This work finds that rapid retrograde signaling from the photosynthesizing chloroplast to alter nuclear gene expression involves a metabolite-linked pathway that depends on triose phosphate/phosphate translocator.
Abstract
Regulation of the expression of nuclear genes encoding chloroplast proteins allows for metabolic adjustment in response to changing environmental conditions. This regulation is linked to retrograde signals that transmit information on the metabolic state of the chloroplast to the nucleus. Transcripts of several APETALA2/ETHYLENE RESPONSE FACTOR transcription factors (AP2/ERF-TFs) were found to respond within 10 min after transfer of low-light-acclimated Arabidopsis thaliana plants to high light. Initiation of this transcriptional response was completed within 1 min after transfer to high light. The fast responses of four AP2/ERF genes, ERF6, RRTF1, ERF104, and ERF105, were entirely deregulated in triose phosphate/phosphate translocator (tpt) mutants. Similarly, activation of MITOGEN-ACTIVATED PROTEIN KINASE6 (MPK6) was upregulated after 1 min in the wild type but not in the tpt mutant. Based on this, together with altered transcript regulation in mpk6 and erf6 mutants, a retrograde signal transmission model is proposed starting with metabolite export through the triose phosphate/phosphate translocator with subsequent MPK6 activation leading to initiation of AP2/ERF-TF gene expression and other downstream gene targets. The results show that operational retrograde signaling in response to high light involves a metabolite-linked pathway in addition to previously described redox and hormonal pathways.
INTRODUCTION
Appropriate acclimation to a changing environment is necessary to optimize plant growth and fitness. A range of biotic and abiotic factors may influence the acclimation process during the plant life cycle. In addition to long-term acclimation to usually slowly changing stressors like salinity or drought, plants need to respond to fast-acting stressors within rather short time periods (e.g., to heat, cold, or changing light intensities). In high light, overreduction of the photosynthetic electron transport chain is linked to increased release of reactive oxygen species (ROS) (Walters et al., 1996). In order to minimize photooxidative damage caused by ROS (Mittler, 2002) nonphotochemical quenching allows for efficient dissipation of excitation pressure (Niyogi, 1999). In addition, plants use several mechanisms to counteract adverse effects of ROS. ROS are detoxified by the water-water cycle in an either ascorbate-dependent enzymatic process (Asada 1999, 2000) or ascorbate-independent pathway with peroxiredoxins (Dietz et al., 2006; Muthuramalingam et al., 2009). Like the majority of chloroplast proteins, all antioxidant enzymes are encoded in the nucleus (Abdallah et al., 2000; Sakamoto et al., 2008). To coordinate gene expression with metabolic needs, a retrograde signaling network originates from the chloroplast and transmits information on the metabolic state to the nucleus. Singlet oxygen, the redox state of plastoquinone, hydrogen peroxide, metabolites, abscisic acid, and salicylic acid are examples of signaling cues that have been discussed as possible cues for operational and developmental retrograde signaling (reviewed in Baier and Dietz, 2005; Nott et al., 2006; Pogson et al., 2008; Szechyńska-Hebda and Karpiński, 2013). The processes involved in acclimation to excess light intensities in Arabidopsis thaliana were investigated by Karpinski et al. (1997, 1999), who observed rapid transcriptional upregulation of ROS scavengers like ASCORBATE PEROXIDASE1 (APX1) and APX2 within 7 min after a 10-fold increase in light intensity. Moreover, H2O2 released from the bundle sheaths was proposed to be a key player in signal transmission. H2O2 accumulates to amounts that can be detected in the apoplast mostly near the veins by 3,3′-diaminobenzidine staining within 10 to 20 min (Fryer et al., 2003; Galvez-Valdivieso et al., 2009). In a previous study, low (L) or normal (N) light-acclimated Arabidopsis plants were transferred to high light corresponding to a 10- or 100-fold light shift. Transcript levels of the water-water cycle enzymes reached similar levels after 6 h despite the very different starting points and physiological conditions (Oelze et al., 2012). Changes in transcript levels indicate either altered transcription or mRNA turnover. Following the light intensity increase, retrograde signaling pathways rapidly transmit information and allow for adjustment of gene expression by activation of existing transcription factors (TFs) (Kadonaga, 2004). In a subsequent step, downstream secondary TFs may be upregulated transcriptionally. Davletova et al. (2005) observed expression of the zinc finger protein (ZAT12) after application of H2O2. op den Camp et al. (2003) investigated transcript regulation in the fluorescent (flu) mutant in order to differentiate between singlet oxygen and ROS signaling. Furthermore, ROS were shown to activate phosphorylation cascades that involved mitogen-activated protein kinases (MAPK) and activation of TFs (Nakagami et al., 2005). The MAPK kinase kinase, MEKK1, mediates the H2O2-induced MAPK activation, regulates ROS homeostasis, and, hence, plays a key role in cellular redox control (Nakagami et al., 2006).
One of the biggest TF families containing 146 members is the APETALA2/ETHYLENE RESPONSE FACTORs (AP2/ERFs) (Riechmann and Meyerowitz, 1998; Licausi et al., 2013). Besides their role in developmental processes like flower organ development (Jofuku et al., 1994), they strongly respond to various abiotic stressors and are involved in stress acclimation (Dietz et al., 2010; Lata and Prasad, 2011). Involved in responses to cold, salinity, or drought stress, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN1A (DREB1A), 2A, 1B, and 2B probably are the best studied representatives of stress-responsive AP2/ERF-TFs (Sakuma et al., 2006). Expression profiles and comparison of dreb mutants and DREB overexpressors proved the important and specific function of DREBs in acclimation to abiotic stresses. Treatments with abscisic acid, DCMU, Suc, and low or high light resulted in transcript regulation of different clusters of AP2/ERF-TFs with either a broad response to all or most treatments or specific changes to selected effectors (Vogel et al., 2012). Furthermore, regulation of AP2/ERF transcript accumulation upon combinatorial treatments deviated strongly from additive effects in response to single stresses (Vogel et al., 2012). In addition to this general evidence for a role of AP2/ERF-TFs in retrograde signaling, the AP2/ERF-TF ABSCISIC ACID INSENSITIVE4 acts downstream of the master switch GENOME UNCOUPLED1 (Richly et al., 2003), which mediates signal transmission by pathways originating from protochlorophyllide, photosynthetic electron transport, and oxidative stress in seedlings (Koussevitzky et al., 2007). Khandelwal et al. (2008) reported expressional upregulation of REDOX RESPONSIVE TRANSCRIPTION FACTOR1 (RRTF1) in response to high light and DCMU and placed it in the central position of a redox responsive coexpression network. Leaves of a rrtf1 line bleach within 48 h of high-light treatment and the coexpression network of redox-responsive genes collapses. Moreover, RRTF1 is upregulated in response to high light in a systemic manner, which enables acquired acclimation of distal leaves unexposed to high light conditions (Gordon et al., 2012). RELATED TO APETALA 2.4a (RAP2.4a), another AP2/ERF-related TF, displays redox-dependent binding to the promoter of 2-CYSTEINE PEROXIREDOXIN A in protoplasts (Shaikhali et al., 2008). The conformation of RAP2.4a changes with redox status, and it does not bind to the 2CPA promoter under highly reducing or oxidizing conditions in vitro (Shaikhali et al., 2008). These data define AP2/ERF-TFs as interesting candidates to link retrograde signaling and nuclear gene expression.
This work aimed to unravel the order of events that induce the activation of the acclimation response following a sudden and strong light intensity jump. Special attention was given to the starting hypothesis that an AP2/ERF-TF coexpression network, identified by bioinformatics analyses as being light responsive, plays a central role in retrograde signaling. The peculiar time-dependent regulation of transcript levels of these AP2/ERF-TFs motivated us to search for the trigger that initiates their expression. By analysis of mutants defective in specific retrograde signaling pathways, the triose phosphate/phosphate translocator and MITOGEN-ACTIVATED PROTEIN KINASE6 (MPK6) were identified as key players in this signal initiation. Further signal transmission in response to high light by AP2/ERF-TFs was addressed by comparison of wild-type plants with an AP2/ERF-TF T-DNA insertion line.
RESULTS
A Set of AP2/ERF TFs Rapidly Responds to High Light
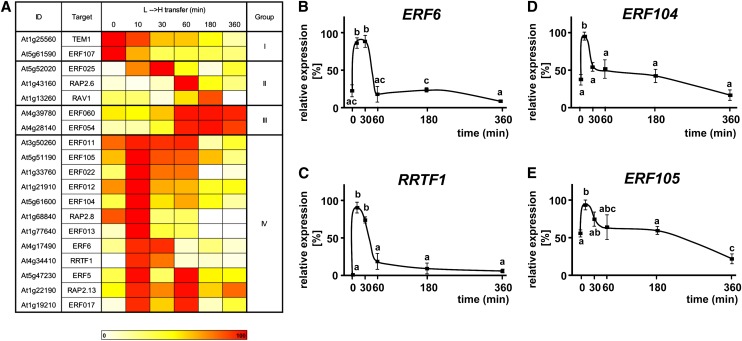
We analyzed the transcriptional responses to high (H) light of members of a potential AP2/ERF-TF coexpression network (Dietz et al., 2010) using plants that were fully acclimated to L-light, as previously shown based on chloroplast ultrastructure and electron transport rate (Oelze et al., 2012). Upon transfer to H-light, transcript levels of antioxidant defense and marker genes of retrograde signaling profoundly changed in these plants within 6 h (Oelze et al., 2012; Alsharafa et al., 2014). We enlarged the network starting with RRTF1 using a second tool for coexpression analysis (Expression Angler; Toufighi et al., 2005); the obtained network consisted of 19 AP2/ERF-TFs with a total of 50 edges between the different members (Supplemental Figure 1). The transcript levels of these AP2/ERF-TFs were quantified during a time course following the transfer from L-light to H-light (8 to 800 µmol quanta m−2 s−1) and from N-light (80 µmol quanta m−2 s−1) to H-light. Based on their time-dependent expressional profiles upon L-light to H-light (L→H) transfer, the 19 different AP2/ERF-TFs were divided into four groups (Figure 1A). Twelve TFs (Group IV) showed a fast upregulation at t = 10 min after L→H shift, mostly followed by a fast decrease of expression, which resulted in very low mRNA levels after 60 min in H-light. The transcriptional changes were similar in plants transferred from N→H-light (Supplemental Figure 2) but with lower magnitude. This indicates a specific role of these TFs in H-light acclimation irrespective of the previous acclimation to L- or N-light.
Figure 1.
Transcript Levels of AP2/ERF-TFs in Response to High-Light Treatment.
(A) The AP2/ERF-TFs from the coexpression network (Supplemental Figure 1) were classified in four groups according to their time-dependent transcript regulation during L→H shift. Expression patterns in ecotype Col-0 were analyzed using nonquantitative RT-PCR. Validity of RT-PCR results was confirmed by comparison with quantitative PCR for four transcripts and gave correlation coefficients of r = 0.94 to 1.00 for the increases during the first 30 min. Twelve AP2/ERF-TFs had an expression maximum at t = 10 min after L→H shift followed by downregulation.
(B) to (E) Transcript regulation of four AP2/ERF-TFs was validated by quantitative PCR (n = 3 independent experiments with duplicate determinations, mean ± sd, letters indicate groups of same significance, Student’s t test: P < 0.05). The highest transcript level for each gene was set to 100%.
[See online article for color version of this figure.]
Due to the higher magnitude of transcriptional changes in the L→H shift, the subsequent experiments were conducted with the 100-fold light increment. Among the members of Group IV, we selected four AP2/ERF-TFs for more detailed analysis: ERF6 showed a 4-fold upregulation with high transcript levels at t = 10 and 30 min of L→H shift followed by a strong decrease and significantly lower mRNA levels after 60 min (Figure 1B). A similar expression profile was detected for RRTF1 but with almost no expression in L-light (Figure 1C). Furthermore, ERF104 and ERF105 displayed the same significant upregulation at t = 10 min but lacked the subsequent fast decrease (Figures 1D and 1E).
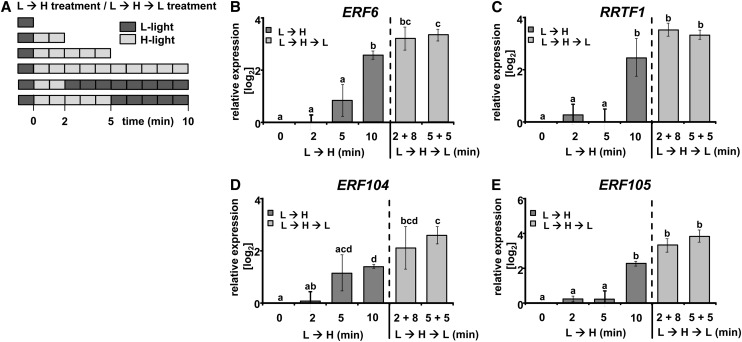
The Transcriptional Response Is Triggered within 2 Min
In order to define the kinetics of the triggering event leading to the fast upregulation of the AP2/ERF-TFs at t = 10 min, we designed an experiment to determine the possible point of no return for the initiation of gene expression of the four selected AP2/ERF-TFs. Whole plants were shifted from L-light to H-light for the durations of 2, 5, and 10 min, respectively. Plants left in L-light were used as a control (Figure 2A). In addition, other plants were shifted to H-light for 2 or 5 min and afterwards transferred back to L-light for 8 min or 5 min (L→H→L treatment), respectively, and thus harvested 10 min after the L→H shift (Figure 2A). All selected AP2/ERF-TFs (ERF6, RRTF1, ERF104, and ERF105) showed significant ∼2.5-fold log2 transcript upregulation at t = 10 min (Figures 2B to 2E, dark-gray columns), while no significant upregulation was seen at t = 2 or t = 5 min. By contrast, upregulation similar to that at t = 10 min of continuous H-light was seen for all four AP2/ERF-TFs in the L→H→L treatments, i.e., 2 min L→H followed by 8 min L-light and 5 min L→H followed by 5 min L-light (Figures 2B to 2E, light-gray columns). These results demonstrate that the trigger responsible for the Group IV response was completed by t = 2 min.
Figure 2.
Point of No Return for Transcript Accumulation of AP2/ERF-TFs.
(A) The length of H-light treatment was varied to define the time point of initiation of transcript regulation. Plants (Col-0) were harvested at indicated time points of H-light (L→H), or plants were transferred back to L-light after 2 or 5 min H-light and harvested at t = 10 min (light-gray bars, H-light; dark-gray bars, L-light).
(B) to (E) All selected AP2/ERF-TFs showed significant transcript accumulation at t = 10 min of L→H (dark-gray columns). A treatment consisting of 2 min H-light followed by 8 min L-light resulted in similar upregulation (light-gray columns) (quantitative PCR, n = 3 independent experiments with duplicate determinations, mean ± se, letters indicate significance groups, Student’s t test P < 0.05).
Temperature as a trigger could be excluded since it was unchanged within the first minutes of the H-light shift when measured with a small thermosensor directly fixed on the leaf surface (Figure 3A). It is well established that ROS are produced under conditions of excess excitation energy and act as signaling molecules in a concentration-dependent manner (Møller et al., 2007). Following the L→H shift, H2O2 accumulated in the leaves from 2 µmol/g fresh weight up to 4.2 µmol/g fresh weight at t = 30 min (Figure 3B). Subsequently, H2O2 concentration declined and had similar levels in L→H as in L-light at t = 60 min to t = 360 min.
Figure 3.
Heat and H2O2 as Possible Signal Triggers.
(A) Temperature measurements by a thermoelement (n = 3, ±sd) showed no change in leaf temperature during the first minutes of treatment.
(B) The H2O2 concentration doubled at t = 30 min of L→H shift from ∼2 to more than 4 µmol/g fresh weight. Within 60 min of treatment, the concentration decreased back to L-light levels (n = 3 independent experiments, mean ± sd, letters indicate significance groups, Student’s t test P < 0.05).
Triose Phosphate/Phosphate Translocator Is Involved in Early Transcriptional Light Response
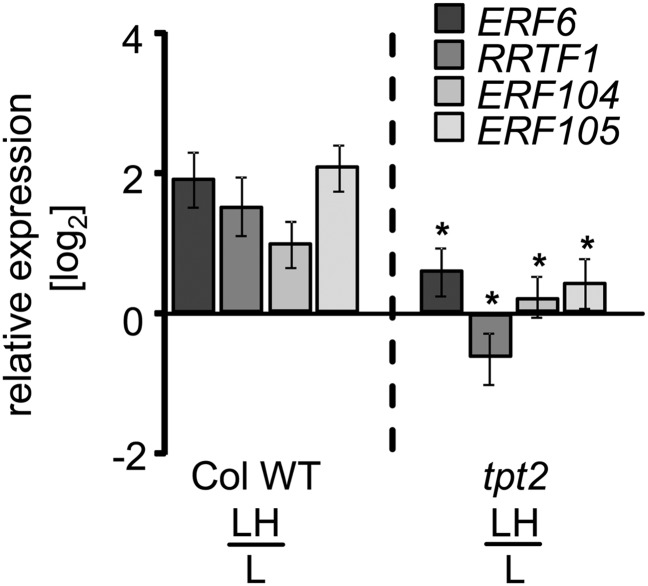
In an attempt to link AP2/ERF-TF transcript accumulation to retrograde signaling, we analyzed Arabidopsis mutants with defects in proposed chloroplast-to-nucleus signaling pathways (Figure 4; Supplemental Figure 3). Transcript levels of the four TFs were quantified in the mutants and presented as ratio L→Ht=10 to L-light controls. We focused on identifying possible components of retrograde signaling that could be involved in metabolite signaling. The assumption was that metabolite changes are among the very fast and first changes that occur in light shifts. Triose phosphate translocator (TPT) is of prime importance for the export of triose phosphate from the chloroplast in exchange for cytosolic phosphate (Flügge, 1999) and appeared to be an ideal candidate for transmission of metabolite signals from the chloroplast to the nucleus since metabolites of the Calvin cycle are known to be autocatalytically built up immediately with activation of photosynthesis (Dietz and Heber, 1984). Two different insertion mutants of the triose phosphate/phosphate translocator (tpt1, Schneider et al., 2002; tpt2, Schmitz et al., 2012; see Supplemental Figure 4A for schematics of insertion site) lack the activity of the TPT. In contrast to the wild type, none of the investigated AP2/ERF-TFs showed strong upregulation of transcript in either tpt mutant at t = 10 min of L→H (Figure 4; Supplemental Figure 3A). It is important to note that deregulation of AP2/ERF-TF transcript accumulation in tpt2 relative to the wild type was also observed following a transfer from N→H (Supplemental Figure 5). Again, significant deregulation was detected for all investigated TFs.
Figure 4.
Comparison of AP2/ERF-TF Transcript Levels in Wild-Type Plants and tpt2 Mutant (Col-0).
ERF6, RRTF1, ERF104, and ERF105 transcript levels were quantified in wild-type plants and tpt2 mutant defective in TPT 10 min following the L→H transfer (quantitative PCR, n = 3 independent experiments with duplicate determinations, mean ± se, asterisks indicate significant difference from the wild-type transcript level, Student’s t test P < 0.05).
Chloroplast thylakoid protein kinase STN7 mediates short-term and long-term acclimation and senses the redox state of the plastoquinone pool and thus links nuclear gene expression to intersystem electron transport (Allen and Pfannschmidt, 2000; Pesaresi et al., 2009). The stn7 mutant, which carries a T-DNA insertion in the fourth exon (Supplemental Figure 4A), is defective in a thylakoid-bound kinase responsible for state transition and might unravel a connection between the expression of AP2/ERF-TFs and plastoquinone-dependent redox signaling. Although the transcriptional responses of the four AP2/ERF-TFs ERF6, RRTF1, ERF104, and ERF105 in the mutant did not entirely match the responses in the wild type, significant differences were not observed (Supplemental Figure 3B). H-light will lead to excess reducing capacity in chloroplasts (Krömer and Scheibe, 1996). Excess reduction capacity can be released to the cytosol via the malate valve using the chloroplast malate dehydrogenase (MDH). The mdh mutant is defective in the chloroplast MDH (Hebbelmann et al., 2012; Supplemental Figure 4A) and unable to activate the malate valve (Hameister et al., 2007). For ERF6, ERF104, and ERF105, similar reaction patterns were observed in the mdh mutants and the wild type (Supplemental Figure 3B). Only for RRTF1 was an opposite response seen, with a decrease upon H-light transfer in the mdh mutant, which resulted in a negative ratio of the transcript level (Supplemental Figure 3B). In order to investigate the possible involvement of chloroplast H2O2, we analyzed the transcript levels of the four selected AP2/ERF-TF in an Arabidopsis line expressing Escherichia coli catalase inside the chloroplast (KatE2; Oelze et al., 2012). Except for higher transcript accumulation of RRTF1, significant differences in the ratios did not exist (Supplemental Figure 3B).
Mitogen-Activated Kinases Function Downstream of Metabolite Export
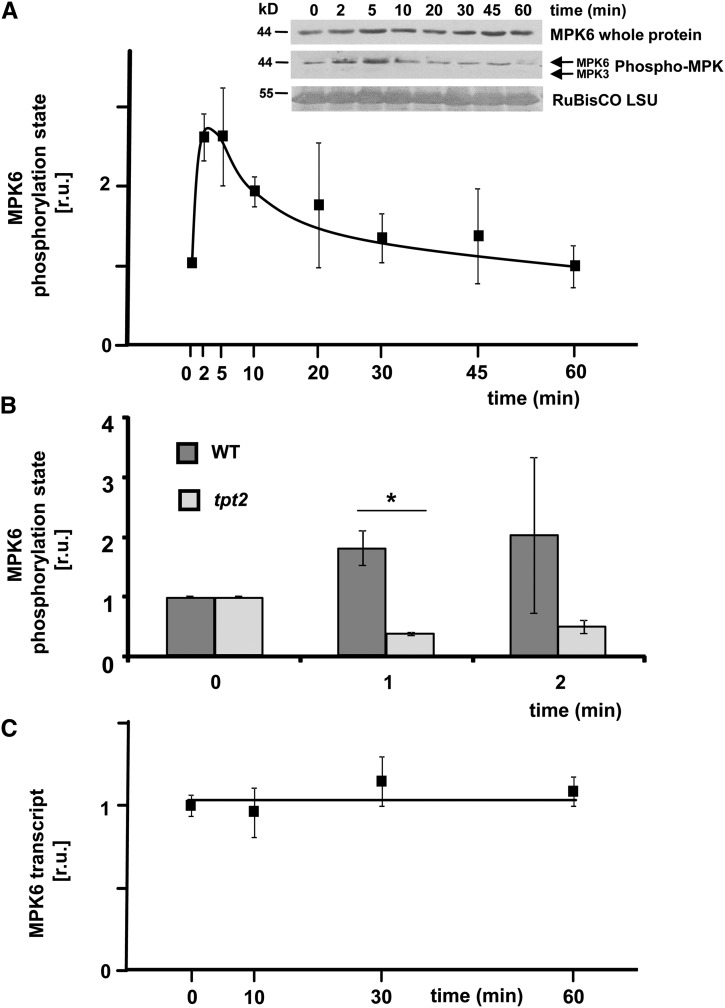
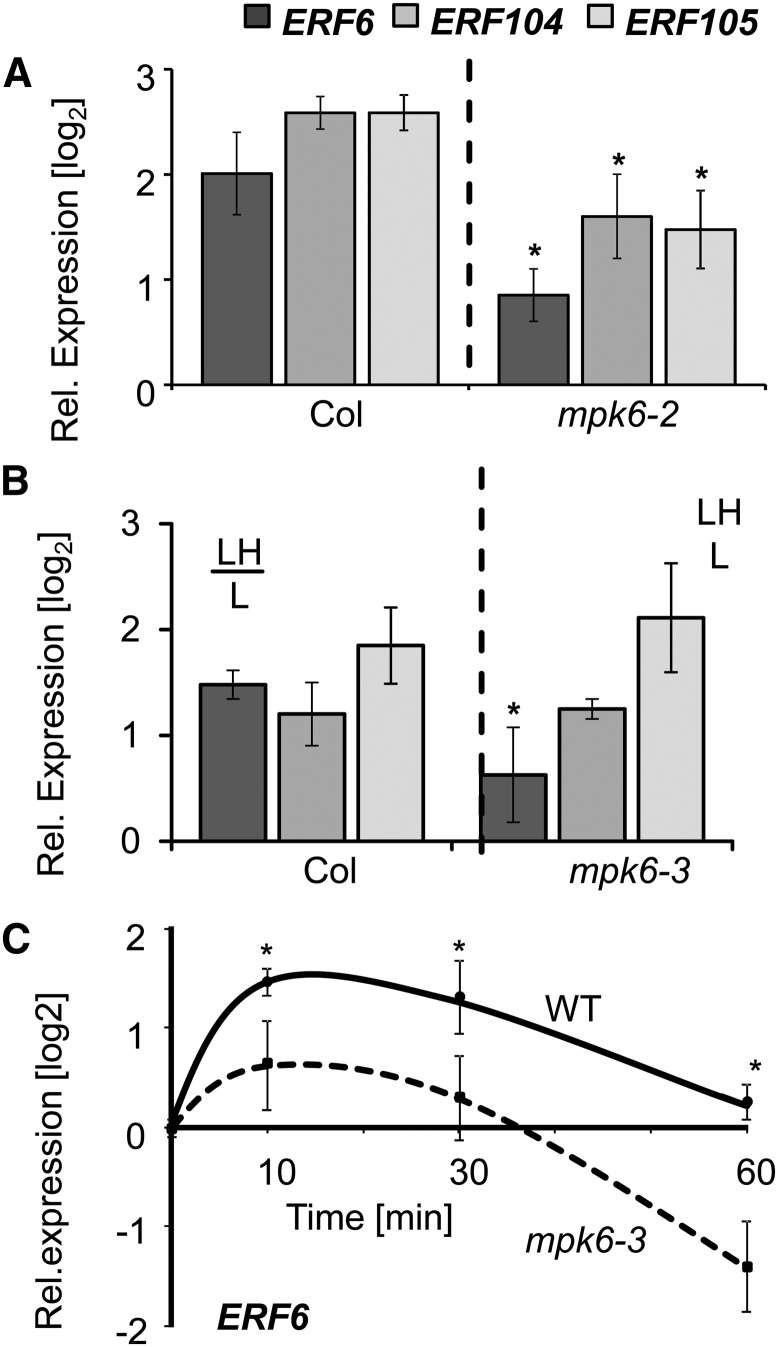
Activation of MAPK cascades is a well-established process in response to abiotic and biotic stresses (Rodriguez et al., 2010) but its involvement in retrograde signaling has not been shown to date. Bethke et al. (2009) reported on an interaction between ERF104 and MPK6, which is disrupted after flagellin peptide (FLG22) application. This prompted us to investigate the phosphorylation state of MAPKs in our L→H shift experiment. Using antibodies that recognize phosphorylated (and, hence, activated forms of) MAPKs (Bethke et al., 2009), phosphorylation of a MAPK of the expected size for MPK6 was detected (Figure 5A). MPK6 phosphorylation showed a very fast and ∼2-fold upregulation at t = 2 min and t = 5 min of L→H treatment, which subsequently decreased, reaching a similar level at t = 60 min in L→H as in L-light (Figure 5A). While the MAPK phosphorylation was much lower than that observed in FLG22-treated plants, it could be reproduced in numerous experiments and hence represents strong evidence that there is H-light-mediated MPK6 activation. By contrast, the activation was absent in mpk6 mutants transferred from L-light to H-light and, although the antibody can also detect activation of MPK3, 4, and 11, phosphorylation of none of these could be observed during the H-light shift, neither in the wild type nor in mpk6 mutants (Supplemental Figure 6). Interestingly, the early MPK6 activation at t = 1 and 2 min was not seen in the tpt2 mutant (Figure 5B). In contrast to MPK6 activation, the MPK6 transcript level was unaltered in the wild type during 60 min of L→H shift (Figure 5C). Taken together, these results suggest that a TPT-dependent retrograde signal transduction pathway leads to MPK6 activation upon shifting from L→H light. To test for a link between such a MAPK cascade and the initiation of expression of the AP2/ERF-TFs at t = 10 min, the transcript levels of ERF6, ERF104, and ERF105 were analyzed in two different mutants of MPK6 at t = 10 min of L→H transfer (Figure 6). ERF6 showed significantly less transcript accumulation at t = 10 min in both investigated mpk6 mutants. Based on this result of deregulated ERF6 in mpk6, we focused on ERF6 in the subsequent analyses.
Figure 5.
H-Light Response of MPK6.
(A) The phosphorylation state of MPK6 in response to the 100-fold light shift was determined in wild-type Col-0 plants by immunoblotting with antibodies recognizing phosphorylated forms of MAPKs as well as MPK6-specific antibodies (inset shows a representative immunoblot). Three independent experiments were performed, and the relative MPK6 activation was calculated on the basis of the band intensities as determined by densitometry (±se). A 2-fold increase occurred after t = 2 min of L→H followed by a decrease to L-light levels at t = 60 min.
(B) Comparison of MPK6-activity between the wild type and tpt2 mutants. The MAPK6 phosphorylation state increased in the wild type as before, but not in the mutant (n = 3 independent experiments, mean ± se, asterisk indicates significant difference between the wild type and mutant, Student’s t test, P < 0.05).
(C) Transcript level of MPK6 (n = 3, with duplicate quantitative PCR determinations, mean ± se).
Figure 6.
Expression of ERF6, ERF104, and ERF105 in mpk6 Mutants (Col-0).
(A) and (B) Transcript levels of ERF6, ERF104, and ERF105 were quantified by quantitative PCR in the wild type and two different mpk6 mutants: (A) mpk6-2 and (B) mpk6-3 in response to L→H shift at t = 10 min.
(C) Kinetics of ERF6 transcript regulation in the wild type and mpk6-3 mutant upon L→H light transfer (n = 3 independent experiments with duplicate determinations, mean ± se, asterisks indicate significant difference, Student’s t test P < 0.05).
Certain cis-Elements Are Enriched in Targeted Promoters
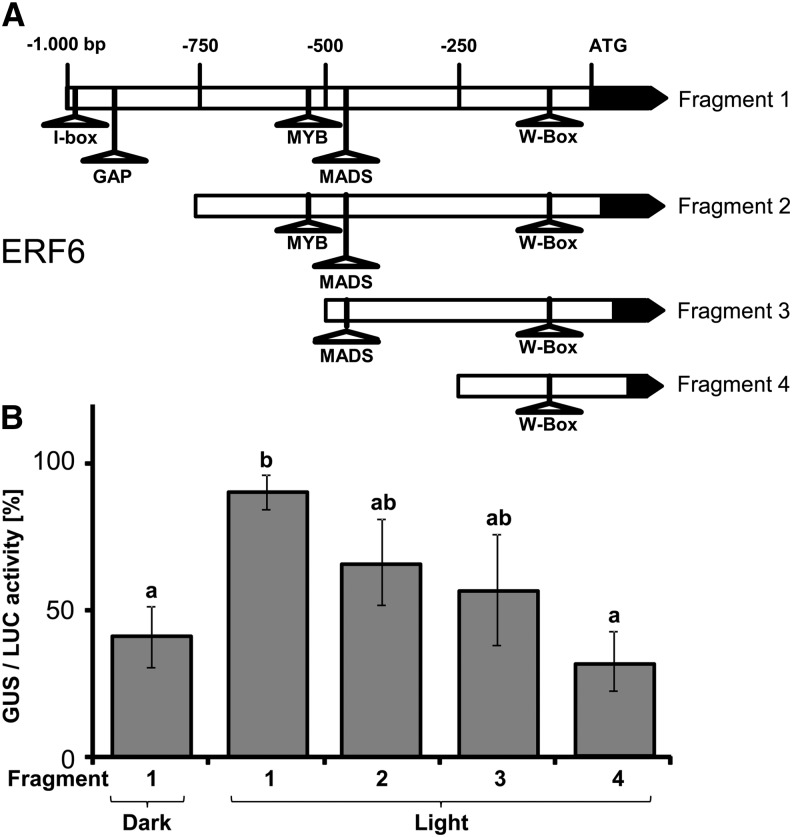
The rapid initiation of AP2/ERF-TF gene expression suggests the existence of TFs that are inactive under L-light conditions and rapidly activated after transfer to H-light. In order to search for possible TF candidates, we screened for common cis-elements in the promoter regions of Group IV members of the coexpression network. Overall, 193 cis-elements were found in the 1000-bp upstream regions of the 12 selected AP2/ERF-TFs (Table 1). Further promoter analysis for common motifs using MEME (Bailey et al., 2006) identified two motifs of 6 to 8 bp length that are common to four promoters, three sites common to three promoters, and >45 sites common to two promoters. No motif that was represented in all 12 promoters could be found, indicating that several TFs act downstream of MPK6 and activate early AP2/ERF gene expression upon the shift to H-light. To explore possible autoregulation within the AP2/ERF-TF network itself, the expression patterns of the two closest neighbors in the AP2/ERF-TF coexpression network (ERF104 and ERF105) were investigated after the L→H shift compared in wild-type and erf6 mutant plants. The T-DNA insertion in the erf6 mutant was confirmed at the genomic DNA level, as was its lack of expression at the transcript level (Supplemental Figures 4B to 4D). The transcriptional alterations in response to the L→H shift of both ERF104 and ERF105 were similar in the wild type and the erf6 mutant (Supplemental Figure 7). To confirm the light responsiveness of erf6 expression, to exclude transcript stabilization as cause for ERF6 accumulation and to further investigate the initiation of the AP2/ERF-TF expression, fragments of the ERF6 promoter were fused to a β-glucuronidase (GUS) reporter gene (Figure 7A). Protoplasts were transfected in parallel with the pERF6:GUS and a pUBIQUITIN:LUCIFERASE (LUC) constructs. GUS and LUC activities were quantified in a H-light shift experiment. The complete 1000-bp fragment could be activated significantly by the light shift as shown by the higher GUS/LUC ratio relative to the dark control. The GUS/LUC ratio of fragment 4 was reduced in comparison to fragment 1 and showed no difference from the dark control (Figure 7B).
Table 1. Overview of cis-Elements within the Promoter Regions of Group IV AP2/ERF Genes.
| AP2/ERF (AtID) | Name | MYB/MYB-Related | WRKY | MADS | MYC | bZIP | AP2/ERF | GAP | I | Other |
|---|---|---|---|---|---|---|---|---|---|---|
| At1g19210 | ERF017 | 5 | 2 | – | – | 3 | – | – | – | 6 |
| At1g21910 | ERF012 | 3 | 3 | 1 | – | – | 1 | 1 | – | 1 |
| At1g22190 | RAP2.13 | 3 | – | 1 | – | – | - | - | 1 | 3 |
| At1g33760 | ERF022 | 6 | 2 | 2 | 1 | 1 | 1 | 1 | ||
| At1g68840 | RAP2.8 | 6 | 6 | – | – | 1 | – | – | – | 3 |
| At1g77640 | ERF013 | 5 | 2 | 2 | – | – | 2 | 1 | – | 1 |
| At3g50260 | ERF011 | 4 | 1 | 1 | 1 | 2 | ||||
| At4g17490 | ERF6 | 1 | 1 | 1 | – | 1 | – | 1 | 1 | 1 |
| At4g34410 | RRTF1 | 1 | 1 | 2 | 4 | 1 | – | – | 1 | 8 |
| At5g47230 | ERF5 | 5 | 4 | – | – | – | 1 | – | – | 2 |
| At5g51190 | ERF105 | 4 | 3 | – | – | – | – | – | – | 1 |
| At5g61600 | ERF104 | 5 | – | 2 | 8 | 1 | 1 | – | 1 | 4 |
| Total | 48 | 24 | 12 | 13 | 7 | 7 | 4 | 5 | 33 |
To analyze the total number and type of cis-elements within Group IV members, the 1000 bp upstream of the ATG region was searched using the ATHENA database (O’Connor et al., 2005).
Figure 7.
GUS Activity of ERF6 Promoter Fragments.
(A) The 1000-bp upstream region of the ERF6 promoter was truncated to obtain four fragments that were fused to GUS reporter gene. cis-elements that are potential binding sites for light-responsive TFs (I- and GAP-box) were detected in the promoters of the network; however, MYB, MADS, and WRKY boxes were present in higher numbers.
(B) GUS/LUC activity of the different fragments measured after protoplast transfection. Fragment 1 showed the highest activity after light shift. Fragment 4 showed no differential activity compared with darkness control (n = 5 independent experiments, mean ± se, letters indicate significance groups, Student’s t test P < 0.05).
Downstream Targets of ERF6 Are Deregulated
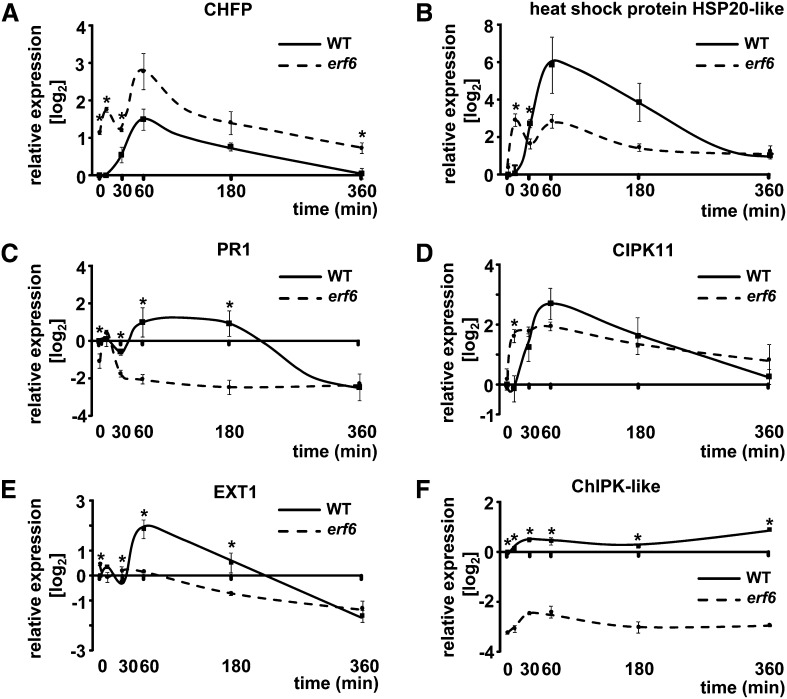
To find downstream targets of ERF6, a single ATH1 Gene-Chip was hybridized with probes synthesized from total RNA from erf6 mutants and the wild type after a L→H treatment of t = 30 min. Eighteen genes were tentatively identified as being differentially regulated (Supplemental Table 1). Six transcripts from this list were selected for confirmation by quantitative PCR. The expression of genes encoding chloroplast protein kinase ChlPK-like, a chitinase family protein (CHFP), the heat shock protein HSP20-like (HSP20-like), pathogenesis-related 1 (PR1), and extensin1 (EXT1) appeared to be downregulated in the erf6 mutant, whereas the gene for calcineurin B-like protein interacting protein kinase 11 (CIPK) was upregulated in erf6. Changes in transcript levels were quantified during the L→H shift at different time points in erf6 mutants and wild-type plants (Figure 8).
Figure 8.
Transcript Profiling of Possible ERF6 Target Genes.
On the basis of a single microarray experiment, putative target genes of the ERF6 TF were selected and RNA levels quantified by quantitative PCR. All selected genes showed significantly altered expression levels in the erf6 mutant (quantitative PCR, n = 3 independent experiments with duplicate determinations, mean ± se, asterisks indicate significant difference, Student’s t test P < 0.05).
In summary, the exploratory microarray results from the L→H experiment at t = 30 min could be confirmed for three of six selected targets. However, each of the other three targets also revealed distinctly different transcript kinetics in erf6 and the wild type albeit at other time points and partly in the opposite direction.
DISCUSSION
Successful acclimation to H-light depends on fast molecular responses to avoid photooxidative damage (Karpinski et al., 1997; Niyogi, 1999; Li et al., 2009). Here, it is shown that at least one type of signaling initiation after L→H transfer is detectable as MPK6 phosphorylation within a minute and by subsequent transcriptional responses within 5 min. Thus, the pace of these responses is in the same time range as activation of established fast regulatory responses such as nonphotochemical quenching (Niyogi,1999), redox-dependent activation of reductive metabolism (Oelze et al., 2008), stimulation of the malate valve for export of excess reducing power (Hebbelmann et al., 2012), and dissipation of excess energy in mitochondria (Xu et al., 2011).
The processes investigated herein are involved in operational control of nuclear gene expression via retrograde adjustment (Pogson et al., 2008). AP2/ERF-TFs are known to be involved in abiotic stress response (Agarwal et al., 2006; Dietz et al., 2010) and light acclimation (Koussevitzky et al., 2007; Khandelwal et al., 2008; Gordon et al., 2012). The identified AP2/ERF-TF coexpression network of 19 members revealed H-light responsiveness (Figure 1), 12 members of which were upregulated at t = 10 min following the L→H shift and upon N→H shift. The similar type of regulation excludes a major influence of starvation of L-light plants on TF expression since N-light plants were subjected to normal growth conditions, and the H-light treatment was started 6 h after beginning of light phase. The fast response suggests a role in very early signaling of excess light and possibly in signal amplification. Despite the fact that L-light plants develop fewer and smaller leaves and display lower electron transport rate, L-light plants are vital and perfectly acclimate to the 100-fold light increment (Oelze et al., 2012). Photoinhibition is fully reversible and the transcriptome of L→H-shifted plants highly resembles that of N→H-shifted plants after 6 h H-light (Alsharafa et al., 2014). The regulation of early responding coexpressed AP2/ERF-TFs was considered as an interesting starting point for further investigation of upstream and downstream events in the early H-light response.
Triggering the AP2/ERF-TF Coexpression Network
The strong upregulation of the 12 AP2/ERFs in H-light implies that upstream signaling events precede this regulation. The time window of the activation of this trigger could be narrowed down to ∼1 to 2 min, which proved to be a point of no return for close to maximal upregulation of AP2/ERF transcript levels after 10 min (Figure 2) even when returned back to L-light, and also enabled MPK6 activation (Figure 5). The short time period makes it unlikely that de novo–synthesized TFs are involved in transcriptional control of AP2/ERFs of Group IV. Thus, existing TFs present in L-light plants likely are activated ≤2 min following the L→H transfer. The promoter analysis (Table 1) indicates the presence of a high number of cis-elements for MYB-TFs. These MYB-TFs might be able to activate the network upon H-light treatment based on the involvement of MYB-TFs in many stress-responsive signal transduction pathways (Stracke et al., 2001; Dubos et al., 2010). The highest GUS/LUC activity was seen only in the 1000-bp erf6 promoter region (Figure 7). An I-box and a GAP-box were found in the region between −750 and −1000 bp (Figure 7), which gives hints to transcription initiation. The I-box plays a role in the response to different light conditions in tomato (Solanum lycopersicum) (Giuliano et al., 1988), whereas the GAP-box was found in the promoter region of the gene for chloroplast glyceraldehyde-3-phosphate dehydrogenase and shown to be light regulated (Jeong and Shih, 2003). WRKY TFs can be excluded at least as sole activators since the smallest promoter fragment containing the conserved W-box did not mediate light-dependent reporter gene activation. Since the I- and GAP-box are present only in about half of the 12 AP/ERF-TFs, other motifs must be involved in the H-light response as well.
The unaltered leaf temperature excludes heat stress as a trigger for the upregulation of AP2/ERF transcripts within the first minutes of L→H shift (Figure 3). When leaves were pretreated with H2O2 prior to the shift to excess excitation conditions, they developed less photooxidative damage and APX2 transcript levels, a stress marker, accumulated to lower levels (Karpinski et al., 1999). Based on these findings, H2O2 was suggested to function as messenger in local and systemic signaling in response to excess light (Bechtold et al., 2008). In line with these results, H2O2 transiently accumulated at t = 30 min in the L→H experiment (Figure 3), supporting its suggested role in signal transmission. However, the kinetics of H2O2 accumulation appeared to be too slow and the maximum of accumulation at t = 30 min was reached too late to function as trigger for the signaling pathway leading to expression of the early responding AP2/ERF-TFs. The point of no return for the transcriptional activation was determined at t ≤ 2 min of L→H shift. This kinetic discrepancy and the dependency on TPT make it unlikely that H2O2 acts as the immediate and first trigger for the induction of MPK6 phosphorylation 1 or 2 min after transfer to H-light. This is tentatively supported by the fact that the transcriptional response of the AP2/ERF-TFs was unaltered in the KatE2 line that overexpresses the E. coli catalase in the chloroplast (Supplemental Figure 3). The activity of E. coli catalase in chloroplasts upon H-light treatment is difficult to judge since catalase is known to be light sensitive under certain conditions and to have a low Km for H2O2 (Feierabend and Engel, 1986). However, there exists experimental evidence that E. coli catalase is active since inhibition of photosynthesis in transgenic plants expressing glycolate oxidase in the chloroplast was overcome by coexpression of KatE (Maurino and Flügge, 2009), suggesting efficient decomposition of glycolate oxidase–generated H2O2. Thus, H2O2 signals appear to be of subordinate importance both at the very early time point of H-light acclimation and at later time points as reported for the same experiment at t = 6 h by Oelze et al. (2012). There remains the likelihood that H2O2 participates in retrograde signaling on intermediate time scale of 15 to 45 min (Karpinski et al., 1997, 1999; Galvez-Valdivieso et al., 2009). Furthermore, other ROS like singlet oxygen could be involved in these signaling pathways. Apel and coworkers investigated the singlet oxygen–dependent pathways using flu and executer in single and triple mutants (op den Camp et al., 2003; Wagner et al., 2004; Kim et al., 2012). Singlet oxygen signaling triggers expression of marker genes, such as BAP1. However, Galvez-Valdivieso et al. (2009) observed downregulation of BAP1 following illumination with 5-fold higher intensity than growth light. This study also included early time points of 5 and 10 min. Likewise, redox changes of the photosynthetic electron transport chain and signal transmission mediated by STN7 can be excluded as trigger for the AP2/ERF-TF expression in excess light since the accumulation of transcript levels were similar in the wild type and stn7 at t = 10 min (Supplemental Figure 3). In addition, the chloroplast MDH and alterations in the NADPH-to-NADP+ ratio mediated by the malate valve are not involved in the initiation of ERF6, ERF104, and ERF105 accumulation (Supplemental Figure 3). The different expression profile of RRTF1 in the mdh mutant (Supplemental Figure 3) where the redox balance between the chloroplast and cytosol is changed could be due to the previously described involvement of RRTF1 in redox regulation (Khandelwal et al., 2008). The function of RRTF1 in redox homeostasis still remains elusive. AP2/ERF-TF transcript regulation depends on interfering signaling pathways. This hypothesis is supported by results from transcript analysis where mRNA levels of 53 AP2/ERF-TFs expressed in leaves were assessed for dependence on treatments inducing retrograde signals (Vogel et al., 2012). Some of these AP2/ERF-TFs share highly similar regions in their DNA binding domains. Nevertheless, their expression profiles were completely different, leading to the assumption of involvement in different signaling pathways (Vogel et al., 2012).
TPT enables the export of dihydroxyacetone phosphate (DHAP) into the cytosol in exchange for phosphate (Fliege et al., 1978; Flügge, 1987, 1992) but is only functional in photosynthetically active tissues (Schulz et al., 1993; Flügge, 1995). The expression change of selected AP2/ERF-TFs at t = 10 min was strongly and significantly suppressed in both tpt mutants (Figure 4; Supplemental Figure 3A). Therefore, the initiation of AP2/ERF-TF expression appears to rely on altered metabolite levels linked to triose phosphate export in the very early phase of the light shift. DHAP increases after illumination of darkened leaves within seconds as shown in nonaqueously fractionated chloroplasts that reflect the metabolic situation of the chloroplasts in vivo (Dietz and Heber, 1984). Exported triose phosphate is converted to 3-phosphoglycerate with simultaneous generation of ATP and NADPH and subsequent Suc biosynthesis (Flügge, 1999). Thus, concentration changes of DHAP, ATP, and NAD(P)H are connected and may activate protein phosphorylation cascades. To test this model, subcellular metabolite levels would need to be determined, for example after nonaqueous fractionation of leaf tissue (Gerhardt and Heldt, 1984). Since plants were exposed to L-light prior to transfer to H-light, the Calvin cycle is preactivated. Thus, upon transfer to H-light, the activation state should allow for instantaneous buildup of metabolites in the stroma (Dietz and Heber, 1986) and the cytosol mediated by TPT.
Linking MAPK Cascades and AP2/ERF-TF Expression
Protein phosphorylation by MAPK cascades is a well-known process in eukaryotic signal transmission (Tena et al., 2001; Rodriguez et al., 2010). The doubling of MPK6 activation within 1 min after the L→H shift indicates that MAPK cascades involving MPK6 are involved in the very fast response to the 100-fold light shift (Figure 5A). Thus, expression initiation of the AP2/ERF-TFs, which precedes the transcript maximum at t = 10 min, correlates with activation of MAPK cascades as indicated by MPK6 phosphorylation. This activation was absent in H-light shifted mpk6 mutants, proving activation of MPK6 in the wild type. Moreover, none of the other detected MAPKs showed a higher phosphorylation state in the wild type or mpk6 mutants, ruling out compensatory redundancy of MPK3 and MPK6 in the early phase of the H-light shift (Supplemental Figure 6). The absence of MPK6 activation in the tpt2 mutant connects triose phosphate export to MAPK cascades in the early H-light response (Figure 5B). Bethke et al. (2009) observed phosphorylation of ERF104 by MPK6 upon flg22 application. MPK6 is involved in rapid biotic stress responses (Asai et al., 2002). Likewise, ERF6 was described as substrate of MPK3/MPK6, and this activation was linked to defense gene expression (Meng et al., 2013). Teige et al. (2004) showed upregulation of MPK6 activity by MKK2 phosphorylation after cold or salinity treatment. In addition, MPK6 activation after application of ethylene precursors induced ETHYLENE INSENSITIVE3, which acts as positive regulator in ethylene signaling (Yoo et al., 2008). The transcript accumulation of ERF6 at t = 10 min of L→H was significantly reduced but not completely abolished in both mpk6 mutants, whereas ERF104 and ERF105 showed a decrease only in the mpk6-2 mutant (Figure 6). Despite these differences, the results support the hypothesis of TPT and MPK6 involvement in rapid ERF transcript accumulation. The mentioned reports show that MPK6 interacts with many proteins. This allows for two interesting conclusions: First, the TPT-dependent retrograde signaling likely addresses different downstream pathways that then adjust gene expression to H-light conditions; second, the TPT pathway likely has crosstalk with other pathogen and abiotic stress–related signaling pathways that involve MPK6.
Signal Amplification by ERF6
The unchanged transcript levels of ERF104 and ERF105 in erf6 mutants compared with wild-type plants demonstrate that regulation of these AP2/ERF-TFs occurs independently of the network itself and excludes short term feedback regulation. It is suggested that the AP2/ERF-TF coexpression network acts as downstream signal amplification module in the H-light acclimation response. Several putative targets of ERF6 were identified by altered transcript levels in the mutant. Transcript accumulation of chitinase genes in Arabidopsis in H-light response was already observed by Takenaka et al. (2009). The higher transcript levels of CHITINASE FAMILY PROTEIN (CHFP) suggest a repressive function of ERF6 in the signaling cascade of CHFP expression initiation (Figure 8A). However, an ERF-associated amphiphilic repression motif (EAR-motif; Dong and Liu, 2010) is not present in the sequence of CHFP. Thus, the repressive function must be indirect. In general, chitinases are involved in pathogen defense (Shin et al., 2008) and are known to be developmentally regulated (Kwon et al., 2005). The response of PR1, which is a known marker for biotic stress (Van Loon and Van Strien, 1999), was significantly lower in erf6 mutants (Figure 8C). However, besides the involvement in biotic stress, it was shown that mRNA levels of PR1 also accumulated in response to salicylic acid or jasmonic acid signaling in the absence of biotic stress, both of which are involved in retrograde signaling processes (Ward et al., 1991; Reinbothe et al., 1993; Niki et al., 1998). In addition, Qiu et al. (2008) demonstrated that PR1 expression is downregulated in mkk1 and mkk2 mutants where MKK2 serves as the kinase upstream of MPK6 (Teige et al., 2004). Hence, PR1 expression might depend on MPK cascades and ERF6. EXT1 is involved in developmental processes since its expression alters the thickness of root cell walls (Merkouropoulos et al., 1999). In addition EXT1 levels are upregulated in response to abiotic stress in all plant tissues (Merkouropoulos and Shirsat, 2003).
The lower expression of HSP20-like after 30 min of L→H transfer characterizes this gene as a possible downstream target of ERF6 (Figure 8C). Heat shock proteins participate in signal transduction in H-light acclimation and in hardening responses. Rossel et al. (2002) reported transcript accumulation of HSP20 and HSP70 after 60 and 120 min of a 10-fold light intensity shift. The enhanced expression of HSP20-like in wild-type plants is in line with the accumulation of H2O2 at t = 30 min in this work. HSP transcripts are upregulated after H2O2 application (Desikan et al., 2001) as well as in catalase-deficient mutants in Arabidopsis (Vandenabeele et al., 2004). CIPK11 is part of calcium-dependent signaling and activates Na/H+ antiporters (Halfter et al., 2000). The strong upregulation in the wild type at t = 60 min suggests involvement of calcium-dependent signaling in H-light response (Figure 8D). In erf6 mutants, CIPK11 mRNA accumulation occurred earlier (t = 10 min), but the peak was less pronounced compared with the wild type peak at t = 60 min. Investigation of Ca2+ levels and changes in cytosolic Ca2+ activities might provide insight into Ca2+ signaling responses and probable alterations in the mutants. Chloroplast protein kinase-like (ChlPK-like) is possibly involved in tocopherol biosynthesis (Vidi et al., 2006). Tocopherol, as a low molecular mass antioxidant, counteracts membrane oxidation and protects photosystem II (Gill and Tuteja, 2010; Traber and Stevens, 2011). Hence, measurements of tocopherol levels during a L→H transition might provide more information about involved signaling pathways and acclimation processes to H-light.
Proposed Model of Early H-Light Signaling
The results of this study in concert with data from recent publications support a hypothetical model for the time-dependent regulation processes in response to H-light. Within a few seconds after the light shift, the cytosolic level of DHAP, and subsequently of linked metabolites, such as ATP and NADPH, change as a consequence of translocation of DHAP from the chloroplast to the cytosol by the TPT. In turn, these changes activate protein phosphorylation cascades leading to MPK6 phosphorylation peaking 1 to 2 min after L→H transfer. It is suggested that phosphorylation of constitutive but inactive TFs initiates expression of the AP2/ERF-TFs. Accumulating transcript levels for AP2/ERF-TFs and their subsequent translation together with higher H2O2 levels and H2O2-dependent signaling amplify the H-light acclimation response through HSPs and kinases. Thus, within minutes, cascades of signaling pathways lead to the activation and amplification of several target genes and contribute to global acclimation. In parallel or at slightly longer time scales, other retrograde signals such as H2O2 (Galvez-Valdivieso et al., 2009), singlet oxygen, possibly via EXECUTER (op den Camp et al., 2003; Wagner et al., 2004), and hormones (Park et al., 2013) also may contribute to the operational retrograde signaling leading to efficient H-light acclimation.
METHODS
Plant Growth and Treatments
Arabidopsis thaliana wild-type Columbia-0 (Col-0) and wild-type Wassilewskija plants and the mutants KatE2 (Oelze et al., 2012), erf6 (SAIL_1236_H11), mdh (Hameister et al., 2007), mpk6-2 (Müller et al., 2010), mpk6-3 (Bethke et al., 2009), stn7 (Pesaresi et al., 2009), tpt1 (Schneider et al., 2002), and tpt2 (Schmitz et al., 2012) were grown in soil culture (1:1:1 mixture of Frühsdorfer Erde Klocke P, perlite, and vermiculite) for 4.5 weeks under the following conditions: 10 h of light, 80 μmol quanta m−2 s−1, 23°C, and 14 h darkness at 18°C; 55% relative humidity. Except for the KatE2 mutant, all mutants used in this study are T-DNA exon insertion mutants (Supplemental Figure 4). For L-light acclimation 3-week-old N-light-grown plants were transferred to L-light (8 μmol quanta m−2 s−1) and further grown for 10 d. These plants have been shown to be fully shade acclimated (Oelze et al., 2012). For the treatments, whole plants were illuminated with H-light (800 μmol quanta m−2 s−1) for different time periods or left in low light as control. Harvest time was always 3 pm. Whole rosettes were harvested and immediately frozen in liquid nitrogen for all subsequent analyses.
Network Construction
The network was constructed as previously published (Dietz et al., 2010). Moreover, a second database (Expression Angler; Toufighi et al., 2005) was employed to resolve the coexpression network in more detail. The cutoff for possible coexpressed AP2/ERF-TFs was set to Pearson correlation coefficient r = 0.4.
Promoter Analysis and Cloning of Constructs
To analyze the promoter regions of the genes for AP2/ERF-TFs, the 1000-bp region upstream of the ATG start codon of each gene was analyzed for common cis-elements. To this end, the ATHENA database (O’Connor et al., 2005) was used. The promoter regions (up to 1000 bp upstream) of the rapidly responding AP2/ERFs were also analyzed for common cis motifs using MEME (Bailey et al., 2006). The 1000-bp promoter region of the AP2/ERF-TF ERF6 (At4g17490) was amplified using specific primers (5′-AAAAGGATCCTTCACTTACAAATGAAGC GA-3′: 5′-AAAACTCGAGTTTGGAGGAAACAGAGAATT-3′) in a RoboCycler (Stratagene) and visualized in agarose gels. The constructs with shortened fragments were generated in the same way using the following specific forward primers: Fragment 2, 5′-AAAAGGATCCACTGGTTGAACT-3′; fragment 3, 5′-AAAAGGATCCAACGTGTGTTTATC-3′; fragment 4, 5′-AAAAGGATCCGCAGCCTAATTAAA-3′; while the reverse primer was in each case the same as for fragment 1. After purification using the SV Gel and PCR Clean Up System (Promega), these fragments were cloned into the pBT10-GUS vector (Sprenger-Haussels and Weisshaar, 2000) using BamHI and XhoI (NEB). The plasmids were sequenced (MWG Ebersberg) using the specific primers 5′-CTGGCCTTTTGCTCACATG-3′ or 5′-GTATAAAGACTTCGCGCTGATAC-3′ and verified plasmids were used for activity measurements.
Transfection and GUS Activity Measurements
Twenty micrograms of the various pBT10-ERF6-Promoter-GUS constructs was transfected into protoplasts together with a pBT10-UBI-LUC vector (Sprenger-Haussels and Weisshaar, 2000). As a positive control the pBT10-35S-GUS vector (Sprenger-Haussels and Weisshaar, 2000) was used. The transfection protocol was described earlier by Seidel et al. (2004). The protoplasts were treated for 10 min with H-light (800 μmol quanta m−2 s−1) and afterwards incubated for 4 h in darkness for optimal protein expression of GUS or LUC. The protoplast solution was centrifuged for 5 min at 800 rpm (SIGMA 3-18K; Osterode). The supernatant was discarded and the pellet again briefly centrifuged (Biofuge fresco; Hereaus). After shock freezing in liquid nitrogen, the pellets were dissolved in Beetle-Lysis Juice (PJK) and centrifuged for 10 min at 10,000g. The resulting supernatant was immediately used for measuring luciferase activity (Firefly Beetle Juice; PJK) or GUS activity (Beta GUS Juice PLUS; PJK) according to the manufacturer’s instructions. To eliminate differences between the samples according the transfection efficiency, the GUS activity was normalized as ratio of the fluorescence value and the measured luciferase activity for each sample. Thus GUS activities are presented as GUS/LUC ratios with maximum value set to 100%.
T-DNA Insertion Mutant Analysis
The analysis of the T-DNA insertion line SAIL_1236_H11 of ERF6 was done as described by Vogel et al. (2012) following the DNA extraction protocol of Edwards et al. (1991).
Transcript Profiling
RNA isolation and cDNA synthesis were performed as described by Wormuth et al. (2006). Transcript levels were quantified with RT-PCR as previously published (Finkemeier et al., 2005). Quantitative real-time PCR was performed as described by Oelze et al. (2012) with the single difference using MESA Blue MasterMix Plus SYBR Assay (Eurogentec). ACTIN2 was used as reference gene. Comparable transcript levels of ACTIN2 during H-light treatment and L-light or N-light controls were checked. Primers are given in Supplemental Table 2.
Microarray Hybridization and Analysis
Whole plants of Arabidopsis wild type and erf6 mutants were treated for 30 min with a 100-fold light shift (L→H). Total RNA was extracted by the Qiagen Plant RNA kit from whole rosettes following the manufacturer’s instructions. The purity was checked by Nanodrop analysis (ND-1000; Peqlab) and gel electrophoresis. A total amount of 1 µg in 10 μL water was sent to the Nottingham Arabidopsis Stock Centre for hybridization (Craigon et al., 2004). The data were further analyzed with Flex Array v1.6.1 (Blazejczyk et al., 2007) using RMA normalization. The difference between the normalized log2 values of the erf6 mutant and wild-type plants was calculated.
Temperature Measurement
A thermoelement (nickel/chromate [Ø0.05 mm], nickel [Ø0.06 mm]) was used for measuring leaf temperature. The small thermoelement was fixed on the upper side of the leaf to monitor the temperature with a voltmeter while illuminating the plant with H-light.
Determination of H2O2 Concentrations and MPK Activity
Leaves were frozen in liquid nitrogen, and H2O2 was measured by chemiluminescence according to Pérez and Rubio (2006). Protein extraction and quantification as well as SDS-PAGE and immunoblotting were done as described by Ahlfors et al. (2004).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACTIN 2 (At3g18780), ChlPK-like (At5g05200), CHFP (At2g43620), CIPK11 (At2g30360), ERF5 (At5g47230), ERF6 (At4g17490), ERF011 (At3g50260), ERF012 (At1g21910), ERF013 (At1g77640), ERF017 (At1g19210), ERF022 (At1g33760), ERF025 (At5g52020), ERF054 (At4g28140), ERF060 (At4g39780), ERF104 (At5g61600), ERF105 (At5g51190), ERF107 (At5g61590), EXT1 (At1g76930), HSP20-like (At1g53540), MDH (At5g58330), MPK6 (At2g43790), PR1 (At2g14610), RAP2.6 (At1g43160), RAP2.8 (At1g68840), RAP2.13 (At1g22190), RAV1 (At1g13260), RRTF1 (At4g34410), STN7 (At1g68830), TEM1 (At1g25560), and TPT (At5g46110).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Potential H-Light-Responsive Coexpression Network of AP2/ERF-TFs.
Supplemental Figure 2. Transcript Regulation of AP2/ERF-TFs in N-Light-Grown Plants in Response to H-Light Treatment.
Supplemental Figure 3. Comparison of AP2/ERF-TF Transcript Levels in Wild-Type Plants and Mutants.
Supplemental Figure 4. Schematics of T-DNA Insertion Sites of Mutants Used in This Paper and Genetic Confirmation of the erf6 Insertion Mutant.
Supplemental Figure 5. Transcript Abundances of AP2/ERF-TFs after 10 Min of N→H in the Wild Type and tpt2 Mutant.
Supplemental Figure 6.: Comparison of Phosphorylation State of Different MAPKs in Wild-Type and mpk6 Mutant Plants.
Supplemental Figure 7. Expression of ERF104 and ERF105 in the erf6 Mutant.
Supplemental Table 1. Results of the Single-Chip Hybridization.
Supplemental Table 2. Primers.
Supplementary Material
Acknowledgments
We acknowledge support by the Deutsche Forschungsgemeinschaft (FOR 804; P.P. and J.L., SFB648). We thank Ulf-Ingo Flügge (Koeln), Dario Leister (Munich), Renate Scheibe (Osnabrück), and Veronica Maurino (Duesseldorf) for providing us with mutant Arabidopsis.
AUTHOR CONTRIBUTIONS
M.O.V. designed the research, performed the transcript profiling, microarray data, and bioinformatic analyses, and wrote the article. M.M. performed the research, in particular, the MPK assays and part of the transcript quantifications. K.K., P.P., and K.A. performed the research. J.L. supervised and discussed data. K.-J.D. designed the project, supervised and discussed the results, and wrote the article.
Glossary
- ROS
reactive oxygen species
- TF
transcription factor
- MAPK
mitogen-activated protein kinase
- TPT
triose phosphate translocator
- MDH
malate dehydrogenase
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abdallah F., Salamini F., Leister D. (2000). A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 5: 141–142. [DOI] [PubMed] [Google Scholar]
- Agarwal P.K., Agarwal P., Reddy M.K., Sopory S.K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25: 1263–1274. [DOI] [PubMed] [Google Scholar]
- Ahlfors R., Macioszek V., Rudd J., Brosché M., Schlichting R., Scheel D., Kangasjärvi J. (2004). Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J. 40: 512–522. [DOI] [PubMed] [Google Scholar]
- Allen J.F., Pfannschmidt T. (2000). Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsharafa K., Vogel M.O., Oelze M.L., Moore M., Stingl N., König K., Friedman H., Mueller M.J., Dietz K.J. (2014). Kinetics of retrograde signalling initiation in the high light response of Arabidopsis thaliana. Philos. Trans. R. Soc. Lond. B Biol. Sci. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess light. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 601–639. [DOI] [PubMed] [Google Scholar]
- Asada K. (2000). The water-water cycle as alternative photon and electron sinks. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1419–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Baier M., Dietz K.J. (2005). Chloroplasts as source and target of cellular redox regulation: A discussion on chloroplast redox signals in the context of plant physiology. J. Exp. Bot. 56: 1449–1462. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U., Richard O., Zamboni A., Gapper C., Geisler M., Pogson B., Karpinski S., Mullineaux P.M. (2008). Impact of chloroplastic- and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. J. Exp. Bot. 59: 121–133. [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazejczyk, M., Miron, M., and Nadon, R. (2007). FlexArray: A Statistical Data Analysis Software for Fene Expression Microarrays. (Montreal, Canada: Genome Quebec), http://genomequebec.mcgill.ca/FlexArray.
- Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S. (2004). NASCArrays: A repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 32: D575–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S., Rizhsky L., Liang H.J., Shengqiang Z., Oliver D.J., Coutu J., Shulaev V., Schlauch K., Mittler R. (2005). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., A-H-Mackerness S., Hancock J.T., Neill S.J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K.J., Heber U. (1984). Rate-limiting factors in leaf photosynthesis. I. Carbon fluxes in the Calvin cycle. Biochim. Biophys. Acta Bioenergetics 767: 432–443. [Google Scholar]
- Dietz K.J., Heber U. (1986). Light and CO2 limitation of photosynthesis and states of the reactions regenerating ribulose-1,5-bisphosphate or reducing 3-phosphoglycerate. Biochim. Biophys. Acta Bioenergetics 848: 392–401. [Google Scholar]
- Dietz K.J., Jacob S., Oelze M.L., Laxa M., Tognetti V., de Miranda S.M., Baier M., Finkemeier I. (2006). The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 57: 1697–1709. [DOI] [PubMed] [Google Scholar]
- Dietz K.J., Vogel M.O., Viehhauser A. (2010). AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245: 3–14. [DOI] [PubMed] [Google Scholar]
- Dong C.J., Liu J.Y. (2010). The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 10: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581. [DOI] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feierabend J., Engel S. (1986). Photoinactivation of catalase in vitro and in leaves. Arch. Biochem. Biophys. 251: 567–576. [DOI] [PubMed] [Google Scholar]
- Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L.J., Dietz K.J. (2005). The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 280: 12168–12180. [DOI] [PubMed] [Google Scholar]
- Fliege R., Flügge U.I., Werdan K., Heldt H.W. (1978). Specific transport of inorganic phosphate, 3-phosphoglycerate and triosephosphates across the inner membrane of the envelope in spinach chloroplasts. Biochim. Biophys. Acta 502: 232–247. [DOI] [PubMed] [Google Scholar]
- Flügge, U.I. (1987). Physiological function and physical characteristics of the chloroplast phosphate translocator. In Progress in Photosynthesis Research, J Biggins, ed (The Hague, The Netherlands: Nijhoff), pp. 739–746.. [Google Scholar]
- Flügge U.I. (1992). Reaction mechanism and asymmetric orientation of the reconstituted chloroplast phosphate translocator. Biochim. Biophys. Acta 1110: 112–118. [DOI] [PubMed] [Google Scholar]
- Flügge U.I. (1995). Phosphate translocation in the regulation of photosynthesis. J. Exp. Bot. 46: 1317–1323. [Google Scholar]
- Flügge U.I. (1999). Phosphate translocators in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 27–45. [DOI] [PubMed] [Google Scholar]
- Fryer M.J., Ball L., Oxborough K., Karpinski S., Mullineaux P.M., Baker N.R. (2003). Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 33: 691–705. [DOI] [PubMed] [Google Scholar]
- Galvez-Valdivieso G., Fryer M.J., Lawson T., Slattery K., Truman W., Smirnoff N., Asami T., Davies W.J., Jones A.M., Baker N.R., Mullineaux P.M. (2009). The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21: 2143–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt R., Heldt H.W. (1984). Measurement of subcellular metabolite levels in leaves by fractionation of freeze-stopped material in nonaqueous media. Plant Physiol. 75: 542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.S., Tuteja N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48: 909–930. [DOI] [PubMed] [Google Scholar]
- Giuliano G., Pichersky E., Malik V.S., Timko M.P., Scolnik P.A., Cashmore A.R. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 85: 7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M.J., Carmody M.E., Albrecht V., Pogson B.J. (2012). Systemic and local responses to repeated HL stress-induced retrograde signaling in Arabidopsis. Front. Plant Sci. 3: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U., Ishitani M., Zhu J.K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97: 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameister S., Becker B., Holtgrefe S., Strodtkötter I., Linke V., Backhausen J.E., Scheibe R. (2007). Transcriptional regulation of NADP-dependent malate dehydrogenase: Comparative genetics and identification of DNA-binding proteins. J. Mol. Evol. 65: 437–455. [DOI] [PubMed] [Google Scholar]
- Hebbelmann I., et al. (2012). Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J. Exp. Bot. 63: 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong M.J., Shih M.C. (2003). Interaction of a GATA factor with cis-acting elements involved in light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Biochem. Biophys. Res. Commun. 300: 555–562. Erratum. Biochem. Biophys. Res. Commun. 333: 1385. [DOI] [PubMed] [Google Scholar]
- Jofuku K.D., den Boer B.G.W., Van Montagu M., Okamuro J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J.T. (2004). Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116: 247–257. [DOI] [PubMed] [Google Scholar]
- Karpinski S., Escobar C., Karpinska B., Creissen G., Mullineaux P.M. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9: 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S., Reynolds H., Karpinska B., Wingsle G., Creissen G., Mullineaux P. (1999). Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284: 654–657. [DOI] [PubMed] [Google Scholar]
- Khandelwal A., Elvitigala T., Ghosh B., Quatrano R.S. (2008). Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiol. 148: 2050–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Meskauskiene R., Zhang S., Lee K.P., Lakshmanan Ashok M., Blajecka K., Herrfurth C., Feussner I., Apel K. (2012). Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719. [PubMed] [Google Scholar]
- Krömer S., Scheibe R. (1996). Function of the chloroplastic malate valve for respiration during photosynthesis. Biochem. Soc. Trans. 24: 761–766. [DOI] [PubMed] [Google Scholar]
- Kwon H.K., Yokoyama R., Nishitani K. (2005). A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol. 46: 843–857. [DOI] [PubMed] [Google Scholar]
- Lata C., Prasad M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62: 4731–4748. [DOI] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260. [DOI] [PubMed] [Google Scholar]
- Licausi F., Ohme-Takagi M., Perata P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 199: 639–649. [DOI] [PubMed] [Google Scholar]
- Maurino, V.G., and Flügge, U.I. (2009). Means for improving agrobiological traits in a plant by providing a plant cell comprising in its chloroplasts enzymatic activities for converting glycolate into malate. Patent Application No. WO2009103782, August 27, 2009.
- Merkouropoulos G., Shirsat A.H. (2003). The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217: 356–366. [DOI] [PubMed] [Google Scholar]
- Merkouropoulos G., Barnett D.C., Shirsat A.H. (1999). The Arabidopsis extensin gene is developmentally regulated, is induced by wounding, methyl jasmonate, abscisic and salicylic acid, and codes for a protein with unusual motifs. Planta 208: 212–219. [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7: 405–410. [DOI] [PubMed] [Google Scholar]
- Meng X., Xu J., He Y., Yang K.Y., Mordorski B., Liu Y., Zhang S. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller I.M., Jensen P.E., Hansson A. (2007). Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58: 459–481. [DOI] [PubMed] [Google Scholar]
- Müller J., Beck M., Mettbach U., Komis G., Hause G., Menzel D., Samaj J. (2010). Arabidopsis MPK6 is involved in cell division plane control during early root development, and localizes to the pre-prophase band, phragmoplast, trans-Golgi network and plasma membrane. Plant J. 61: 234–248. [DOI] [PubMed] [Google Scholar]
- Muthuramalingam M., Seidel T., Laxa M., Nunes de Miranda S.M., Gärtner F., Ströher E., Kandlbinder A., Dietz K.J. (2009). Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Mol. Plant 2: 1273–1288. [DOI] [PubMed] [Google Scholar]
- Nakagami H., Pitzschke A., Hirt H. (2005). Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 10: 339–346. [DOI] [PubMed] [Google Scholar]
- Nakagami H., Soukupová H., Schikora A., Zárský V., Hirt H. (2006). A Mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281: 38697–38704. [DOI] [PubMed] [Google Scholar]
- Niki T., Mitsuhara I., Seo S., Ohtsubo N., Ohashi Y. (1998). Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 39: 500–507. [Google Scholar]
- Niyogi K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 333–359. [DOI] [PubMed] [Google Scholar]
- Nott A., Jung H.S., Koussevitzky S., Chory J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57: 739–759. [DOI] [PubMed] [Google Scholar]
- O’Connor T.R., Dyreson C., Wyrick J.J. (2005). Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413. [DOI] [PubMed] [Google Scholar]
- Oelze M.L., Kandlbinder A., Dietz K.J. (2008). Redox regulation and overreduction control in the photosynthesizing cell: Complexity in redox regulatory networks. Biochim. Biophys. Acta 1780: 1261–1272. [DOI] [PubMed] [Google Scholar]
- Oelze M.L., Vogel M.O., Alsharafa K., Kahmann U., Viehhauser A., Maurino V.G., Dietz K.J. (2012). Efficient acclimation of the chloroplast antioxidant defence of Arabidopsis thaliana leaves in response to a 10- or 100-fold light increment and the possible involvement of retrograde signals. J. Exp. Bot. 63: 1297–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp R.G.L., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon A., Wagner D., Hideg E., Göbel C., Feussner I., Nater M., Apel K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-W., et al. (2013). Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 110: 9559–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez F.J., Rubio S. (2006). An improved chemiluminescence method for hydrogen peroxide determination in plants. Plant Growth Regul. 48: 89–95. [Google Scholar]
- Pesaresi P., Hertle A., Pribil M., Kleine T., Wagner R., Strissel H., Ihnatowicz A., Bonardi V., Scharfenberg M., Schneider A., Pfannschmidt T., Leister D. (2009). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21: 2402–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Woo N.S., Förster B., Small I.D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13: 602–609. [DOI] [PubMed] [Google Scholar]
- Qiu J.L., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S., Reinbothe C., Parthier B. (1993). Methyl jasmonate-regulated translation of nuclear-encoded chloroplast proteins in barley (Hordeum vulgare L. cv. salome). J. Biol. Chem. 268: 10606–10611. [PubMed] [Google Scholar]
- Richly E., Dietzmann A., Biehl A., Kurth J., Laloi C., Apel K., Salamini F., Leister D. (2003). Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO Rep. 4: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Meyerowitz E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379: 633–646. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Rossel J.B., Wilson I.W., Pogson B.J. (2002). Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 130: 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, W., Miyagishima, S.Y., and Jarvis, P. (2008). Chloroplast biogenesis: Control of plastid development, protein import, division and inheritance. The Arabidopsis Book 6: e0110, doi/10.1199/tab.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Osakabe Y., Qin F., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Schöttler M.A., Krueger S., Geimer S., Schneider A., Kleine T., Leister D., Bell K., Flügge U.I., Häusler R.E. (2012). Defects in leaf carbohydrate metabolism compromise acclimation to high light and lead to a high chlorophyll fluorescence phenotype in Arabidopsis thaliana. BMC Plant Biol. 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Häusler R.E., Kolukisaoglu U., Kunze R., van der Graaff E., Schwacke R., Catoni E., Desimone M., Flügge U.I. (2002). An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilisation is abolished. Plant J. 32: 685–699. [DOI] [PubMed] [Google Scholar]
- Schulz B., Frommer W.B., Flügge U.I., Hummel S., Fischer K., Willmitzer L. (1993). Expression of the triose phosphate translocator gene from potato is light dependent and restricted to green tissues. Mol. Gen. Genet. 238: 357–361. [DOI] [PubMed] [Google Scholar]
- Seidel T., Kluge C., Hanitzsch M., Ross J., Sauer M., Dietz K.J., Golldack D. (2004). Colocalization and FRET-analysis of subunits c and a of the vacuolar H+-ATPase in living plant cells. J. Biotechnol. 112: 165–175. [DOI] [PubMed] [Google Scholar]
- Shaikhali J., Heiber I., Seidel T., Ströher E., Hiltscher H., Birkmann S., Dietz K.J., Baier M. (2008). The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 8: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Mackintosh C.A., Lewis J., Heinen S.J., Radmer L., Dill-Macky R., Baldridge G.D., Zeyen R.J., Muehlbauer G.J. (2008). Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J. Exp. Bot. 59: 2371–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger-Haussels M., Weisshaar B. (2000). Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 22: 1–8. [DOI] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456. [DOI] [PubMed] [Google Scholar]
- Szechyńska-Hebda M., Karpiński S. (2013). Light intensity-dependent retrograde signalling in higher plants. J. Plant Physiol. 170: 1501–1516. [DOI] [PubMed] [Google Scholar]
- Takenaka Y., Nakano S., Tamoi M., Sakuda S., Fukamizo T. (2009). Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: Chitinase inhibitor allosamidin enhances stress tolerance. Biosci. Biotechnol. Biochem. 73: 1066–1071. [DOI] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15: 141–152. [DOI] [PubMed] [Google Scholar]
- Tena G., Asai T., Chiu W.L., Sheen J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4: 392–400. [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J. 43: 153–163. [DOI] [PubMed] [Google Scholar]
- Traber M.G., Stevens J.F. (2011). Vitamins C and E: Beneficial effects from a mechanistic perspective. Free Radic. Biol. Med. 51: 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H.K., Zabeau M., Van Montagu M., Inzé D., Van Breusegem F. (2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39: 45–58. [DOI] [PubMed] [Google Scholar]
- Van Loon L.C., Van Strien E.A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55: 85–97. [Google Scholar]
- Vidi P.A., Kanwischer M., Baginsky S., Austin J.R., Csucs G., Dörmann P., Kessler F., Bréhélin C. (2006). Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281: 11225–11234. [DOI] [PubMed] [Google Scholar]
- Vogel M.O., Gomez-Perez D., Probst N., Dietz K.J. (2012). Combinatorial signal integration by APETALA2/Ethylene Response Factor (ERF)-transcription factors and the involvement of AP2-2 in starvation response. Int. J. Mol. Sci. 13: 5933–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., Lee K.P., Würsch M., Laloi C., Nater M., Hideg E., Apel K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185. [DOI] [PubMed] [Google Scholar]
- Walters R.G., Ruban A.V., Horton P. (1996). Identification of proton-active residues in a higher plant light-harvesting complex. Proc. Natl. Acad. Sci. USA 93: 14204–14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E.R., Uknes S.J., Williams S.C., Dincher S.S., Wiederhold D.L., Alexander D.C., Ahl-Goy P., Metraux J.P., Ryals J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormuth D., Baier M., Kandlbinder A., Scheibe R., Hartung W., Dietz K.J. (2006). Regulation of gene expression by photosynthetic signals triggered through modified CO2 availability. BMC Plant Biol. 6: 15–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Yuan S., Lin H.H. (2011). Response of mitochondrial alternative oxidase (AOX) to light signals. Plant Signal. Behav. 6: 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.