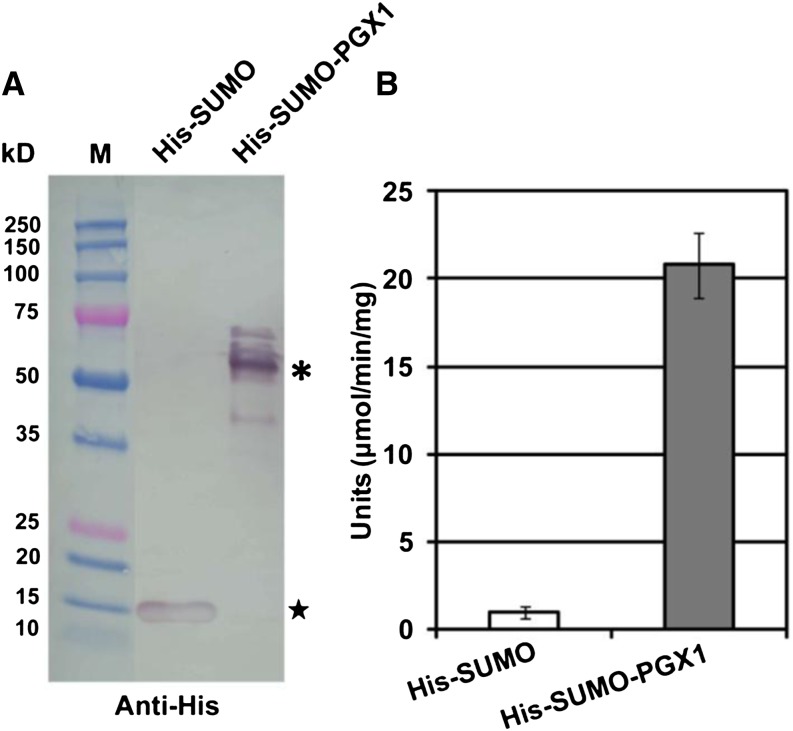
To date, few Arabidopsis putative PGs have been experimentally confirmed to display bona fide PG activity (Rhee et al., 2003; Ogawa et al., 2009). To determine whether PGX1 encodes an active PG, its full coding sequence was synthesized in a codon-optimized form, cloned into a bacterial expression vector as a 6XHis-tagged fusion with SUMO to enhance protein solubility (Peroutka et al., 2011), transformed into Escherichia coli strain BL21-DE3, and purified using nickel-NTA agarose. Immunoblotting against purified supernatant from E. coli expressing the full-length fusion protein detected the presence of a major band corresponding to the predicted 65 kD size of the His-SUMO-PGX1 fusion (Figure 7A). Supernatant containing the partially purified protein was tested for in vitro PG activity using a polygalacturonic acid substrate (Gross, 1982) and was found to contain 20.8 ± 1.9 (sd) units of PG activity, whereas supernatants from bacteria transformed with the empty vector did not display significant PG activity (Figure 7B). These results indicate that heterologously expressed His-SUMO-PGX1 can cleave polygalacturonans in vitro.
Figure 7.
Purified PGX1 Exhibits PG Activity in Vitro.
(A) Immunoblotting using anti-His antibody shows that soluble His-tagged PGX1 is expressed in E. coli after induction. M, prestained protein standard (Bio-Rad). Star indicates the size of His-SUMO alone, and asterisk indicates the predicted size of His-SUMO-PGX1.
(B) The specific activity of PGX1 was calculated from three different samples. PG activity units are defined as the amount of enzyme that releases 1 μmol of reducing end groups per minute per milligram of protein at 30°C. Purified protein from empty vector cultures was used as a control. Error bars indicate sd.
[See online article for color version of this figure.]
Altered PGX1 Expression Results in Changes in Total PG Activity, Pectin Molecular Mass, and Cell Wall Makeup
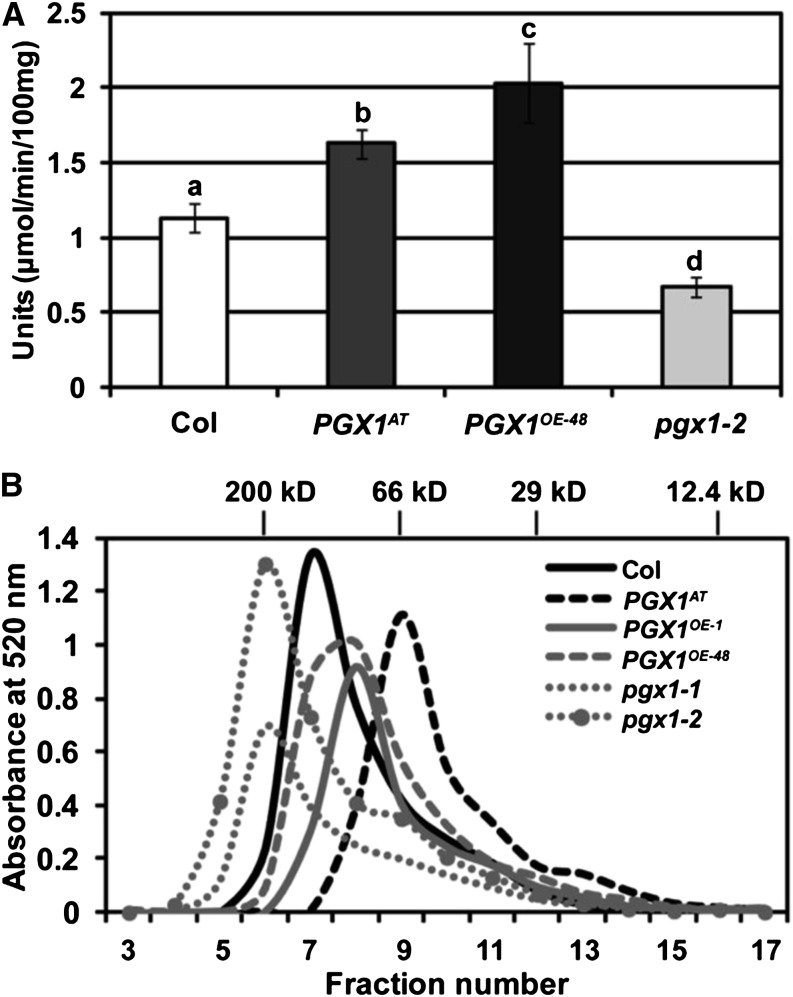
Given that PGX1 cleaves polygalacturonic acid in vitro, we next wanted to test whether changes in its expression affected total PG activity in planta. Total proteins were extracted from leaves of 40-d-old Col, PGX1AT, PGX1OE-48, and pgx1-2 plants and assayed for PG activity. We found that total protein from PGX1AT and PGX1OE-48 plants possessed higher PG activity than Col controls, whereas total protein from pgx1-2 plants possessed lower PG activity (Figure 8A). These results suggest that PGX1 contributes significantly to total PG activity in leaves. We next examined the molecular mass distribution of pectins isolated from 6-d-old etiolated seedling walls by 1,2-cyclohexylenedinitrilo-tetraacetic acid (CDTA) extraction using size-exclusion chromatography and found that pectins extracted from PGX1AT, PGX1OE-1, and PGX1OE-48 walls had smaller average molecular masses than those extracted from Col walls, whereas pectins extracted from pgx1-1 and pgx1-2 cell walls had larger average molecular masses (Figure 8B). Together, these data indicate that increased PGX1 expression results in higher total PG activity to generate smaller HG molecules, whereas reduced PGX1 expression results in less PG activity and therefore larger HG molecules.
Figure 8.
Measurement of Total PG Activity and Pectin MW in PGX1 Mutant and Overexpression Lines.
(A) PG activity in vivo is lower in pgx1-2 and higher in PGX1AT and PGX1OE-48 plants. Leaves of 40-d-old plants of Col, pgx1-2, PGX1AT, and PGX1OE48 grown in 16 h light/8 h dark were used to isolate total plant proteins. Total protein amounts were measured by Bradford assay. PG activity units are expressed as the amount of enzyme releasing 1 μmol reducing end groups per 1 min per 100 mg protein at 30°C. Error bars indicate sd (n = 3 replicates per genotype). Lowercase letters indicate significantly different groups (P < 0.05, one-way ANOVA and Tukey test).
(B) Molecular mass profile of CDTA-soluble pectin from Col, PGX1AT, PGX1OE-1, PGX1OE-48, pgx1-1, and pgx1-2, 6-d-old etiolated seedlings. Molecular distribution was analyzed by size-exclusion chromatography on a Superdex 75 5/150 GL column with cross-linked agarose and dextran. Fractions 3 to 17, which contained the only significant uronic acid contents of 30 total fractions, are shown. The column was calibrated using molecular standards including β-amylase, 200 kD; BSA, 66 kD; carbonic anhydrase, 29 kD; and cytochrome c, 12.4 kD. Uronic acid was measured by absorbance at 520 nm (Blumenkrantz and Asboe-Hansen, 1973).
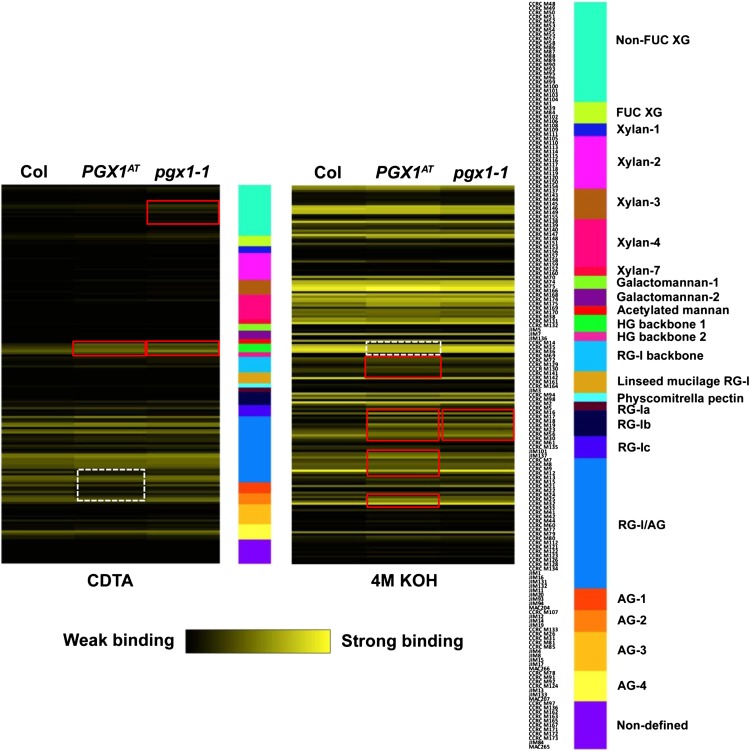
To examine the molecular phenotypes of cell walls in PGX1 overexpression and mutant plants, we performed monosaccharide composition analysis of cell walls derived from 6-d-old etiolated seedlings and destarched leaves of all six genotypes (Col, PGX1AT, PGX1OE-1, PGX1OE-48, pgx1-1, and pgx1-2), focusing on changes that were consistent between at least two overexpression and/or both mutant lines (Table 1). In etiolated seedlings, the relative amount of Fuc was higher in PGX1AT, PGX1OE-1, and PGX1OE-48 walls, whereas the relative amount of galacturonic acid was lower in PGX1OE-1 and PGX1OE-48 walls and the relative amount of glucuronic acid was lower in pgx1-1 and pgx1-2 walls. In leaves, Fuc and Gal were lower in pgx1-1 and pgx1-2 walls, rhamnose, Ara, and Xyl were lower in PGX1AT, PGX1OE, and pgx1 walls, Glc was higher in PGX1OE and pgx1 walls, and galacturonic acid was lower in PGX1OE-1 and PGX1OE-48 walls. We also performed glycome profiling (Pattathil et al., 2010, 2012) for walls derived from 6-d-old etiolated Col, PGX1AT, and pgx1-1 seedlings that were sequentially extracted with CDTA, which extracts mainly pectins, and 4 M KOH, which extracts additional pectins and hemicelluloses (Fry, 1988) (Figure 9). Glycome profiling revealed increased CDTA-extractable HG, reduced CDTA-extractable arabinogalactan proteins (AG), reduced KOH-extractable HG, and increased KOH-extractable rhamnogalacturonan I (RG-I) and AG in PGX1AT walls compared with controls, whereas pgx1-1 walls contained increased CDTA-extractable xyloglucan, CDTA-extractable HG, and KOH-extractable RG-I/AG. These results indicate that the molecular composition and/or structure of the cell wall are altered when PGX1 expression is either enhanced or reduced.
Table 1. Monosaccharide Composition of Wild-Type and PGX1 Mutant Cell Walls.
| Tissue | Genotype | Fuc | Rhamnose | Ara | Gal | Glc | Xyl | GalA | GlcA |
|---|---|---|---|---|---|---|---|---|---|
| ES | Col | 5.74 ± 0.42 | 5.37 ± 0.90 | 35.33 ± 3.10 | 16.38 ± 1.02 | 5.55 ± 0.91 | 8.61 ± 0.59 | 23.03 ± 2.98 | 3.73 ± 0.59 |
| PGX1AT | 6.81 ± 0.75 | 5.81 ± 0.64 | 32.80 ± 1.35 | 16.96 ± 1.09 | 6.01 ± 0.60 | 8.49 ± 0.66 | 22.02 ± 2.67 | 3.34 ± 0.98 | |
| PGX1OE-1 | 6.88 ± 0.55 | 5.72 ± 0.45 | 34.69 ± 3.22 | 18.15 ± 1.11 | 5.10 ± 0.62 | 8.42 ± 0.22 | 19.29 ± 2.28 | 3.51 ± 0.70 | |
| PGX1OE-48 | 7.14 ± 0.42 | 5.27 ± 0.65 | 30.76 ± 2.05 | 21.33 ± 2.74 | 5.92 ± 0.69 | 8.80 ± 0.70 | 19.07 ± 2.72 | 3.40 ± 0.66 | |
| pgx1-1 | 6.33 ± 0.55 | 4.69 ± 0.59 | 33.97 ± 1.87 | 16.60 ± 1.07 | 5.60 ± 0.76 | 8.99 ± 0.67 | 23.82 ± 4.07 | 0.54 ± 0.20 | |
| pgx1-2 | 6.56 ± 0.59 | 5.77 ± 1.38 | 30.39 ± 1.79 | 19.69 ± 1.38 | 6.10 ± 0.21 | 7.83 ± 0.39 | 22.36 ± 3.16 | 2.60 ± 0.93 | |
| L | Col | 2.84 ± 0.56 | 1.93 ± 0.15 | 8.01 ± 0.50 | 2.67 ± 0.11 | 50.10 ± 1.51 | 4.00 ± 0.20 | 30.41 ± 2.05 | |
| PGX1AT | 2.77 ± 0.32 | 1.43 ± 0.06 | 5.94 ± 0.25 | 1.97 ± 0.12 | 53.90 ± 3.71 | 3.23 ± 0.05 | 30.79 ± 3.90 | ||
| PGX1OE-1 | 2.50 ± 0.77 | 1.53 ± 0.11 | 5.57 ± 0.23 | 2.32 ± 0.18 | 67.80 ± 1.00 | 4.16 ± 0.42 | 16.14 ± 1.40 | ||
| PGX1OE-48 | 2.93 ± 0.21 | 1.66 ± 0.04 | 6.04 ± 0.20 | 2.58 ± 0.14 | 61.10 ± 1.49 | 3.38 ± 0.10 | 22.33 ± 1.78 | ||
| pgx1-1 | 2.24 ± 0.22 | 1.16 ± 0.05 | 4.84 ± 0.39 | 1.46 ± 0.08 | 64.10 ± 1.74 | 3.39 ± 0.17 | 22.83 ± 1.87 | ||
| pgx1-2 | 2.10 ± 0.29 | 1.26 ± 0.08 | 5.01 ± 0.44 | 1.57 ± 0.14 | 59.00 ± 6.63 | 3.33 ± 0.10 | 27.76 ± 7.35 |
Sugar composition (GalA, galacturonic acid; GlcA, glucuronic acid) from 6-d-old etiolated seedlings (ES) and leaves from 40-d-old plants (L) of the wild type, PGX1AT, PGX1OE-1, PGX1OE-48, pgx1-1, and pgx1-2 genotypes. Sugar amounts are presented as mean percentages ± sd (n = 6). Bold values indicate mutant glycosyl residue compositions that are at least ±15% and are significantly different (P < 0.05, t test) compared to the wild type for at least two overexpression and/or for both mutant lines.
Figure 9.
Glycome Profiling of Sequential Extractions of Cell Walls from 6-d-Old Etiolated Col, PGX1AT, and pgx1-1 Seedlings.
Cell wall extracts were probed by ELISA with 172 glycan-directed monoclonal antibodies (Pattathil et al., 2010). Data (A450 − A655) are presented as heat maps with black to bright yellow, indicating weakest to strongest binding. The panel on the right lists the array of monoclonal antibodies in 24 groups according to the glycan recognized by the antibodies. Altered patterns of monoclonal antibody binding PGX1AT and/or pgx1-1 fractions compared with wild-type fractions are highlighted by red (increased binding) or dashed white (decreased binding) rectangles.
PGX1 T-DNA Insertion and Activation Tag Plants Possess a Higher Number of Flowers with Extra Petals and More Variable Phyllotactic Patterning in Floral Meristems
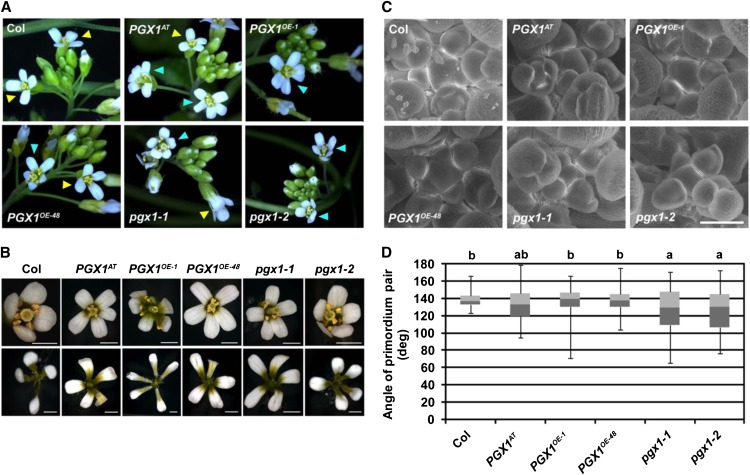
When growing PGX1 overexpression or mutant plants, we noticed that their flowers sometimes possessed five or six petals, rather than the four petals usually seen in Col flowers (Figures 10A and 10B). The extra petals did not appear to arise from splitting of these organs after initiation, since each petal contained an independent base in the abnormal flowers (Figure 10B). We quantified petal number for flowers in Col controls and mutant plants grown in the same growth containers and found that for both the overexpression and T-DNA lines, the proportion of flowers with extra petals was significantly higher than in Col plants (Table 2; P < 0.01, χ2 test). For flowers with more than four petals, we often observed additional sepals (46/53 flowers) but fewer stamens (35/53 flowers). To investigate floral development more closely, we imaged floral meristems of all genotypes using scanning electron microscopy and observed more variable angles between flower primordia, which normally arise at a consistent angle of 137 degrees relative to the previous primordium (Hamant et al., 2010), in PGX1AT, PGX1OE-1, PGX1OE-48, pgx1-1, and pgx1-2 floral meristems, with pgx1-1 and pgx1-2 meristems producing organs at significantly smaller relative angles than Col meristems (Figure 10D). These data indicate that changes in PGX1 expression perturb normal floral organ patterning.
Figure 10.
PGX1AT, PGX1OE, and pgx1 Mutants Possess Flowers with Extra Petals.
(A) and (B) Flowers of 6-week-old Col, PGX1AT, PGX1OE-1, PGX1OE-48, pgx1-1, and pgx1-2 plants grown in a 16-h-light/8-h-dark photoperiod. In (A), yellow arrowheads indicate flowers with four petals and cyan arrowheads indicate flowers with extra petals. In (B), flowers were isolated (top panels) and dissected to reveal petal bases (bottom panels). Bars = 1 mm.
(C) Scanning electron micrographs of Col, PGX1AT, PGX1OE, and pgx1 floral meristems. Bar = 100 μm.
(D) Box plot distributions of floral primordium pair angles from Col, PGX1AT, PGX1OE, and pgx1 plants. Whiskers define data range, dark-gray boxes define second quartiles, and light-gray bars define third quartiles (n ≥ 39 measurements per genotype). Lowercase letters indicate significantly different groups (P < 0.05, one-way ANOVA and Tukey test).
Table 2. Quantification and Statistical Analysis of Flower Numbers with Additional Petals in PGX1 Mutant and Overexpression Lines.
| Genotype | Col | PGX1AT | Col | PGX1OE-1 | Col | PGX1OE-48 | Col | pgx1-1 | Col | pgx1-2 |
|---|---|---|---|---|---|---|---|---|---|---|
| No. flowers w/four petals | 895 | 844 | 244 | 255 | 829 | 832 | 1187 | 1412 | 1011 | 887 |
| No. flowers w/five petals | 39 | 85 | 6 | 13 | 20 | 39 | 35 | 66 | 24 | 54 |
| Percentage w/extra petals | 4.18 | 9.15 | 2.40 | 4.85 | 2.36 | 4.48 | 2.86 | 4.47 | 2.32 | 5.74 |
| P (χ2) | 4.0 × 10−14 | 3.2 × 10−3 | 2.1 × 10−5 | 5.6 × 10−17 | 3.9 × 10−13 | |||||
| No. replicates | 3 | 1 | 2 | 4 | 3 | |||||
All completely open flowers were counted for each experiment. The percentage indicates the ratio of flowers with five or more petals to total completely open flowers.
DISCUSSION
In this work, we used activation tag screening to identify genes involved in cell expansion whose loss-of-function phenotypes might be masked by redundancy or that might be involved in essential processes. Based on our initial screening results, we anticipate that many additional candidate genes among the over 1000 estimated to be involved in cell wall biosynthesis and/or modification (Somerville et al., 2004) can be isolated and characterized using this approach, and we are characterizing several wall-related candidates from among the initial hits. However, the potential complications associated with loss-of-function analyses for members of large gene families makes the functional characterization of these candidates via reverse genetic approaches (Alonso et al., 2003) a challenge. Additional approaches, such as biochemical analysis of wall composition (Reiter et al., 1997) in response to gene overexpression and the generation of multiplexed loss-of-function mutants for related gene family members, may be required to fully elucidate the functions of these candidates in cell wall modification during growth.
The finding that PGX1AT and PGX1OE cells throughout etiolated hypocotyls, both in growing cells at the top and middle of the hypocotyl and at the base of the hypocotyl where cell elongation has ceased (Derbyshire et al., 2007a), are longer than wild-type cells at the same position (Figures 1 and 2) suggests that overexpression of PGX1 enhances cell expansion in this organ. These data also imply that the capacity for cell expansion is not maximized in wild-type hypocotyls, possibly due to rigidification of the cell wall by pectin cross-linking (Derbyshire et al., 2007a; Zhao et al., 2008), which we predict should be diminished in PGX1 overexpression lines. Importantly, we did not observe significant changes in cell number along the lengths of hypocotyls in any lines tested (Supplemental Figure 3), suggesting that cell overproliferation at earlier developmental stages does not contribute to the observed changes in hypocotyl length.
Although there are 69 annotated Arabidopsis PG-encoding genes, disrupting PGX1 function by T-DNA insertions results in reduced cell size and hypocotyl elongation in the mutant lines relative to the wild type (Figure 3). This finding is surprising given the potential for redundancy among PG family members, but At3g26610 is the sole member of a subclade of PGs in Arabidopsis (Supplemental Figure 2) (Kim and Patterson, 2006; González-Carranza et al., 2007) and might thus play a unique role in wall loosening and expansion in etiolated hypocotyls and other tissues, including leaves (Supplemental Figure 5). We observed a correlation between PGX1 expression levels (Figures 1 to 3) and total PG activity in leaves (Figure 8A), suggesting that this gene contributes a significant proportion of total PG activity in at least some tissues. In agreement with this interpretation, we observed a higher average molecular mass for pectins isolated from pgx1 mutant walls and a lower average molecular mass for pectins isolated from PGX1AT and PGX1OE walls (Figure 8B), indicating that PGX1 cleaves HG (Atkinson et al., 2002; Posé et al., 2013), possibly reducing the capacity of the pectin network to cross-link and enabling wall expansion. At this point, we do not know how PGX1 activity is limited in cell walls to prevent it from aberrantly degrading pectin, but it is possible that it is regulated by limited mobility in intact walls and/or binding to PG inhibitor proteins (Alexandersson et al., 2011).
Our glycomics profiling experiments (Figure 9) revealed a complex set of relatively subtle changes in the cell walls of PGX1AT and pgx1-1 etiolated seedlings. We posit two possible interpretations of the finding that CDTA-extractable HG is higher in both lines relative to the wild type: First, there might be a relatively narrow molecular mass range within which HG is maximally entangled in the cell wall; and second, the balance between HG cross-linking and its interaction with other wall components is perturbed in either case, leading to increased CDTA extractability. We also noted a slight increase in xyloglucan CDTA extractability in the pgx1-1 mutant, suggesting that altered pectin in the mutant might prevent xyloglucan from interacting normally with the wall. Altered extractability of RG-I/AG in PGX1AT plants might result from compensatory changes in RG-I/AG cross-linking to the wall in response to reduced HG cross-linking. However, we observed an increase in KOH-extractable RG-I/AG in pgx1-1 walls without an accompanying reduction in CDTA-extractable RG-I/AG, suggesting that the overall amount of RGI/AG might be altered in this mutant. The reduction in total GalA content in PGX1OE-1 and PGX1OE-48 etiolated seedlings and leaves (Table 1) might also arise from compensatory changes in pectin biosynthesis in these lines; however, it is also possible that HG solubility was increased in these lines such that significant HG (and, thus, GalA) was lost from these wall samples during alcohol-insoluble residue (AIR) isolation.
Interestingly, the in vitro PG activity of heterologously expressed PGX1 (Figure 7) is more than 9 times that of several enzymatically characterized Arabidopsis PGs (Ogawa et al., 2009), further suggesting that it might play a major role in HG degradation in at least some tissues. However, comparable enzymatic activity measurements for additional PGs within this family will be required to support this hypothesis. PGX1 is predicted to be coexpressed (Mostafavi et al., 2008; Mutwil et al., 2009) with the xyloglucan endotransglycosylase/hydrolase-encoding gene XTH21 as well as other wall polymer-degradation genes, implying that it might be part of a multifunctional expression module involved in cell wall remodeling. Detailed analyses of the transcriptional regulation and functions of this coexpressed network of genes will be required to test this hypothesis.
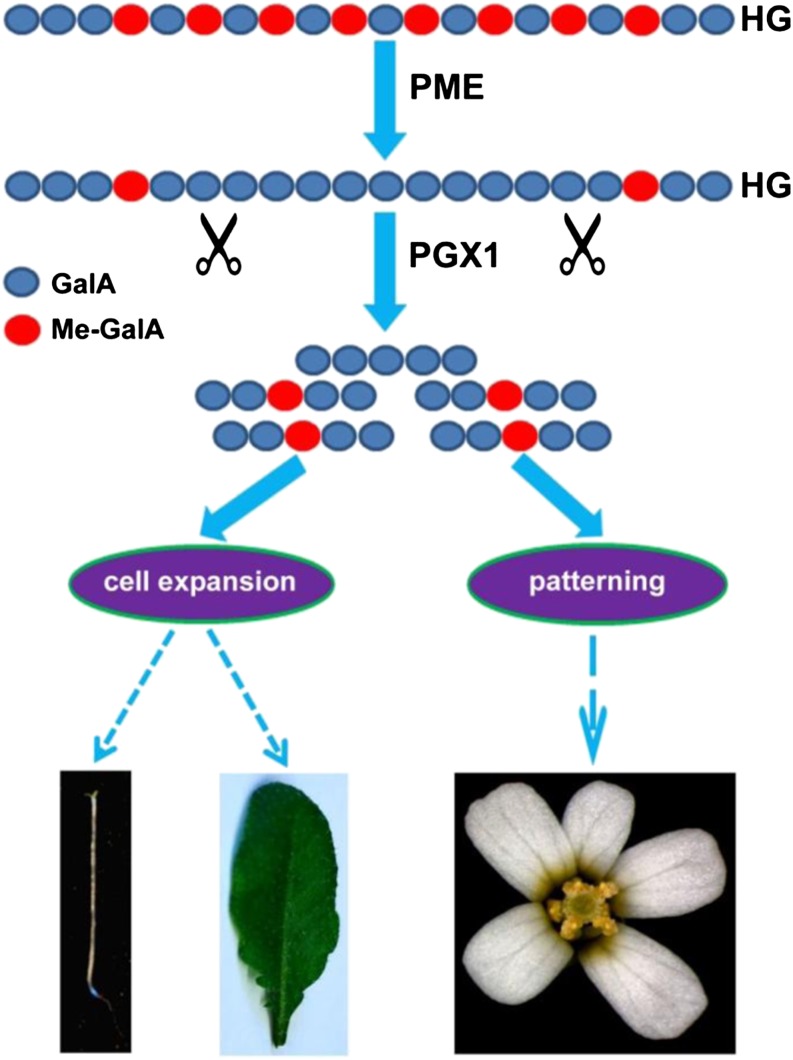
Our finding that plants with either reduced or increased PGX1 mRNA levels and total PG activity display a higher frequency of aberrant floral organ number and higher variability in floral organ primordium angles in comparison to wild-type plants (Figure 10) seems paradoxical at first glance but could potentially be explained by a requirement for a finely tuned balance of PG activity to robustly maintain the patterning mechanisms present in the developing flower, which might mirror those observed in the shoot apical meristem in their dependence upon the stiffness and degradability of the pectin network in cell walls (Peaucelle et al., 2011). We hypothesize that by tuning pectin methylation, cross-linking, and molecular mass, pectin methyl-esterases and PGs might act as downstream components of the regulatory networks that control cell expansion, as well as floral phyllotaxis and organogenesis (Monfared et al., 2013), and present these ideas in a conceptual model in Figure 11.
Figure 11.
A Schematic Model of PGX1 Function in Plant Development.
HG (blue polymer) is secreted into the apoplast in a highly methylated form after synthesis in the Golgi. Pectin can be demethylated in the wall by the activity of pectin methyl-esterases (PMEs). Demethylated HG might be cleaved by PGX1 and other PGs, loosening the wall and enabling normal hypocotyl elongation, leaf expansion, and flower development.
In summary, the evidence presented in this work highlights the function of PGs in cell expansion in addition to their established roles in cell separation (Rhee et al., 2003; Kim and Patterson, 2006; González-Carranza et al., 2007; Ogawa et al., 2009). Intriguingly, cells adjacent to abscission zones often expand after abscission (Patterson, 2001), highlighting a possible connection between the molecular mechanisms that underlie wall expansion and cell separation. Future investigation of how these processes are related should provide additional insights into how plants orchestrate a complex sequence of molecular behaviors to modify their cell walls in order to accomplish growth, reproduction, and senescence.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana plants were of the Col ecotype. Activation tagged lines were generated in the Somerville lab by Wolf Scheible, and seed pools are available from the ABRC. Plants were grown on MS plates containing 2.2 g/L MS salts (Caisson Laboratories), 0.6 g/L MES (Research Organics), 1% (w/v) Suc, and 0.8% (w/v) agar-agar (Research Organics), pH 5.6, under long-day conditions (16 h light/8 h dark) in a 22°C chamber. Seedlings were transferred from MS plates to soil with nutrient solution (Miracle Gro) for growth in a growth room with the same light regime and temperature. Adult plants were imaged using a Nikon D5100 DSLR camera and rosette diameters were measured using ImageJ.
Activation Tagging Mutant Screening and Adapter Ligation PCR
Pools of activation tagged seeds transformed with pSKI015 (Weigel et al., 2000) were surface-sterilized in 30% bleach + 0.1% SDS (w/v) for 20 min, washed four times with sterile MilliQ water, suspended in sterile 0.15% agar (w/v), and stored in the dark at 4°C for 2 to 7 d before sowing on MS without Suc in empty 200-μL pipette tip boxes. Approximately 1000 seeds were sown in each box, and boxes were exposed to light for 4 h, then wrapped in aluminum foil and placed in a 22°C growth chamber for 6 d. Seedlings with long hypocotyls were transferred to MS plates for vertical growth under 24 h light at 22°C for 10 to 14 d, after which they were transplanted to soil. The progeny of each putative mutant were screened for heritable hypocotyl length by growth on vertical MS plates without Suc for 6 d in the dark, scanning on a Scanjet 8300 scanner (HP) at 600 dpi, measuring hypocotyl lengths in ImageJ, and testing for significantly longer hypocotyls (P < 0.01, t test, two replicates). Other progeny were tested for resistance to methionine sulfoximine (MSO; Sigma-Aldrich) (Maughan and Cobbett, 2003) by growth on MS plates containing 5 μM MSO in the light for 5 d; plants with well-developed cotyledons and roots over 0.67 cm long were scored as resistant. Lines containing likely single insertions were identified by χ2 analysis (P > 0.1, two replicates). For adapter ligation PCR, genomic DNA was prepared from rosette leaves using the Genelute Plant Genomic DNA miniprep kit (Sigma-Aldrich) and the protocol of O’Malley et al. (2007) was followed using both HindIII-EcoRI and AseI adapter primer sets. PCR products were extracted from 1% agarose TAE gels and sequenced, and the sequences were aligned to pSKI015 and the Arabidopsis genome using NCBI BLAST and TAIR Seqviewer tools.
Identification of T-DNA Insertion Mutants
T-DNA insertion lines (Alonso et al., 2003) for PGX1 were obtained from the ABRC. WiscDsLox262B06 (intron) and Salk_026818 (exon) lines were designated pgx1-1 and pgx1-2, respectively. Homozygous T-DNA insertion lines were identified using primers designed at http://signal.salk.edu/tdnaprimers.2.html (Supplemental Table 1).
Plasmid Construction and Plant Transformation
For constitutive overexpression, a full-length synthetic PGX1 coding sequence (DNA 2.0) was cloned into pEarleyGate 104 (Earley et al., 2006) using the Gateway cloning system (Life Technologies) to generate the overexpression construct 35S:YFP-PGX1. For complementation experiments, fragments of the region 2.5 kb upstream of the PGX1 translation initiation site from Col genomic DNA and the full coding sequence of PGX1 from Col flower cDNA were amplified by Phusion hot start high-fidelity DNA Polymerase (Finnzyme) with gene-specific primers (Supplemental Table 1) and ligated by overlap PCR (Lu, 2005). After adding adenosine to the 3′ end of PCR product using Taq polymerase, the final PCR fragment was cloned into pCR8 using the pCR8/GW/TOPO TA cloning system (Life Technologies) and recombined into the binary vector pMDC110 (Curtis and Grossniklaus, 2003) to generate PGX1pro:PGX1-GFP. Gel Extraction and Cycle Pure kits (Omega Bio-Tek) were used for gel and PCR purification, and a Plasmid Mini Kit (Omega Bio-Tek) was used for plasmid isolation. All amplified sequences were sequenced (dnaLIMS; Penn State University) to verify that no mutations were introduced. Col and T-DNA mutant plants were transformed with Agrobacterium tumefaciens strain GV3101 using the floral dip method (Clough and Bent, 1998). Positive transformants were selected on MS plates containing 5 μM MSO or 25 μg/mL hygromycin (Omega Scientific).
Hypocotyl and Cell Length Measurements
Seeds were surface-sterilized in 30% bleach + 0.1% (w/v) SDS, washed four times with sterile water, stored at 4°C for 3 d, and sown on MS plates without Suc. Seeds were exposed to light for 4 h to stimulate germination, then wrapped in two layers of aluminum foil and grown for 6 d at 22°C. Plates with etiolated seedlings were scanned using an HP Scanjet 8300 scanner at 600 dpi, and hypocotyl lengths were measured using Image J. To measure cell length, 6-d-old etiolated seedlings were mounted in water and images of epidermal cells (at least 30 seedlings of each genotype) were recorded using a Plan Neofluar ×10 0.3–numerical aperture air objective (Zeiss) on a Zeiss Axio Observer SD spinning disk confocal microscope with a CSU-X1 spinning disk head (Yokogawa). Cell length in recorded images was quantified using ImageJ.
Gene Expression Analysis
Total RNA was extracted from seedlings and adult tissues using a Plant RNA Kit (Omega Bio-Tek). Samples were treated with RNase-free DNase I (NEB) on a column to remove genomic DNA. RNA concentration was measured by spectrophotometer (NanoDrop 2000C), and first-strand cDNA was synthesized using qScript cDNA SuperMix (Quanta Biosciences) from 500 ng DNase I-treated total RNA. RT-PCR was performed using cDNA as a template with gene-specific primers, followed by agarose gel electrophoresis. ACT2 (At3g18780) was amplified as an internal control (Czechowski et al., 2005). qPCR was performed using SYBR Green FastMix (Quanta Biosciences) with cDNA and gene-specific primers on a StepOne Plus Real-Time PCR machine (Applied Biosystems). Data were analyzed using StepOne software (Applied Biosystems) and transcript levels of PGX1 were calculated relative to ACT2 using the ΔΔCT method. Primer sequences are listed in Supplemental Table 1. In addition to measurement of mRNA expression in different tissues by qPCR, the spatial expression pattern of PGX1 was examined by promoter-GUS analysis. A 2.5-kb fragment upstream of the PGX1 start codon was amplified from Col genomic DNA, ligated into pCR8 (Life Technologies), and recombined into pMDC162 (Curtis and Grossniklaus, 2003) using LR Clonase (Life Technologies) to generate a PGX1pro:GUS reporter gene construct. The promoter-GUS construct was transformed into Col plants as above, 20 independent transgenic lines were obtained, and T2 lines were used for expression analysis. For GUS assays, tissues were immersed in buffer containing 50 mM sodium phosphate, pH 7.2, 0.2% (v/v) Triton X-100, 2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, and cyclohexylammonium salt (X-Gluc) and incubated at 37°C in the dark for 2 to 16 h. Tissues were decolored in 70% ethanol, and images of GUS staining were recorded using a Discovery V12 fluorescence dissecting microscope (Zeiss).
Subcellular Localization of PGX1-GFP
Transgenic plants expressing PGX1pro:PGX1-GFP were generated by Agrobacterium-mediated transformation of Col plants. The signal peptide and transmembrane domain of PGX1 were predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP) and TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM), respectively. Five-day-old light-grown PGX1pro:PGX1-GFP seedlings were used to observe GFP fluorescence under a spinning disk confocal microscope with a ×100 1.40–numerical aperture oil-immersion objective (Zeiss), and images were recorded and contrast-enhanced using ImageJ. Some seedlings were subjected to plasmolysis in 1 M mannitol for 5 min and/or stained with 5 μM FM4-64 (Life Technologies) for 10 min at room temperature (Bolte et al., 2004). GFP fluorescence was detected using a 488-nm excitation laser and 525/50-nm emission filter, and FM4-64 was detected using a 561-nm excitation laser and 617/73-nm emission filter.
Expression and Purification of PGX1
Expression and purification of PGX1 followed (Xiao et al., 2009) with modifications. The synthetic PGX1 open reading frame was cloned into pSUMO (gift from Ming Tien) using BsaI and XhoI (NEB) restriction sites. Expression of the 6xHis-SUMO-PGX1 fusion protein was induced by 1 mM isopropyl β-d-thiogalactopyranoside (Gold Biotechnology) in Escherichia coli strain BL21-DE3. After induction for 4 h at 25°C, cells were harvested by centrifugation at 10,000g for 10 min at 4°C and resuspended in 20 mL of protein extraction buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 10 mM imidazole, and 2.5 µg/mL DNase, pH 8.0). The cells were sonicated for 3 × 10 s pulses and supernatants were isolated by centrifugation at 10,000g for 20 min at 4°C. Supernatants were incubated with Ni-NTA Agarose (5 PRIME) with gentle shaking (50 rpm) at 4°C for 1 h, then added to a purification column (Thermo) and washed twice in extraction buffer and twice in washing buffer (50 mM sodium phosphate, 300 mM NaCl, and 20 mM imidazole, pH 8.0). His-tagged proteins were eluted using elution buffer (50 mM sodium phosphate, 300 mM NaCl, and 250 mM imidazole pH 8.0). Purified protein samples were dialyzed in 50 mM sodium acetate, pH 5.0, at 4°C overnight using Spectra/Por MWCO 12-14,000 membrane (Spectrum Labs). For immunoblotting, proteins were separated on 12% Mini-PROTEAN precast gels (Bio-Rad) using a Mini-PROTEAN Tetra Cell (Bio-Rad). Proteins were electrophoretically transferred to Amersham Hybond-ECL membrane (GE Healthcare) using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). Transfer buffer consisted of 48 mM Tris base, 39 mM glycine, 20% methanol, and 0.0375% (w/v) SDS. The membrane was blocked with TBST (25 mM Tris base, 150 mM NaCl, and 0.1% Tween 20, pH 7.5) containing 5% (w/v) skim milk powder. The membrane was incubated with an anti-His primary antibody (1:5000; Sigma-Aldrich) in blocking solution for 1 h at room temperature. After washing in TBST (3 × 5 min), the blot was incubated with alkaline phosphatase–conjugated goat anti-mouse secondary antibody (1:5000; Sigma-Aldrich) for 1 h, washed as above, and developed using nitro-blue tetrazolium chloride, 5-bromo-4-chloro-3′-indolyphosphate solution (Sigma-Aldrich).
PG Activity Assay
Purified PGX1 protein concentration was measured using Bradford dye reagent (Amresco) (Bradford, 1976) with BSA (Amresco) as a standard. For enzymatic activity assays, PG activity was determined by measuring an increase in reducing end groups (Gross, 1982) using d-galacturonic acid (Sigma-Aldrich) as a standard. Protein samples were added to a solution containing 37.5 mM Na-acetate, pH 4.4, and 0.2% (w/v) polygalacturonic acid as a substrate (Sigma-Aldrich) and incubated at 30°C for 3 h. Released reducing end groups were labeled with 2-cyanoacetamide (Sigma-Aldrich), and absorbance at 276 nm was measured using a NanoDrop 2000C spectrophotometer. One unit of PG activity was defined as the amount of enzyme releasing 1 µmol reducing end groups per minute (Ogawa et al., 2009).
Plant Protein Extraction
Plant proteins were isolated according to Peretto et al. (1992) with minor modifications. Five grams of rosette leaves from 40-d-old plants were ground into fine powder in liquid nitrogen by mortar and pestle. The powder was transferred to another cold mortar with 5 mL extraction buffer consisting of 50 mM Tris-HCl, pH 7.5, 3 mM EDTA, 2.5 mM DTT (Sigma-Aldrich), 2 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich), 1 M NaCl, and 10% (v/v) glycerol to continue grinding. Homogenate was transferred into a 15-mL conical tube and kept at 4°C for 1 h. Cell debris was removed by centrifugation at 4500g for 10 min at 4°C. After filtering with a 100-µm cell strainer, the supernatant was collected by centrifugation at 25,000g for 40 min at 4°C and dialyzed against 50 mM Na-acetate buffer (pH 5.0) overnight at 4°C using Spectra/Por MWCO 12-14,000 membrane (Spectrum Labs).
Preparation of AIR Extracts and AIR Fractionation
Rosette leaves of 40-d-old plants and 6-d-old dark-grown seedlings were ground into a fine powder in liquid nitrogen by mortar and pestle. The powder was suspended in 30 mL chloroform/methanol (1:1, v/v) and rocked for 1 h at room temperature. After centrifugation at 4500g for 10 min at room temperature, the supernatant was removed and the residue was resuspended in 30 mL 70% ethanol and rocked for 1 h at room temperature. Centrifugation and resuspension in 70% ethanol were repeated four times. Wall residue was air-dried with 100% acetone at room temperature. Starch was removed by treatment with 90% DMSO (Sigma-Aldrich) overnight with rotation at room temperature. Destarched wall material was rinsed with 90% DMSO once, then washed six times with 70% ethanol (Fry, 1988). The absence of starch was assessed by iodine/potassium iodide. The final residue was air-dried with 100% acetone. AIR was sequentially extracted with 50 mM CDTA (Aqua Solutions), pH 6.0, and 4 M KOH containing 1% (w/v) sodium borohydride (Sigma-Aldrich). Each extraction was performed for 24 h with constant shaking (50 rpm) at room temperature. Samples were centrifuged at 4500g for 10 min to sediment CDTA-insoluble residue and used for KOH extraction after washing the pellet with water. CTDA and KOH fractions from the soluble supernatants were precipitated in 70% ethanol and lyophilized.
Size-Exclusion Chromatography and Uronic Acid Assays
A Superdex 75 5/150 GL (GE Healthcare) gel filtration column was used to fractionate the polymers from CDTA-soluble material. Samples were dissolved by adding 5 mg to 1 mL sodium acetate buffer, vortexing, and filtering through a 0.45-µm filter. Gel media were equilibrated with 0.1 M sodium acetate buffer (pH 5.0) before loading 150 μL of each sample and eluted at a flow rate of 0.25 mL/min (Posé et al., 2013). Thirty fractions of 250 μL were collected and analyzed for uronic acid content. A size standard kit was purchased from Sigma-Aldrich (MWGF200). Uronic acid content was determined as described (Blumenkrantz and Asboe-Hansen, 1973) with modifications. Samples were mixed with 1 mL concentrated sulfuric acid containing 0.0125 M sodium tetraborate (Sigma-Aldrich) and boiled for 5 min in a 100°C water bath. After cooling to room temperature, background absorbance was read at 520 nm using a NanoDrop 2000C spectrophotometer. Twenty microliters of 0.15% (w/v) m-hydroxydiphenyl (Sigma-Aldrich) dissolved in 0.5% (w/v) NaOH was added and pipetted thoroughly before incubating at room temperature for 5 min, and then measuring absorbance at 520 nm again.
Trifluoroacetic Acid Hydrolysis and Sugar Monosaccharide Composition Analysis
Trifluoroacetic acid hydrolysis was performed with the Dionex TN53. Twenty milligrams of AIR was dissolved in 1 mL water, vortexed to resuspend, and 20 μL of the suspension was transferred into a new screw-top tube. BSA (Sigma-Aldrich) and water were used as positive and negative controls, respectively. Hydrolysis was performed with 125 μL each of 13.5 M trifluoroacetic acid (Sigma-Aldrich) and water for 2 h at 100°C, and samples were cooled on ice for 10 min, spun down briefly to pellet condensate, and air-dried. Hydrolyzed samples were resuspended in 200 μL water and vortexed, and insoluble pellets were collected by centrifugation at 4500g for 5 min at room temperature. The supernatant was filtered through a 0.2-µm filter, and 20 μL of each sample was injected into a high-pressure anion exchange chromatography system with pulsed-amperometric detection using a CarboPac PA20 guard (3 × 30 mm) and column (3 × 150 mm; Dionex ICS-5000; Thermo Scientific). Elution steps were 10% of 100 mM NaOH for 15 min, followed by a 0 to 100% gradient of 100 mM NaOH and 100 mM NaOAc for 20 min. Sugar monosaccharide peaks in the samples were quantified using standards with known concentrations (Fuc, rhamnose, Ara, Gal, Glc, and Xyl at 100 μg/mL; galacturonic acid and glucuronic acid at 1 mg/mL). All chemicals were purchased from Sigma-Aldrich, and all eluents supplied by Thermo Scientific. Three technical replicates were performed for each sample.
Total Sugar Estimation and ELISA
Total sugar content was quantified by the phenol-sulfuric acid assay (Dubois et al., 1956). AIR fractions were suspended in deionized water, and 250 μL 5% (w/v) phenol (Sigma-Aldrich) was added and mixed well with sample. Then, 1.25 mL 96% sulfuric acid was jetted into the reaction solution, which was immediately vortexed. After incubation for 30 min at room temperature in dark, absorbance at 490 nm was measured using a spectrophotometer (Evolution 220 UV-Visible; Thermo Scientific). Total sugar content was calculated based on a standard curve of d-Glc.
ELISAs were performed as described (Pattathil et al., 2010, 2012), using 50 μL of 40 μg/mL wall extract per well. After primary and secondary antibody incubation and development, the absorbance of each well was read at 450 nm and 655 nm using a microplate reader (Synergy H1; BioTek). Net optical density values were calculated by subtracting A655 from A450, and glycome profiling data were presented using heat maps generated using the R software package.
Morphological Analysis of Flowers and Floral Meristems
Six-week-old plants were examined for floral organ phenotypes. Col and mutant plants were grown in the same flat (16 plants of each genotype) to ensure that all growth conditions were as similar as possible. All fully opened flowers were scored for petal number. The number of replicates for each genotype is listed in Table 2. χ2 tests were used to calculate the significance of differences between the proportion of normal and abnormal flowers in wild-type and mutant lines. For scanning electron microscopy, inflorescence meristems were dissected under a Discovery V12 fluorescence dissecting microscope (Zeiss) and fixed in fixative solution (2.5% glutaraldehyde in 0.1 M sodium cacodylate) overnight at 4°C. Tissue samples were washed in 0.1 M sodium cacodylate buffer for 3 × 10 min, then dehydrated with an acetone series. Samples were critical-point dried (CPD-030), mounted onto aluminum stubs with carbon tape, and viewed by scanning electron microscopy (ESEM FEI; Quanta 200) using 15-kV accelerating voltage under 80-Pa pressure in low vacuum conditions. Angles of primordium pairs were measured in ImageJ.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative (AGI) or GenBank/EMBL databases under the following accession numbers: PGX1 corresponds to AGI number At3g26610, and ACT2 corresponds to AGI number At3g18780.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Experimental Flow Chart for Activation Tag Screening for Genes Involved in Cell Expansion.
Supplemental Figure 2. Phylogenetic Analysis of Arabidopsis Putative PGs.
Supplemental Figure 3. Cell Number in Hypocotyls of 6-d-Old Etiolated Seedlings.
Supplemental Figure 4. Quantification of PGX1 mRNA Expression in Adult Tissues of Col and PGX1AT Plants.
Supplemental Figure 5. PGX1AT and PGX1OE Rosettes Are Larger Than Wild-Type Rosettes.
Supplemental Figure 6. Forty-Five-Day-Old Plants of Col, pgx1-1, pgx1-2, PGX1AT, PGX1OE-1, and PGX1OE-48 Grown in Long-Day Conditions.
Supplemental Figure 7. Hypocotyls of 6-d-Old Etiolated Seedlings of PGX1pro:PGX1-GFP Complementation Lines Are Comparable in Length to Wild-Type Hypocotyls.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Alignment Used to Generate the Phylogeny Presented in Supplemental Figure 2.
Supplementary Material
Acknowledgments
We thank Wolf-Rudiger Scheible for generating the activation tag lines used in this work, Liza Wilson and Sarah Kiemle for technical assistance with monosaccharide analysis and size-exclusion chromatography, and Wenting Xi for technical assistance with activation tag screening. We also thank Joseph Hill Jr. and Juan Du for assistance with immunoblotting and for providing bacterial expression vectors and Daniel Cosgrove for a critical reading of the article. Activation tag screening and generation of overexpression constructs was supported by the Energy Biosciences Institute; all other work was supported as part of The Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, under Award DE-SC0001090.
AUTHOR CONTRIBUTIONS
C.X., C.S., and C.T.A. designed the research. C.X. and C.T.A. performed research. C.X. and C.T.A. analyzed data. C.T.A. and C.X. wrote the article.
Glossary
- HG
homogalacturonan
- PG
polygalacturonase
- Col
Columbia
- MS
Murashige and Skoog
- qPCR
quantitative real-time RT-PCR
- MSO
methionine sulfoximine
- CDTA
1,2-cyclohexylenedinitrilo-tetraacetic acid
- AIR
alcohol-insoluble residue
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abasolo W., Eder M., Yamauchi K., Obel N., Reinecke A., Neumetzler L., Dunlop J.W., Mouille G., Pauly M., Höfte H., Burgert I. (2009). Pectin may hinder the unfolding of xyloglucan chains during cell deformation: Implications of the mechanical performance of Arabidopsis hypocotyls with pectin alterations. Mol. Plant 2: 990–999. [DOI] [PubMed] [Google Scholar]
- Alexandersson E., Becker J.V., Jacobson D., Nguema-Ona E., Steyn C., Denby K.J., Vivier M.A. (2011). Constitutive expression of a grapevine polygalacturonase-inhibiting protein affects gene expression and cell wall properties in uninfected tobacco. BMC Res. Notes 4: 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson C.T., Carroll A., Akhmetova L., Somerville C. (2010). Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson R.G., Schröder R., Hallett I.C., Cohen D., MacRae E.A. (2002). Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 129: 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson R.G., Sutherland P.W., Johnston S.L., Gunaseelan K., Hallett I.C., Mitra D., Brummell D.A., Schröder R., Johnston J.W., Schaffer R.J. (2012). Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol. 12: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo M.A., Hao Z., Mohnen D. (2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64: 747–779. [DOI] [PubMed] [Google Scholar]
- Biswal A.K., Soeno K., Gandla M.L., Immerzeel P., Pattathil S., Lucenius J., Serimaa R., Hahn M.G., Moritz T., Jönsson L.J., Israelsson-Nordström M., Mellerowicz E.J. (2014). Aspen pectate lyase PtxtPL1-27 mobilizes matrix polysaccharides from woody tissues and improves saccharification yield. Biotechnol. Biofuels 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenkrantz N., Asboe-Hansen G. (1973). New method for quantitative determination of uronic acids. Anal. Biochem. 54: 484–489. [DOI] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Bugbee W.M. (1993). A pectin lyase inhibitor protein from cell walls of sugar beet. Phytopathology 83: 63–68. [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 37: D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P., Findlay K., McCann M.C., Roberts K. (2007b). Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. J. Exp. Bot. 58: 2079–2089. [DOI] [PubMed] [Google Scholar]
- Derbyshire P., McCann M.C., Roberts K. (2007a). Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biol. 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick-Pérez M., Zhang Y., Hayes J., Salazar A., Zabotina O.A., Hong M. (2011). Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50: 989–1000. [DOI] [PubMed] [Google Scholar]
- Dubois M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28: 350–356. [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Ferrari S., Sella L., Janni M., De Lorenzo G., Favaron F., D'Ovidio R. (2012). Transgenic expression of polygalacturonase-inhibiting proteins in Arabidopsis and wheat increases resistance to the flower pathogen Fusarium graminearum. Plant Biol. (Stuttg.) 14: 31–38. [DOI] [PubMed] [Google Scholar]
- Ferrari S., Vairo D., Ausubel F.M., Cervone F., De Lorenzo G. (2003). Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15: 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, S.C. (1988). The Growing Plant Cell Wall: Chemical and Metabolic Analysis. (Harlow, Essex, UK: Burnt Mill; New York: Longman Scientific & Technical; J. Wiley). [Google Scholar]
- González-Carranza Z.H., Elliott K.A., Roberts J.A. (2007). Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 58: 3719–3730. [DOI] [PubMed] [Google Scholar]
- Gross K.C. (1982). A rapid and sensitive spectrophotometric method for assaying polygalacturonase using 2-cyanoacetamide. HortScience 17: 933–934. [Google Scholar]
- Hamant O., Traas J., Boudaoud A. (2010). Regulation of shape and patterning in plant development. Curr. Opin. Genet. Dev. 20: 454–459. [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Verboom R.E., Tonti-Filippini J., Small I., Millar A.H. (2007). SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res. 35: D213–D218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolie R.P., Duvetter T., Van Loey A.M., Hendrickx M.E. (2010). Pectin methylesterase and its proteinaceous inhibitor: a review. Carbohydr. Res. 345: 2583–2595. [DOI] [PubMed] [Google Scholar]
- Jordan D.B., Bowman M.J., Braker J.D., Dien B.S., Hector R.E., Lee C.C., Mertens J.A., Wagschal K. (2012). Plant cell walls to ethanol. Biochem. J. 442: 241–252. [DOI] [PubMed] [Google Scholar]
- Keegstra K. (2010). Plant cell walls. Plant Physiol. 154: 483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Patterson S.E. (2006). Expression divergence and functional redundancy of polygalacturonases in floral organ abscission. Plant Signal. Behav. 1: 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Shiu S.H., Thoma S., Li W.H., Patterson S.E. (2006). Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 7: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Ma Y., Chen N., Guo S., Liu H., Guo X., Chong K., Xu Y. (2013). Overexpression of stress-inducible OsBURP16, the beta-subunit of polygalacturonase 1, decreases pectin contents and cell adhesion, and increases abiotic stress sensitivity in rice. Plant Cell Environ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q. (2005). Seamless cloning and gene fusion. Trends Biotechnol. 23: 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S.E., et al. (2010). Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J. 64: 191–203. [DOI] [PubMed] [Google Scholar]
- Marcus S.E., Verhertbruggen Y., Hervé C., Ordaz-Ortiz J.J., Farkas V., Pedersen H.L., Willats W.G., Knox J.P. (2008). Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Rodríguez M.C., Orchard J., Seymour G.B. (2002). Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 53: 2115–2119. [DOI] [PubMed] [Google Scholar]
- Maughan S.C., Cobbett C.S. (2003). Methionine sulfoximine, an alternative selection for the bar marker in plants. J. Biotechnol. 102: 125–128. [DOI] [PubMed] [Google Scholar]
- Maulik A., Sarkar A.I., Devi S., Basu S. (2012). Polygalacturonase-inhibiting proteins: Leucine-rich repeat proteins in plant defence. Plant Biol. (Stuttg.) 14: 22–30. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S.J., Cosgrove D.J. (1995). Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 107: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F. (2001). Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6: 414–419. [DOI] [PubMed] [Google Scholar]
- Monfared M.M., Carles C.C., Rossignol P., Pires H.R., Fletcher J.C. (2013). The ULT1 and ULT2 trxG genes play overlapping roles in Arabidopsis development and gene regulation. Mol. Plant 6: 1564–1579. [DOI] [PubMed] [Google Scholar]
- Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q. (2008). GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9 (suppl. 1): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Ruprecht C., Giorgi F.M., Bringmann M., Usadel B., Persson S. (2009). Transcriptional wiring of cell wall-related genes in Arabidopsis. Mol. Plant 2: 1015–1024. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Kay P., Wilson S., Swain S.M. (2009). ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21: 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R.C., Alonso J.M., Kim C.J., Leisse T.J., Ecker J.R. (2007). An adapter ligation-mediated PCR method for high-throughput mapping of T-DNA inserts in the Arabidopsis genome. Nat. Protoc. 2: 2910–2917. [DOI] [PubMed] [Google Scholar]
- Palusa S.G., Golovkin M., Shin S.B., Richardson D.N., Reddy A.S. (2007). Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol. 174: 537–550. [DOI] [PubMed] [Google Scholar]
- Park Y.B., Cosgrove D.J. (2012). A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 158: 1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S., et al. (2010). A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153: 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S., Avci U., Miller J.S., Hahn M.G. (2012). Immunological approaches to plant cell wall and biomass characterization: Glycome profiling. Methods Mol. Biol. 908: 61–72. [DOI] [PubMed] [Google Scholar]
- Patterson S.E. (2001). Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 126: 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A., Braybrook S., Höfte H. (2012). Cell wall mechanics and growth control in plants: The role of pectins revisited. Front Plant Sci 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A., Braybrook S.A., Le Guillou L., Bron E., Kuhlemeier C., Höfte H. (2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21: 1720–1726. [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Louvet R., Johansen J.N., Höfte H., Laufs P., Pelloux J., Mouille G. (2008). Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr. Biol. 18: 1943–1948. [DOI] [PubMed] [Google Scholar]
- Peretto R., Favaron F., Bettini V., De Lorenzo G., Marini S., Alghisi P., Cervone F., Bonfante P. (1992). Expression and localization of polygalacturonase during the outgrowth of lateral roots in Allium porrum L. Planta 188: 164–172. [DOI] [PubMed] [Google Scholar]
- Peroutka III, R.J., Orcutt, S.J., Strickler, J.E., and Butt, T.R. (2011). SUMO fusion technology for enhanced protein expression and purification in prokaryotes and eukaryotes. In Heterologous Gene Expression in E. coli, J.T.C. Evans and M.-Q. Xu, eds (New York: Humana Press), pp. 15–30. [DOI] [PubMed] [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C.R. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé S., Paniagua C., Cifuentes M., Blanco-Portales R., Quesada M.A., Mercado J.A. (2013). Insights into the effects of polygalacturonase FaPG1 gene silencing on pectin matrix disassembly, enhanced tissue integrity, and firmness in ripe strawberry fruits. J. Exp. Bot. 64: 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protsenko M.A., Buza N.L., Krinitsyna A.A., Bulantseva E.A., Korableva N.P. (2008). Polygalacturonase-inhibiting protein is a structural component of plant cell wall. Biochemistry (Moscow) 73: 1053–1062. [DOI] [PubMed] [Google Scholar]
- Refrégier G., Pelletier S., Jaillard D., Höfte H. (2004). Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol. 135: 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W.D., Chapple C., Somerville C.R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12: 335–345. [DOI] [PubMed] [Google Scholar]
- Rhee S.Y., Somerville C.R. (1998). Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J. 15: 79–88. [DOI] [PubMed] [Google Scholar]
- Rhee S.Y., Osborne E., Poindexter P.D., Somerville C.R. (2003). Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 133: 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H. (2004). Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211. [DOI] [PubMed] [Google Scholar]
- Sun L., van Nocker S. (2010). Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol. 10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., et al. (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25: 270–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H., Chen X., Nurmberg P., Grant J.J., SantaMaria M., Chini A., Gilroy E., Birch P.R., Loake G.J. (2004). Activation tagging in plants: A tool for gene discovery. Funct. Integr. Genomics 4: 258–266. [DOI] [PubMed] [Google Scholar]
- Tian C., Beeson W.T., Iavarone A.T., Sun J., Marletta M.A., Cate J.H., Glass N.L. (2009). Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. USA 106: 22157–22162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sandt V.S., Suslov D., Verbelen J.P., Vissenberg K. (2007). Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. (Lond.) 100: 1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken J.P., Schols H.A., Oomen R.J., McCann M.C., Ulvskov P., Voragen A.G., Visser R.G. (2003). If homogalacturonan were a side chain of rhamnogalacturonan I. Implications for cell wall architecture. Plant Physiol. 132: 1781–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Chen F., Yu X., Lin C., Fu Y.F. (2009). Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 71: 39–50. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Yuan S., Wang X., Zhang Y., Zhu H., Lu C. (2008). Restoration of mature etiolated cucumber hypocotyl cell wall susceptibility to expansin by pretreatment with fungal pectinases and EGTA in vitro. Plant Physiol. 147: 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.