This work reveals that the photoreceptor CRYPTOCHROME1 (CRY1), but not CRY2, is involved in the perception of blue light in dormant barley grains, which inhibits germination by impeding ABA decline in the embryo.
Abstract
It is well known that abscisic acid (ABA) plays a central role in the regulation of seed dormancy and that transcriptional regulation of genes encoding ABA biosynthetic and degradation enzymes is responsible for determining ABA content. However, little is known about the upstream signaling pathways impinging on transcription to ultimately regulate ABA content or how environmental signals (e.g., light and cold) might direct such expression in grains. Our previous studies indicated that light is a key environmental signal inhibiting germination in dormant grains of barley (Hordeum vulgare), wheat (Triticum aestivum), and Brachypodium distachyon and that this effect attenuates as after-ripening progresses further. We found that the blue component of the light spectrum inhibits completion of germination in barley by inducing the expression of the ABA biosynthetic gene 9-cis-epoxycarotenoid dioxygenase and dampening expression of ABA 8’-hydroxylase, thus increasing ABA content in the grain. We have now created barley transgenic lines downregulating the genes encoding the blue light receptors CRYTOCHROME (CRY1) and CRY2. Our results demonstrate that CRY1 is the key receptor perceiving and transducing the blue light signal in dormant grains.
INTRODUCTION
Completion of germination is a critical step in the plant life cycle, and its timing is strongly determined by dormancy status and environmental cues such as light, moisture, and temperature. Dormancy has an important adaptive role in optimizing seed germination to coordinate seedling establishment, plant growth, and flowering (viz. the rest of the life cycle) to occur under the most suitable conditions for the species. Compared with their wild ancestors, many modern cultivars of bread wheat (Triticum aestivum) and barley (Hordeum vulgare) have low dormancy at grain maturity due to positive selection for rapid and uniform germination during early domestication and modern breeding (Gubler et al., 2005; Meyer and Purugganan, 2013). The loss of dormancy at maturity in combination with exposure to cool wet conditions makes many wheat and barley varieties susceptible to premature germination in the spike, commonly called preharvest sprouting. The effect of environmental cues such as light quality on the genetic mechanisms regulating dormancy initiation, maintenance, and loss in cereals are largely unknown.
Numerous studies of dicot seeds have investigated the role of light, in particular red (R) and far-red (FR) wavelengths, in regulating the progression of seed germination (Seo et al., 2009). In Arabidopsis thaliana seeds, there is also strong evidence that light signaling mediated through the PHYTOCHROME (PHY) family of photoreceptors has a major effect in determining the level of seed dormancy at maturity and the germination conditions required to break dormancy (Heschel et al., 2008). Genetic dissection of the individual roles for the PHYs has shown that they have partially overlapping and diverse functions in defining germination conditions necessary to break or promote dormancy. Our understanding of the role of PHYs in regulating dormancy/germination in cereals is very limited due to a lack of mutants. A recent study of Brachypodium distachyon found that highly dormant Bd18-1 grains showed R/FR photoreversible influences on seed dormancy, implying a role for PHYB or possibly PHYC in breaking dormancy (Barrero et al., 2012). Early studies of wild oats (Avena fatua) demonstrated similar photoreversible dormancy on dehusked seeds (reviewed in Simpson, 1990). It is interesting to note that so far there have not been any reported examples of R/FR reversible effects on germination in domesticated cereals such as wheat or barley, raising the prospect that it may have been lost in domestication (Barrero et al., 2012).
In contrast with the lack of information on R/FR light regulation of cereal germination, there are a number of reports detailing a strong inhibition of wheat and barley grain germination under blue light (BL) at harvest maturity (Chaussat and Zoppolo, 1983; Gubler et al., 2008; Jacobsen et al., 2013). The BL light response appears to be restricted to the Poaceae with reports of BL inhibition of germination in Lolium rigidum and B. distachyon, but none outside the grass family (Goggin et al., 2008; Barrero et al., 2012). The BL response is closely linked to dormancy status. Freshly harvested grains do not complete germination even in darkness but partially after-ripened grains do. These partially after-ripened grains, illuminated with light containing a BL component, are inhibited from completing germination until this sensitivity starts attenuating in strength (Gubler et al., 2008; Barrero et al., 2009, 2012), becoming completely lost in fully after-ripened barley grains that complete germination equally well under BL or darkness (Gubler et al., 2008).
There are two major classes of BL photoreceptors in plants: CRYPTOCHROMES (CRYs) and PHOTOTROPINS (PHOTs) (Christie, 2007; Chaves et al., 2011). CRYs are flavoproteins that are part of the DNA photolyase superfamily and are present in plants, animals, and bacteria (Yu et al., 2010). In those plants studied to date, there are two paralogous proteins, CRY1 and CRY2, which have overlapping roles in the plant circadian clock (Imaizumi and Kay, 2006; Más and Yanovsky, 2009). They also have distinct roles; for example, CRY2 acts specifically in photoperiod regulation of flowering (Guo et al., 1998; Mockler et al., 2003), while both contribute, nonredundantly, to some aspects of photomorphogenesis (Folta and Spalding, 2001). In barley, wheat, and other monocot species, there are two CRY1 genes (CRY1a and CRY1b) and one CRY2 gene (Perrotta et al., 2001; Xu et al., 2010).
PHOTs are light-activated Ser/Thr protein kinases that regulate a range of responses that serve to optimize the photosynthetic efficiency of plants, including chloroplast movement and the size of the stomatal aperture (Briggs and Christie, 2002). In rice (Oryza sativa), PHOTs consist of a small gene family with a duplication of PHOT1 and a single PHOT2 gene (Kanegae et al., 2000; Jain et al., 2007).
There is indirect evidence linking BL perception by the CRYs to photoinhibition of germination in dormant cereal grains, but genetic evidence supporting this hypothesis is missing. It is also unknown if one or both CRY paralogs are involved in the photoinhibition of germination. In dormant barley grains, green light partly reverses the inhibitory effect of BL on germination progression, which implies a role for CRY and not PHOT in the light response (Kennis et al., 2004; Hoang et al., 2013). Green light has been shown to inhibit CRY-mediated effects such as hypocotyl elongation (Bouly et al., 2007; Sellaro et al., 2010), and the CRYs are known to repress some green light–stimulated, shade-associated transcriptome alterations underlying photomorphogenic alterations to leaves (Zhang et al., 2011; Wang and Folta, 2013). So far, there are no reports of PHOT receptors mediating a green light response (Goggin and Steadman, 2012). Further support pointing to the CRYs as being responsible for BL-mediated inhibition of germination can be demonstrated in germination assays of Arabidopsis seeds overexpressing wheat CRYs (Xu et al., 2009). Expression of Ta-CRY1a and Ta-CRY2 in Arabidopsis seeds enhanced their sensitivity to abscisic acid (ABA) and salt during germination compared with the wild type. In Arabidopsis, BL has been shown to partially repress germination, but that effect is dependent on PHYB, not on CRY1 (Poppe et al., 1998).
In order to examine the role of Hv-CRYs in the BL inhibition of germination, we used RNA interference (RNAi) to knock down CRY1a/b or CRY2 in barley grains and examined the effect on seed dormancy, gene expression, and hormone content. We report that RNAi knockdown evidence supports a role for CRY1 but not CRY2 in BL inhibition of germination in barley. In addition, our results demonstrate that CRY1 mediates BL-induced NCED1 expression. This produces an accumulation of ABA in BL-illuminated barley grains that then exhibit stronger grain dormancy.
RESULTS
Dormancy and Germination Phenotypes of Hv-CRY1a/b and Hv-CRY2 RNAi and Null Segregant Grains
To test the role of CRY1 and CRY2 in BL-mediated photo-dormancy in barley, we used an RNAi silencing approach. Given the considerable similarity (81% nucleotide identity) between CRY1a and CRY1b, a RNAi hairpin construct was made that was capable of silencing both genes, while a second RNAi construct was made that would silence Hv-CRY2 specifically (Figure 1; Supplemental Tables 1 and 2). RNAi constructs were introduced into the Golden Promise cultivar of barley using an Agrobacterium tumefaciens–based transformation protocol. Four independent T3 families, showing a decrease in CRY1a/b or CRY2 transcript abundance by quantitative real-time PCR (qRT-PCR; silencing ranging from ∼70 to 20% in different lines) in homozygous RNAi grains compared with sibling null segregants, were selected for phenotyping (Figures 2A and 2B).
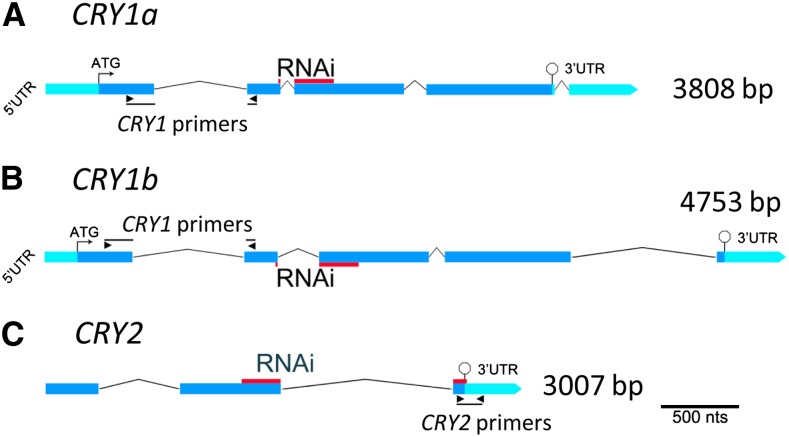
Figure 1.
Schematic Representation of the Barley CRY1a, CRY1b, and CRY2 Genes.
Barley CRY1a and CRY1b gene sequences and the CRY2 partial length gene sequence were retrieved from the National Center for Biotechnology Information and used to design RNAi cassettes (see Methods). CRY1a (A), CRY1b (B), and CRY2 (C) sequences were annotated to reveal the UTRs and coding regions.
(A) and (B) A 355-bp region (red bar) containing portions of the second and third exons of CRY1a (A) and sharing 78% identity with CRY1b (B) over the same region (red bar) was chosen for silencing CRY1a/b.
(C) For silencing CRY2, a 339-nucleotide region spanning the last intron in the coding sequence of that gene (red bar) and showing low similarity to CR1a/b was chosen. The 5′ UTR and 3′ UTRs (when known) are colored light blue, the coding regions of the genes are dark blue, and these are joined by chevrons depicting the introns. The translational start site (when known) is indicated by a bent arrow at the ATG, while the stop codon is denoted with a stop sign. The positions of the primers used for expression analysis are also indicated.
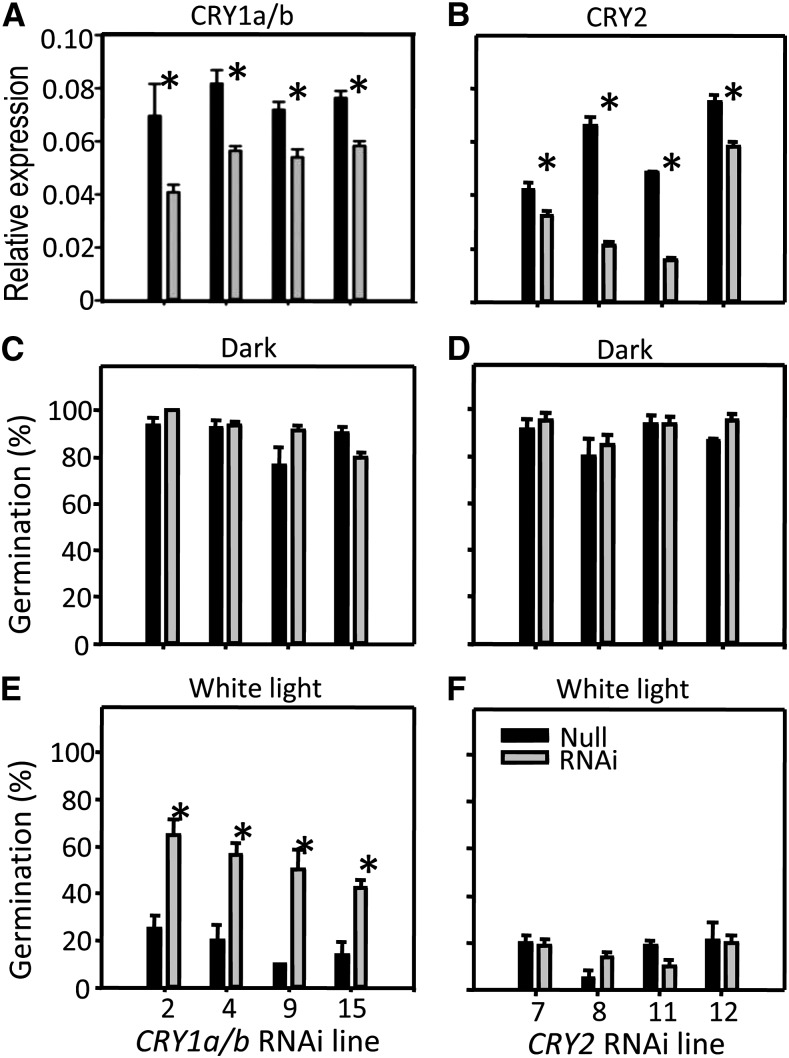
Figure 2.
Silencing Effects in CRY1a/b and CRY2 Transgenic Lines and Their Dormancy.
(A) and (B) Expression of CRY1a/b and CRY2 in barley grains imbibed 18 h in darkness from four independent, homozygous, single-insertion CRY1a/b lines (A), four independent, homozygous, single-insertion CRY2 lines (B), and from their corresponding null segregant siblings was tested by qRT-PCR, revealing a consistent expression reduction of the targeted genes.
(C) to (F) Grains from the same harvest were tested for dormancy in darkness ([C] and [D]) or under WL ([E] and [F]).
Each point is the average ± se of three biological replicates. Statistically significant differences using Dunnett’s test at α = 0.05 are indicated with an asterisk.
Grains from homozygous CRY1a/b and CRY2 RNAi plants and null segregants were harvested at maturity and tested for dormancy after 1 month of after-ripening. Regardless of genotype or line, partially after-ripened grains imbibed in darkness showed >80% germination (Figures 2C and 2D). When dormancy was tested under continuous white light (WL), all four CRY1a/b RNAi lines exhibited reduced sensitivity and reached higher germination percentages (45 to 65% germination) compared with their respective sibling null lines (8 to 25%; Figure 2E). However, all four CRY2 RNAi lines completed germination very similarly to their null segregant controls under continuous WL (∼20% germination; Figure 2F).
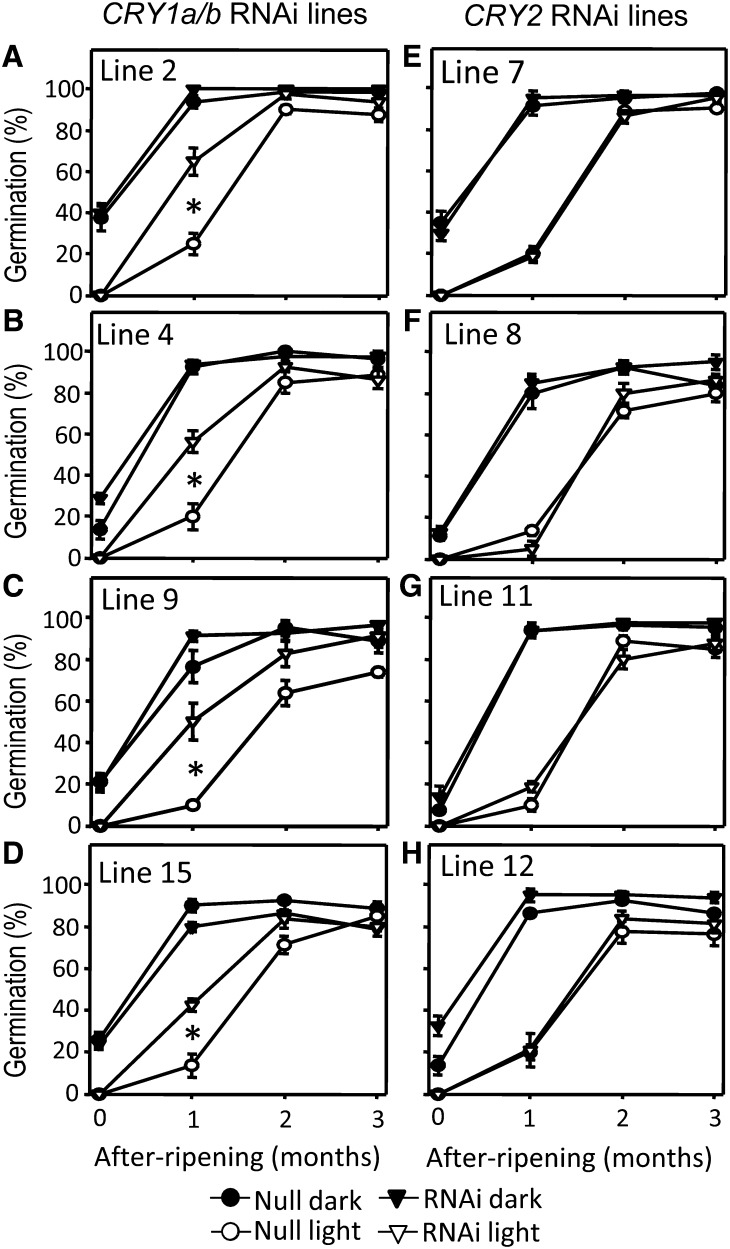
We also tested the germination of CRY1a/b or CRY2 lines during an after-ripening time course (Figure 3). All lines were highly dormant when freshly harvested (no after-ripening) with 0% germination under continuous WL and no more than 40% germination in continuous darkness (Figure 3), and no differences in final percentage germination were observed between any of the RNAi lines and their respective sibling null grains. Upon partial after-ripening (1 month) the percentage germination was altered in all CRY1a/b lines in WL, as reported in the previous paragraph (Figures 3A to 3D). After 2 months of after-ripening, dormancy was greatly reduced even under WL conditions, but small differences between CRY1a/b lines and null segregants were still detectable. After 3 months of after-ripening, dormancy was no longer evident and all lines completed germination almost equally well in darkness and in WL.
Figure 3.
After-Ripening Time Course of the RNAi CRY1a/b and CRY2 Lines.
The four lines carrying the CRY1a/b RNAi construct and the four lines carrying the CRY2 RNAi construct, together with their control null segregants, were after-ripened for various durations before being tested for germination in either continuous WL (open symbols) or darkness (filled symbols). Each point is the average ± se of final percentage germination values after 5 d from three biological replicates. Statistically significant differences between RNAi lines and null segregants using Dunnett’s test at α = 0.05 are indicated with an asterisk.
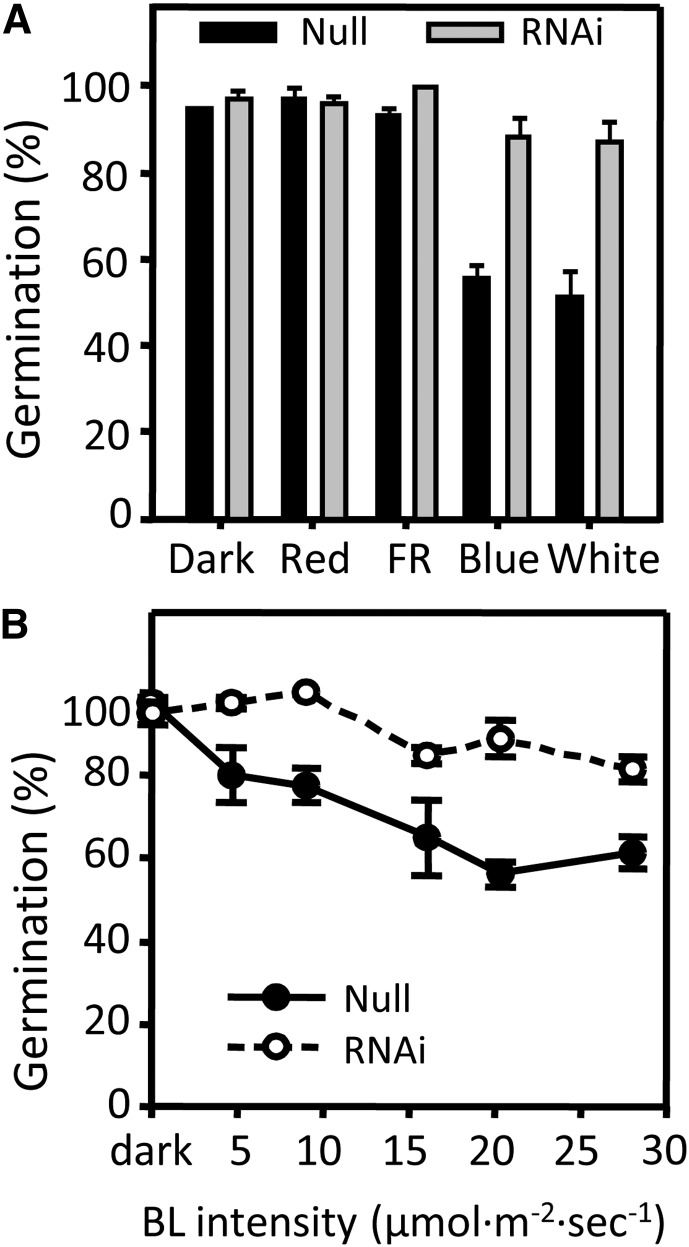
CRY1a/b RNAi line 15 was selected for further analyses to test the germination response to different light qualities. None of the partially after-ripened CRY1a/b RNAi grains, when compared with the sibling null grains, demonstrated a significant difference in germination following imbibition in darkness or under R or FR light (Figure 4A). However, when germinated under 20 μmol m−2 s−1 BL, CRY1a/b RNAi showed greater germination than did its null segregant, indicating that CRY1a/b RNAi was specifically defective in BL-mediated repression of germination (Figure 4A). Altering the BL fluence revealed that the RNAi repression of CRY1a/b caused a lessening of BL responses between 5 and 28 μmol m−2 s−1 (Figure 4B), which is the highest fluence rate technically possible with the device used. At 5 and 9 μmol m−2 s−1, the response of the CRY1a/b RNAi line was almost identical to that in darkness, indicating that the RNAi line was not capable of perceiving such low BL fluence rates, in contrast with the null segregant.
Figure 4.
CRY1a/b RNAi Lines Show a Germination Phenotype in Response to BL.
Grains of CRY1a/b RNAi line 15 and its null segregant were tested for dormancy in various light qualities following partial after-ripening at 37°C.
(A) Grains were germinated in darkness or under constant R, FR, BL, or WL.
(B) CRY1a/b RNAi line 15 and null segregant grains were placed under constant BL at various fluence rates. Percentage of germination after 5 d was recorded.
We did not observe any other phenotype in the CRY1a/b and CRY2 RNAi plants, and even the coleoptile length (Supplemental Figure 1), which is a phenotype influenced by BL in both monocots and dicots, did not show differences between RNAi and null segregants.
BL-Regulated Gene Expression in CRY1a/b RNAi Grains and ABA Analysis
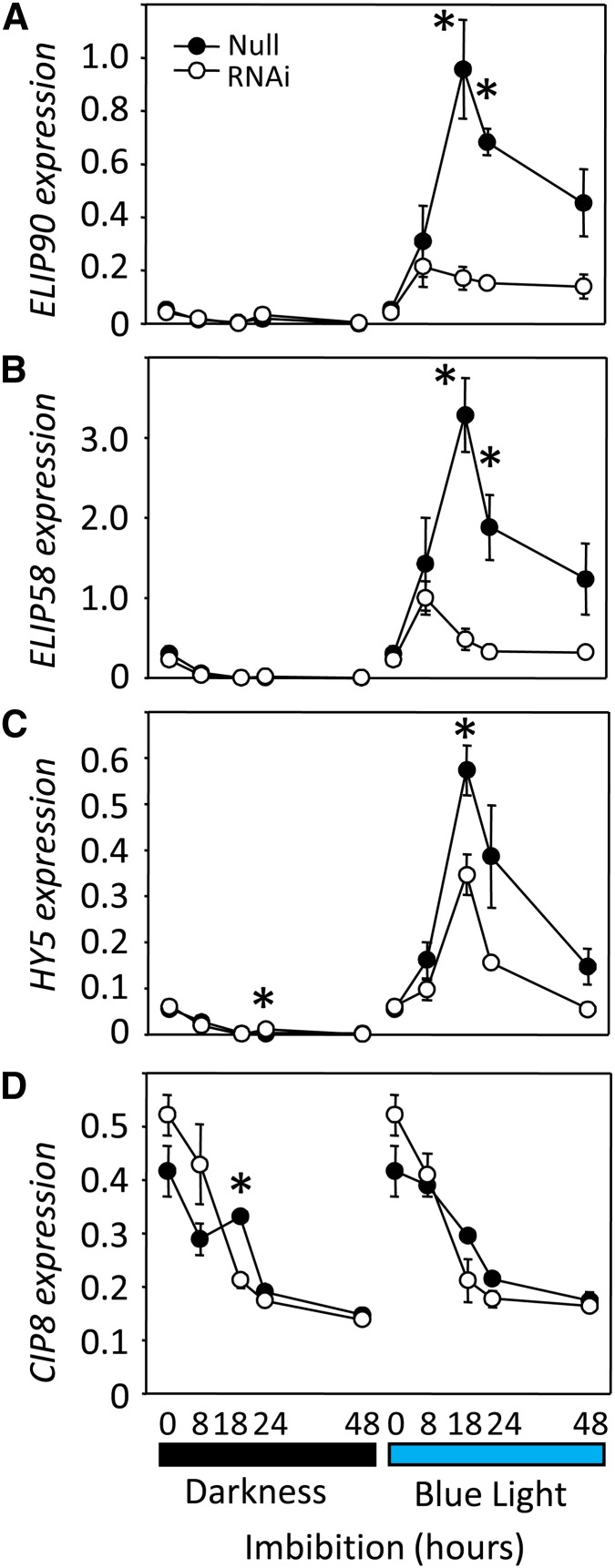
Partially after-ripened grains (19 d after-ripened; see Methods) from CRY1a/b RNAi line 15 were imbibed for various periods under continuous BL or darkness. RNA samples were purified from embryos and expression of selected genes monitored by qRT-PCR (Figure 5). To test the efficacy of the RNAi repression of CRY1 function, the expression of reported BL-inducible genes EARLY-LIGHT INDUCED PROTEIN58 (ELIP58) and ELIP90 (Adamska et al., 1992; Harari-Steinberg et al., 2001) and LONG HYPOCOTYL5 (HY5; Holm et al., 2002; Fittinghoff et al., 2006; Sellaro et al., 2009) was compared in the CRY1a/b RNAi grains and null segregant grains. In the null grains imbibed on water, ELIP90 and ELIP58 were induced rapidly in response to BL and reached a maximum at 18 h, declining thereafter (Figures 5A and 5B). By contrast, expression of ELIP90 and ELIP58 in CRY1a/b RNAi grains was almost abolished and increased only slightly in response to BL. In addition, the expression of HY5, a BL-regulated transcription factor, followed a similar pattern to the ELIP genes with lower fold induction in response to BL in the CRY1a/b RNAi grains compared with the null segregant (Figure 5C). We also investigated the expression of another gene that mediates the BL signaling pathway in Arabidopsis, COP1-interacting protein 8 (CIP8), which is involved in HY5 degradation (Hardtke et al., 2002). In agreement with earlier studies in Arabidopsis, Hv-CIP8 expression did not show any BL response, and the time course showed very little difference in expression patterns between imbibed CRY1a/b RNAi grains and null grains (Figure 5D).
Figure 5.
Relative Expression of Genes Involved in the BL Signaling Pathway in the CRY1a/b RNAi and Null Segregant Embryos.
Reverse-transcribed mRNA from embryos from partially after-ripened grains imbibed in darkness or under BL for different durations was tested using gene-specific primers for transcript abundance of ELIP90 (A), ELIP58 (B), HY5 (C), or CIP8 (D). Each point is the average ± se of relative transcript abundances from three biological replicates of 15 embryos each. Statistically significant differences between RNAi lines and null segregants using Dunnett’s two-tailed test at α = 0.05 are indicated with an asterisk.
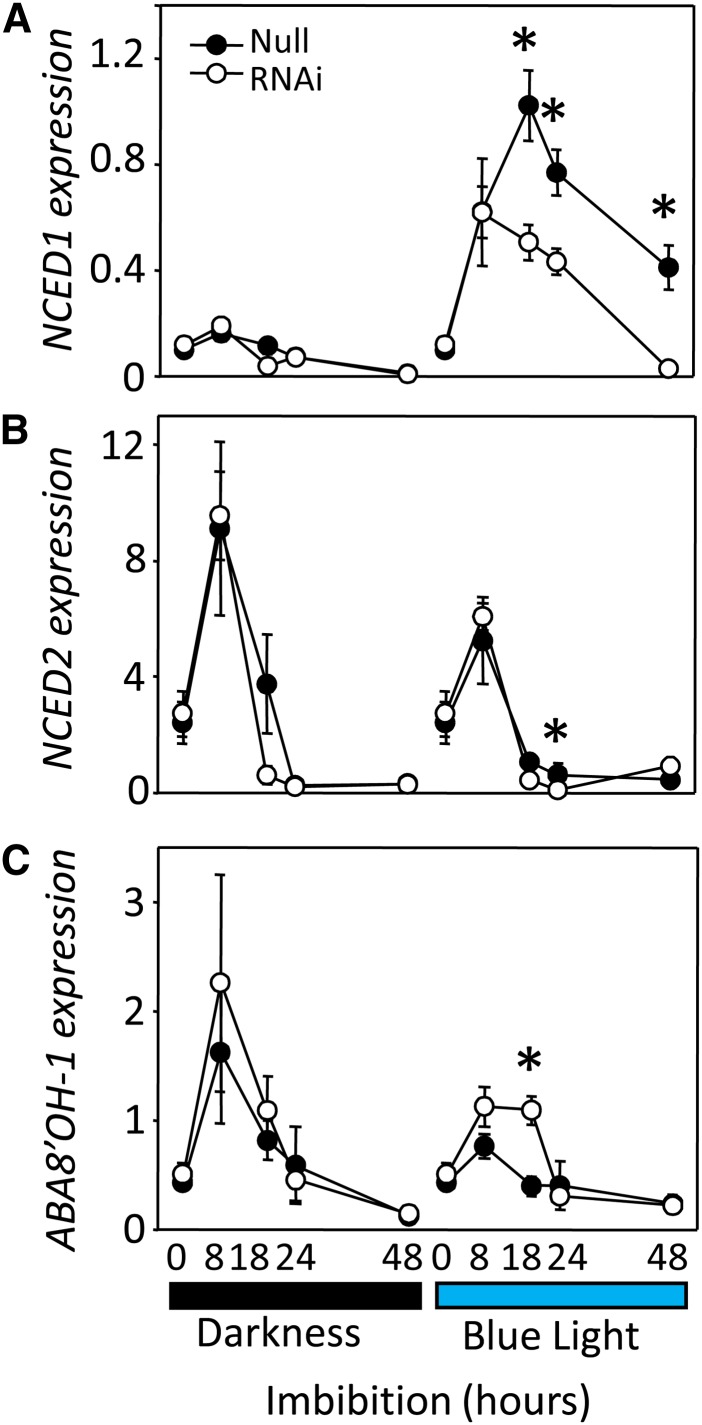
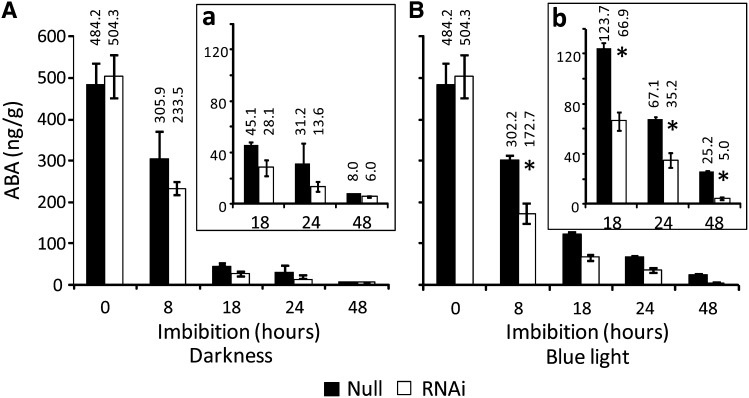
We previously demonstrated that BL inhibition of germination in barley grains is correlated with BL enhancement of NCED1 expression and ABA content in embryos (Gubler et al., 2008). To test whether the BL regulation of NCED1 is dependent on CRY1, we compared expression of NCED1 in the embryos of CRY1a/b RNAi and null grains. As shown in Figure 6A, NCED1 was strongly induced over 5-fold in null grains imbibed under BL compared with dark imbibed grains. No differences in the BL response were observed between CRY1a/b RNAi and null grains in the first 8 h of imbibition, but by 18 h of imbibition, the expression of NCED1 in the RNAi grains was <50% that in the null grains. Thereafter, the BL induction of NCED1 in the RNAi grains remained lower than in the null grains up to 48 h of imbibition. In agreement with previous results (Gubler et al., 2008), other ABA metabolic genes, such as NCED2, did not show any, or at least no sustained, BL response in the RNAi or null imbibed grain (Figure 6B). However, the ABA catabolic gene ABA8’OH-1 showed a small increase in expression in the CRY1a/b RNAi line in comparison with the null grains at 8 and 18 h after imbibition under blue light (Figure 6C). Thus, reduced CRY1a/b expression in the RNAi grains increases ABA8’OH-1 expression early after imbibition and strongly reduces NCED1 after 18 h imbibition in embryos of grains imbibed up to 48 h. In addition to gene expression analyses, we measured ABA content in embryos of RNAi and null grains imbibed under continuous BL and darkness. Reduced CRY1a/b expression did not affect embryo ABA content when the grains were imbibed in the dark (Figure 7A) with ABA content in both CRY1a/b RNAi and null grains declining at a similar rate from 500 ng·g−1 fresh weight in dry embryos to 6 to 7 ng·g−1 fresh weight after 48 h imbibition. By contrast, the decline in embryo ABA content in null grains imbibed under BL was lower, with over 2-fold greater ABA content compared with dark-imbibed grains between 8 and 48 h (Figure 7B). As deduced from the gene expression changes, the BL-mediated, greater embryo ABA content observed in the null embryos at the end of 48-h imbibition was not observed in embryos from the CRY1a/b RNAi grains, indicating that CRY1 is required for BL-mediated promotion of ABA content in dormant barley embryos.
Figure 6.
Relative Expression of Genes Involved in ABA Metabolism in the CRY1a/b RNAi and Null Segregant Embryos.
Reverse-transcribed mRNA from embryos from partially after-ripened grains imbibed in darkness or under BL for different durations was tested using gene-specific primers for transcript abundance of NCED1 (A), NCED2 (B), and ABA8’OH1 (C). Each point is the average ± se of relative transcript abundances from three biological replicates of 15 embryos each. Statistically significant differences between RNAi lines and null segregants using Dunnett’s two-tailed test at α = 0.05 are indicated with an asterisk.
Figure 7.
ABA Quantification on Embryos from the HvCRY1a/b RNAi Grains and Null Segregant Grains.
ABA was extracted from embryos from dry and imbibed partially after-ripened grains. Imbibition occurred in darkness (A) or under BL (B). ABA was quantified using liquid chromatography–tandem mass spectrometry. Numbers above columns indicate the average ABA concentration for each sample. Subpanels ([a] and [b]) are shown to magnify the latest time points with a rescaled axis. Each point is the average ± se of hormone amounts from four biological replicates. Statistically significant differences between RNAi lines and null segregants using Dunnett’s test at α = 0.05 are indicated with an asterisk.
DISCUSSION
Light quality plays a key role in regulating dormancy and germination in seeds. R and FR reversibility of germination has been demonstrated in many plant species and appears to be a widely conserved mechanism acting through the PHYB photoreceptor. Recent genetic analysis has revealed that PHY photoreceptors are also implicated in defining conditions to break or maintain dormancy (Heschel et al., 2008). Our work and that of others have previously demonstrated that BL is also a key wavelength in regulating germination of freshly harvested seeds of the grass family. To date, grains of barley, wheat, B. distachyon, and L. rigidum have been shown to exhibit BL-mediated inhibition of germination (Chaussat and Zoppolo, 1983; Goggin et al., 2008; Gubler et al., 2008; Barrero et al., 2012; Jacobsen et al., 2013). The BL response is linked to grain dormancy. Initially, and if the grains are highly dormant, no BL effect can be measured because grains will not germinate in darkness or under BL. As the grains after-ripen, the BL response is made manifest as the difference in percentage germination between dark-imbibed and light-imbibed grains increases. Subsequently, as after-ripening progresses, the BL inhibition of germination attenuates until the grains are no longer responsive and can be considered nondormant. This work provides genetic evidence that the BL response in barley grains is mediated by the CRY1 photoreceptor. RNAi knockdown of the CRY1a/b mRNA led to reduced BL inhibition of germination of barley grains, while knockdown of CRY2 mRNA had no effect. These results are in agreement with earlier evidence based on green light inhibition of the BL response in barley, which implies a role for CRY in light inhibition of germination (Hoang et al., 2013). This work demonstrates that Hv-CRY1 is a repressor of seed germination, and it is interesting to note that BL repression of spore germination has been observed in the fern Adiantum capillus-veneris (Sugai and Furuya, 1985), indicating that the role of CRY1 in repressing germination may have arisen early in the evolution of plants. There are also reports of BL increasing the production of ABA in the fungus Botrytis cinerea (Marumo et al., 1982) and that would indicate that the basic light mechanism by which germination is inhibited existed even earlier. The phenotype displayed by the Hv-CRY1a/b RNAi lines can be seen only in partially after-ripened grains, but in neither freshly harvested, nor in nondormant grains. This observation indicates that the role of Hv-CRY1 is revealed during after-ripening. Other photoreceptors might transduce the light inhibition of germination initially when grains are very dormant, but their function seems to dissipate with short after-ripening. The initial regulation of dormancy and germination could involve other photoreceptors, such as PHYB, the influence of which may be short lived in dry grains during after-ripening. This suggestion is supported by previous results in B. distachyon where two light pathways regulate dormancy and germination, one short-lasting and involving R/FR light and the other long-lasting and involving BL, have been described (Figure 8; Barrero et al., 2012). Our results in barley indicate the long-lasting BL inhibition of germination must be driven by CRY1, which attenuates to nothing during 2 to 3 months of after-ripening. In Arabidopsis, considered a photoblastic species, BL can partially inhibit seed germination but this effect is dependent on PHYB (Poppe et al., 1998). To date, there are no reports linking CRY1 with the maintenance of dormancy status of Arabidopsis seeds. So, while similar to B. distachyon in using multiple light qualities to regulate seed germination, including BL, Arabidopsis apparently does not use the same photoreceptor to do this.
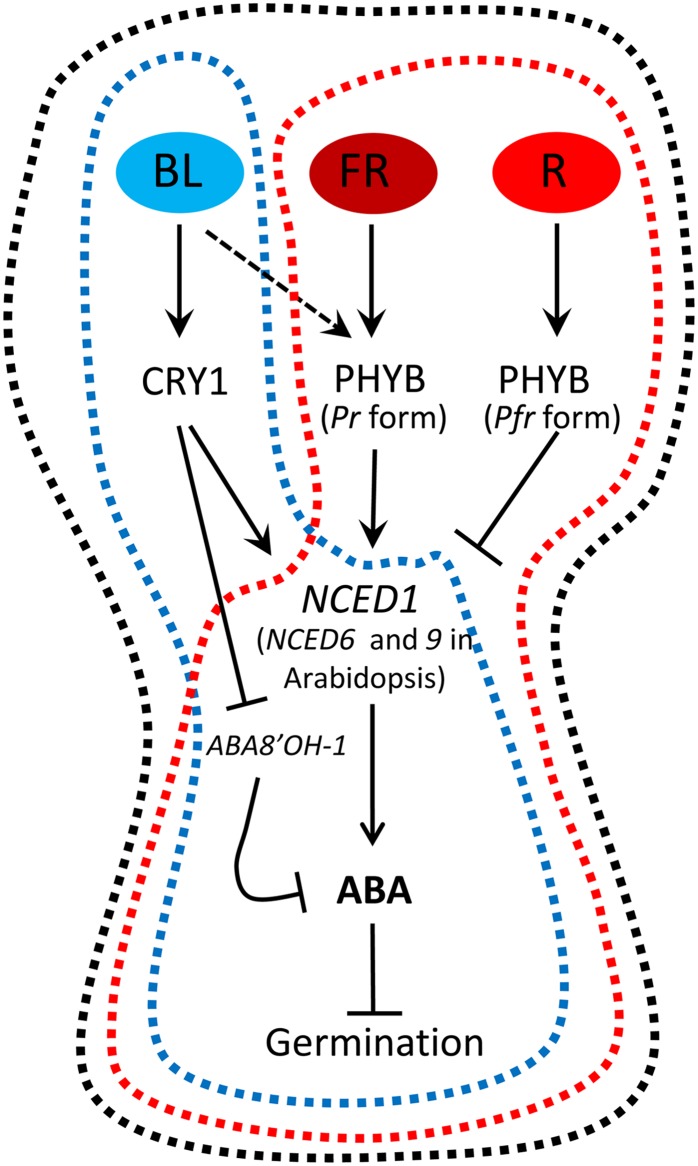
Figure 8.
A Model of the Light Regulation of Germination in the Grasses and Arabidopsis.
In grasses BL, FR, and R wavelengths are able to regulate germination in dormant grains. In Arabidopsis, R and FR wavelengths have a key role in regulating germination, and a PHYB-mediated BL effect has been also detected, although no relationship with seed dormancy has been reported. Black-dotted line: In wild grasses such as B. distachyon, strong effects of R (promoting germination) and of FR and BL (inhibiting germination) are detectable in dormant grains. Blue-dotted line: In domesticated grasses such as wheat and barley, the existence of R and FR effects has not been reported, while the BL inhibitory effect is conserved. Red-dotted line: In Arabidopsis and other dicots, R and FR had a reversible effect on seed germination driven by changes in the PHYB conformation (Pfr and Pr forms), and BL can partially mimic the FR effect. In cereal grains, BL is perceived by CRY1, and possibly also by other photoreceptors such as PHYB, inducing the expression of the NCED1 gene, while repressing ABA8’OH-1, which stabilizes the ABA content in the embryo, blocking germination. FR and R wavelengths are perceived by PHYB and by other PHY family members. In grasses, the targets of R and FR are not known, but in Arabidopsis, FR induces the expression of NCED6 and NCED9, while R blocks their expression, increasing or decreasing ABA levels, respectively.
In dicots, CRY1 regulates many aspects of plant development, including hypocotyl elongation, leaflet expansion, circadian clock entrainment, root elongation, and anthocyanin production (Li and Yang, 2007). In the case of monocots, the role of CRY1 is poorly defined due to the limited number of functional analyses of monocot CRY genes. This is due in part to the duplication of CRY1 genes in monocots presumably making it difficult to identify mutants in genetic screens (Perrotta et al., 2001; Szucs et al., 2006; Zhang et al., 2006; Xu et al., 2010). Coleoptile elongation in rice, maize (Zea mays), oats, and now barley (Supplemental Figure 1) is known to be inhibited by BL, and there is genetic evidence that BL inhibition is mediated by both CRY1 and CRY2. Overexpression of Os-CRY1b in rice results in shorter coleoptiles in a BL-dependant manner (Zhang et al., 2006). Similarly, Ta-CRY1a and Ta-CRY2 overexpression in Arabidopsis seedlings leads to a shorter hypocotyl phenotype in response to BL (Xu et al., 2009). Since our study failed to detect any clear coleoptile phenotype in the Hv-CRY1 and Hv-CRY2 RNAi lines (Supplemental Figure 1), it appears likely that both Hv-CRY1 and Hv-CRY2 function redundantly in regulating coleoptile elongation. Apart from the hypocotyl phenotype, it was found that overexpression of Ta-CRY genes in Arabidopsis seeds resulted in altered stress and ABA responses (Xu et al., 2009). Arabidopsis seeds overexpressing Ta-CRY1a and Ta-CRY2 showed a normal germination phenotype compared with the wild type when imbibed in water but when imbibed in the presence of ABA exhibited lower germination percentages. It is not clear if the altered response to ABA is dependent on BL illumination as the seeds were imbibed in 12-h-WL\12-h-dark regime. In the same study, based on semiquantitative analysis of the expression pattern in different tissues, it is proposed that Ta-CRY2 may have a more important role during germination than Ta-CRY1a, but our results in barley demonstrate that it is actually the Hv-CRY1 paralogs that are the principal BL receptors regulating germination in cereal grains. One possibility for this discrepancy is that the semiquantitative RT-PCR used in the wheat study does not reflect the real CRY protein levels of the grain embryo, but we cannot rule out the possibility that these two classes of CRY photoreceptor have different roles in wheat and barley. In any case, the mechanism linking CRYs and stress responses could be the same as that regulating the BL inhibition of germination of dormant grains.
ELIPs are nucleus-encoded thylakoid membrane proteins that are rapidly induced in response to light and have been associated with protection of the photosynthetic apparatus in green tissues and with the development of photosynthetic units in the first hours of the greening process. The use of photoreceptor mutants in Arabidopsis has shown that ELIP1 and ELIP2 are transcriptionally induced by R, FR, and blue light (Harari-Steinberg et al., 2001). In other species like pea (Pisum sativum), ELIP expression is specifically regulated by BL (Adamska et al., 1992). ELIPs are also involved in stress mechanisms and are produced in response to desiccation in fern spores (Raghavan and Kamalay, 1993) and in the resurrection plants Craterostigma plantagineum and Tortula ruralis (Bartels et al., 1992; Zeng et al., 2002). In those plants, ELIP expression is induced by ABA but only in the presence of light. In barley, two ELIPs have been characterized, ELIP58 and ELIP90 (Grimm and Kloppstech, 1987), and both of them are strongly expressed in dormant (but not in after-ripened) grains in response to light (Barrero et al., 2009). We demonstrated that Hv-CRY1 is the major BL receptor mediating the transcriptional upregulation of these ELIP genes. In barley, the BL induction of ELIP90 and ELIP58 is almost abolished in the CRY1a/b RNAi grains. Despite the strong connection between ELIP expression and dormancy status or with stress and ABA, no demonstration of a direct involvement of ELIPs in seed dormancy has been reported, although in Arabidopsis their expression seems to be positively correlated with germination under conditions of abiotic stress (Rizza et al., 2011).
On the other hand, there has been a number of reports of interactions between light and hormone signaling in seeds, in particular highlighting PHY regulation of endogenous levels of gibberellins (GAs) and ABA in germinating Arabidopsis and lettuce (Lactuca sativa) seeds (Seo et al., 2009). In Arabidopsis seeds, the decrease in ABA content during germination can be modulated by R and FR treatments initially (Seo et al., 2006; Oh et al., 2007). By 24 h imbibition, FR-treated seeds retain greater ABA contents than FR/R-treated seeds. Expression of genes encoding ABA biosynthetic enzymes such as NCED6 and NCED9 is strongly promoted by FR and inhibited by R in imbibed Arabidopsis seeds. By contrast, GA content is enhanced by R light through the activation of a number of genes encoding biosynthetic enzymes (At-GA3ox1 and At-GA3ox2) and repression of at least one gene (At-GA2ox2) whose product catabolizes bioactive GA. Our results indicate that Hv-CRY1 inhibition of germination through BL is a striking parallel in this respect to the action of PHYB when illuminated by FR light in imbibed Arabidopsis seeds (Figure 8). It has been previously shown that NCED1 is upregulated by BL in dormant barley grains and that the increased expression is associated with a greater ABA content compared with dark-imbibed grains after 24 h of imbibition (Figure 8). Our results show that the BL induction of Hv-NCED1 and promotion of ABA content is mediated at least in part by Hv-CRY1. We have also found that BL, sensed through Hv-CRY1, can repress the expression of Hv-ABA8’OH-1 during early imbibition, which can be important for explaining changes in ABA content early during imbibition. It is interesting to note that NCED1 expression and the expression of HY5 were downregulated in illuminated RNAi grains but not as much as other genes responsive to BL, such as the ELIPs. This may indicate that HY5 and NCED1 expression may be under the partial regulation of other BL photoreceptors such as PHOTs. We also found previously that Hv-ELIP expression is strongly affected by after-ripening, while the expression of Hv-NCED1 is not (Barrero et al., 2009), which again indicates that the light-regulated pathway controlling the expression of ELIPs and NCED1 must have common but also different elements.
In summary, we identified the key light receptor involved in the inhibition of germination by BL in partially after-ripened grains of barley. We demonstrated that in partially after-ripened barley grains exposed to BL, CRY1 plays a major role in the induction of NCED1 and also in the repression of ABA8’OH-1, the genes encoding the key ABA biosynthetic and catabolic enzymes. Those changes are correlated with the stabilization of the ABA content in the embryo, thus preventing the completion of germination (Figure 8). We have shown using a transgenic approach that the level of expression of CRY1a/b can affect the response to BL in partially after-ripened grains, modifying the percentage germination. However, the molecular basis of the loss of BL sensitivity during after-ripening remains unknown. Natural variability in the levels of expression or stability of the CRY1 photoreceptor may contribute to the differences seen in grain dormancy and in the germination behavior displayed by different barley or wheat varieties. The manipulation of CRY1 gene expression or CRY1 light sensitivity, stability, or signaling potency in grains could be the first steps in molecular re-engineering centered around light-based strategies to modify the germination responses in cereals prone to preharvest sprouting.
METHODS
RNAi Cassette Construction, Barley Transformation, and Grain Production
Primers based on published barley (Hordeum vulgare) CRY sequences (Szucs et al., 2006; Matsumoto et al., 2011) were used to amplify cDNA regions from cv Golden Promise (Supplemental Table 1). The CRY1a/b hairpin construct was designed to target a 355-bp (Supplemental Table 2) cDNA region of CRY1a that was 78% identical to the same region in CRY1b, but had low identity (54%) with the equivalent region in CRY2. A separate construct of 339 bp (Supplemental Table 2) was used to target the 3′ end of the CRY2 coding region, extending 12 nucleotides beyond the stop codon into the 3′ untranslated region (UTR). The CRY2 target sequence selected for silencing had low identity with the CRY1a (48%) and CRY1b (42%; alignments not shown). The 3′ UTR of CRY2 was truncated in the gene (accession number DQ201156), necessitating the use of the 3′ UTR sequence from the cDNA from cultivar Haruna Nijo (accession number AK353828) for primer design. Hairpin RNAi constructs targeting CRY1a/b or CRY2 were made by inserting the amplicons spanning the regions reported above in both orientations (using the BamHI and SmaI sites for one orientation and the KpnI and SpeI for the other) into the hairpin RNAi vector pStarling (Gubler et al., 2008). The hairpin RNAi constructs were subcloned into the NotI site of the binary vector pWBVec8 (Wang et al., 1998) before being transferred into Agrobacterium tumefaciens strain AGL0 by triparental mating. Transformation of Golden Promise barley was performed using the Agrobacterium-mediated technique as described by Jacobsen et al. (2006). The selected plants were grown in naturally lit phytotron glasshouses with air temperature set at 17°C/9°C day/night cycle as previously described (Jacobsen et al., 2002; Millar et al., 2006).
T3 grains from homozygous T2 plant populations from four independently transformed CRY1a/b RNAi and four CRY2 RNAi barley (cv Golden Promise) lines, along with their corresponding null segregant lines (which were derived from the same T0 line) were harvested in August, 2011 (Harvest 1). Heads were harvested at maturity, dried for 7 d, and threshed by hand to prevent damage to the husk and embryo. Sufficient quantities of the threshed grains were stored at −20°C to preserve dormancy, and still others were incubated at 37°C for various periods to promote after-ripening. At intervals, seeds were removed from 37°C and aliquots tested for the completion of germination in darkness or WL while the remaining seeds were placed back at 37°C to continue dry after-ripening. Line CRY1a/b RNAi 15 was grown again to obtain greater number of grains and was harvested in February, 2012 (Harvest 2). Grains from Harvest 2 had a lower level of dormancy than Harvest 1 and had to be after-ripened for only 19 d to reduce any dark dormancy to achieve a maximum response to light, when contrasted with the high germination percentages in darkness. At 19 d of after-ripening, line CRY1a/b 15 showed 55% germination under BL, while its null segregant showed 87% germination. In darkness, the germination of these lines was 95 and 100%, respectively.
Germination Assays
Four replications of 20 seeds each (from Harvest 1) were placed on Whatman filter paper (No. 598) with 5 mL of distilled water in a Petri dish to assess germination. Plates were sealed with Parafilm and wrapped in two layers of aluminum foil for germination in darkness or exposed to continuous WL (130 μmol m−2 s−1; Philips TLD 36W/865 fluorescent tubes) or to 11 μmol m−2 s−1 R, 63 μmol m−2 s−1 FR, or 20 μmol m−2 s−1 BL emitting diodes in custom-built chambers (Gubler et al., 2008). Additionally, a BL dose–response curve was prepared for line 15 using BL fluence rates from 0 (darkness) to 28 μmol m−2 s−1. The number of seeds from which the coleorhiza had protruded beyond the husk (germination completed) was assessed each day for 5 d. For those seeds germinating under illumination conditions preventing assessment of coleorhiza protrusion, a green safe light was employed (Barrero et al., 2012).
qRT-PCR Analysis of Gene Expression
Embryos from partially after-ripened grains of four different RNAi lines of CRY1a/b, CRY2, and null segregant controls (from Harvest 1) were collected after 18 h in darkness or under 130 μmol m−2 s−1 WL. Embryos from partially after-ripened grains (from Harvest 2) were collected at various time points following incubation in darkness or under 10 μmol m−2 s−1 BL. For RNA extraction, frozen embryos were pulverized in liquid nitrogen using a mortar and pestle, and the total RNA contained within them was extracted (Chang et al., 1993). The quality and quantity of total RNA were assessed spectrophotometrically before an aliquot was treated with DNase I using a kit (Qiagen) and the quantity of RNA remaining reassessed using a spectrophotometer (NanoDrop Products). Equal amounts of DNA-free, total RNA was reverse transcribed (SuperScript III; Invitrogen), and qRT-PCR reactions utilizing Platinum TAQ (Invitrogen) were performed on a Corbett Rotor-Gene 6000 qRT-PCR machine (Qiagen) or a 7900HT Fast Real-Time PCR system (Applied Biosystems). Results were imported into Excel, and the expression of Hv-Actin (AY145451; Trevaskis et al., 2006) was used as an internal control to normalize gene expression. The sequences of primers used for gene expression analyses are listed in Supplemental Table 3. For CRY1a/b, primers were designed that could amplify both Hv-CRY1a and Hv-CRY1b genes. Primers for detecting the expression of Hv-CRY1a/b and Hv-CRY2 were positioned outside the fragment chosen for making the hairpin construct.
ABA Analysis
Samples of 15 embryos were dissected from dry or imbibed grains (from Harvest 2) using a dissection scalpel under a microscope and were frozen in dry ice immediately. ABA analysis was performed using the liquid chromatography–mass spectrometry method described by Dave et al. (2011). Embryo samples were ground to powder in a tissue lyser, ABA was extracted in 1.9 mL of 70:30 (v/v) acetone:50 mM citric acid solution, and 20 ng [2H6]-ABA (Olchemim) was added as an internal standard. Samples were incubated with continuous shaking for over 3 h at 4°C. After incubation, the acetone layer was evaporated overnight before the samples were re-extracted with diethyl ether. Samples were centrifuged for 10 min, and the upper phase was collected in a glass vial and evaporated to dryness in a speed vacuum centrifuge. Dry samples were resuspended in 50 μL 100% methanol and filtered through a 0.45-μm GHP membrane (Pall). Finally, samples were analyzed on an liquid chromatography–tandem mass spectrometry system (Agilent 6530 Accurate-Mass Q-TOF LC/MS; Agilent Technologies) with a jet stream ion source. ABA and its internal standard were quantified by tandem mass spectrometry using the scan mode at m/z 263.13 and 269.16, respectively, in negative ionization mode.
Coleoptile Length Assessment
Rockwool prefabricated plug sheets were wetted overnight in excess water in large polypropylene containers. The plug sheets were transferred to similar containers holding 3 cm of water. Whatman 3-mm paper strips (3 × 30 cm) were folded in half lengthwise and placed as an inverted “v” between the rows of adjacent plugs from which they absorbed water. Nine barley grains per genotype were placed, crease-side-down, between adjacent plugs, resting on the moist 3-mm paper apex. Grains were moist chilled at 4°C in darkness for 3 d prior to being removed to 25°C for 14 d, either in darkness or in a chamber illuminated by constant blue light (23 μmol m−2 s−1). The coleoptile lengths of the resulting seedlings were measured thereafter.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Hv-CRY1a, DQ201150; Hv-CRY1b, DQ201153; and Hv-CRY2, DQ201156.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Coleoptile Lengths of RNAi CRY1a/b and CRY2 Seedlings.
Supplemental Table 1. Primers Used for Construction of Hv-CRY1a/b and Hv-CRY2 RNAI Hairpins.
Supplemental Table 2. Sequences Used in the Hv-CRY1a/b and Hv-CRY2 RNAI Hairpin Constructs.
Supplemental Table 3. Primer Sequences Used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank Kerrie Ramm and Trijntje Hughes for their excellent and dedicated technical assistance. We also thank Steve Swain help with coleoptile analysis and for comments on the article and Scott Boden and John Jacobsen for their critical reviews. This work was supported by the CSIRO, by the Grains and Research Corporation, by a CSIRO McMaster Fellowship to A.B.D., and by a USDA–National Institute of Food and Agriculture sabbatical grant (2012-67014-19421) to A.B.D.
AUTHOR CONTRIBUTIONS
F.G. and J.M.B. designed the research. A.B.D. and J.M.B performed the experiments and analyzed the data. Q.X. contributed new analytic tools. F.G., A.B.D., and J.M.B wrote the article.
Glossary
- R
red
- FR
far-red
- BL
blue light
- RNAi
RNA interference
- qRT-PCR
quantitative real-time PCR
- WL
white light
- ABA
abscisic acid
- GA
gibberellin
- UTR
untranslated region
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adamska I., Ohad I., Kloppstech K. (1992). Synthesis of the early light-inducible protein is controlled by blue light and related to light stress. Proc. Natl. Acad. Sci. USA 89: 2610–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero J.M., Jacobsen J.V., Talbot M.J., White R.G., Swain S.M., Garvin D.F., Gubler F. (2012). Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytol. 193: 376–386. [DOI] [PubMed] [Google Scholar]
- Barrero J.M., Talbot M.J., White R.G., Jacobsen J.V., Gubler F. (2009). Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol. 150: 1006–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D., Hanke C., Schneider K., Michel D., Salamini F. (1992). A desiccation-related Elip-like gene from the resurrection plant Craterostigma plantagineum is regulated by light and ABA. EMBO J. 11: 2771–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouly J.P., Schleicher E., Dionisio-Sese M., Vandenbussche F., Van Der Straeten D., Bakrim N., Meier S., Batschauer A., Galland P., Bittl R., Ahmad M. (2007). Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem. 282: 9383–9391. [DOI] [PubMed] [Google Scholar]
- Briggs W.R., Christie J.M. (2002). Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 7: 204–210. [DOI] [PubMed] [Google Scholar]
- Chang S., Puryear J., Cairney J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11: 113–116. [Google Scholar]
- Chaussat R., Zoppolo J. (1983). Lumière et germination de l'orge. Bios. 14: 30–32. [Google Scholar]
- Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L.O., van der Horst G.T., Batschauer A., Ahmad M. (2011). The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62: 335–364. [DOI] [PubMed] [Google Scholar]
- Christie J.M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58: 21–45. [DOI] [PubMed] [Google Scholar]
- Dave A., Hernández L., He Z., Andriotis V.M.E., Vaistij F.E., Larson T.R., Graham I.A. (2011). 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23: 583–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittinghoff K., Laubinger S., Nixdorf M., Fackendahl P., Baumgardt R.L., Batschauer A., Hoecker U. (2006). Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 47: 577–590. [DOI] [PubMed] [Google Scholar]
- Folta K.M., Spalding E.P. (2001). Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 26: 471–478. [DOI] [PubMed] [Google Scholar]
- Goggin D.E., Steadman K.J. (2012). Blue and green are frequently seen: Responses of seeds to short- and mid-wavelength light. Seed Sci. Res. 22: 27–35. [Google Scholar]
- Goggin D.E., Steadman K.J., Powles S.B. (2008). Green and blue light photoreceptors are involved in maintenance of dormancy in imbibed annual ryegrass (Lolium rigidum) seeds. New Phytol. 180: 81–89. [DOI] [PubMed] [Google Scholar]
- Gubler F., Hughes T., Waterhouse P., Jacobsen J. (2008). Regulation of dormancy in barley by blue light and after-ripening: effects on abscisic acid and gibberellin metabolism. Plant Physiol. 147: 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Millar A.A., Jacobsen J.V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8: 183–187. [DOI] [PubMed] [Google Scholar]
- Grimm B., Kloppstech K. (1987). The early light-inducible proteins of barley. Characterization of two families of 2-h-specific nuclear-coded chloroplast proteins. Eur. J. Biochem. 167: 493–499. [DOI] [PubMed] [Google Scholar]
- Guo H., Yang H., Mockler T.C., Lin C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363. [DOI] [PubMed] [Google Scholar]
- Harari-Steinberg O., Ohad I., Chamovitz D.A. (2001). Dissection of the light signal transduction pathways regulating the two early light-induced protein genes in Arabidopsis. Plant Physiol. 127: 986–997. [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Okamoto H., Stoop-Myer C., Deng X.W. (2002). Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30: 385–394. [DOI] [PubMed] [Google Scholar]
- Heschel M.S., Butler C.M., Barua D., Chiang G.C.K., Wheeler A., Sharrock R.A., Whitelam G.C., Donohue K. (2008). New roles of phytochromes during seed germination. Int. J. Plant Sci. 169: 531–540. [Google Scholar]
- Hoang H.H., Sechet J., Bailly C., Leymarie J., Corbineau F. (November 21, 2013). Inhibition of germination of dormant barley (Hordeum vulgare L.) grains by blue light as related to oxygen and hormonal regulation. Plant Cell Environ. http://dx.doi.org/10.1111/pce.12239. [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Kay S.A. (2006). Photoperiodic control of flowering: Not only by coincidence. Trends Plant Sci. 11: 550–558. [DOI] [PubMed] [Google Scholar]
- Jacobsen J.V., Barrero J.M., Hughes T., Julkowska M., Taylor J.M., Xu Q., Gubler F. (2013). Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain (Triticum aestivum L.). Planta 238: 121–138. [DOI] [PubMed] [Google Scholar]
- Jacobsen J.V., Pearce D.W., Poole A.T., Pharis R.P., Mander L.N. (2002). Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant. 115: 428–441. [DOI] [PubMed] [Google Scholar]
- Jacobsen, J.V., Venables, I., Wang, M.B., Matthews, P., Ayliffe, M., and Gubler, F. (2006). Barley (Hordeum vulgare L.). In Agrobacterium Protocols, 2nd ed, Vol. 1, K. Wang, ed (Totowa, NJ: Humana Press), pp. 171–183. [DOI] [PubMed] [Google Scholar]
- Jain M., Sharma P., Tyagi S.B., Tyagi A.K., Khurana J.P. (2007). Light regulation and differential tissue-specific expression of phototropin homologues from rice (Oryza sativa ssp. indica). Plant Sci. 172: 164–171. [Google Scholar]
- Kanegae H., Tahir M., Savazzini F., Yamamoto K., Yano M., Sasaki T., Kanegae T., Wada M., Takano M. (2000). Rice NPH1 homologues, OsNPH1a and OsNPH1b, are differently photoregulated. Plant Cell Physiol. 41: 415–423. [DOI] [PubMed] [Google Scholar]
- Kennis J.T., van Stokkum I.H., Crosson S., Gauden M., Moffat K., van Grondelle R. (2004). The LOV2 domain of phototropin: A reversible photochromic switch. J. Am. Chem. Soc. 126: 4512–4513. [DOI] [PubMed] [Google Scholar]
- Li Q.-H., Yang H.-Q. (2007). Cryptochrome signaling in plants. Photochem. Photobiol. 83: 94–101. [DOI] [PubMed] [Google Scholar]
- Marumo S., Katayama M., Komori E., Ozaki Y., Natsume M., Kondo S. (1982). Microbial production of abscisic acid by Botrytis cinerea. Agric. Biol. Chem. 46: 1967–1968. [Google Scholar]
- Más P., Yanovsky M.J. (2009). Time for circadian rhythms: Plants get synchronized. Curr. Opin. Plant Biol. 12: 574–579. [DOI] [PubMed] [Google Scholar]
- Matsumoto T., et al. (2011). Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol. 156: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R.S., Purugganan M.D. (2013). Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 14: 840–852. [DOI] [PubMed] [Google Scholar]
- Millar A.A., Jacobsen J.V., Ross J.J., Helliwell C.A., Poole A.T., Scofield G., Reid J.B., Gubler F. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 45: 942–954. [DOI] [PubMed] [Google Scholar]
- Mockler T., Yang H., Yu X., Parikh D., Cheng Y.C., Dolan S., Lin C. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 100: 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta G., Yahoubyan G., Nebuloso E., Renzi L., Giuliano G. (2001). Tomato and barley contain duplicated copies of cryptochrome 1. Plant Cell Environ. 24: 991–997. [Google Scholar]
- Poppe C., Sweere U., Drumm-Herrel H., Schäfer E. (1998). The blue light receptor cryptochrome 1 can act independently of phytochrome A and B in Arabidopsis thaliana. Plant J. 16: 465–471. [DOI] [PubMed] [Google Scholar]
- Raghavan V., Kamalay J.C. (1993). Expression of two cloned mRNA sequences during development and germination of spores of the sensitive, Onoclea sensibilis L. Planta 189: 1–9. [DOI] [PubMed] [Google Scholar]
- Rizza A., Boccaccini A., Lopez-Vidriero I., Costantino P., Vittorioso P. (2011). Inactivation of the ELIP1 and ELIP2 genes affects Arabidopsis seed germination. New Phytol. 190: 896–905. [DOI] [PubMed] [Google Scholar]
- Sellaro R., Crepy M., Trupkin S.A., Karayekov E., Buchovsky A.S., Rossi C., Casal J.J. (2010). Cryptochrome as a sensor of the blue/green ratio of natural radiation in Arabidopsis. Plant Physiol. 154: 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R., Hoecker U., Yanovsky M., Chory J., Casal J.J. (2009). Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr. Biol. 19: 1216–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48: 354–366. [DOI] [PubMed] [Google Scholar]
- Seo M., Nambara E., Choi G., Yamaguchi S. (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69: 463–472. [DOI] [PubMed] [Google Scholar]
- Sugai M., Furuya M. (1985). Action spectrum in ultraviolet and blue light region for the inhibition of red-light-induced spore germination in Adiantum capillus-veneris L. Plant Cell Physiol. 26: 953–956. [Google Scholar]
- Simpson, G.M. (1990). Seed Dormancy in Grasses. (Cambridge, New York: Cambridge University Press). [Google Scholar]
- Szucs P., Karsai I., von Zitzewitz J., Mészáros K., Cooper L.L., Gu Y.Q., Chen T.H., Hayes P.M., Skinner J.S. (2006). Positional relationships between photoperiod response QTL and photoreceptor and vernalization genes in barley. Theor. Appl. Genet. 112: 1277–1285. [DOI] [PubMed] [Google Scholar]
- Trevaskis B., Hemming M.N., Peacock W.J., Dennis E.S. (2006). HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 140: 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Li Z., Matthews P.R., Upadhyaya N.M., Waterhouse P.M., Wang M.B., Li Z.Y. (1998). Improved vectors for Agrobacterium-mediated transformation of monocot plants. Acta Hortic. 461: 401–407. [Google Scholar]
- Wang Y., Folta K.M. (2013). Contributions of green light to plant growth and development. Am. J. Bot. 100: 70–78. [DOI] [PubMed] [Google Scholar]
- Xu P., Xiang Y., Zhu H., Xu H., Zhang Z., Zhang C., Zhang L., Ma Z. (2009). Wheat cryptochromes: Subcellular localization and involvement in photomorphogenesis and osmotic stress responses. Plant Physiol. 149: 760–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Zhu H.L., Xu H.B., Zhang Z.Z., Zhang C.Q., Zhang L.X., Ma Z.Q. (2010). Composition and phylogenetic analysis of wheat cryptochrome gene family. Mol. Biol. Rep. 37: 825–832. [DOI] [PubMed] [Google Scholar]
- Yu X., Liu H., Klejnot J., Lin C. (2010). The cryptochrome blue light receptors. The Arabidopsis Book 8: e0135, /10.1199/tab.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q., Chen X.B., Wood A.J. (2002). Two early light-inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. J. Exp. Bot. 53: 1197–1205. [DOI] [PubMed] [Google Scholar]
- Zhang T., Maruhnich S.A., Folta K.M. (2011). Green light induces shade avoidance symptoms. Plant Physiol. 157: 1528–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-C., Gong S.-F., Li Q.-H., Sang Y., Yang H.Q. (2006). Functional and signaling mechanism analysis of rice CRYPTOCHROME 1. Plant J. 46: 971–983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.