A mechanism was identified by which degradation of D14, the putative receptor of the plant hormone strigolactone, is promoted by this hormone. This mechanism, which is mediated by protein degradation via the proteasome machinery and requires the activity of the F-box protein MAX2, should cause a substantial drop in strigolactone perception, thereby limiting hormone signaling duration and intensity.
Abstract
Strigolactones (SLs) are phytohormones that play a central role in regulating shoot branching. SL perception and signaling involves the F-box protein MAX2 and the hydrolase DWARF14 (D14), proposed to act as an SL receptor. We used strong loss-of-function alleles of the Arabidopsis thaliana D14 gene to characterize D14 function from early axillary bud development through to lateral shoot outgrowth and demonstrated a role of this gene in the control of flowering time. Our data show that D14 distribution in vivo overlaps with that reported for MAX2 at both the tissue and subcellular levels, allowing physical interactions between these proteins. Our grafting studies indicate that neither D14 mRNA nor the protein move over a long range upwards in the plant. Like MAX2, D14 is required locally in the aerial part of the plant to suppress shoot branching. We also identified a mechanism of SL-induced, MAX2-dependent proteasome-mediated degradation of D14. This negative feedback loop would cause a substantial drop in SL perception, which would effectively limit SL signaling duration and intensity.
INTRODUCTION
Plant architecture is determined in great part by branching patterns. Branches develop from axillary meristems (AMs) that form at the base of leaves. AMs undergo a growth period during which they initiate leaf primordia (and sometimes flower meristems) with no internode elongation and give rise to axillary buds. These buds can be developmentally arrested for long time periods or continue to grow to generate lateral shoots. The great diversity of branching patterns among flowering plants is determined both by the arrangement of leaves in the stem (phyllotaxis), where axillary buds are formed, and by the decision of buds to grow out to give rise to a branch or remain dormant. Phyllotaxis is mostly unaffected by external cues, whereas axillary bud outgrowth shows great plasticity, as it is regulated by developmental and environmental stimuli, such as apical dominance, nutrient availability, and light quality.
Study of the genetic pathways that regulate branch development shows a large degree of conservation over a wide range of angiosperm species, such as maize (Zea mays), rice (Oryza sativa), petunia (Petunia hybrida), tomato (Solanum lycopersicum), pea (Pisum sativum), and thale cress (Arabidopsis thaliana). Genetic and regulatory divergence have nonetheless been found (Drummond et al., 2011; Delaux et al., 2012; Challis et al., 2013), which could help to account for the variations in branching patterns between groups. In all species analyzed, long-distance signaling and local gene activity participate in the regulation of axillary bud growth. Systemic signaling that prevents branch outgrowth involves auxin and the recently identified hormone strigolactone (SL; Gomez-Roldan et al., 2008; Umehara et al., 2008).
Auxin is synthesized in the shoot apex and transported basipetally (toward the root) through the polar auxin transport stream (PATS) (Thimann and Skoog, 1933; Skoog and Thimann, 1934). SL is synthesized in the root and other plant organs and is transported acropetally (toward the shoot) in the xylem (Foo et al., 2001; Kohlen et al., 2011). Auxin is thought to suppress branching indirectly through two mechanisms, by modulating the activity of second messengers (i.e., SL and cytokinin; Nordström et al., 2004; Brewer et al., 2009) and due to competition between shoot apices for auxin export into the PATS (Domagalska and Leyser, 2011). SL is also proposed to have two roles in the regulation of shoot branching, including dampening of auxin transport, thus enhancing competition between buds for their common auxin sink (Bennett et al., 2006; Prusinkiewicz et al., 2009; Crawford et al., 2010; Balla et al., 2011; Shinohara et al., 2013), and transcriptional activation of the growth repressor BRANCHED1 (BRC1) (Braun et al., 2012; Dun et al., 2012, 2013). BRC1-like genes, expressed inside axillary buds, encode class II TCP (for TEOSINTE BRANCHED1, CYCLOIDEA, and PCF) transcription factors (Martín-Trillo and Cubas, 2010) that delay bud growth and development and promote bud dormancy (Aguilar-Martínez et al., 2007; Finlayson, 2007; Martín-Trillo et al., 2011; Braun et al., 2012; Dun et al., 2012; González-Grandío et al., 2013).
Several genes involved in SL synthesis and perception, whose mutants show increased branching and reduced stature, have been characterized in a number of species. Active SL is synthesized from β-carotene by a set of proteins conserved in mono- and dicotyledons. The DWARF27 (D27) gene, first characterized in rice, encodes an isomerase that acts at the initial steps of this pathway by transforming trans-β-carotene to 9-cis-β-carotene (Lin et al., 2009; Alder et al., 2012; Waters et al., 2012a). CAROTENOID CLEAVAGE DIOXYGENASE7 (CCD7) and CCD8 (HTD1/D17/DAD3/RMS5/MAX3 and D10/DAD1/RMS1/MAX4, respectively) (Sorefan et al., 2003; Booker et al., 2005; Snowden et al., 2005; Johnson et al., 2006; Zou et al., 2006; Arite et al., 2007; Drummond et al., 2009; Pasare et al., 2013) produce carlactone, a putative intermediate compound in the pathway (Alder et al., 2012; Seto et al., 2014). The MORE AXILLARY GROWTH1 (MAX1) gene, so far identified only in Arabidopsis, encodes a cytochrome P450 monooxygenase that acts downstream of CCD7 and CCD8 (Booker et al., 2005). The phenotype of excessive branching in mutants for all these SL synthesis genes can be rescued by application of SL.
A second group of genes whose mutants are at least partially insensitive to SL are thought to be involved in SL perception and signaling. MAX2 genes (Stirnberg et al., 2002; Ishikawa et al., 2005; Johnson et al., 2006) encode F-box proteins that participate in SCF (for Skp1, Cullin, RBX1, F-box protein) complexes of E3 ubiquitin ligases. D14/DECREASED APICAL DOMINANCE2/HTD2 (D14/DAD2/HTD2) code for α/β-fold hydrolases that bind and hydrolyze SL in vitro (Arite et al., 2009; Gao et al., 2009; Liu et al., 2009; Gaiji et al., 2012; Hamiaux et al., 2012; Waters et al., 2012b; Kagiyama et al., 2013; Nakamura et al., 2013). In yeast two-hybrid assays, the petunia DAD2 and MAX2b proteins interact in an SL concentration–dependent manner (Hamiaux et al., 2012). This interaction has been confirmed in rice for D14 and D3 (Jiang et al., 2013; Zhou et al., 2013). These findings have led to the proposal that DAD2/D14 is the SL receptor that, after interaction with SL, binds MAX2/D3 to select target proteins for degradation. Rice proteins D53 and SLENDER RICE1 (SLR1) (Jiang et al., 2013; Zhou et al., 2013; Nakamura et al., 2013) and Arabidopsis BRI1-EMS-SUPPRESSOR1 (BES1) (Wang et al., 2013) are likely targets for degradation through this pathway. A protein closely related to D14/DAD2, D14-like/KARRIKIN-INSENSITIVE2 (KAI2), genetically interacts with MAX2 (Waters et al., 2012b). KAI2 binds a second hormone, karrikin (KAR), a compound present in bushfire smoke that triggers seed germination (Bythell-Douglas et al., 2013; Guo et al., 2013b; Kagiyama et al., 2013). These observations suggested that D14 and KAI2 mediate SL and KAR signaling, respectively, through interaction with MAX2 (Waters et al., 2012b, 2013).
In this study, we screened for mutants with excessive branching at high-planting density and identified seto5, an allele of the Arabidopsis D14 gene. Seto5 carries a point mutation in a conserved residue of the protein that reveals an amino acid position essential for D14 function. We compared D14 promoter activity and D14 protein distribution and found that they are not identical, which might indicate that D14 moves between cells within a short range. Nonetheless, it cannot move acropetally from the root to rescue the shoot branching phenotype of d14 mutants. Most notably, we discovered a mechanism of negative feedback regulation by which SL induces rapid degradation of D14, which could effectively limit the duration and intensity of SL signaling.
RESULTS
Identification and Phenotypic Characterization of the seto5 Mutant
To identify genes involved in the regulation of shoot branching, we performed a genetic screen to search for mutants with increased numbers of lateral shoots. We grew ethyl methanesulfonate–mutagenized Columbia-0 (Col-0) plants at a density of nine plants/36 cm2 pot, a condition that leads to complete branch suppression in wild-type plants (Aguilar-Martínez et al., 2007). We screened for individuals with four or more lateral branches and termed them seto (bush in Spanish) mutants. One plant bred true (seto5), yielding plants that consistently displayed a bushy phenotype. We backcrossed seto5 to wild-type Col-0 plants and confirmed 3:1 wild type:mutant segregation in the F2 population, indicating that a single locus is responsible for the phenotype.
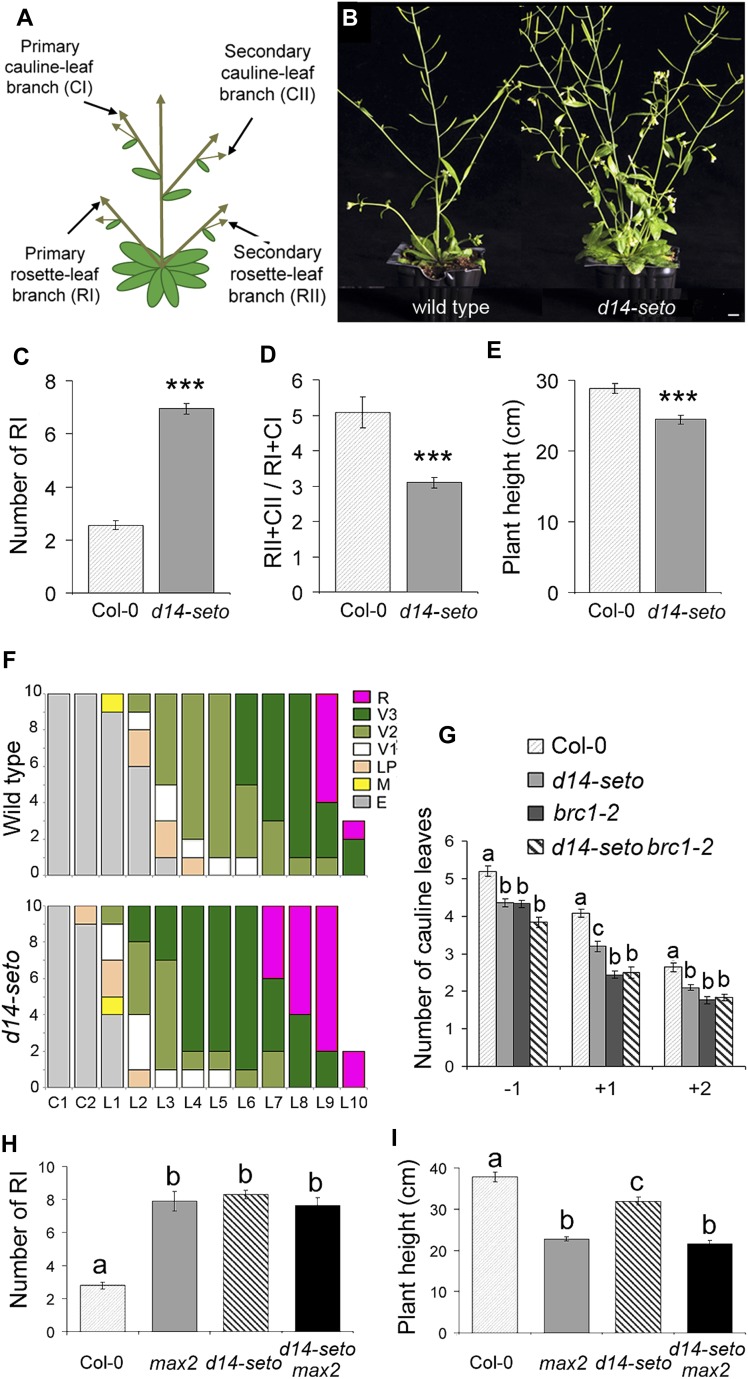
seto5 mutants, backcrossed twice to Col-0, had a significantly larger number of primary rosette branches (RI, Figure 1) than the wild type (Figures 1B and 1C). In growth conditions in which wild-type plants had approximately two primary rosette branches at maturity, mutants had more than six primary rosette branches. seto5 plants had fewer secondary branches (RII and CII, Figure 1A) relative to primary branch number (Figure 1D). In addition, mutant plants were slightly shorter than controls (Figure 1E).
Figure 1.
Phenotypes of d14-seto Single and Double mutants.
(A) Arabidopsis branching structure.
(B) Close-up of mature wild-type Col-0 (left) and seto5 (here termed d14-seto) (right) rosettes showing their lateral shoot phenotype. Bar = 1 cm.
(C) Number of primary rosette branches (RI) of wild-type and d14-seto plants.
(D) Number of secondary branches (RII+CII) relative to the number of primary branches (RI+CI).
(E) Height of the main inflorescence of the same set of plants.
(F) Developmental stages of buds in the axils of cotyledons (C1 and C2) and rosette leaves (L1 to L10) of wild-type (top) and d14-seto (bottom) individuals. R, reproductive stage, V1 to V3, vegetative stages; LP, leaf primordium stages; M, meristem; E, empty axil. Developmental stages are as defined (Aguilar-Martínez et al., 2007) (n = 10).
(G) Flowering time, expressed as number of leaves, of lateral inflorescences of wild-type, d14-seto, and brc1-2 mutants and d14-seto brc1-2 double mutants. −1, uppermost RI; +1 and +2, first and second basal-most CI branches.
(H) and (I) Number of RI branches (H) and height of the main inflorescence (I) of d14-seto max2-1 double mutants. Asterisks denote significant differences in Student’s t tests (P < 0.0001). Letters denote significant differences in one-way ANOVA test (Tukey test P < 0.05). Data shown as mean ± se (n = 20).
To study the seto5 mutant phenotype during early bud development, we compared the developmental stage of wild-type and seto5 axillary buds formed at identical node positions in plants grown for 28 d in long photoperiods. At this stage, all plants had undergone flowering and were starting to bolt. In wild-type plants, buds nearest the apex were more developmentally advanced than those farther from the apex (Figure 1F, top). This gradient was also observed in seto5 mutants, but all buds were developmentally more advanced than those of the wild type (Figure 1F, bottom).
To determine whether flowering time of lateral inflorescences was also accelerated, we counted, in lateral shoots, the number of leaves formed before emergence of the first flower. In wild-type and seto5 plants, we studied the branch formed in the most apical rosette leaf (−1) and those in the two most basal cauline leaves (+1 and +2) (Figure 1G). seto5 mutant lateral inflorescences had one fewer leaf than the wild type, as reported for mutants in the BRC1 locus (Niwa et al., 2013; Figure 1G). Double mutant seto5 brc1-2 plants resembled brc1-2 mutants, indicating that brc1-2 is epistatic for this character (Figure 1G).
These results suggest that, in wild-type plants, the SETO5 locus delays axillary bud development, lateral shoot outgrowth, and flowering time of lateral inflorescences.
Cloning of seto5
High-resolution mapping (Supplemental Figure 1) combined with whole-genome sequencing of seto5 individuals allowed the identification of a single homozygous nonsynonymous nucleotide substitution localized within the 163-kb interval of chromosome 3 that included seto5. It affected the At3g03990 gene and was a C→T transition at position 506 relative to the predicted translation start site (TSS). This caused a Pro→Leu substitution at position 169 of the encoded protein (Supplemental Figure 1). The gene At3g03990 is At-D14 (Waters et al., 2012b), the ortholog of rice D14 (Arite et al., 2009; Gao et al., 2009; Liu et al., 2009) and petunia DAD2 (Hamiaux et al., 2012). These loci are required for SL-dependent inhibition of shoot branching.
The F1 progeny from crosses between seto5 mutants and the strong mutant d14-1 in which full-length transcripts are undetectable (Waters et al., 2012b; Supplemental Figure 2A) were phenotypically mutant: They showed an increased number of primary rosette branches and reduced stature relative to wild-type plants (Supplemental Figures 2B and 2C). This confirmed that the phenotype of seto5 was caused by the point mutation in D14, and we renamed this mutant d14-seto. A 2.3-kb genomic region comprising 553 bp 5′ of the TSS, the 804-bp coding sequence (CDS), and a 916-bp region 3′of the stop codon (Supplemental Figure 2A) of D14 was sufficient to complement the d14-seto mutation (Supplemental Figures 2D and 2E), indicating that this sequence contained the regulatory regions necessary for D14 function. Constructs carrying only the 553 bp 5′ of the TSS and the 804-bp CDS also rescued the d14-seto mutant phenotype (Supplemental Figure 2F).
We analyzed the phenotype of two additional mutant lines with T-DNA insertions at 153 and 71 bp 5′ of the D14 TSS, termed d14-3 and d14-4, respectively (SALK_057876 and GABI-KAT-759C03; Supplemental Figure 2A). Neither showed excess of branching or reduced stature in the homozygous condition nor in heteroallelic combination with d14-seto (Supplemental Figure 2B) despite d14-3 showing significantly reduced levels of D14 mRNA (Supplemental Figure 3). This suggests that D14 mRNA levels are not limiting in Arabidopsis. We also generated d14-seto max2-1 double mutants whose phenotype resembled that of max2-1 plants (Figures 1H and 1I), confirming D14 involvement in the SL pathway.
In summary, the d14-seto mutant is a strong loss-of-function allele of D14 that carries a point mutation in the CDS of the gene.
Relationship between D14 and BRC1
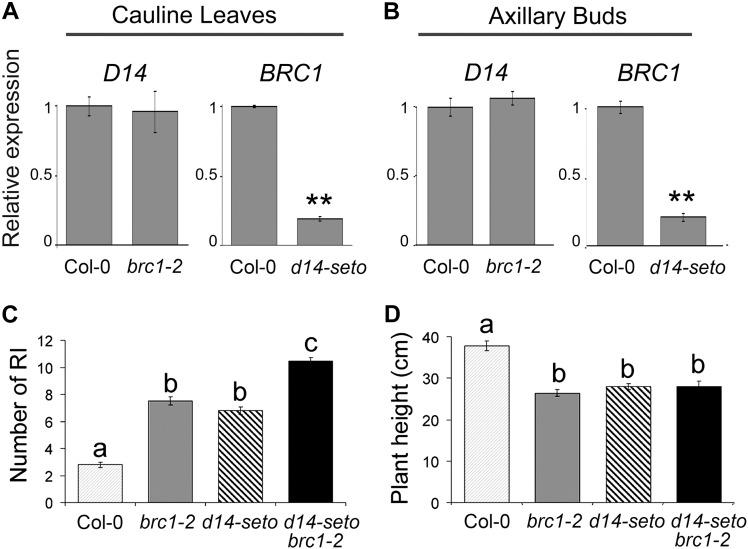
It has been proposed that BRC1 is downstream of the SL pathway in pea, Arabidopsis, and rice (Aguilar-Martínez et al., 2007; Finlayson, 2007; Brewer et al., 2009; Braun et al., 2012; Dun et al., 2012, 2013; Minakuchi et al., 2010). To further test this relationship, we studied transcript levels of each gene in cauline leaves and axillary buds of the reciprocal single mutant. D14 mRNA levels were unaltered in brc1-2 mutants (Figures 2A and 2B), whereas BRC1 mRNA levels were greatly reduced in d14-seto mutants, in both axillary buds and cauline leaves (Figures 2A and 2B). Then, we studied the branching and height phenotypes of wild-type, d14-seto, and brc1-2 plants as well as double mutant d14-seto brc1-2 plants. Double mutants had significantly more branches than the single mutants, indicating additivity of the phenotypes (Figure 2C). This was in contrast with the flowering time and height phenotypes, which showed no additivity (Figures 1G and 2D).
Figure 2.
Genetic Relationship between BRC1 and D14.
(A) and (B) Transcript abundance of D14 and BRC1 in cauline leaves (A) and axillary buds (B) of wild-type, d14-seto, and brc1-2 quantified by qPCR. Data shown as mean ±se (n = 3 to 5 biological replicates). Asterisks denote significant differences in Student’s t test (**P < 0.01).
(C) Number of RI (primary rosette branches) of wild-type, brc1-2, d14-seto, and d14-seto brc1-2 F3 plant siblings.
(D) Height of the main inflorescence of the same plants. Letters denote significant differences in one-way ANOVA test (Tukey test P < 0.05). Data shown as mean ±se (n = 20).
These results support both the transcriptional regulation of BRC1 by the SL pathway and also partially nonoverlapping roles for BRC1 and SL-related genes in the regulation of shoot branching (see Discussion).
The d14-seto Protein
D14 belongs to the α/β-fold hydrolase superfamily (Ishikawa et al., 2005; Arite et al., 2009). The crystal structures of petunia DAD2, rice D14, Arabidopsis D14, and KAI2 show that they have a canonical α/β-fold hydrolase domain with a substrate binding pocket and the Ser-His-Asp catalytic triad necessary for hydrolase activity. A cap formed by four helices partially covers the active site with nonpolar residues (Hamiaux et al., 2012; Bythell-Douglas et al., 2013; Guo et al., 2013b; Kagiyama et al., 2013; Zhao et al., 2013; Figure 3C). In D14 proteins, the pocket can bind the synthetic SL analog GR24 (Hamiaux et al., 2012; Kagiyama et al., 2013; Nakamura et al., 2013). In KAI2, the pocket binds the synthetic KAR KAR1 (Bythell-Douglas et al., 2013; Guo et al., 2013b; Kagiyama et al., 2013). Protein destabilization and conformational changes have been detected after ligand binding in both protein types (Hamiaux et al., 2012; Guo et al., 2013b; Nakamura et al., 2013).
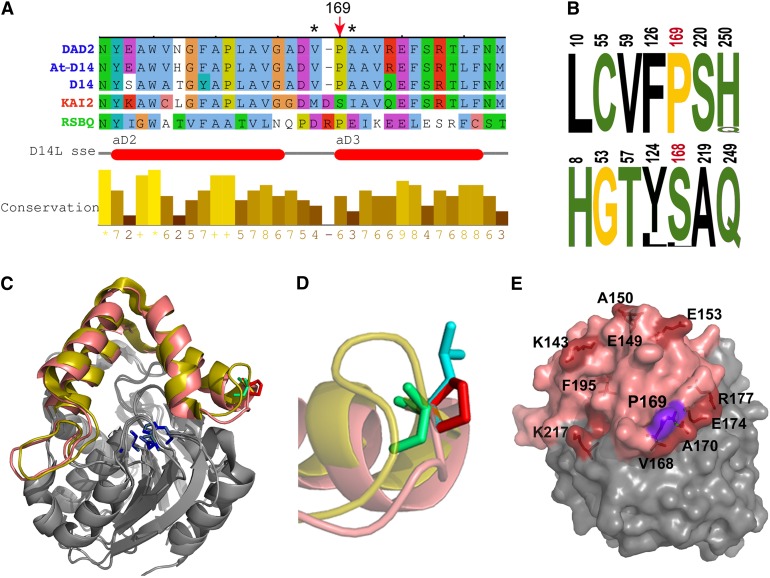
Figure 3.
The d14-seto Protein.
(A) Sequence alignment of the Arabidopsis D14 segment comprising the Pro169Leu mutation, with ortholog sequences petunia DAD2 (Hamiaux et al., 2012), rice D14 (Arite et al., 2009; Gao et al., 2009; Liu et al., 2009; Hamiaux et al., 2012), paralog KAI2 (Waters et al., 2012b), and related bacterial protein RsbQ (Brody et al., 2001). Red arrow indicates Pro-169 and corresponding amino acid Ser-168 in KAI2. Asterisks indicate residues Met-166 and Ile-169, which undergo conformational changes in KAI2 after KAR1 binding (Guo et al., 2013b). Horizontal red bars indicate the position of two of the KAI2 cap α-helices (Kagiyama et al., 2013).
(B) Logos of SDPs that differ in D14 (top) and KAI2 (bottom) ortholog sequences. Numbering corresponds to D14 and KAI2 protein sequences. Pro-169 and Ser-168 are shown in red. Hydrophobic residues are indicated in black, polar residues in green, and Gly and Pro in yellow. Letter size represents percentage of conservation within protein classes.
(C) Front view of D14 (PDB:4ih4) and KAI2 (PDB:3w06) structural alignment. Helical caps of D14 and KAI2 are highlighted in salmon pink and yellow, respectively. Active site residues are in blue, D14 Pro-169 is in red, and KAI2 S168 is in green.
(D) Close-up view and side chain superposition of wild-type D14 P169 (red), mutant Leu-169 (blue), and KAI2 Ser-168 (green). Note that the Pro-169 side chain is exposed to the solvent and that KAI2 loop (yellow) is longer than that of D14.
(E) D14 structure in surface representation. Residues corresponding to residues in KAI2 that undergo side-chain movement after KAR1 binding are labeled and highlighted in red, Pro-169 in purple and cap domain in pink.
The strong phenotype of d14-seto, which has a single Pro169Leu substitution, suggested an important role for this residue. We mapped this position in the 3D protein structure using PyMOL (www.pymol.org) and found that it is located at the N terminus of cap helix αD3, with the side chain exposed to the solvent (Figures 3A and 3C to 3E; Supplemental Figure 4). To study potential destabilizing effects of this mutation, we compared the differences in free energy of unfolding (ΔΔG) between the wild-type and mutant proteins. No large destabilizing effects were predicted for this mutation (Supplemental Table 1).
Sequence comparison of a large collection of D14-related proteins revealed that Pro-169 is fully conserved among D14-related proteins, while KAI2-related proteins have a highly conserved Ser in this position (Figure 3A; Supplemental Figure 4). Indeed, Pro-169/Ser-168 is a specificity-determining position (SDP; de Juan et al., 2013) that may provide functional specificity to D14 and KAI2 (Figure 3B). This position is adjacent to a loop whose length and composition also differ between D14-type and KAI2-type proteins (Figures 3A, 3C, and 3D; Supplemental Figure 4).
KAR1 binding to KAI2 causes conformational changes in the side chains of 11 KAI2 protein residues (Guo et al., 2013b). Four of them (Met-166, Ile-169, Glu-173, and Arg-176) are in close proximity to Ser-168 (Supplemental Figure 5). Mapping of the equivalent residues in D14 (Val-168, Ala-170, Glu-174, and Arg-177; Supplemental Figure 4) confirmed that they also cluster around Pro-169 (Figure 3E).
In summary, the d14-seto allele has a Pro→Leu mutation in an SDP located not in the active site, but in the external surface of one of the D14 protein cap helices. Calculations of free energy of unfolding (ΔΔG) indicate that this mutation should not destabilize the 3D structure. However, this mutation is located in a protein surface region that, by analogy with KAI2, could undergo conformational changes after SL binding and interfere with protein–protein interactions essential for functional SL signaling.
D14 Expression Patterns and Protein Distribution during Arabidopsis Development
To identify the plant tissues in which D14 has an important role, we studied its mRNA and protein distribution during plant development. First, we analyzed its mRNA levels in different tissues by quantitative real-time PCR (qPCR). In 28-d-old plants grown in long days (which had undergone flowering), D14 was transcribed at high levels in rosette and cauline leaves and at lower levels in axillary buds, inflorescences, stems, and roots (Supplemental Figure 6).
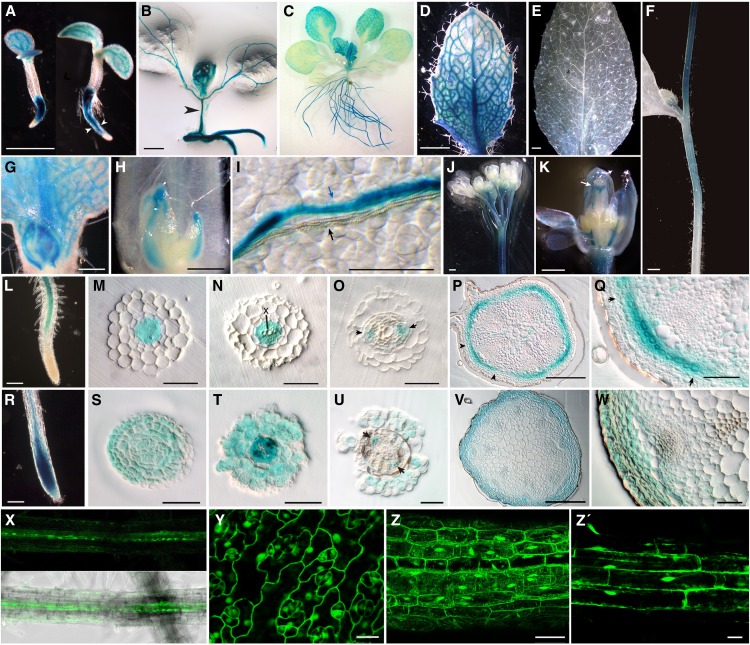
We then generated transcriptional and translational fusions lines of D14. For this, we made three constructs carrying the 540 bp upstream of the D14 TSS fused to the CDS of the β-GLUCURONIDASE (GUS) gene (D14pro:GUS) or the same D14 promoter region fused to the CDSs of D14 and GUS or GREEN FLUORESCENT PROTEIN (GFP; D14pro:D14:GUS or D14pro:D14:GFP). These two types of lines could complement the phenotype of d14-seto mutants (Supplemental Figure 2). We studied GUS/GFP expression in nine representative T3 homozygous lines. In 5-d-old D14pro:GUS seedlings, GUS staining accumulated in the root and in the developing vascular tissue of cotyledons. Root expression became progressively restricted to the vascular cylinder (Figure 4A). In 10-d-old seedlings, the vascular tissue of the hypocotyl also showed GUS activity (Figure 4B). In general, root expression was undetectable in the meristematic zone but was strong in the differentiation zone and detectable in the elongation zone of some lines (Figure 4L). Analysis of thin sections of plastic-embedded roots confirmed that GUS expression was progressively restricted to phloem strands (Figures 4M to 4O). The lack of expression in the root meristem and GUS accumulation in root phloem cells was consistent with previous high-resolution expression profilings (Brady et al., 2007). In the aerial part, the D14 promoter was active throughout leaf primordia and young leaves (Figures 4B and 4C). Expression was progressively restricted to the phloem in expanding cotyledons, leaves, sepals, petals, and stamen filaments (Figures 4B, 4D, 4I, and 4K) and was almost undetectable in mature leaves (Figure 4E). GUS signal was also strong in the style (Figure 4K), flower pedicels (Figure 4J), and the apical-most region of the inflorescence stems but not in the basal-most region (Figures 4F and 4J). Thin sections showed that GUS accumulated in a ring of cortex cells in the stem (Figures 4P and 4Q). Cortex sectors adjacent to the vascular bundles showed more GUS expression that those next to interfascicular regions (Figures 4P and 4Q). In addition, the D14 promoter was active throughout axillary buds (Figures 4G and 4H). The general distribution of D14:GUS in D14pro:D14:GUS lines paralleled that of the promoter activity but D14:GUS was more widespread: In nine independent homozygous lines, the protein was also detected in the root meristem (Figures 4R and 4S), root and shoot epidermis (Figures 4T to 4W), and shoot vasculature (Figures 4U and 4W).
Figure 4.
D14 Promoter Activity and D14 Protein Distribution during Arabidopsis Development.
GUS histochemical activity of Arabidopsis D14pro:GUS ([A] to [Q]) and D14pro:D14:GUS ([R] to [W]) transgenic plants.
(A) Five-day-old transgenic seedlings. The plant on the right, more advanced in development, shows expression in the root more restricted to the vascular cylinder (arrowheads) than that of the less developmentally advanced (left).
(B) Ten-day-old seedling with GUS activity in the vascular tissue of the hypocotyl (arrowhead).
(C) Eighteen-day-old vegetative rosette.
(D) Young rosette leaf from plant in (C).
(E) Mature cauline leaf from 30-day-old plant.
(F) Stem of the main inflorescence showing a gradient of GUS activity with a maximum near the apex.
(G) Bud in the axil of a young rosette leaf.
(H) Bud in the axil of a mature rosette leaf.
(I) Detail of a rosette leaf surface. Note the separation between the xylem (white) bundle (black arrow) and the phloem (blue) bundle expressing GUS (blue arrow).
(J) Main inflorescence. GUS accumulates in the apical-most stem region and in flower pedicels.
(K) Close-up of a developing flower. Signal in the style is indicated (white arrow).
(L) Root tip.
(M) to (O) The 3-μm transverse plastic-embedded sections of root similar to that in (L).
(M) Distal section showing GUS staining in procambium cells.
(N) GUS is excluded from xylem cells (X).
(O) More proximal section showing promoter activity in phloem cells (arrows).
(P) Transverse plastic-embedded section of a stem internode of the primary inflorescence.
(Q) Close-up of a section similar to that shown in (M), with cortex cells but not epidermis cells expressing GUS. Notice the stronger signal in the vascular bundle sector flanked by the arrows in (P) and (Q).
(R) Root tip similar to that in (L). D14:GUS is present in the root tip.
(S) to (U) The 3-μm transverse plastic-embedded sections of root tips. (S) shows the meristematic zone, and (T) and (U) are sections similar to those in (N) and (O). GUS signal is widespread in (S) and (T) and accumulates in the epidermis, cortex, and phloem (arrowheads) in (U).
(V) and (W) Stem transverse plastic-embedded sections comparable to those in (P) to (Q). GUS is detectable throughout the cortex, epidermis, and phloem (arrowheads).
(X) GFP fluorescence image (top) and fluorescence merged with bright-field image (bottom) of a transgenic D14pro:D14:GFP root.
(Y) to (Z’) Leaf (Y), hypocotyl (Z), and root (Z’) cells of CaMV35Spro:D14:GFP transgenic plants. GFP is detected in nucleus and cytoplasm.
Bars = 1 mm in (A), (D) to (F), and (K), 500 μm in (L), 200 μm in (B), (M), and (P), 100 μm in (G) and (N), 50 μm in (H) to (J), (Q), and (Z), 15 μm in (Y) and (Z’).
D14:GFP subcellular localization, analyzed in D14pro:D14:GFP and CaMV35Spro:D14:GFP lines, was both cytoplasmic and nuclear in all tissues studied (Figures 4X to 4Z), consistent with WoLF PSORT predictions (Horton et al., 2007).
D14 Does Not Move Long Distances Acropetally in the Plant to Regulate Shoot Branching
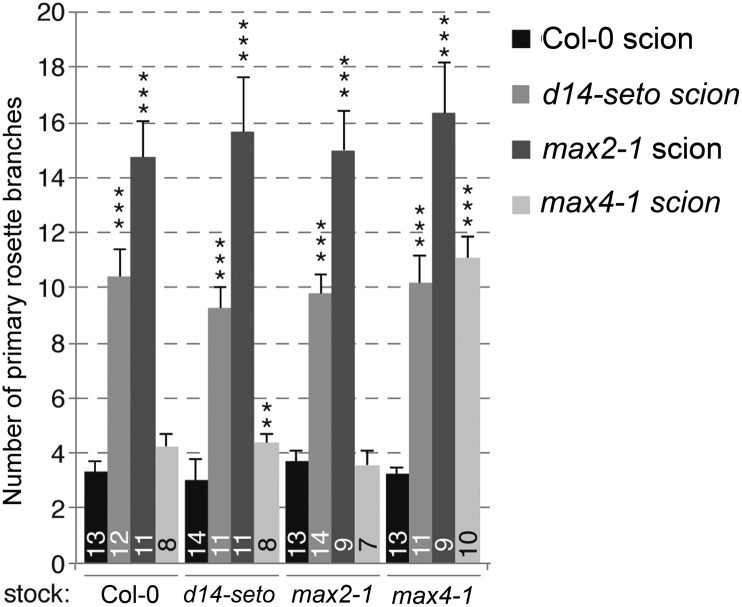
To test whether D14 could act non-cell autonomously in the regulation of shoot branching, we performed reciprocal micrografting between wild-type, d14-seto, max2-1, and max4-1 rootstocks and scions and studied their branching phenotypes (Figure 5). If the D14 mRNA or protein moved long distance acropetally or if D14 was exclusively involved in the synthesis of a bioactive compound transported upwards in the plant, d14-seto scions grafted to wild-type stocks would have a wild-type, or at least a partially rescued, branching phenotype. Instead, we found that d14-seto scions grafted to either wild-type, max2-1, or max4-1 rootstocks remained bushy, like max2-1 scions (Figure 5). This was in agreement with grafting experiments performed with dad2 in petunia (Simons et al., 2007) and indicated that, like MAX2, D14 is required locally in the aerial part of the plant to suppress shoot branching. This is also consistent with the proposed interaction between D14 and MAX2 and with a role for D14 in SL perception and signaling.
Figure 5.
Spatial Requirement of D14 to Suppress Branching.
Number of rosette branches in plants obtained from the micrografts indicated, quantified 14 d after bolting. Data shown as mean ± se. Numbers on bars indicate the number of individuals analyzed. **P < 0.01; ***P < 0.001.
D14 Transcription Is Neither Responsive to SL nor Auxin Signaling nor to Bud Growth Status
To analyze D14 transcriptional regulation, we tested whether expression of this gene was affected by SL or auxin signaling, as described for other SL-related genes (Sorefan et al., 2003; Bainbridge et al., 2005; Foo et al., 2005; Snowden et al., 2005; Johnson et al., 2006; Zou et al., 2006; Arite et al., 2007; Simons et al., 2007; Hayward et al., 2009; Mashiguchi et al., 2009; Guan et al., 2012; Waters et al., 2012b) or by bud growth status, like petunia DAD2 (Hamiaux et al., 2012).
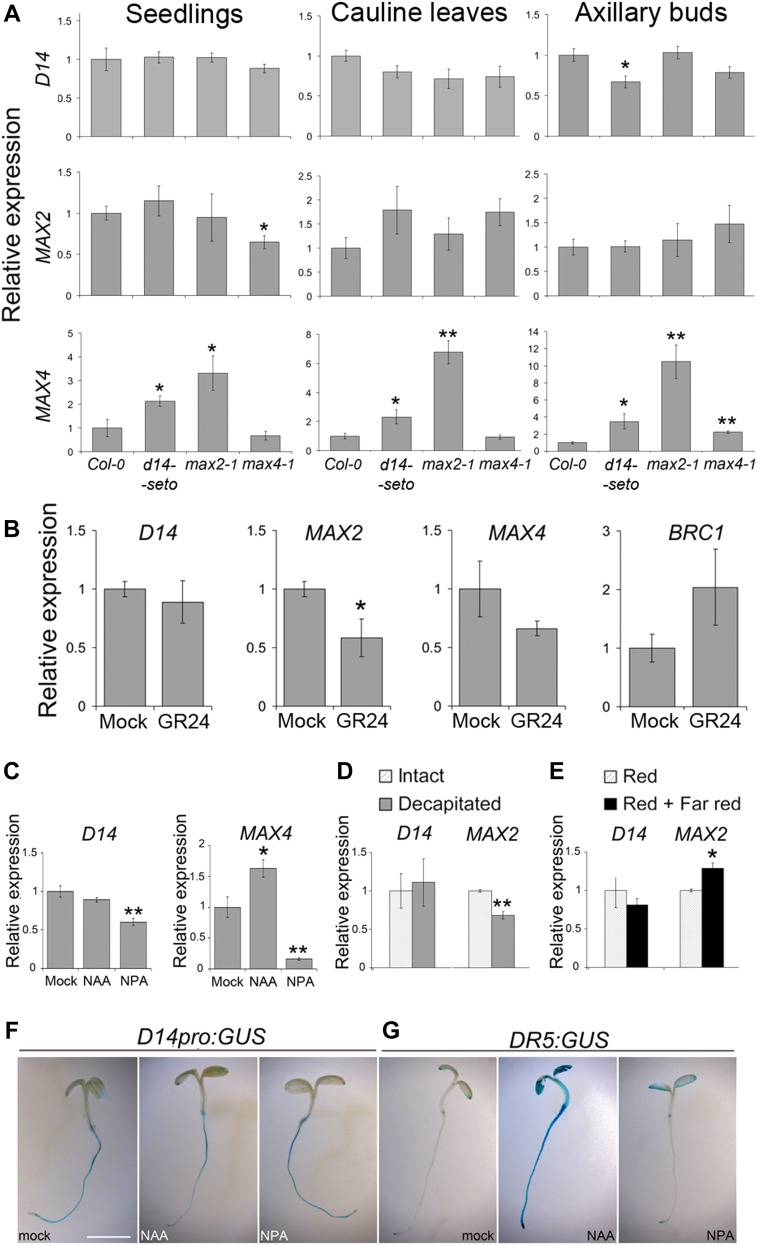
We quantified D14 transcript levels in young seedlings, cauline leaves, and axillary buds of max2-1, max4-1, and d14-seto mutants. Expression of D14 did not change significantly in any background or plant tissue, except for a slight reduction in transcript levels in d14-seto mutant axillary buds (Figure 6A). We then treated young wild-type seedlings with 5 μM synthetic SL GR24 (24 h), which confirmed that D14 mRNA levels were unresponsive to GR24. MAX2, by contrast, responded negatively to GR24 (Figure 6B). To test whether D14 transcription is positively regulated by auxin levels or transport, we compared D14 mRNA levels of young seedlings treated with synthetic auxin 1-naphthaleneacetic acid (NAA), the auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA), or the mock control (24 h). D14 transcript levels showed only moderate reduction in NPA-treated plants (Figure 6C). In a second experiment, we monitored the response of the D14 promoter to NAA or NPA treatment (24 h) in D14pro:GUS seedlings; there were no significant changes in GUS activity in treated relative to mock-treated seedlings (Figure 6F), although NAA effectively activated the synthetic auxin response promoter DR5 (Figure 6G).
Figure 6.
Transcriptional Regulation of D14.
(A) qPCR quantification of D14, MAX2, and MAX4 transcript abundance in seedlings, cauline leaves, and axillary buds of d14-seto, max2-1, or max4-1 plants relative to levels in wild-type Col-0 plants.
(B) Transcript abundance of D14, MAX2, and MAX4 and BRC1 in seedlings treated for 24 h with 5 μM GR24 (synthetic SL analog) compared with mock-treated plants.
(C) D14 and MAX4 transcript levels in 10-d-old seedlings treated with 10 μM NAA or 10 μM NPA (auxin transport inhibitor) relative to levels in mock-treated plants.
(D) and (E) D14 and MAX2 mRNA levels measured in axillary buds of decapitated plants 8 h after treatment relative to levels in intact plants grown in parallel (D) and of plants treated with red + far-red light for 8 h relative to levels in red light–treated plants grown in parallel (E).
(F) and (G) Histochemical assays of D14pro:GUS (F) and DR5:GUS (G) in 4-d-old seedlings treated for 24 h with 10 μM NAA, 10 μM NPA, or mock. Bar = 1 mm for (F) and (G). Data shown as mean ± se (n = 3 to 4 biological replicates). Asterisks denote significant differences in Student’s t test (*P < 0.05; **P < 0.01).
Finally, in axillary bud tissue, we studied the transcriptional response of D14 to treatments affecting bud growth status, including decapitation of the main shoot, which promotes bud activation, and 8-h exposure to far-red-rich light, which promotes bud dormancy. D14 mRNA levels were unaffected by both treatments (Figures 6D and 6E), whereas MAX2 mRNA levels correlated positively with bud dormancy in both cases (Figures 6D and 6E).
These results show that D14 mRNA levels are thus not responsive to changes in SL or auxin signaling, nor do they correlate positively with bud growth arrest in Arabidopsis.
The D14 Protein Is Rapidly Degraded in the Presence of SL
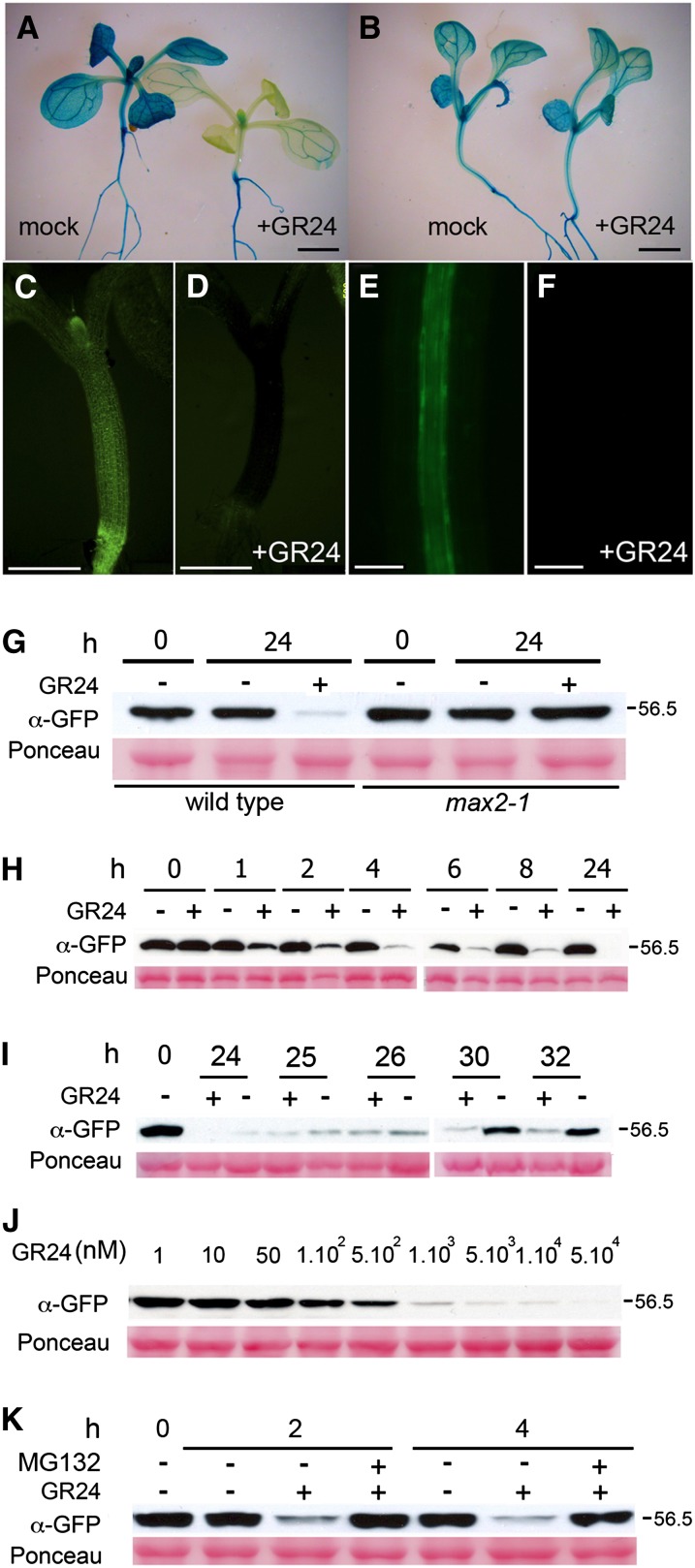
As D14 did not appear to be regulated transcriptionally, we investigated a potential posttranslational regulation. To test whether the D14 protein was regulated by SL, we treated young seedlings carrying either D14pro:D14:GUS/GFP or CaMV35Spro:D14:GFP constructs with GR24 and studied D14:GUS and D14:GFP protein levels at different times. When incubated for 24 h with 5 μM GR24, plants showed a striking reduction in GUS and GFP signal especially in rosette leaves and hypocotyl, although the effect was also noticeable in roots (Figures 7A and 7C to 7G; Supplemental Figure 7). By contrast, max2-1 mutant plants carrying the same constructs did not show reduced levels of GUS/GFP when treated with GR24 (Figures 7B and 7G; Supplemental Figure 8). GR24-treated D14pro:GUS control plants did not display reduced GUS protein levels (Supplemental Figure 9).
Figure 7.
SL Destabilizes the D14 Protein.
(A) to (F) GUS histochemical activity of D14pro:D14:GUS plants in Col-0 (A) or max2-1 (B) background after 24-h treatment with mock (left) or GR24 (right). Fluorescence image of young CaMV35Spro:D14:GFP seedlings after 24-h treatment with mock (C) or GR24 (D). Fluorescence image of D14pro:D14:GFP seedlings after 24-h treatment with mock (E) or GR24 (F). Bars = 500 μm.
(G) D14:GFP response to GR24 analyzed by immunoblot using α-GFP antibody in D14pro:D14:GUS plants in Col-0 (wild type) or max2-1.
(H) Time course of D14:GFP accumulation in response to GR24. Protein extracts (10 μg) from 5-d-old D14pro:D14:GFP seedlings treated with GR24 (+) or mock (−) for 0 to 24 h were separated by 10% SDS-PAGE and identified as a 56.5-kD band.
(I) After a 24-h GR24 treatment, seedlings were maintained in MS with (+) or without GR24 (−) for 1 to 8 h.
(J) Dose response of D14:GFP to 24-h treatments with increasingly higher concentrations of GR24.
(K) D14:GFP response to GR24 in the presence of MG132. In (G) to (K), Ponceau stainings are included for loading reference. All GR24 treatments used a 5 μM concentration unless indicated.
We then performed a time-course analysis of the response in Arabidopsis. We did immunoblots with α-GFP in D14pro:D14:GFP seedlings treated 1, 2, 4, 6, 8, and 24 h with 5 μM GR24 and detected a negative effect of GR24 on D14:GFP accumulation as early as 1 to 2 h after the beginning of the GR24 treatment (Figure 7H). Protein levels begun to recover 26 h later, even in the presence of GR24, perhaps due to GR24 hydrolysis by D14 and new protein synthesis. Removal of GR24 after 24 h led to accelerated protein accumulation (Figure 7I). The effect of GR24 was dose dependent and still detectable at GR24 concentrations of 50 to 100 nM (Figure 7J). This response was strongly suppressed by simultaneous treatment of plants with the proteasome inhibitor MG132 (Figure 7K).
All these results indicate that SL promotes rapid degradation of D14 and that this response requires a functional MAX2 gene. If D14 is the SL receptor, as proposed, this negative feedback regulation would cause a substantial drop in SL perception that could very effectively limit the extent of SL signaling.
DISCUSSION
D14 as an SL Receptor
Two alternative roles have been proposed for D14-type proteins in plants, as enzymes that transform SL into bioactive compounds and as SL receptors. Growing evidence supports the latter possibility and suggests that D14 links SL perception and signaling through protein–protein interactions with MAX2-type proteins (Arite et al., 2009; Hamiaux et al., 2012; Waters et al., 2012b; Kagiyama et al., 2013). Our grafting experiments, which show that the bushy phenotype of d14-seto mutants cannot be rescued by grafting their shoots to wild-type rootstocks, are consistent with grafting experiments performed with dad2-1 in petunia (Simons et al., 2007) and confirm a local requirement for D14 in the aerial part of the plant. This is in agreement with the proposal that D14 and MAX2 (also required in the shoot for branch suppression; Booker et al., 2005; Stirnberg et al., 2007) must interact to trigger SL signaling.
Spatial Regulation of SL Signaling
According to the recent model, D14 and MAX2 interact in the presence of SL (Hamiaux et al., 2012; Zhou et al., 2013; Jiang et al., 2013), although in Arabidopsis, this interaction may not be direct (Wang et al., 2013). As MAX2, D14, and SL are not distributed ubiquitously, the SL response would only take place in tissues in which these three factors coincide. MAX2 is widely distributed in axillary buds, young leaves, hypocotyl, vasculature, and flower organs (Shen et al., 2007; Stirnberg et al., 2007). D14 expression patterns and protein distribution largely overlap with those of MAX2 in axillary buds, root vasculature, and carpel. Moreover, the wider distribution of GUS in D14pro:D14:GUS relative to D14pro:GUS lines raises the possibility that the D14 protein moves, at least within a short range, and is perhaps unloaded from the root phloem. Alternatively, regulatory motifs within the open reading frame of D14 could, in D14pro:D14:GUS lines, regulate transcription in tissues adjacent to those identified using the D14pro:GUS lines. In situ hybridization of D14 mRNA and immunolocalization of the D14 protein will help elucidate these possibilities. In some other tissues, such as the shoot cambium, where MAX2 has been shown to play an essential role (Agusti and Greb, 2013), the D14 protein does not accumulate measurably. One possibility is that MAX2 does not interact with D14 for this function; alternatively, small, below-detection amounts of D14, which perhaps is degraded rapidly after the interaction, are sufficient to trigger SL signaling in this tissue. At the subcellular level, the nuclear localization of MAX2 (Shen et al., 2007; Stirnberg et al., 2007) and D14 (which is nuclear and cytoplasmic) would allow their physical interactions in the nucleus without special translocation events.
By contrast, the interaction between SL and D14 may need to be promoted. SL has been shown to be transported in the xylem sap (Foo et al., 2001; Kohlen et al., 2011) where D14 is undetectable. Moreover, some D14- and MAX2-expressing tissues, such as very young axillary buds, whose development responds to SL signaling, are not yet connected to the vasculature. Although SL accumulation in phloem and other tissues is yet unknown, this seems to imply that a system of cell-to-cell SL delivery may be required to regulate SL–D14 interactions. Such a system has been described in petunia, where it is regulated by the ABC transporter PDR1 (Kretzschmar et al., 2012). Arabidopsis SL transporters have not yet been identified, but transgenic plants carrying the PDR1pro:GUS construct show promoter activity in the vascular tissue, stem nodes, and regions that subtend axillary buds (Kretzschmar et al., 2012). SL might be exported from the xylem to the basal region of buds, thus promoting bud arrest.
The d14-seto Protein
We identified d14-seto, a loss-of-function mutant allele in the D14 locus, which has a single Pro169Leu amino acid substitution and causes a consistently strong, bushy phenotype. The Pro-169 position is probably not involved in ligand binding or hydrolytic activity, nor is the Pro169Leu mutation predicted to cause large destabilizing effects in the protein structure. Pro-169 is located in the external surface of the cap domain with the side chain exposed to the solvent, suggesting that the mutation affects critical protein–protein interactions. This position is one of the seven SDPs (Rausell et al., 2010) found between D14 and KAI2 proteins, which have either conserved Pro or Ser, respectively, suggesting that these residues help determine the functional specificity of each protein type. It is also unlikely that they are involved in D14–MAX2 interactions, as both D14 and KAI2 seem to interact with MAX2, based on molecular and genetic evidence (Nelson et al., 2011; Hamiaux et al., 2012; Waters et al., 2012b; Kagiyama et al., 2013).
Pro-169 could participate in the recognition of sets of proteins to be targeted for degradation through the SL pathway, such as D53-, BES1-, or DELLA-related proteins (Jiang et al., 2013; Nakamura et al., 2013; Wang et al., 2013; Zhou et al., 2013). The equivalent position in KAI2, Ser-168, is surrounded by amino acids that undergo measurable conformational changes upon KAR1-KAI2 binding (Guo et al., 2013b). If this situation is analogous for D14, SL binding and/or hydrolysis could cause allosteric changes in the area surrounding Pro-169, creating or modifying a protein-interacting surface. This model would resemble that of the gibberellin receptor GID1, an α/β-fold hydrolase superfamily protein, in which gibberellin acts as an allosteric effector and induces conformational changes in the cap that folds back to generate a DELLA binding surface (Murase et al., 2008; Shimada et al., 2008). Further experiments will help evaluate this model; second-site mutagenesis and search for d14-seto suppressors could reveal additional components of the pathway acting in close proximity to the SL-D14 complex.
Relation of BRC1 and the SL Pathway
A positive transcriptional regulation of BRC1 by the SL pathway has been proposed based on the strong downregulation of BRC1 in axillary buds of SL mutants and its upregulation in buds in response to SL application (Aguilar-Martínez et al., 2007; Finlayson, 2007; Braun et al., 2012; Dun et al., 2012, 2013). However, these transcriptional changes could simply reflect SL-dependent bud dormancy or activity, to which BRC1 expression is tightly associated. Now, we have also observed a strong transcriptional downregulation of BRC1 in d14-seto cauline leaves, organs in which no obvious phenotypic effects are detected in brc1 mutants. This supports a direct transcriptional regulation of BRC1 by the SL pathway in organs different from axillary buds. Moreover, the late flowering phenotype of d14-seto brc1-2 double mutants, identical to that of brc1-2 mutants, suggests that in the control of flowering time, SLs could act entirely through regulation of BRC1, whose protein could in turn interact with FT, as described by Niwa et al. (2013). By contrast, the additive shoot branching phenotypes of d14-seto and brc1-2 supports that SLs act not only by upregulating BRC1 but also by other mechanisms not directly related to BRC1 (i.e., PATS dampening and degradation of branching-promoting factors, such as D53 and BES1; Jiang et al., 2013; Wang et al., 2013; Zhou et al., 2013).
Conservation and Evolution of D14-Like Gene Regulation
Mutants bearing loss-of-function alleles of D14-like genes in rice (Arite et al., 2009; Gao et al., 2009; Liu et al., 2009), petunia (Hamiaux et al., 2012), and Arabidopsis (Waters et al., 2012b; this study) show increased branching and reduced stature, indicating functional conservation of these genes in the determination of plant architecture. Some degree of divergence in their regulation is nonetheless evident. For instance, the rice D14 gene was reported to be transcribed in parenchyma cells surrounding the xylem (Arite et al., 2009), whereas in Arabidopsis, we found the strongest transcriptional activity of D14 in the phloem, young organs, and cortex cells of elongating stems. Petunia DAD2 and rice D14 mRNA levels correlate positively with bud dormancy (Arite et al., 2009; Hamiaux et al., 2012), while this is not the case for Arabidopsis D14. In rice, TB1 was proposed to be a transcriptional activator of D14. In protoplast assays, TB1 interacts with MADS57, a transcriptional repressor of D14, thus reducing the inhibitory effect on D14 transcription (Guo et al., 2013a). If this regulatory pathway was conserved in Arabidopsis, we would predict that mutants in the Os-Tb1 homolog, BRC1, would have reduced D14 mRNA levels. We have found that this is not the case. Indeed, D14 transcription levels are not greatly altered by any stimulus studied so far, with the exception of darkness (Waters and Smith, 2013). Effective regulation of protein stability in this group might have rendered its transcriptional regulation irrelevant, leading to evolutionary loss.
Negative Feedback Loops in SL Signaling
We observed that SL destabilizes D14, probably by promoting its proteasome-mediated degradation. If D14 is confirmed as the SL receptor, this would represent an example of an interesting phenomenon whereby a plant hormone triggers degradation of its own receptor. This response requires a functional MAX2 gene, as we found that D14 is resistant to SL in max2-1 mutants. MAX2 could participate directly in the SCF complex that tags D14 for degradation, implying that D14 could be a MAX2 substrate, like SLR1, D53, and BES1 (Jiang et al., 2013; Nakamura et al., 2013; Wang et al., 2013; Zhou et al., 2013). Alternatively, other proteins targeted by the SCFMAX2 complex might prevent D14 degradation, so that their removal during SL signal transduction could lead to D14 destabilization through other ubiquitin-related systems. Interestingly, while D53 and BES1 degradation occurs within 8 to 30 min after SL treatment, D14 degradation is slower, 1 to 2 h after SL treatment. This is in agreement with Jiang et al. (2013) and Zhou et al. (2013), who did not find SL-mediated destabilization of D14 in 1 h. One possible scenario is that once the SL-dependent substrates are degraded via the SL:D14:SCFMAX2 complex, D14 itself becomes destabilized. This negative feedback regulation to modulate D14 protein levels would cause a drop in SL perception that could effectively limit the extent of SL signaling. This scenario resembles that of the clock-associated protein EARLY FLOWERING3 that, after acting as substrate adaptor to promote recognition of GIGANTEA by the E3-ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), is in turn ubiquitinated and degraded via COP1, leading to a cycling signaling (Yu et al., 2008).
In summary, SL signaling homeostasis seems to be modulated by a strong negative feedback regulation that affects not only the transcription of SL synthesis genes (Bainbridge et al., 2005; Foo et al., 2005; Snowden et al., 2005; Johnson et al., 2006; Arite et al., 2007; Drummond et al., 2009; Dun et al., 2009; Hayward et al., 2009) but also the stability of the D14 protein. The transcriptional regulatory mechanisms require functional SL signaling. This posttranslational regulation of D14 requires at least functional MAX2, but it remains unknown whether SL signal transduction is necessary. Assays performed using the d14-seto protein will help clarify this point.
Additional mechanisms that regulate this pathway might involve regulation of D14 cell-to-cell movement, modulation of SL transport, and/or availability and regulation of D14 nuclear localization to modulate the intensity of D14-MAX2 interactions. Further work is still needed to understand the stoichiometry of this process and the regulatory bottlenecks through which the signaling pathway is mostly regulated.
METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis thaliana was the Col-0 ecotype. The d14-seto allele was an ethyl methanesulfonate mutant generated by Lehle Seeds (www.arabidopsis.com). Homozygote seeds from plants backcrossed twice to wild type Col-0 were used. d14-1 (NASC ID: N913109), d14-3 (NASC ID: N678534), and d14-4 (NASC ID: N557876) were obtained from the Nottingham Arabidopsis Stock Centre. brc1-2 has been described (Aguilar-Martínez et al., 2007). max2-1 and max4-1 mutants were provided by O. Leyser. Wild-type and mutant seeds were sown on commercial soil and vermiculite at a 3:1 proportion, stratified in darkness (4°C, 3 d), and grown in a 16-h-light/8-h-dark photoperiod at 22°C in white light (W; PAR, 100 μmol m−2 s−1). For the experiment in Figure 2D, seeds were grown in conditions as above and exposed for 8 h to white light (W; red [R]:far red [FR] ratio = 11.7) or W supplemented with FR (W+FR, R:FR ratio = 0.2) as described (González-Grandío et al., 2013). For auxin response assays, seedlings were germinated in vertical plates in Murashige and Skoog (MS) medium, 0.7% agar, 1% Suc (MAS) and grown as above for 15 d and then transferred to MAS + 10 μm NAA or MAS + mock (24 h).
Light Sources
White light was provided by cool-white 20-W F20T12/CW tubes (Phillips). Supplemental far-red light was provided by lamp tubes carrying far-red 735-nm LEDs (L735-03AU; EPITEX).
Protein Structure Analyses
Homolog sequences for D14, KAI2, and RsbQ were obtained by BLAST searches against the UniProt database. Sequence hits were further aligned with MUSCLE (Edgar, 2004). SDPs were analyzed with JDet (Muth et al., 2012) using the S3det method (Rausell et al., 2010) implemented in the JDet software. D14 (PDB:4ih4) and KAI2 (PDB:3w06) structures were aligned with FATCAT (Ye and Godzik, 2004). Mutant side chain orientation (Figure 3) was generated using FoldX (Schymkowitz et al., 2005). We identified residues of D14 corresponding to those in KAI2 undergoing side-chain movement after KAR1 binding (Guo et al., 2013b). These residues were mapped in D14 structure (PDB:4ih4) using PyMOL (www.pymol.org). To assess the change in protein stability after mutation (ΔΔG), we used the D14 structure (PDB:4ih4) with the prediction methods FoldX (Schymkowitz et al., 2005), I-Mutant (Capriotti et al., 2005), SDM (Worth et al., 2011), Eris (Yin et al., 2007), and Concoord/PBSA (Benedix et al., 2009).
Micrografting
Grafting was performed in tissue culture by joining shoot scions and root stocks of young, 6-d-old seedlings (grown on plates at 25°C in 16-h light days) at the level of their hypocotyls as described (Ragni et al., 2011). Successful grafts were transferred onto soil 7 d after grafting and grown in 16-h-light days. Phenotypes were scored 14 d after bolting.
Decapitation Assay
Col-0 plants were grown until main inflorescences were 2 to 3 cm in length. In half of the plants, the main shoot, including the cauline nodes, was removed; six to eight decapitated and nondecapitated rosettes were collected for each biological replicate 24 h after decapitation. RNA was extracted as described in González-Grandío et al. (2013), and qPCR was performed with three biological replicates.
Positional Cloning
The seto5 mutation was mapped by the Instituto de Bioingeniería–Universidad Miguel Hernández Gene Mapping Facility (Elche, Spain) as described (Ponce et al., 1999, 2006). In brief, for low-resolution mapping, the DNA of 50 F2 phenotypically mutant plants was extracted individually and used as a template to multiplex PCR coamplify 32 SSLP and insertion/deletion molecular markers using fluorescently labeled oligonucleotides as primers. For fine mapping, 400 additional F2 plants were used to assess linkage iteratively between seto5 and molecular markers designed according to the polymorphisms between Landsberg erecta and Col-0 described at the Monsanto Arabidopsis Polymorphism Collection database (http://www.arabidopsis.org).
Genome Sequencing
Whole-genome sequencing of 40 μg of pooled genomic DNA from six seto5 individuals was performed by BGI (www.genomics.cn); 2.79 Gb of clean data was analyzed with an average effective depth of 22.75 X.
Phenotypic Analysis
Branches (shoots >1 cm) were counted 2 weeks after the main inflorescence became visible. AM initiation and early bud development phenotype analyses were performed as described (Aguilar-Martínez et al., 2007). Flowering time of cauline and rosette branches was analyzed by counting the number of rosette and cauline leaves.
Plastic Embedding of Stem Sections
Stem fragments (5 to 10 mm) from immediately above the uppermost rosette leaf were fixed in 4% glutaraldehyde and 0.1% Tween (20 to 24 h), dehydrated in ethanol series up to 100% ethanol, washed in preinfiltration solution (50% ethanol and 50% infiltration solution), and passed to the infiltration solution of the Historesin Standard kit (Leica). Subsequent steps were done according to the manufacturer’s instructions. Sections (3 μm) of resin-embedded specimens were prepared with a Leica microtome and tungsten carbide blades, floated in a 50°C water bath, collected on a slide, and allowed to dry. Nontransgenic lines were stained with 1% cresyl violet (Sigma-Aldrich C-5042). Sections were mounted in Surgipath micromount (Leica).
GUS Histochemistry
GUS staining was performed as described (Sessions et al., 1999).
GR24 Treatments
Seedlings were grown in vertical MS + agar plates and were then transferred to liquid MS + 1% Suc medium with 5 µM GR24 (from a stock solution in acetone) or the corresponding acetone volume (mock) and grown in normal growth conditions for different times. When specified, other concentrations of GR24 were used. For the MG132 treatment, MG132 was added to a concentration of 50 µM at T0, and identical amounts were added to the incubation solution every hour. For the immunoblot experiments, 1-week-old seedlings were used.
Protein Extraction and Immunoblots
Five-day-old seedlings were frozen in liquid N2, and total protein was extracted in PBS buffer, 0.1% SDS, 0.1% Triton X-100, 1 mM PMSF, 5 mM β-mercaptoethanol, and protease inhibitors (5 µM E-64, 50 µM leupeptin, 1 µM pepstatin, and 10 µg/mL aprotinin). The extract was centrifuged (15 min, 16,000g, 4°C), and the supernatant was collected. Protein concentration was determined in a Bradford assay. Protein extracts (10 μg) were denatured by boiling (5 min, 95°C), separated by 10% SDS-PAGE, and transferred to polyvinylidene difluoride membranes (Bio-Rad). To confirm equivalent protein loading, membranes were stained with Ponceau reagent. Membranes were probed with anti-GFP-horseradish peroxidase antibody (1:1000; Milteny Biotec), and signal was detected using ECL reagent (Invitrogen).
D14 Constructs
The CDS of D14 and d14-seto and the 2.3-kb genomic fragment of D14 and the D14 promoter were amplified from genomic DNA using Phusion polymerase (Finnzymes) with indicated primers (Supplemental Table 2). The CDS of MAX2 was amplified using indicated primers (Supplemental Table 2). PCR fragments were BP cloned into the entry vector pDONR207 (Gateway; Invitrogen). For the CaMV35Spro:D14:GFP construct, the CDS was LR cloned into the destination vector pGWB5. For D14pro:GFP:D14, the genomic fragment was cloned into pGWB4. For D14pro:GUS, the promoter was cloned into pGWB3. All pGW vectors were provided by Tsuyoshi Nakagawa (Shimane University; Nakagawa et al., 2007).
Arabidopsis Transgenic Plants
Transgenic plants (Col-0) were generated by agroinfiltration using the floral dip method (Clough and Bent, 1998). T3 homozygous lines generated from T1 individuals carrying a single transgene insertion were analyzed.
RNA Preparation and qPCR Analyses
Material was harvested from at least eight individuals and a minimum of three biological replicates per genotype/treatment and stored in liquid N2. RNA was isolated with the RNeasy plant mini kit (Qiagen). Possible traces of DNA were eliminated with an RNase-Free DNase set (Qiagen). RNA was used to make cDNA with the High-Capacity cDNA archive kit (Applied Biosystems). The qPCR reactions were performed with Power SYBR Green (Applied Biosystems) and the Applied Biosystems 7300 real-time PCR system, according to the manufacturer’s instructions. Three technical replicates were done for each biological replicate. Primers pairs are described in Supplemental Table 2. The SAND gene was used as reference (Czechowski et al., 2005).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: D14 (At3g03990), NM_111270.2; GI:30679007.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mapping and Cloning of the seto5 Locus.
Supplemental Figure 2. Complementation of the d14-seto Mutant and D14 Allelic Series.
Supplemental Figure 3. Relative D14 and BRC1 mRNA Levels in Axillary Buds of the d14-3 Mutant, Quantified by qPCR.
Supplemental Figure 4. Multiple Sequence Alignment of the D14 and KAI2 Orthologs.
Supplemental Figure 5. 3D Structure of D14 and KAI2 and Residues Affected upon KAR1 Binding.
Supplemental Figure 6. D14 Relative mRNA Levels in Different Tissues Analyzed by qPCR.
Supplemental Figure 7. The D14:GUS Protein Is Destabilized by SL.
Supplemental Figure 8. D14:GUS Is Not Destabilized by SL in a max2-1 Background.
Supplemental Figure 9. GUS Is Not Destabilized by SL.
Supplemental Table 1. Difference in Free Energy of Unfolding between the Wild-Type D14 Protein and Mutant d14-seto Protein.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Desmond Bradley, Eduardo González, and Michael Nicolas for helpful comments on the article, María Rosa Ponce, José Luis Micol, and the TRANSPLANTA Consortium for mapping the seto5 mutant, Concepción Manzano for help with the mutant screening, Catherine Rameau and Binne Zwanenburg for GR24, Ottoline Leyser for seed stocks, Chidi Afamefule for qPCR of the d14-3 mutants, and Catherine Mark for editorial assistance. This work was supported by the Spanish Ministerio de Educación y Ciencia (BIO2008-00581 and CSD2007-00057), Ministerio de Ciencia y Tecnología (BIO2011-25687), and the Fundación Ramón Areces.
AUTHOR CONTRIBUTIONS
F.C. and P.C. designed the research. F.C., K.N., C.S.H., M.L.R., J.C.S.-F., M.C., and P.C. performed the experiments. P.C., C.S.H., M.C., and F.C. analyzed the data. M.C. and J.C.S.-F. contributed analytical tools. P.C. wrote the article.
Glossary
- AM
axillary meristem
- SL
strigolactone
- PATS
polar auxin transport stream
- KAR
karrikin
- Col-0
Columbia-0
- TSS
translation start site
- CDS
coding sequence
- SDP
specificity-determining position
- qPCR
quantitative real-time PCR
- NAA
1-naphthaleneacetic acid
- NPA
1-N-naphthylphthalamic acid
- MS
Murashige and Skoog
Footnotes
Online version contains Web-only data.
References
- Aguilar-Martínez J.A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti J., Greb T. (2013). Going with the wind—Adaptive dynamics of plant secondary meristems. Mech. Dev. 130: 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351. [DOI] [PubMed] [Google Scholar]
- Arite T., Iwata H., Ohshima K., Maekawa M., Nakajima M., Kojima M., Sakakibara H., Kyozuka J. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite T., Umehara M., Ishikawa S., Hanada A., Maekawa M., Yamaguchi S., Kyozuka J. (2009). d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Bainbridge K., Sorefan K., Ward S., Leyser O. (2005). Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J. 44: 569–580. [DOI] [PubMed] [Google Scholar]
- Balla J., Kalousek P., Reinöhl V., Friml J., Procházka S. (2011). Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J. 65: 571–577. [DOI] [PubMed] [Google Scholar]
- Benedix A., Becker C.M., de Groot B.L., Caflisch A., Böckmann R.A. (2009). Predicting free energy changes using structural ensembles. Nat. Methods 6: 3–4. [DOI] [PubMed] [Google Scholar]
- Bennett T., Sieberer T., Willett B., Booker J., Luschnig C., Leyser O. (2006). The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16: 553–563. [DOI] [PubMed] [Google Scholar]
- Booker J., Sieberer T., Wright W., Williamson L., Willett B., Stirnberg P., Turnbull C., Srinivasan M., Goddard P., Leyser O. (2005). MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8: 443–449. [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806. [DOI] [PubMed] [Google Scholar]
- Braun N., et al. (2012). The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer P.B., Dun E.A., Ferguson B.J., Rameau C., Beveridge C.A. (2009). Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 150: 482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody M.S., Vijay K., Price C.W. (2001). Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the sigma(B) transcription factor in Bacillus subtilis. J. Bacteriol. 183: 6422–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bythell-Douglas R., Waters M.T., Scaffidi A., Flematti G.R., Smith S.M., Bond C.S. (2013). The structure of the karrikin-insensitive protein (KAI2) in Arabidopsis thaliana. PLoS ONE 8: e54758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Casadio R. (2005). I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 33: W306–W310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R.J., Hepworth J., Mouchel C., Waites R., Leyser O. (2013). A role for more axillary growth1 (MAX1) in evolutionary diversity in strigolactone signaling upstream of MAX2. Plant Physiol. 161: 1885–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crawford S., Shinohara N., Sieberer T., Williamson L., George G., Hepworth J., Müller D., Domagalska M.A., Leyser O. (2010). Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan D., Pazos F., Valencia A. (2013). Emerging methods in protein co-evolution. Nat. Rev. Genet. 14: 249–261. [DOI] [PubMed] [Google Scholar]
- Delaux P.M., Xie X., Timme R.E., Puech-Pages V., Dunand C., Lecompte E., Delwiche C.F., Yoneyama K., Bécard G., Séjalon-Delmas N. (2012). Origin of strigolactones in the green lineage. New Phytol. 195: 857–871. [DOI] [PubMed] [Google Scholar]
- Domagalska M.A., Leyser O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12: 211–221. [DOI] [PubMed] [Google Scholar]
- Drummond R.S., Martínez-Sánchez N.M., Janssen B.J., Templeton K.R., Simons J.L., Quinn B.D., Karunairetnam S., Snowden K.C. (2009). Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol. 151: 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond R.S., Sheehan H., Simons J.L., Martínez-Sánchez N.M., Turner R.M., Putterill J., Snowden K.C. (2011). The expression of petunia strigolactone pathway genes is altered as part of the endogenous developmental program. Front. Plant Sci. 2: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun E.A., Brewer P.B., Beveridge C.A. (2009). Strigolactones: Discovery of the elusive shoot branching hormone. Trends Plant Sci. 14: 364–372. [DOI] [PubMed] [Google Scholar]
- Dun E.A., de Saint Germain A., Rameau C., Beveridge C.A. (2012). Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 158: 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun E.A., de Saint Germain A., Rameau C., Beveridge C.A. (2013). Dynamics of strigolactone function and shoot branching responses in Pisum sativum. Mol. Plant 6: 128–140. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson S.A. (2007). Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol. 48: 667–677. [DOI] [PubMed] [Google Scholar]
- Foo E., Bullier E., Goussot M., Foucher F., Rameau C., Beveridge C.A. (2005). The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17: 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E., Turnbull C.G., Beveridge C.A. (2001). Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol. 126: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiji N., Cardinale F., Prandi C., Bonfante P., Ranghino G. (2012). The computational-based structure of Dwarf14 provides evidence for its role as potential strigolactone receptor in plants. BMC Res. Notes 5: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Qian Q., Liu X., Yan M., Feng Q., Dong G., Liu J., Han B. (2009). Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol. Biol. 71: 265–276. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194. [DOI] [PubMed] [Google Scholar]
- González-Grandío E., Poza-Carrión C., Sorzano C.O., Cubas P. (2013). BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25: 834–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J.C., Koch K.E., Suzuki M., Wu S., Latshaw S., Petruff T., Goulet C., Klee H.J., McCarty D.R. (2012). Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol. 160: 1303–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Xu Y., Liu H., Mao Z., Zhang C., Ma Y., Zhang Q., Meng Z., Chong K. (2013a). The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Zheng Z., La Clair J.J., Chory J., Noel J.P. (2013b). Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc. Natl. Acad. Sci. USA 110: 8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C., Drummond R.S., Janssen B.J., Ledger S.E., Cooney J.M., Newcomb R.D., Snowden K.C. (2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22: 2032–2036. [DOI] [PubMed] [Google Scholar]
- Hayward A., Stirnberg P., Beveridge C., Leyser O. (2009). Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 151: 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 35: W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S., Maekawa M., Arite T., Onishi K., Takamure I., Kyozuka J. (2005). Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46: 79–86. [DOI] [PubMed] [Google Scholar]
- Jiang L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X., Brcich T., Dun E.A., Goussot M., Haurogné K., Beveridge C.A., Rameau C. (2006). Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 142: 1014–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M., Hirano Y., Mori T., Kim S.Y., Kyozuka J., Seto Y., Yamaguchi S., Hakoshima T. (2013). Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18: 147–160. [DOI] [PubMed] [Google Scholar]
- Kohlen W., Charnikhova T., Liu Q., Bours R., Domagalska M.A., Beguerie S., Verstappen F., Leyser O., Bouwmeester H., Ruyter-Spira C. (2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 155: 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T., Kohlen W., Sasse J., Borghi L., Schlegel M., Bachelier J.B., Reinhardt D., Bours R., Bouwmeester H.J., Martinoia E. (2012). A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344. [DOI] [PubMed] [Google Scholar]
- Lin H., Wang R., Qian Q., Yan M., Meng X., Fu Z., Yan C., Jiang B., Su Z., Li J., Wang Y. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Wu C., Fu Y., Hu G., Si H., Zhu L., Luan W., He Z., Sun Z. (2009). Identification and characterization of HTD2: A novel gene negatively regulating tiller bud outgrowth in rice. Planta 230: 649–658. [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15: 31–39. [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Grandío E.G., Serra F., Marcel F., Rodríguez-Buey M.L., Schmitz G., Theres K., Bendahmane A., Dopazo H., Cubas P. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67: 701–714. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., Sasaki E., Shimada Y., Nagae M., Ueno K., Nakano T., Yoneyama K., Suzuki Y., Asami T. (2009). Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci. Biotechnol. Biochem. 73: 2460–2465. [DOI] [PubMed] [Google Scholar]
- Minakuchi K., Kameoka H., Yasuno N., Umehara M., Luo L., Kobayashi K., Hanada A., Ueno K., Asami T., Yamaguchi S., Kyozuka J. (2010). FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 7: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463. [DOI] [PubMed] [Google Scholar]
- Muth T., García-Martín J.A., Rausell A., Juan D., Valencia A., Pazos F. (2012). JDet: Interactive calculation and visualization of function-related conservation patterns in multiple sequence alignments and structures. Bioinformatics 28: 584–586. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakamura H., et al. (2013). Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4: 2613. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Scaffidi A., Dun E.A., Waters M.T., Flematti G.R., Dixon K.W., Beveridge C.A., Ghisalberti E.L., Smith S.M. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M., Daimon Y., Kurotani K., Higo A., Pruneda-Paz J.L., Breton G., Mitsuda N., Kay S.A., Ohme-Takagi M., Endo M., Araki T. (2013). BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25: 1228–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A., Tarkowski P., Tarkowska D., Norbaek R., Astot C., Dolezal K., Sandberg G. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 101: 8039–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasare S.A., Ducreux L.J., Morris W.L., Campbell R., Sharma S.K., Roumeliotis E., Kohlen W., van der Krol S., Bramley P.M., Roberts A.G., Fraser P.D., Taylor M.A. (2013). The role of the potato (Solanum tuberosum) CCD8 gene in stolon and tuber development. New Phytol. 198: 1108–1120. [DOI] [PubMed] [Google Scholar]
- Ponce M.R., Robles P., Micol J.L. (1999). High-throughput genetic mapping in Arabidopsis thaliana. Mol. Gen. Genet. 261: 408–415. [DOI] [PubMed] [Google Scholar]
- Ponce M.R., Robles P., Lozano F.M., Brotóns M.A., Micol J.L. (2006). Low-resolution mapping of untagged mutations. Methods Mol. Biol. 323: 105–113. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P., Crawford S., Smith R.S., Ljung K., Bennett T., Ongaro V., Leyser O. (2009). Control of bud activation by an auxin transport switch. Proc. Natl. Acad. Sci. USA 106: 17431–17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni L., Nieminen K., Pacheco-Villalobos D., Sibout R., Schwechheimer C., Hardtke C.S. (2011). Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion. Plant Cell 23: 1322–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausell A., Juan D., Pazos F., Valencia A. (2010). Protein interactions and ligand binding: From protein subfamilies to functional specificity. Proc. Natl. Acad. Sci. USA 107: 1995–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. (2005). The FoldX web server: An online force field. Nucleic Acids Res. 33: W382–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20: 259–263. [DOI] [PubMed] [Google Scholar]
- Seto, Y., Sado, A., Asami, K., Hanada, A., Umehara, M., Akiyama, K., and Yamaguchi, S. (2014). Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci USA 11: 1640–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Luong P., Huq E. (2007). The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol. 145: 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Nakatsu T., Nakajima M., Naoe Y., Ohmiya H., Kato H., Matsuoka M. (2008). Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523. [DOI] [PubMed] [Google Scholar]
- Shinohara N., Taylor C., Leyser O. (2013). Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons J.L., Napoli C.A., Janssen B.J., Plummer K.M., Snowden K.C. (2007). Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol. 143: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F., Thimann K.V. (1934). Further experiments on the inhibition of the development of lateral buds by growth hormone. Proc. Natl. Acad. Sci. USA 20: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden K.C., Simkin A.J., Janssen B.J., Templeton K.R., Loucas H.M., Simons J.L., Karunairetnam S., Gleave A.P., Clark D.G., Klee H.J. (2005). The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K., Booker J., Haurogné K., Goussot M., Bainbridge K., Foo E., Chatfield S., Ward S., Beveridge C., Rameau C., Leyser O. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17: 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., Furner I.J., Ottoline Leyser H.M. (2007). MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50: 80–94. [DOI] [PubMed] [Google Scholar]
- Stirnberg P., van De Sande K., Leyser H.M. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Thimann K.V., Skoog F. (1933). Studies on the growth hormone of plants: III. The inhibiting action of the growth substance on bud development. Proc. Natl. Acad. Sci. USA 19: 714–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., Yamaguchi S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun S., Zhu W., Jia K., Yang H., Wang X. (2013). Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 27: 681–688. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Smith S.M. (2013). KAI2- and MAX2-mediated responses to karrikins and strigolactones are largely independent of HY5 in Arabidopsis seedlings. Mol. Plant 6: 63–75. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Brewer P.B., Bussell J.D., Smith S.M., Beveridge C.A. (2012a). The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 159: 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K., Dixon K.W., Smith S.M. (2012b). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Scaffidi A., Flematti G.R., Smith S.M. (2013). The origins and mechanisms of karrikin signalling. Curr. Opin. Plant Biol. 16: 667–673. [DOI] [PubMed] [Google Scholar]
- Worth C.L., Preissner R., Blundell T.L. (2011). SDM—A server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 39: W215–W222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Godzik A. (2004). FATCAT: A web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 32: W582–W585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S., Ding F., Dokholyan N.V. (2007). Eris: An automated estimator of protein stability. Nat. Methods 4: 466–467. [DOI] [PubMed] [Google Scholar]
- Yu J.W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.H., et al. (2013). Crystal structures of two phytohormone signal-transducing α/β hydrolases: Karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., et al. (2013). D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Zhang S., Zhang W., Li G., Chen Z., Zhai W., Zhao X., Pan X., Xie Q., Zhu L. (2006). The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48: 687–698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.