This study demonstrates that the jasmonate-activated transcription factor MYC2 modulates both the quantity and quality of ethylene-activated transcription factor EIN3, by directly inducing F-box gene EBF1 expression to promote EIN3 protein degradation and interacting with EIN3 to inhibit its transcriptional activity, thus negatively regulating HLS1 expression and apical hook development.
Abstract
The apical hook is an essential structure that enables epigeal plants to protrude through the soil. Arabidopsis thaliana HOOKLESS1 (HLS1) is reported to be a key regulator of hook development and a direct target gene of the ethylene (ET)-activated transcription factors ETHYLENE INSENSITIVE3 (EIN3) and its close homolog EIN3-Like1. Previous research has shown that the phytohormones jasmonate (JA) and ET antagonistically regulate apical hook development, although the underlying molecular mechanism is largely unknown. Here, we report that JA represses hook formation by reducing HLS1 expression. Our results further reveal that the JA-activated transcription factor MYC2 represses EIN3 function to reduce HLS1 expression through at least the following two layers of regulation: (1) MYC2 binds to the promoter of an F-box gene, EIN3 BINDING F-BOX PROTEIN1, to induce its expression and thus promote EIN3 degradation; and (2) MYC2 physically interacts with EIN3 and inhibits its DNA binding activity. Collectively, our findings shed light on the molecular mechanism underlying the antagonism between JA and ET during apical hook development and provide insight into the coaction of multiple phytohormones in the regulation of plant growth and development.
INTRODUCTION
To protect apical meristematic tissues and cotyledons (embryonic leaves) from damage during germination, many epigeal plants have evolved an elegant organ named the apical hook. Etiolated Arabidopsis thaliana seedlings form an apical hook with closed cotyledons, which open upon exposure to light to facilitate photosynthesis (Liscum and Hangarter, 1993). Light and several plant hormones have been found to regulate apical hook development. Ethylene (ET) and gibberellins (GAs) are two major positive regulators of hook formation, while light, brassinosteroids, and jasmonate (JA) are negative regulators (Liscum and Hangarter, 1993; Turner et al., 2002; Alabadí et al., 2004; Li et al., 2004; Vriezen et al., 2004; De Grauwe et al., 2005).
The formation of an apical hook is a consequence of differential cell growth (Raz and Ecker, 1999), in which the plant hormone auxin is reported to play a major role. HOOKLESS1 (HLS1), a protein with similarity to N-acetyltransferase, has been identified as a key modulator of auxin distribution and responses during apical hook formation of Arabidopsis seedlings (Roman et al., 1995; Lehman et al., 1996). The loss-of-function mutant hls1-1 fails to form an apical hook when grown in darkness, and such a hookless phenotype is also observed upon inhibition of auxin transport or alteration of auxin distribution (Lehman et al., 1996). ET promotes HLS1 transcript accumulation via ETHYLENE INSENSITIVE3 (EIN3), which directly binds to its promoter, thus leading to exaggerated hook curvature (Lehman et al., 1996; Chang et al., 2013). Our recent work demonstrates that GA promotes hook formation partly by inducing the expression of HLS1 via relieving the repression of DELLA proteins on EIN3 (An et al., 2012). These results indicate that HLS1 is a central regulator of multiple signaling pathways in the control of auxin-induced differential cell growth during apical hook development.
ET is a gaseous hormone that widely regulates plant growth and development (Johnson and Ecker, 1998). A typical ET response is the so-called “triple response,” including shortened root and hypocotyl as well as exaggerated hook curvature of etiolated seedlings (Roman et al., 1995). Several ET signaling components have been uncovered through forward genetics approaches (Roman et al., 1995; Alonso et al., 2003). The ethylene receptors (ETR1, ETR2, ERS1, ERS2, and EIN4) and CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) are negative regulators of ET signaling, whereas EIN2, EIN3, and EIN3-Like1 (EIL1) are positive regulators. EIN3 and EIL1 are two crucial transcription factors that regulate most, if not all, of the ET-responsive phenotypes (Chao et al., 1997; Alonso et al., 2003; An et al., 2010). ET activates EIN3 and EIL1 by increasing their protein stability. In the absence of ET, EIN3 and EIL1 are subject to proteasomal degradation mediated by two F-box proteins, EIN3 BINDING F-BOX PROTEIN1 (EBF1) and EBF2 (Guo and Ecker, 2003; Potuschak et al., 2003; Olmedo et al., 2006). ET treatment reduces the stability of EBF1/2, which results in EIN3/EIL1 accumulation (An et al., 2010). EIN3 binds to the EBF2 promoter and activates EBF2 transcription (Konishi and Yanagisawa, 2008), which forms a negative feedback loop that fine-tunes the accumulation of EIN3/EIL1.
JA is another plant hormone that regulates myriad developmental processes, the wound response, and pathogen defense (Browse, 2009). After synthesis, JA is conjugated with Ile to form JA-Ile, which is the bioactive form of JA in plants (Staswick et al., 2002; Staswick and Tiryaki, 2004; Fonseca et al., 2009). CORONATINE INSENSITIVE1 (COI1), an F-box protein, has been identified through JA-insensitive mutant screening (Benedetti et al., 1995; Xie et al., 1998). JASMONATE ZIM-DOMAIN PROTEINS (JAZs) are the direct targets of COI1 and are degraded very quickly upon JA treatment (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). A number of JAZ-interacting transcription factors have been isolated, including MYC2/MYC3/MYC4 (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011), R2R3-MYB TRANSCRIPTION FACTOR21/24 (MYB21/MYB24) (Song et al., 2011), EIN3/EIL1 (Zhu et al., 2011), and TRANSPARENT TESTA8/GLABRA3/ENHANCER OF GLABRA3/MYB75/GLABRA1 (Qi et al., 2011) complexes and INDUCER OF CBF EXPRESSION1 (ICE1) and ICE2 (Hu et al., 2013). JAZs repress their target transcription factors through directly or indirectly recruiting TOPLESS corepressor protein or interacting with HISTONE DEACETYLASE6 (HDA6) to inhibit transcription (Pauwels et al., 2010; Zhu et al., 2011; Shyu et al., 2012). Crystallographic analysis shows that COI1 and JAZs together constitute the coreceptor for JA-Ile (Yan et al., 2009; Sheard et al., 2010). Binding of JA-Ile to this coreceptor stimulates COI1-JAZs interaction via a “molecular glue” mechanism and thus promotes JAZ degradation (Sheard et al., 2010). The removal of JAZs thus derepresses the above-mentioned transcription factors to activate their downstream genes and produce different JA responses.
ET and JA are found to coordinately (cooperatively or antagonistically) regulate plant growth, development, and pathogen defense responses (Dong, 1998; Li and Guo, 2007). Both ET and JA treatment induce the expression of pathogen-responsive genes, such as ETHYLENE RESPONSE FACTOR1 (ERF1), a direct target of EIN3/EIL1 (Solano et al., 1998; Lorenzo et al., 2003). We previously identified EIN3/EIL1 as a novel class of JAZ-interacting proteins and further demonstrated that both JA and ET signaling are required for activating EIN3/EIL1 to integrate jasmonate-ethylene (JA-ET) coaction in the plant defense responses and root hair development (Zhu et al., 2011).
Nonetheless, the molecular basis for JA-ET antagonism is largely unclear. Here, we show that HLS1 is a necessary component for JA-ET antagonism in hook development. JA reduces HLS1 expression and hook curvature angles even in the presence of ET. We next reveal that JA attenuates HLS1 expression through repressing EIN3/EIL1 activity. JA treatment promotes EIN3/EIL1 proteolysis, which is dependent on SCFEBF1. We further find that the basic/helix-loop-helix transcription factor MYC2 is necessary for this antagonistic effect and that JA activates MYC2 to positively regulate EBF1 expression by directly binding to its promoter. Besides this layer of regulation, MYC2 can also physically interact with EIN3 and directly inhibit its transcriptional activity.
RESULTS
JA Antagonizes ET-Induced Hook Formation in an HLS1-Dependent Manner
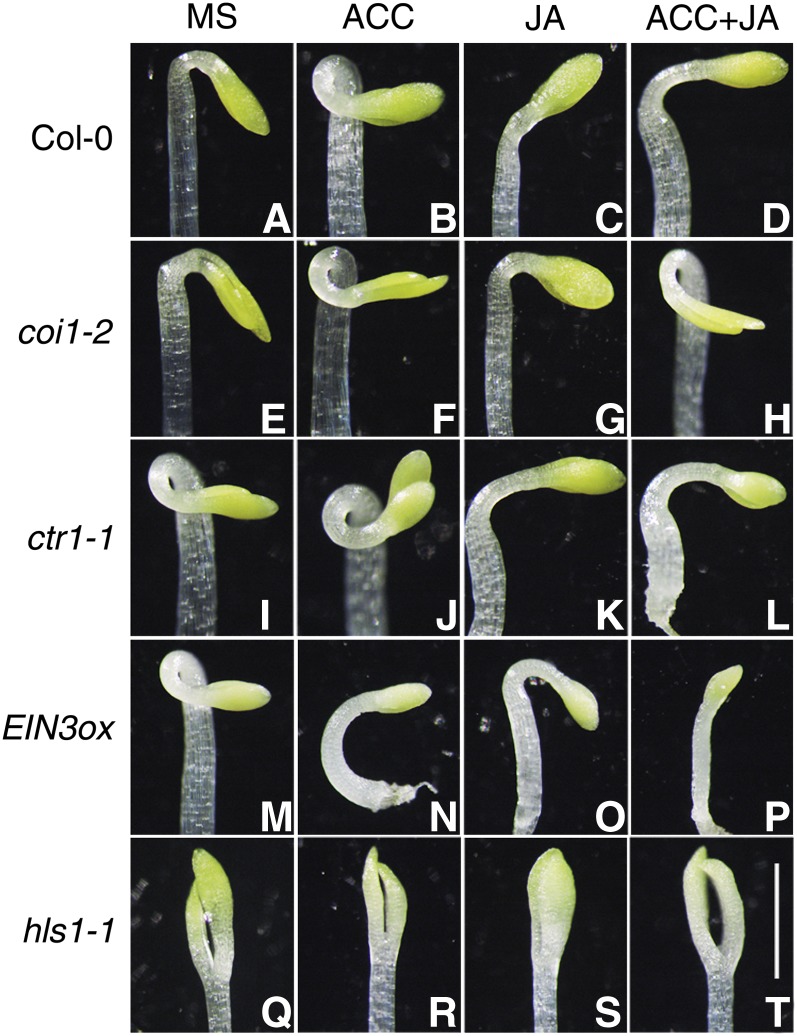
It has been reported that JA represses the ET-induced exaggerated hook in ecotype Columbia-0 (Col-0) and ctr1-1 (Turner et al., 2002). To illustrate the molecular framework of the JA-ET interaction in regulating hook development, we first examined the hook curvature phenotypes in various ET or JA response mutants. As shown in Figure 1, 3-d-old etiolated wild-type (Col-0) seedlings showed exaggerated hooks when grown on 1-aminocyclopropane-1-carboxylic acid (ACC; an ET biosynthesis precursor) medium compared with normal growth conditions (Murashige and Skoog [MS] medium), while the apical hook was dramatically inhibited when plants were grown on JA medium (Figures 1A to 1C). Moreover, JA partly repressed the ET-induced exaggerated hooks when plants were grown on ET plus JA medium (Figure 1D). The JA receptor–defective mutants (coi1-2) formed exaggerated hooks on ACC medium (Figures 1E and 1F) but were not responsive to JA, as expected (Figure 1G). Simultaneous treatment with JA and ET still resulted in coi1-2 forming exaggerated hooks (Figure 1H; also see the quantitative results in Supplemental Figure 1), suggesting that the JA repression of hook formation is COI1 dependent. We next examined the hook phenotypes in the constitutively activated ethylene signaling mutants ctr1-1 and 35S:EIN3 (referred to as EIN3ox). We found that JA treatment repressed the exaggerated hook of ctr1-1 etiolated seedlings grown on MS medium supplemented with or without ET (Figures 1K and 1L). EIN3ox showed exaggerated hooks in the absence of ET treatment (Figure 1M) and formed an extremely short, bent hypocotyl in the presence of ET (Figure 1N). This unusual hook phenotype is probably due to the fact that overactivated ET signaling leads to a distorted hypocotyl and seedling structure, which is also observed in the ebf1 ebf2 double mutant (Potuschak et al., 2003; Olmedo et al., 2006). When compared with seedlings grown under normal growth conditions, JA was also able to partially repress the exaggerated hook of EIN3ox (Figures 1M and 1O; Supplemental Figure 1). These results suggest that a COI1-mediated JA pathway antagonizes an EIN3-mediated ET pathway in the regulation of hook curvature.
Figure 1.
JA Represses ET-Induced Hook Formation.
Three-day-old etiolated seedlings were grown on MS, 10 μM ACC, 50 μM JA, or 10 μM ACC plus 50 μM JA medium. Representative images of Col-0 ([A] to [D]), coi1-2 ([E] to [H]), ctr1-1 ([I] to [L]), EIN3ox ([M] to [P]), and hls1-1 ([Q] to [T]) hooks are shown. Bar = 1 mm.
[See online article for color version of this figure.]
Given that HLS1 is an essential regulator integrating multiple signaling pathways involved in hook formation, we then tested whether HLS1 is involved in JA-ET antagonism in hook development. Our results showed that, compared with wild-type seedlings, hls1-1 was not responsive to ET, JA, or their combined treatment with regard to the apical hook phenotype (Figures 1Q to 1T). This result suggests that HLS1 acts as a common regulator of the JA-ET antagonism in hook formation.
JA Downregulates HLS1 Expression through Inhibiting EIN3/EIL1 Functions
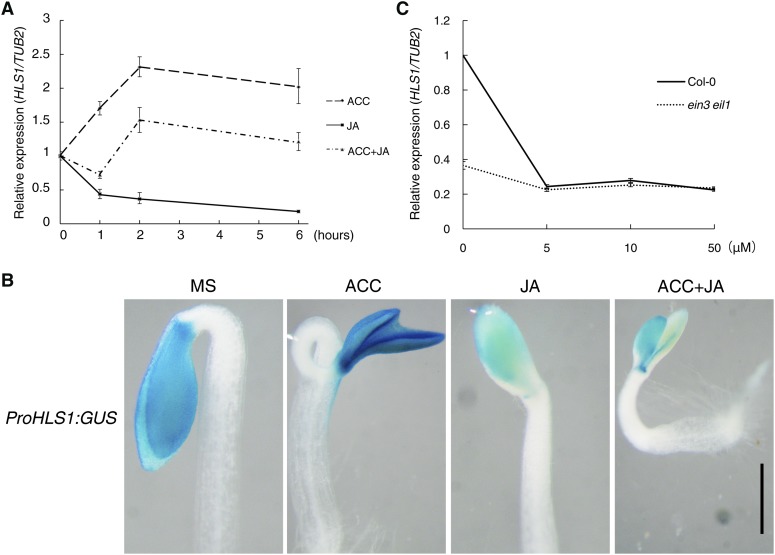
Next, we sought to explore how JA represses hook formation. Given that HLS1 is necessary for the JA-ET antagonism (Figures 1Q to 1T) and that ET treatment induces HLS1 expression (Lehman et al., 1996), we investigated whether JA treatment alters the expression of HLS1. By quantitative RT-PCR (qRT-PCR) analysis, we found that ET rapidly induced HLS1 expression (within 1 h), which reached a maximal steady level after 2 h and declined slightly after 6 h of ET treatment. By contrast, JA constantly reduced HLS1 expression and also suppressed ET-induced HLS1 expression (Figure 2A). Additionally, we generated transgenic plants harboring ProHLS1:GUS (in which β-glucuronidase gene expression is driven by the HLS1 promoter) and detected the GUS activity. Consistent with the qRT-PCR results, ET induced GUS expression in the apical hook and cotyledons, while JA strongly repressed it (Figure 2B).
Figure 2.
JA Represses HLS1 Expression through EIN3/EIL1.
(A) qRT-PCR shows that JA suppresses ET-induced HLS1 expression. Three-day-old etiolated Col-0 was treated with 100 μM ACC, 100 μM JA, or 100 μM ACC plus 100 μM JA for the indicated number of hours. HLS1 expression level was detected and normalized to TUB2. Values shown are means ± sd; n = 3.
(B) ProHLS1:GUS expression assay. Three-day-old etiolated ProHLS1:GUS seedlings grown on MS, 10 μM ACC, 50 μM JA, or 10 μM ACC plus 50 μM JA medium were stained with GUS staining solution for 4 h. Representative images were taken to record expression. Bar = 0.5 mm.
(C) qRT-PCR analysis of HLS1 expression. Three-day-old etiolated Col-0 or ein3 eil1 seedlings were treated with the indicated concentrations of JA for 4 h. HLS1 expression was normalized with TUB2. Values shown are means ± sd; n = 3.
[See online article for color version of this figure.]
Our previous study and a recent EIN3 chromatin immunoprecipitation sequence analysis revealed that HLS1 is a direct target of EIN3/EIL1 (An et al., 2012; Chang et al., 2013). Therefore, we tested whether JA-repressed HLS1 expression is also mediated by EIN3/EIL1. We found that JA-repressed HLS1 expression was largely dependent on EIN3/EIL1, as its expression declined to a much smaller extent in ein3 eil1 than in Col-0 after JA treatment (Figure 2C). This result indicates that EIN3/EIL1 are crucial transcription factors in the JA-mediated repression of HLS1 expression.
JA Promotes EIN3/EIL1 Degradation via SCFEBF1/2
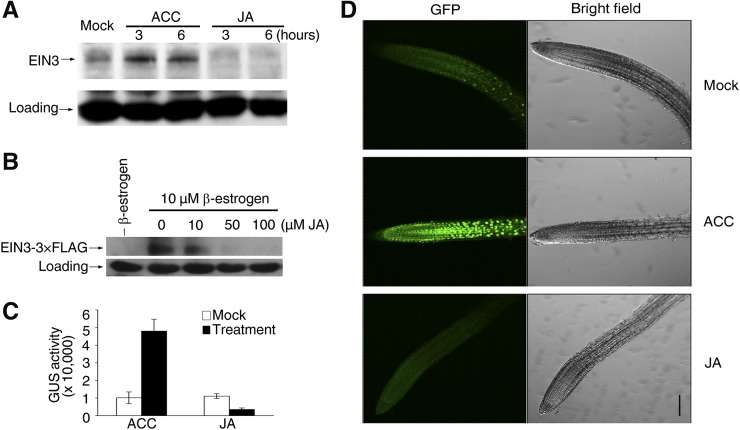
We further assessed whether JA represses HLS1 expression through modulating EIN3/EIL1 function. A principal regulatory mechanism of EIN3/EIL1 function is to alter their protein stability, which can be triggered by environmental stimuli or plant hormones (Guo and Ecker, 2003; Potuschak et al., 2003; Yanagisawa et al., 2003; Lee et al., 2006; Zhong et al., 2009). We then examined whether JA represses the function of EIN3/EIL1 by affecting the stability of these proteins. We first detected the endogenous EIN3 protein abundance in Arabidopsis suspension cell cultures derived from the wild type and found that ET augmented EIN3 accumulation, as reported previously, whereas JA decreased EIN3 abundance (Figure 3A). We also found that JA repressed estradiol-induced EIN3-3×FLAG protein accumulation in a dosage-dependent manner in pER8:EIN3-3×FLAG/Col-0 transgenic plants (Figure 3B). Moreover, we generated transgenic plants expressing an EIN3-GUS fusion driven by its native promoter or an EIN3-FLAG fusion driven by the constitutive 35S promoter in the ein3 eil1 background (ProEIN3:EIN3-GUS/ein3 eil1 and 35S:EIN3-3×FLAG/ein3 eil1, respectively). We found that ACC treatment enhanced GUS activity or EIN3-FLAG accumulation but JA treatment decreased it (Figure 3C; Supplemental Figure 2A). Finally, we examined the level of EIL1 after JA treatment by detecting EIL1-GFP fluorescence in EIL1-GFP transgenic plants and found that ACC strongly promoted GFP signal accumulation, whereas JA reduced it (Figure 3D). Collectively, these results indicate that JA promotes EIN3/EIL1 degradation.
Figure 3.
JA Promotes EIN3/EIL1 Degradation.
(A) and (B) The levels of EIN3 are downregulated by JA treatment. Either cultured suspension cells (A) or transgenic plants harboring inducible EIN3-3×FLAG (with 10 μM β-estradiol preinduction for 4 h before JA treatment) (B) were treated with JA for 4 h. Protein extracts were probed with either anti-EIN3 (A) or anti-FLAG (B) antibody, respectively. Cross-reacting nonspecific bands were used as loading controls. Arrows define the corresponding proteins.
(C) GUS activity in ProEIN3:EIN3-GUS/ein3 eil1. Seedlings were treated with 100 μM ACC or 100 μM JA for 4 h. Values shown are means ± sd; n = 5.
(D) Green fluorescence was observed in 7-d-old EIL1-GFP seedlings after 4 h of treatment with 100 μM ACC or 100 μM JA. Bar = 100 μm.
[See online article for color version of this figure.]
EIN3/EIL1 are subjected to proteasomal degradation mediated by two F-box proteins, EBF1 and EBF2, in the ET signaling pathway. To determine whether JA-triggered EIN3/EIL1 degradation also occurs via the EBF1/2-proteasome pathway, we introduced inducible EIN3 into the ein3 eil1 ebf1 ebf2 background, which lacks both EBF1 and EBF2 functions (An et al., 2010). After estradiol induction, we treated plants with JA at different time points and found that the level of EIN3 remained unchanged (Supplemental Figure 2B), in contrast with the evident decline in the wild-type background (Figure 3B). Therefore, we conclude that JA promotes EIN3/EIL1 protein degradation through SCFEBF1/2.
JA Activates MYC2 to Induce EBF1 Expression
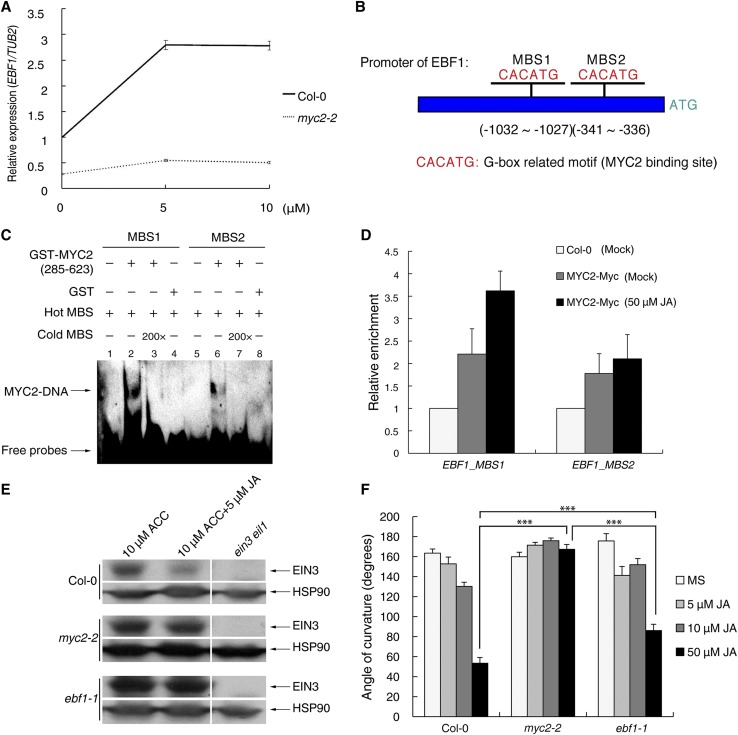
To unravel how JA regulates EIN3/EIL1 degradation through SCFEBF1/2, we first determined whether JA regulates the expression of EBF1 and EBF2. Using qRT-PCR, we observed the induction of EBF1 but not EBF2 by JA and found that this induction was largely diminished in myc2-2 (Figure 4A; Supplemental Figure 3). MYC2 is an important transcription factor mediating multiple JA responses (Kazan and Manners, 2013). Sequence analysis revealed that two putative MYC2 binding sites (MBS1 and MBS2; CACATG) were present in the promoter of EBF1, suggesting that MYC2 may directly bind to the promoter of EBF1 (Figure 4B). To test this possibility, we expressed and purified GST-MYC2 (amino acids 285 to 623; the DNA binding domain of MYC2) and performed an electrophoretic mobility shift assay (EMSA). Our results showed that MYC2 (amino acids 285 to 623) bound to both MBS1 and MBS2 in vitro (Figure 4C) and that the affinity for MBS1 was stronger than that for MBS2. Further in vivo chromatin immunoprecipitation-PCR (ChIP-PCR) analysis showed that Myc-tagged MYC2 (35S:MYC2-4×Myc transgenic plants, referred to as MYC2-myc) preferentially bound to MBS1 after JA treatment (Figure 4D). In addition, we found that the protein stability of EBF1 is not affected by JA treatment (Supplemental Figure 4). Combining the results of in vitro EMSA and in vivo ChIP-PCR assays, we conclude that EBF1 is targeted directly by MYC2.
Figure 4.
MYC2 Directly Regulates EBF1 Expression upon JA Treatment.
(A) Three-day-old etiolated seedlings of Col-0 and myc2-2 were treated with the indicated concentrations of JA for 1 h. The relative expression of EBF1 was detected and normalized to TUB2. Values shown are means ± sd; n = 3.
(B) Schematic illustration of the two putative MYC2 binding sites (CACATG) in the promoter region of EBF1.
(C) EMSA results show that MYC2 binds to the promoter of EBF1. Hot MBS is biotin-labeled MBS probe, while cold MBS is nonlabeled probe for competition (200-fold that of hot MBS). A MYC2 fragment (amino acids 285 to 623) containing the DNA binding domain was purified from E. coli and used for DNA binding assays.
(D) ChIP-PCR shows the in vivo binding of MYC2 with MBS. Cross-linked chromatins extracted from MYC2-Myc were precipitated with anti-MYC antibody. Eluted DNA was subjected to amplification of the neighboring sequences of MBS1 and MBS2 by quantitative PCR. Col-0 plants were used as negative controls.
(E) Protein extracts of 3-d-old etiolated Col-0, ebf1-1, or myc2-2 seedlings treated with the indicated hormones for 4 h were probed with anti-EIN3 or anti-HSP90 antibody (loading control). Arrows define the corresponding proteins.
(F) Quantification of the hook curvature phenotype. Etiolated Col-0, myc2-2, and ebf1-1 seedlings were grown on the indicated media for 3 d, and then the angles of curvature were measured using ImageJ software. Statistical significance was determined using Student’s t test (***P < 0.001). Values shown are means ± se; n = 20.
[See online article for color version of this figure.]
To explore whether MYC2 or EBF1 is required for JA-mediated EIN3 turnover, we examined EIN3 abundance in Col-0, ebf1-1, and myc2-2 seedlings and found that EIN3 was degraded in Col-0 after JA treatment even in the presence of ACC, while it remained unchanged in ebf1-1 or myc2-2 after JA treatment (Figure 4E). Based on these results, we conclude that JA promotes EIN3/EIL1 degradation dependent on both MYC2 and EBF1, likely by activating MYC2 and thereby inducing EBF1 expression.
Since JA-induced EIN3 degradation is dependent on MYC2 and EBF1, we next examined whether the myc2-2 or ebf1-1 mutant is insensitive to JA treatment in terms of the JA-repressed hook curvature phenotype. Statistical analysis of hook curvature showed that myc2-2 was insensitive to various concentrations of JA while ebf1-1 was still responsive to JA, although to a lesser extent (Figure 4F). This result implies the existence of an alternative pathway downstream of MYC2 that bypasses the SCFEBF1-EIN3 degradation module to repress EIN3 function.
MYC2 Physically Interacts with EIN3
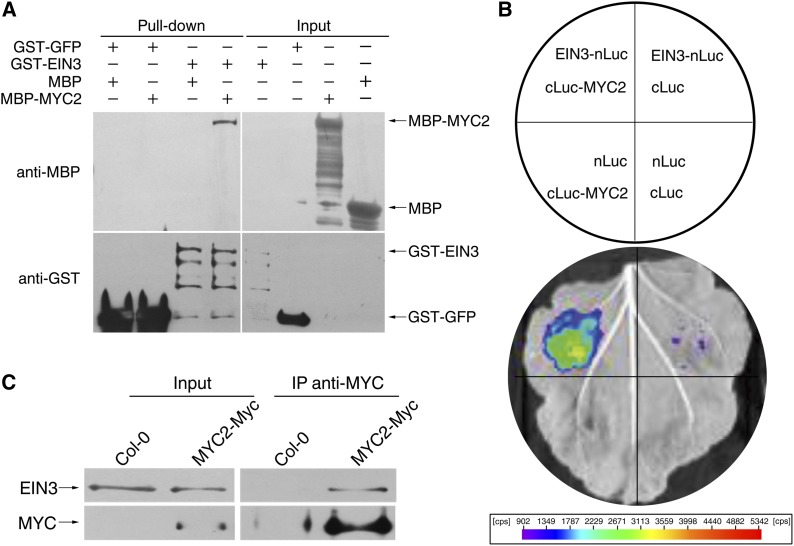
Mutual interactions between transcription factors widely exist in mediating signal transduction. It has been reported that MYC2 interacts with several MYB transcription factors to regulate glucosinolate synthesis (Schweizer et al., 2013), while EIN3 interacts with the basic/helix-loop-helix transcription factor FER-LIKE FE DEFICIENCY-INDUCED TRANSCRIPTION FACTOR to regulate plant iron acquisition (Lingam et al., 2011). We propose that MYC2 physically interacts with EIN3. To test this possibility, we expressed and purified glutathione S-transferase–tagged EIN3 (GST-EIN3) and maltose binding protein–tagged MYC2 (MBP-MYC2) and performed a pull-down assay using these protein fusions. After immobilization of GST-EIN3, MBP-MYC2 can be detected in the pull-down product, indicating that EIN3 and MYC2 interact in vitro (Figure 5A). We next conducted a firefly luciferase (LUC) complementation imaging assay to demonstrate this interaction in vivo. Constructs harboring EIN3 fused with the N terminus of LUC (EIN3-nLUC) and the C terminus of LUC fused with MYC2 (cLUC-MYC2) were coinfiltrated into Nicotiana benthamiana leaves to transiently coexpress these two fusion proteins. A luminescence signal was only detected in EIN3-nLUC/cLUC-MYC2 coexpression regions but not in the negative controls (Figure 5B). Consistent with this transient interaction assay, we also detected the EIN3-MYC2 interaction in a coimmunoprecipitation (co-IP) experiment (Figure 5C). In this experiment, we utilized MYC2-Myc transgenic plants and attempted to immunoprecipitate MYC2-Myc and then detected EIN3 in the immunoprecipitation products. Because the endogenous EIN3 level is very low (Guo and Ecker, 2003) and MYC2 tends to interact with JAZ proteins in the absence of JA treatment, we pretreated plant tissues with ACC (100 μM) to stabilize EIN3 and JA (10 μM) to activate MYC2 and then performed co-IP experiments. Our results showed that EIN3 could be detected in the co-IP products. Taken together, we conclude that MYC2 physically interacts with EIN3.
Figure 5.
MYC2 Physically Interacts with EIN3.
(A) A pull-down assay shows that EIN3 interacts with MYC2. GST fusion proteins were immobilized with Glutathione Sepharose 4B resin, and then MBP fusion proteins were mixed with the corresponding resin. The precipitated products were separated by 10% SDS-PAGE and further blotted with anti-MBP or anti-GST antibody, respectively. Arrows define the corresponding proteins.
(B) A luciferase complementation imaging assay shows that EIN3 and MYC2 interact with each other in N. benthamiana leaves. Agrobacterium strain GV3101 harboring different construct combinations was infiltrated into different N. benthamiana leaf regions. After 3 d of infiltration, luciferase activities were recorded in these regions. cps indicates signal counts per second.
(C) A co-IP assay shows the interaction between EIN3 and MYC2 in vivo. Three-day-old seedlings of Col-0 and MYC2-Myc were pretreated with 10 μM JA and 100 μM ACC. Plant extracts were then immunoprecipitated using anti-MYC antibody, separated on a 10% SDS-PAGE gel, and blotted with anti-MYC or anti-EIN3 antibody. Arrows define the corresponding proteins.
MYC2 Inhibits EIN3 DNA Binding Activity
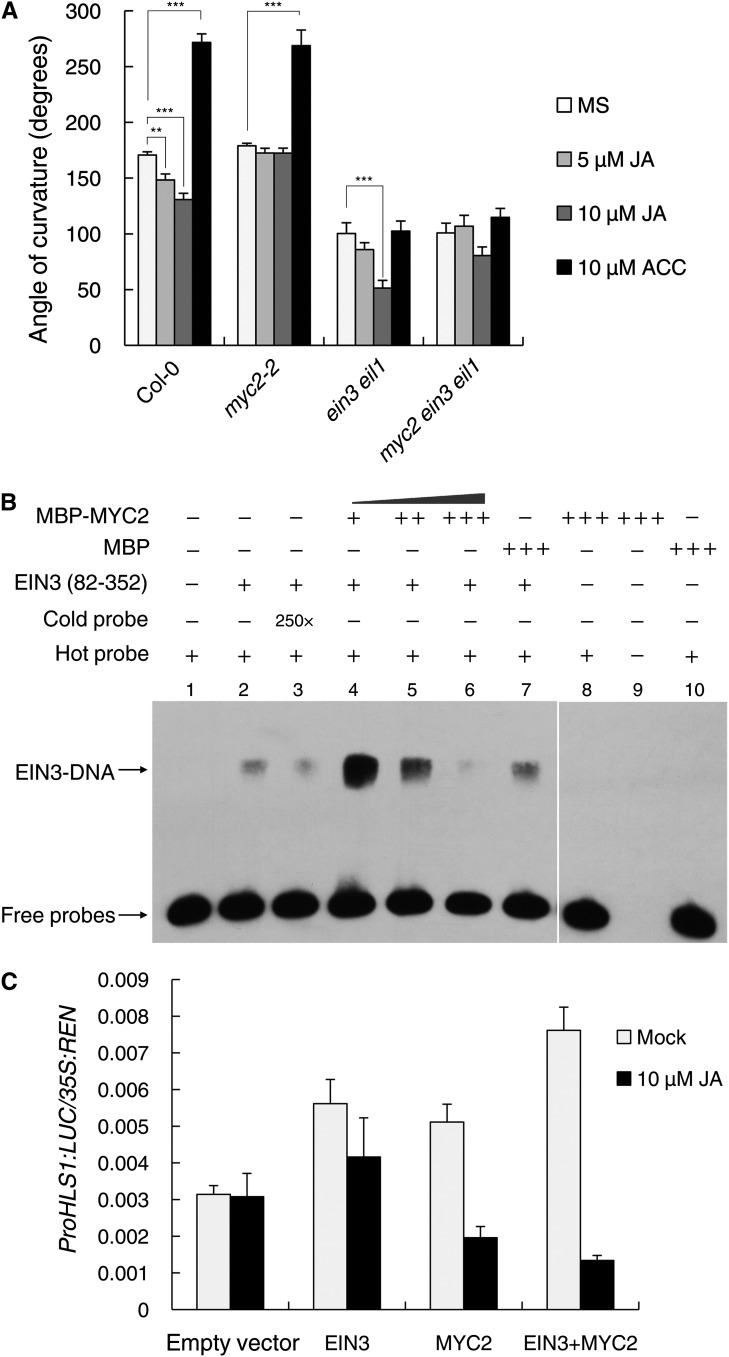
To establish the consequences of the MYC2-EIN3 interaction, we first analyzed the hook phenotypes of the mutants lacking MYC2 and/or EIN3/EIL1 activity. By comparing the myc2-2, ein3 eil1, and myc2 ein3 eil1 mutants upon ET or JA treatment, we found that the hook curvature of myc2-2 was insensitive to JA but responsive to ET (Figure 6A), indicative of the essential role of MYC2 in JA regulation. Although ein3 eil1 was completely insensitive to ET, it still responded to JA in a dosage-dependent manner (Figure 6A). Together with the finding that JA was still able to inhibit HLS1 expression in ein3 eil1 (Figure 2C), this observation suggests the existence of an alternative pathway mediating the repressive effect of JA on hook development when EIN3/EIL1 function is lacking. Furthermore, we found that myc2 ein3 eil1 showed almost an identical hook phenotype regardless of ET or JA treatment (Figure 6A), demonstrating that MYC2 and EIN3/EIL1 are essential signaling components in the regulation of hook curvature by JA and ET.
Figure 6.
MYC2 Interferes with EIN3 DNA Binding and Transcriptional Activities.
(A) Quantification of the hook curvature phenotype. Etiolated Col-0, myc2-2, ein3-1 eil1-1, and myc2 ein3 eil1 seedlings were grown on the indicated media for 3 d, and then the angle of curvature was measured. Statistical significance was calculated between different media and MS medium within the same genotype and was determined using Student’s t test (**P < 0.01, ***P < 0.001). Values shown are means ± se; n = 20.
(B) EMSA results show that MYC2 affects the binding of EIN3 to the HLS1 promoter. Forty picomoles of EIN3 (amino acids 82 to 352) was added to the reaction (lanes 2 to 7). A gradient concentration of MBP-MYC2 was applied (20 pmol for lane 4, 40 pmol for lane 5, and 80 pmol for lanes 6, 8, and 9). Eighty picomoles of MBP was added to the reaction as a negative control.
(C) Transient dual-luciferase reporter assay. Agrobacterium strain GV3101 carrying the reporter plasmid together with different combinations of effector plasmids was infiltrated into N. benthamiana leaves, and the luciferase activity at the sites of infiltration was measured 3 d after infiltration. The activities of firefly luciferase and Renilla luciferase were measured sequentially, and the LUC:REN ratio was calculated as the final transcriptional activity. Values shown are means ± sd; n = 5.
Because EIN3 associates with the promoter of HLS1 to induce its transcription (An et al., 2012), we next examined whether MYC2-EIN3 binding affects EIN3 association with the HLS1 promoter. Using competition EMSA experiments, we found that the addition of a small amount of MYC2 enhanced EIN3 binding to the HLS1 promoter sequence (Figure 6B). We speculate that a low dosage of MYC2 may facilitate the formation of EIN3 homodimers and thus enhance its DNA binding ability, although further evidence is needed to verify this speculation and its possible biological meaning. Nonetheless, with the increase in MYC2 amount, the EIN3 DNA binding ability drastically decreased (Figure 6B). We failed to detect any binding of MYC2 to the HLS1 promoter sequence used, even in the presence of a large amount of MYC2 (Figure 6B), suggesting that MYC2 likely modulates in vitro EIN3 DNA binding via the MYC2-EIN3 interaction rather than through direct binding. Further support came from MYC2 ChIP-PCR experiments demonstrating that none of the HLS1 fragments throughout its promoter region were enriched by MYC2 (Supplemental Figure 5B).
To further investigate the biological consequence of the MYC2-EIN3 interaction, we employed a dual-luciferase reporter approach (Hellens et al., 2005) to determine the effect of MYC2 on EIN3 activity in vivo. In this experiment, the HLS1 promoter-driven firefly luciferase reporter (ProHLS1:LUC) and 35S promoter-driven Renilla luciferase (35S:REN; as an internal control) were constructed in the same plasmid and transiently expressed in N. benthamiana leaves. We monitored the LUC:REN ratio, which reflects in vivo EIN3 activity, when EIN3 and/or MYC2 were coexpressed. As expected, coexpression of EIN3 with ProHLS1:LUC was able to increase the LUC:REN ratio (Figure 6C). Strikingly, coexpression of MYC2 also increased the LUC:REN ratio in the absence of JA treatment, and the increase was more evident upon coexpression of MYC2 plus EIN3 (Figure 6C), which was in accordance with the enhancement of EIN3 DNA binding ability upon the addition of a small amount MYC2 in the EMSA experiment (Figure 6B). Nevertheless, JA treatment markedly repressed EIN3 activity, especially when MYC2 was coexpressed, conditions that inhibited the association between MYC2 and JAZ (Figure 6C).
Taken together, we conclude that JA-activated MYC2 represses EIN3 transcriptional activity through directly interacting with EIN3.
DISCUSSION
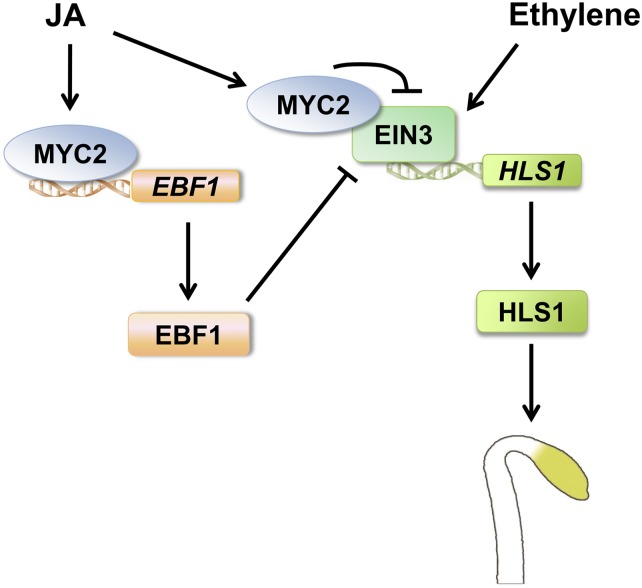
Plants have evolved several classes of phytohormones to regulate growth, development, and tolerance to environmental stresses. Two classes of plant stress hormones, ET and JA, are widely studied for their coactions. For instance, they interdependently and synergistically regulate pathogen-responsive gene expression and plant defense against fungal pathogens through a derepression mechanism (Lorenzo et al., 2003; Zhu et al., 2011). However, the antagonism between these two hormones is poorly understood. In this work, we present a model that provides two mechanistic explanations for the JA-ET antagonism in hook development (Figure 7). In this model, ET initiates a signaling pathway to stabilize EIN3/EIL1 proteins, which directly induce the transcription of HLS1. HLS1 regulates the asymmetric auxin distribution in an unknown manner and ultimately leads to differential cell growth on the two sides of the apical hook (Lehman et al., 1996; Raz and Ecker, 1999; Li et al., 2004). In contrast with ET, JA represses the functions of EIN3/EIL1 in two different ways. On the one hand, JA promotes EIN3/EIL1 degradation by inducing the expression of the F-box gene EBF1 via the activation of MYC2, which results in decreased expression of HLS1. On the other hand, JA-activated MYC2 can directly interact with EIN3 and repress its activity. These two layers of regulation on EIN3 activity contribute to the repression of ET-induced hook formation.
Figure 7.
A Working Model Depicting the Molecular Framework of JA-ET Antagonism in Hook Development.
ET stabilizes EIN3/EIL1 proteins, which directly induce the transcription of HLS1, an essential positive regulator of hook development. By contrast, JA promotes EIN3/EIL1 degradation by inducing EBF1 expression via the activation of MYC2, an important transcription factor in the JA pathway. Meanwhile, JA-activated MYC2 represses EIN3 transcriptional activity via direct protein–protein interaction. These two layers of regulation finally repress ET-mediated hook development.
[See online article for color version of this figure.]
One of the most critical regulators of hook formation is HLS1, which integrates several signaling pathways, including auxin, GA, ET, and light. Our previous studies showed that ET and GA coordinately regulate hook curvature by inducing HLS1 expression (An et al., 2012). This work demonstrates that the JA-mediated repression of hook formation is also dependent on HLS1. We further show that JA reduces HLS1 expression largely by repressing EIN3/EIL1 functions, including both quantity (protein level) and quality (transcriptional activity). Several lines of evidence enabled us to conclude that JA promotes EIN3/EIL1 degradation in an SCFEBF1-dependent manner. Therefore, the JA-ET antagonism in hook development is partly due to the opposite effect that these two hormones have on EIN3/EIL1 protein stability: ET stabilizes their accumulation, whereas JA promotes their degradation. Therefore, this work reinforces the role of HLS1 as a key regulator of hook development and provides a molecular framework for the antagonistic control of HLS1 function by ET and JA.
Our study demonstrates that a JA-activated transcription factor (MYC2) represses ET-activated transcription factors (EIN3/EIL1) in the regulation of HLS1 expression and hook development (Figure 7). It has been reported that the expression level of JA-induced pathogen-responsive genes (ERF1 and its target gene PDF1.2) is much higher in myc2 than in the wild type (Lorenzo et al., 2004; Dombrecht et al., 2007). Because ERF1 is also a direct target of EIN3 (Solano et al., 1998), these results suggest that MYC2 antagonizes EIN3/EIL1 function probably not only with respect to HLS1 expression (in hook development) but also with respect to ERF1 expression (in pathogen defense). In agreement with our study, a recent report also demonstrated that MYC2/3/4 interacts with EIN3/EIL1 and represses EIN3/EIL1 transcriptional activity, and vice versa, in the regulation of hook development and defense against insects (Song et al., 2014).
Interestingly, ERF1 was reported to be induced by both ET and JA and to serve as the integration node for JA-ET coaction in fungal resistance (Solano et al., 1998; Lorenzo et al., 2003). We previously revealed that both ET- and JA-induced ERF1 expression is EIN3/EIL1 dependent and that JA is able to enhance EIN3/EIL1 function by removing JAZs-HDA6 repressors (Zhu et al., 2011). However, as we show in this study, JA negatively regulates EIN3/EIL1 stability, which is in contrast with its positive regulation of EIN3/EIL1 function. To reconcile these seemingly contradictory findings, we hypothesize that JA exerts a dual effect on the function of EIN3/EIL1: the initial or low JA signal activates EIN3/EIL1 through a derepression mechanism (by removing JAZs/HDA6-mediated repression) (Zhu et al., 2011), while the sustained or strong JA signal starts to promote EIN3/EIL1 proteolysis by inducing the expression of the F-box protein (EBF1) via the MYC2 pathway. By analogy, ET also has a dual effect on EIN3/EIL1 function: the initial or low ET signal activates EIN3/EIL1 by destabilizing EBF1/2 proteins (An et al., 2010), while the sustained or strong ET signal suppresses EIN3/EIL1 accumulation by inducing EBF2 transcription (Konishi and Yanagisawa, 2008). The latter negative regulations executed by JA or ET are expected to either prevent the overaccumulation of EIN3/EIL1 or desensitize the activated pathway for the next round of responses.
The mode of JA-ET coaction (either cooperative or antagonistic) is thus determined by the overall effect of JA, which depends on the kinetics or strength of hormone treatment as well as the expression and/or activity of MYC2 and JAZs/HDA6 in different tissue types or at different developmental stages. In agreement with this hypothesis, ERF1 expression is rapidly induced by JA treatment (within 30 min), peaks after 6 h, and declines after 10 h (Lorenzo et al., 2003). Nevertheless, JA constantly downregulates HLS1 expression even after a brief treatment (Figure 2A), which is distinct from its action on ERF1 expression. Since HLS1 is expressed only in the apical hook region, as shown by ProHLS:GUS expression analysis (Figure 2B), we assume that the different expression patterns of these two genes is probably caused by a tissue-specific mechanism. For instance, it is plausible that the derepression of EIN3/EIL1 by JA is trivial, while the proteolysis mechanism predominates in the apical hook region. Further investigations are needed to determine the expression/activity of JAZs/HDA6 and MYC2/EBF1 in the hook region. Taken together, we propose that complicated modes of JA-ET interaction exist in the regulation of diverse processes and that the final output of the two hormones largely depends on the temporal features of hormone signals as well as the cellular context of hormone perception.
METHODS
Plant Materials, Genetic Manipulation, and Growth Conditions
Arabidopsis thaliana ein3-1 eil1-1 (Alonso et al., 2003), ctr1-1 (Kieber et al., 1993), EIN3ox (Chao et al., 1997), coi1-2 (Xu et al., 2002), myc2-2 (Lorenzo et al., 2004), 35S:MYC2-4×MYC (Chen et al., 2011), pER8:EIN3-3×FLAG (Chen et al., 2009), ProHLS1:GUS, 35S:EIN3-FLAG/ein3-1 eil1-1, 35S:EIL1-GFP, 35S:EIN3-GFP/ein3 ei1l, and pER8:EIN3-3×FLAG/ein3-1 eil1-1 ebf1-1 ebf2-1 (An et al., 2010) were described previously. The myc2 ein3 eil1 triple mutants were generated by genetic crosses between myc2-2 and ein3-1 eil1-1 and further characterized by PCR-based genotyping of the F2 population. Seeds were surface-sterilized with 10% bleach and 0.1% Triton X-100 for 5 min and washed with sterile water five times, then placed on MS medium (4.4 g/L MS salt, 1.5% Suc, pH 5.7, and 0.8% agar) with the indicated hormone treatment. After stratification for 3 d, these plates were irradiated with white light for 3 h to promote germination and then kept in darkness at 22°C for 3 d for observing the hook. Images of hook phenotypes were recorded with a dissecting microscope (Olympus).
Solution Preparation
ACC and methyl jasmonate (MeJA) were purchased from Sigma-Aldrich. ACC was dissolved in water to prepare 10 mM stock solution, while MeJA was dissolved in absolute ethanol to prepare 100 mM stock solution. The working solution was diluted from the stock solution. For the mock treatment, ethanol was diluted in water at the same dilution fold as JA. MeJA was used as the JA treatment in all studies here.
Hook Curvature Measurement
Images of individual hooks were acquired using a Canon DSLR camera with a macro lens, and hook angles were then measured using the ImageJ program (http://rsbweb.nih.gov/ij/). The bending angles were scored as described in the literature (Vandenbussche et al., 2010).
Protein Extraction and Immunoblotting
Seedlings were ground in liquid nitrogen and suspended in protein extraction buffer (50 mM Tris-Cl, pH 7.5, 1 M NaCl, 10% glycerol, 0.1% Tween 20, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail) by vortex. Extracts were kept on ice for 30 min and separated via centrifugation (13,000 rpm, 15 min, 4°C). The supernatant was collected and mixed with 5× sample buffer (60 mM Tris-HCl, pH 6.8, 25% glycerol, 2% SDS, 14.4 M mercaptoethanol, and 0.1% bromophenol) and 1 M DTT and boiled for loading on 10 or 12% SDS-PAGE gels to separate proteins. Proteins were transferred to polyvinylidene fluoride membranes (Millipore) following standard procedures. Anti-FLAG (Sigma), anti-MYC (TDY Biotech), anti-HSP90 (Beijing Protein Innovation), and anti-EIN3 (Guo and Ecker, 2003) antibodies were diluted 1000-fold with TBST buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% Tween 20) for incubation with membranes. Goat anti-mouse (or anti-rabbit) horseradish peroxidase–conjugated secondary antibody (Promega) was diluted 2500-fold with TBST buffer.
RNA Extraction, Reverse Transcription, and Real-Time PCR
Total RNA was extracted from etiolated seedlings in Trizol reagent (Invitrogen). After digestion with DNase 1 (TaKaRa), total RNA was subjected to reverse transcription at 42°C for 1 h using M-MLV reverse transcriptase (Promega). The oligonucleotide sequences for all detected genes are listed in Supplemental Table 1. Real-time PCR was performed on a Light Cycler 480 system (Roche) with SYBR Premix Ex Taq reagents (TaKaRa).
GUS Analysis
GUS activity was determined using 4-methylumbelliferyl-β-d-glucuronide (200 μM) as described (Zander et al., 2010). For GUS staining, seedlings were incubated with GUS staining solution (100 mM Na3PO4, pH 7.0, 1 mM EDTA, 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, 1% Triton X-100, and 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) for 4 to 6 h (depending on the reporter line) and washed with washing solution (100 mM Na3PO4 and 1 mM EDTA), and photographs were taken with a dissecting microscope (Olympus).
Protein Expression and Purification
The coding sequences of MYC2 (amino acids 285 to 623) and GFP were digested with EcoRI-SalI and BamHI-EcoRI, respectively, and then inserted into pGEX-5X-1 vectors (GE Healthcare) for GST fusion and transformed into Escherichia coli BL21 (DE3)–competent cells. Protein expression was induced by 0.1 mM isopropyl-β-d-thiogalactopyranoside, and proteins were purified by Glutathione Sepharose 4B (GE Healthcare) following the manufacturer’s instructions. MBP-MYC2 and MBP proteins were expressed and purified as described (Chen et al., 2011). GST-EIN3 protein was expressed and purified using the Bac-to-Bac Baculovirus Expression System (Invitrogen) and purified as described (Wen et al., 2012).
EMSA
Oligonucleotide probes (MBS1, 5′-AAGAATTTGTATGTTCATC-3′ and 5′-CGACTGATGACAAATTTTG-3′; MBS2, 5′-TGATCTTGCGTACCCAATTG-3′ and 5′-GTCAGCATCGTTTATGTTG-3′) were synthesized and labeled with the Biotin 3′ End DNA Labeling Kit (Pierce). Probe sequences of HLS1 were 5′-AATACGTTGAAGCCCACTATTTCAAAATTTACTAGGAGTATTTATN-3′ and 5′-TAAATACTCCTAGTAAATTTTGAAATAGTGGGCTTCAACGTATT-3′ as described (An et al., 2012). EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Pierce). Briefly, 20 fmol of labeled probe was incubated in 1× binding buffer, 2.5% glycerol, 50 mM KCl, 5 mM MgCl2, and 10 mM EDTA with or without proteins at room temperature for 20 min. For nonlabeled probe competition, 4 pmol of nonlabeled probe was added to the reactions.
ChIP-PCR
ChIP-PCR was performed following the literature with minor modifications (Gendrel et al., 2005). Two grams of etiolated seedlings was cross-linked in 1% formaldehyde, and the chromatin was isolated. The indicated antibodies were added to the sonicated chromatin followed by incubation overnight to precipitate bound DNA fragments. Anti-MYC antibodies were purchased from TDY Biotech. After immobilization using Recombinant Protein G-Sepharose 4B (Invitrogen), bound DNA was eluted and amplified by primers corresponding to sequences neighboring the MYC2 binding sites in the EBF1 promoter.
Pull-Down Assay
In vitro–expressed and purified GST fusion proteins (GST-EIN3 and the negative control GST-GFP) were incubated with Glutathione Sepharose 4B (GE Healthcare) in pull-down buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 10% glycerol, 0.5 mM EDTA, 0.1% Triton X-100, 5 mM mercaptoethanol, and 1× protease inhibitor cocktail) for 4 h at 4°C, and then MBP fusion proteins were added and incubated for another 3 h at 4°C. After washing five times with pull-down buffer, precipitated Sepharose beads were collected by brief centrifugation (2000g, 2 min) and then resuspended in protein extraction buffer. Proteins were separated by SDS-PAGE and detected with the corresponding antibody.
Co-IP Assay
Three-day-old etiolated seedlings were treated with 10 μM JA and 100 μM ACC for 3 h before harvest. Proteins were extracted with co-IP buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 10% glycerol, 0.5 mM EDTA, 0.1% Triton X-100, 5 mM mercaptoethanol, and 1× protease inhibitor cocktail). After incubation on ice for 30 min, plant extracts were sonicated and then centrifuged. Cleared extract was combined with anti-MYC antibody (TDY Biotech), together with Recombinant Protein G-Sepharose 4B (Invitrogen), and incubated for 3 h at 4°C. After washing five times with co-IP buffer, agarose beads were collected by centrifugation (2000g, 2 min) and then resuspended in protein extraction buffer. Proteins were separated by SDS-PAGE and detected with the corresponding antibody.
Firefly Luciferase Complementation Imaging Assay
The coding sequences of EIN3 and MYC2 were inserted into the multiple cloning sites of pCAMBIA1300-nLUC (SacI-SalI) and pCAMBIA1300-cLUC (KpnI-XbaI), respectively (Chen et al., 2008). Agrobacterium tumefaciens GV3101 carrying the indicated constructs was cultured to OD600 = 0.5, combined with equal volumes of the adjusted culture for specific groups as shown in the figure legends, and incubated at room temperature without shaking for 3 h and infiltrated into Nicotiana benthamiana leaves. Luciferase activity was detected 3 d after infiltration with the LB 985 NightSHADE system (Berthold Technologies).
Dual-Luciferase Reporter System
Two kilobases of HLS1 promoter was amplified from the Arabidopsis genomic DNA, and the fragment was digested with KpnI and NcoI, inserted into the pGreen II 0800-LUC vector, and used as a reporter plasmid (Hellens et al., 2005).
The coding sequences of EIN3 and MYC2 were amplified by PCR, and both were digested with SpeI-KpnI, inserted into pGreen II 62-SK, and used as effector plasmids (Hellens et al., 2005).
Agrobacterium strain GV3101 carrying the reporter plasmid (ProHLS1:LUC) and specific effector plasmids (empty vector, EIN3, MYC2, or EIN3+MYC2) was cultured to OD600 = 0.5, combined at equal volumes of the indicated combinations, incubated at room temperature without shaking for 3 h, and infiltrated into N. benthamiana leaves. Ten micromoles of JA was infiltrated into the corresponding spots of N. benthamiana leaves 3 h before sample collection. The dual-luciferase reporter system (Promega) was used to analyze the transient expression in N. benthamiana leaves 3 d after infiltration. The activities of firefly (Photinus pyralis) and Renilla reniformis luciferases were measured sequentially from a single sample on a GLO-MAX 20/20 luminometer (Promega). The ratio of LUC to REN was calculated to indicate the final transcriptional activity. Five biological repeats were measured for each sample.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: EIN3 (At3g20770), EIL1 (At2g27050), EBF1 (At2g25490), EBF2 (At5g25350), CTR1 (At5g03730), COI1 (At2g39940), MYC2 (At1g32640), HLS1 (At4g37580), ERF1 (At3g23240), and TUB2 (At5g62690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. JA Represses ET-Induced Hook Formation.
Supplemental Figure 2. JA Represses EIN3 Accumulation in a Manner That Is Dependent on SCFEBF1/2.
Supplemental Figure 3. JA Induces EBF1 Expression but Not EBF2 Expression.
Supplemental Figure 4. The Protein Stability of EBF1 Is Not Affected by JA Treatment.
Supplemental Figure 5. MYC2 Does Not Bind to the HLS1 Promoter in Vivo.
Supplemental Table 1. Primer Sequences Used in This Study.
Supplementary Material
Acknowledgments
We thank Chuanyou Li and Jian-Min Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for kindly providing research materials. We also thank all members of the Guo laboratory for stimulating discussions and suggestions. This work was supported by the National Natural Science Foundation of China (Grants 91217305 and 91017010), the National Basic Research Program of China (973 Program; Grant 2012CB910902), and the Ministry of Agriculture of China (Grant 2010ZX08010-002) to H.G.
AUTHOR CONTRIBUTIONS
H.G., X.Z., and Z.Z. conceived and designed this study. Z.Z. and X.Z. performed all the phenotype observations, genetic studies, EMSA, chromatin immunoprecipitation, and immunoblot experiments. F.A. provided transgenic materials. D.H. performed the transient expression assay. P.L. performed GUS measurements. C.Y. and J.S. performed EIN3 protein expression and purification. Z.Z., X.Z., and H.G. wrote the article. All authors analyzed the data and discussed the article.
Glossary
- ET
ethylene
- GA
gibberellin
- JA
jasmonate
- JA-ET
jasmonate-ethylene
- Col-0
Columbia-0
- ACC
1-aminocyclopropane-1-carboxylic acid
- MS
Murashige and Skoog
- qRT-PCR
quantitative RT-PCR
- EMSA
electrophoretic mobility shift assay
- ChIP-PCR
chromatin immunoprecipitation-PCR
- co-IP
coimmunoprecipitation
- MeJA
methyl jasmonate
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Alabadí D., Gil J., Blázquez M.A., García-Martínez J.L. (2004). Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 134: 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Solano R., Wisman E., Ferrari S., Ausubel F.M., Ecker J.R. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F., Zhang X., Zhu Z., Ji Y., He W., Jiang Z., Li M., Guo H. (2012). Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22: 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C.E., Xie D., Turner J.G. (1995). Coi1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 109: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205. [DOI] [PubMed] [Google Scholar]
- Chang K.N., et al. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144. [DOI] [PubMed] [Google Scholar]
- Chen H., Xue L., Chintamanani S., Germain H., Lin H., Cui H., Cai R., Zuo J., Tang X., Li X., Guo H., Zhou J.M. (2009). ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Sun L., Qi T., Zhang B., Peng W., Liu Y., Xie D. (2011). The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4: 279–288. [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- De Grauwe L., Vandenbussche F., Tietz O., Palme K., Van Der Straeten D. (2005). Auxin, ethylene and brassinosteroids: Tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol. 46: 827–836. [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. (1998). SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1: 316–323. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Jiang L., Wang F., Yu D. (2013). Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.R., Ecker J.R. (1998). The ethylene gas signal transduction pathway: A molecular perspective. Annu. Rev. Genet. 32: 227–254. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2013). MYC2: The master in action. Mol. Plant 6: 686–703. [DOI] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441. [DOI] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2008). Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 55: 821–831. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Deng X.W., Kim W.T. (2006). Possible role of light in the maintenance of EIN3/EIL1 stability in Arabidopsis seedlings. Biochem. Biophys. Res. Commun. 350: 484–491. [DOI] [PubMed] [Google Scholar]
- Lehman A., Black R., Ecker J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85: 183–194. [DOI] [PubMed] [Google Scholar]
- Li H., Guo H. (2007). Molecular basis of the ethylene signaling and response pathway in Arabidopsis. J. Plant Growth Regul. 26: 106–117. [Google Scholar]
- Li H., Johnson P., Stepanova A., Alonso J.M., Ecker J.R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7: 193–204. [DOI] [PubMed] [Google Scholar]
- Lingam S., Mohrbacher J., Brumbarova T., Potuschak T., Fink-Straube C., Blondet E., Genschik P., Bauer P. (2011). Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23: 1815–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E., Hangarter R.P. (1993). Light-stimulated apical hook opening in wild-type Arabidopsis thaliana seedlings. Plant Physiol. 101: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo G., Guo H., Gregory B.D., Nourizadeh S.D., Aguilar-Henonin L., Li H., An F., Guzman P., Ecker J.R. (2006). ETHYLENE-INSENSITIVE5 encodes a 5′→3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. USA 103: 13286–13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689. [DOI] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V., Ecker J.R. (1999). Regulation of differential growth in the apical hook of Arabidopsis. Development 126: 3661–3668. [DOI] [PubMed] [Google Scholar]
- Roman G., Lubarsky B., Kieber J.J., Rothenberg M., Ecker J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139: 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F., Fernández-Calvo P., Zander M., Diez-Diaz M., Fonseca S., Glauser G., Lewsey M.G., Ecker J.R., Solano R., Reymond P. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu C., Figueroa P., Depew C.L., Cooke T.F., Sheard L.B., Moreno J.E., Katsir L., Zheng N., Browse J., Howe G.A. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24: 536–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Huang H., Gao H., Wang J., Wu D., Liu X., Yang S., Zhai Q., Li C., Qi T., Xie D. (2014). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I., Rowe M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Turner J.G., Ellis C., Devoto A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.): S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Petrásek J., Zádníková P., Hoyerová K., Pesek B., Raz V., Swarup R., Bennett M., Zazímalová E., Benková E., Van Der Straeten D. (2010). The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606. [DOI] [PubMed] [Google Scholar]
- Vriezen W.H., Achard P., Harberd N.P., Van Der Straeten D. (2004). Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J. 37: 505–516. [DOI] [PubMed] [Google Scholar]
- Wen X., Zhang C., Ji Y., Zhao Q., He W., An F., Jiang L., Guo H. (2012). Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22: 1613–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S., Yoo S.D., Sheen J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525. [DOI] [PubMed] [Google Scholar]
- Zander M., La Camera S., Lamotte O., Métraux J.P., Gatz C. (2010). Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 61: 200–210. [DOI] [PubMed] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., Guo H. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 106: 21431–21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.