This study demonstrates that 14-3-3 proteins negatively regulate a pathway conferring salt tolerance in plants by repressing the activity of a participating kinase in the absence of salt stress.
Abstract
The Salt Overly Sensitive (SOS) pathway regulates intracellular sodium ion (Na+) homeostasis and salt tolerance in plants. Until recently, little was known about the mechanisms that inhibit the SOS pathway when plants are grown in the absence of salt stress. In this study, we report that the Arabidopsis thaliana 14-3-3 proteins λ and κ interact with SOS2 and repress its kinase activity. Growth in the presence of salt decreases the interaction between SOS2 and the 14-3-3 proteins, leading to kinase activation in planta. 14-3-3 λ interacts with the SOS2 junction domain, which is important for its kinase activity. A phosphorylation site (Ser-294) is identified within this domain by mass spectrometry. Mutation of Ser-294 to Ala or Asp does not affect SOS2 kinase activity in the absence of the 14-3-3 proteins. However, in the presence of 14-3-3 proteins, the inhibition of SOS2 activity is decreased by the Ser-to-Ala mutation and enhanced by the Ser-to-Asp exchange. These results identify 14-3-3 λ and κ as important regulators of salt tolerance. The inhibition of SOS2 mediated by the binding of 14-3-3 proteins represents a novel mechanism that confers basal repression of the SOS pathway in the absence of salt stress.
INTRODUCTION
Soil salinity is a widespread abiotic stress with significant agricultural impact, as it severely reduces plant growth and crop productivity worldwide. The evolutionarily conserved Salt Overly Sensitive (SOS) pathway regulates sodium ion homeostasis during salt stress. SOS1, SOS2, and SOS3, the three major components of the pathway, were initially identified in Arabidopsis thaliana using forward genetic screens to isolate mutants with increased sensitivity to salt (Zhu et al., 1998). Cloning of the genes and characterization of their protein activities have identified SOS3 as a calcium binding protein with four EF hands (Liu and Zhu, 1998), SOS2 as a protein kinase (Liu et al., 2000), and SOS1 as a plasma membrane (PM)-localized Na+/H+ antiporter (Shi et al., 2000; Qiu et al., 2002). SOS3 physically interacts with SOS2 via a FISL/NAF motif in the SOS2 C-terminal regulatory domain and recruits SOS2 to the PM, activating SOS2 in a calcium-dependent manner (Halfter et al., 2000; Albrecht et al., 2001; Guo et al., 2001; Quintero et al., 2002). Localization of the SOS3-SOS2 complex to the PM requires the N-myristoylation of SOS3 at a seven–amino acid consensus motif, MGXXXS/T(K) (Ishitani et al., 2000; Batistič et al., 2008). This complex activates SOS1 by directly phosphorylating its C-terminal autoinhibitory domain (Qiu et al., 2002; Quintero et al., 2002, 2011).
Numerous studies have revealed how components of the SOS pathway are regulated during growth during salt stress exposure. SOS3-Like Calcium Binding Protein8 (SCaBP8)/Calcineurin B-Like10 (CBL10) is also critical for SOS2 activation (Kim et al., 2007; Quan et al., 2007). SCaBP8 is mainly expressed in and functions in the shoot, while SOS3 functions in the root. Both proteins share properties in terms of the regulation of SOS2: they interact with the SOS2 FISL motif, activate SOS2 in a calcium-dependent manner, recruit SOS2 to the PM, and activate SOS1. One difference between the two proteins is the absence of an N-myristoylation motif in SCaBP8 compared with the N-myristoylation of SOS3 (Lin et al., 2009). The phosphorylation of SCaBP8 takes place at the PM and enhances the interaction of SCaBP8 and SOS2 in planta (Lin et al., 2009; Du et al., 2011).
In addition to SOS1, downstream targets of SOS2 include an H+/Ca2+ antiporter, a tonoplast H+-ATPase, and a tonoplast Na+/H+ antiporter (Cheng et al., 2004; Qiu et al., 2004; Batelli et al., 2007). Furthermore, SOS2 has been shown to interact with a protein phosphatase 2C, Nucleoside Diphosphate Kinase2, the catalases CAT2 and CAT3, and the flowering time regulator GIGANTEA (GI) (Ohta et al., 2003; Verslues et al., 2007; Kim et al., 2013), linking SOS2 to diverse signaling pathways.
Additional proteins have also been implicated in the regulation of the SOS1 Na+/H+ antiport activity. For example, a recent study reported that salt (NaCl)-induced phosphatidic acid binds to and activates Mitogen-Activated Protein Kinase6, which in turn phosphorylates the C terminus of SOS1 (Yu et al., 2010). In addition, a reactive oxygen species (ROS) signal has been shown to be required for the regulation of Na+/H+ antiport activity (Leshem et al., 2006; Verslues et al., 2007; Jiang et al., 2012; Zhou et al., 2012). Consistent with these observations, ROS modulates SOS1 mRNA stability (Chung et al., 2008) and SOS1 interacts with a ROS regulator, Radical-Induced Cell Death1 (Katiyar-Agarwal et al., 2006). Moreover, the transporter HKT1 has also been reported to be important for regulating Na+/H+ antiport activity (Rus et al., 2001; Berthomieu et al., 2003; Horie et al., 2006; Møller et al., 2009).
Together, these studies consistently support a model in which the SOS pathway is specifically activated when plants are challenged by salt stress (Zhu, 2003; Lin et al., 2009). However, the mechanisms that underlie SOS pathway regulation or repression in the absence of salt are only partially understood. Recently, it was reported that GI prevents the phosphorylation of SOS1 by SOS2 in the absence of salt stress (Kim et al., 2013).
General Regulatory Factor/14-3-3 proteins are highly conserved in eukaryotes and function in almost all aspects of plant growth and development, including abiotic and biotic stress responses, stomatal opening, primary metabolism, hormone signaling, growth, and cell division (reviewed in Oecking and Jaspert, 2009; Denison et al., 2011; Tseng et al., 2012). The Arabidopsis genome encodes 13 14-3-3 proteins that interact with various target proteins, including kinases, transcription factors, structural proteins, ion channels, and other enzymes (Denison et al., 2011). 14-3-3 proteins bind to phosphorylated proteins and transduce the phosphorylation signals to targets by affecting protein–protein interactions, protein activity, stability, conformation, and localization. Three sequence-specific motifs, RSxpSxP, RSxxpSxP, and YpT, are common phosphorylated peptides for 14-3-3 binding; however, 14-3-3 proteins also interact with proteins without these conserved motifs (Paul et al., 2012). Phosphorylation-independent interactions with 14-3-3 proteins have also been reported (Wang et al., 1999; Fuglsang et al., 2003).
In this study, we report that two 14-3-3 proteins interact with and repress SOS2 activity to inhibit the SOS pathway in plants grown in the absence of salt stress and that sodium reduces the interaction between the 14-3-3 proteins and SOS2, leading to activation of the SOS pathway for salt tolerance.
RESULTS
SOS2 Interacts with 14-3-3 λ in Planta
We previously reported that salt stress induces the phosphorylation of SCaBP8 by SOS2 in Arabidopsis (Lin et al., 2009). To investigate if SOS2 kinase activity is regulated by salt stress, Pro35S:6×Myc-SOS2 transgenic plants in the sos2-2 mutant background (Lin et al., 2009) were left untreated (control) or treated with 100 mM NaCl for 3 h. Myc-SOS2 was immunoprecipitated with anti-Myc antibody–conjugated agarose and then used for in vitro kinase assays with recombinant GLUTATHIONE S-TRANSFERASE (GST)-tagged SCaBP8 as the substrate. SOS2 kinase activity was low in the control-treated sample but was induced upon exposure to NaCl (Figure 1A). To elucidate the mechanism by which SOS2 kinase activity is regulated, we used Pro35S:Flag-HA-SOS2 transgenic plants in the sos2-2 background (in which the transgene was previously shown to rescue the sos2 mutant salt-sensitive phenotype) (Lin et al., 2009) to identify SOS2-interacting proteins. A T3 homozygous line was germinated and grown on Murashige and Skoog medium using normal nutrients (14N) or isotopic nutrients (15N). Ten-day-old plants labeled with 15N were treated with 100 mM NaCl for 24 h, and plants labeled with 14N were treated with water. An equal amount of total protein from both treatments was combined, and Flag-HEMAGGLUTININ (HA)-SOS2 was immunoprecipitated with anti-Flag antibody–conjugated agarose. The Flag epitope was then cleaved off, and anti-HA antibody–conjugated agarose was used for HA-SOS2 immunoprecipitation from the resulting supernatant. Agarose-bound HA-SOS2 was eluted from HA agarose using an HA peptide. After this two-step purification, SOS2 and its interacting proteins were enriched and analyzed by mass spectrometry (MS). One putative SOS2-interacting protein, 14-3-3 λ, exhibited higher tandem MS read values without NaCl compared with those with NaCl treatment. To verify this interaction, Pro35S:6×Myc-SOS2 and Pro35S:Flag-HA-14-3-3 λ individually or in combination were expressed in Arabidopsis leaf protoplasts. SOS2 was immunoprecipitated with anti-C-Myc antibody–conjugated agarose, and 14-3-3 λ was detected using immunoblot assays with anti-Flag antibody. These experiments revealed that 14-3-3 λ was immunoprecipitated with SOS2 (Figure 1B). When Pro35S:6×Myc-14-3-3 λ and the Pro35S:Flag-HA-SOS1 C terminus were coexpressed in Arabidopsis leaf protoplasts for coimmunoprecipitation analysis, 14-3-3 λ did not immunoprecipitate the SOS1 C terminus (Supplemental Figure 1A) or vice versa (Supplemental Figure 1B). These data support the specificity of the observed interaction of SOS2 and 14-3-3 λ.
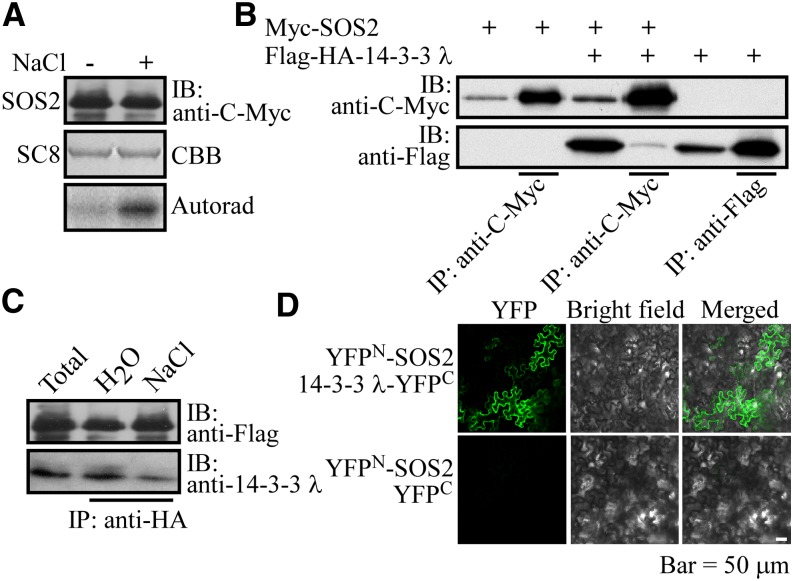
Figure 1.
SOS2 Interacts with 14-3-3 λ in Planta.
(A) SOS2 kinase assay. SOS2 was immunoprecipitated from sos2-2 Pro35S:6×Myc-SOS2 plants grown in the absence (−) or presence (+) of salt and used in in vitro kinase assays with GST-SCaBP8 as substrate. IB, immunoblot; CBB, Coomassie Brilliant Blue; Autorad, autoradiograph. Experimental details are provided in Methods.
(B) Analysis of the SOS2 and 14-3-3 λ interaction in Arabidopsis leaf protoplasts. Purified Pro35S:6×Myc-SOS2 and Pro35S:Flag-HA-14-3-3 λ plasmids were cotransformed into Arabidopsis leaf protoplasts. Transiently expressed proteins were immunoprecipitated with anti-C-Myc antibody–conjugated agarose. Immunoblot assays with anti-C-Myc and anti-Flag antibodies were used to detect Myc-SOS2 and SOS2-interacting 14-3-3 λ, respectively. As a control, Myc-SOS2 or Flag-14-3-3 λ was transiently expressed in Arabidopsis leaf protoplasts and purified with anti-C-Myc or anti-Flag antibody–conjugated agarose, respectively. Anti-C-Myc and anti-Flag antibodies were used to detect immunoprecipitated Myc-SOS2 and Flag-14-3-3 λ, respectively. Experimental details are provided in Methods. IP, immunoprecipitation.
(C) In vivo pull-down assay to analyze the SOS2 and 14-3-3 λ interaction. Ten-day-old Pro35S:Flag-HA-SOS2 transgenic seedlings were treated with water or 100 mM NaCl for 16 h, and then Flag-HA-SOS2 was immunoprecipitated with anti-HA antibody–conjugated agarose. The immunoprecipitated proteins were analyzed by immunoblot using anti-Flag and anti-14-3-3 λ antibodies.
(D) Bimolecular fluorescence complementation analysis in N. benthamiana. Plasmids containing Pro35S:YFPN-SOS2 and Pro35S:YFPC-14-3-3 λ or Pro35S:YFPN-SOS2 and Pro35S:YFPC were transiently cotransformed into N. benthamiana leaves. The YFP fluorescence signal was detected using a Zeiss LSM 510 META confocal microscope. Experimental details are provided in Methods. Bar = 50 μm.
To further confirm this interaction in planta, 14-3-3 λ–specific antibodies were generated by immunizing mice with Escherichia coli–expressed polyhistidine-tagged 14-3-3 λ. To evaluate the specificity of the antibodies, Columbia-0 (Col-0) and the 14-3-3 λ and κ knockdown mutants (14-3-3 κ is the closest homolog of 14-3-3 λ) (Supplemental Figures 2A, 2B, and 2D) were used for immunoblot analysis. Total proteins were extracted from 10-d-old wild-type and single and double mutant seedlings and analyzed by immunoblots with 14-3-3 λ antibodies. Strong cross-reaction was detected in Col-0 and the 14-3-3 κ mutant, weak cross-reaction was detected in the 14-3-3 λ mutant, and no cross-reaction was detected in the 14-3-3 λ κ double mutant (Supplemental Figure 2C). Subsequently, Pro35S:Flag-HA-SOS2 transgenic plants were left untreated or treated with 100 mM NaCl for 16 h, and then Flag-HA-SOS2 was immunoprecipitated with anti-HA antibody–conjugated agarose. The precipitated proteins were analyzed by immunoblot analysis with anti-14-3-3 λ antibodies. Consistent with the tandem MS results, SOS2 pulled down 14-3-3 λ in vivo, and this interaction was reduced with salt treatment, suggesting that salt stress decreases the interaction between SOS2 and 14-3-3 λ (Figure 1C).
Finally, binary constructs YFPN-SOS2 and YFPC-14-3-3 λ were generated for bimolecular fluorescence complementation analysis in Nicotiana benthamiana. Plasmids of YFPN-SOS2 and YFPC-14-3-3 λ or YFPN-SOS2 and the YFPC vector were cotransformed into N. benthamiana leaves by Agrobacterium tumefaciens–mediated transformation. Four days after transformation, the yellow fluorescent protein (YFP) fluorescence signal was detected in leaves transiently expressing the YFPN-SOS2 and YFPC-14-3-3 λ constructs but not in leaves expressing YFPN-SOS2 or the YFPC vector (Figure 1D). Together, these results demonstrate that SOS2 and 14-3-3 λ interact in planta and that this interaction is reduced by salt stress.
14-3-3 λ Represses SOS2 Kinase Activity
Salt stress leads to an increase in SOS2 kinase activity and simultaneously to a reduction in the interaction between SOS2 and 14-3-3 λ (Figures 1A and 1C). Therefore, we reasoned that 14-3-3 λ may repress SOS2 activity in the absence of salt stress and that the addition of sodium may induce disassociation of 14-3-3 λ from SOS2 and allow for SOS2 kinase activation. Since among the 13 members of the 14-3-3 family in Arabidopsis, 14-3-3 λ and 14-3-3 κ share the highest sequence similarity (Supplemental Figure 2D), we also included 14-3-3 κ in our experiments to investigate the potential regulation of SOS2 activity by 14-3-3 proteins. GST-tagged SOS2 and polyhistidine-tagged 14-3-3 λ and 14-3-3 κ were purified from E. coli and used in in vitro kinase assays. Both 14-3-3 λ and 14-3-3 κ partially repressed the SOS2 transphosphorylation activity of Myelin Basic Protein (MBP), while autophosphorylation was not altered (Figure 2A; Supplemental Figure 3A). This reduction in transphosphorylation activity was enhanced with increasing amounts of added 14-3-3 protein (Figure 2B; Supplemental Figure 3B). To confirm that the observed repression is caused by 14-3-3 λ and not simply by an increase in the total amount of protein in the assay or by substances derived from the polyhistidine-tagged protein purification process, we tested the effect of the unrelated cytoplasm-located polyhistidine-tagged Arabidopsis Actin-Depolymerizing Factor4 (ADF4) on SOS2. This protein did not repress SOS2 autophosphorylation or transphosphorylation activity in vitro (Figure 2C; Supplemental Figure 3C). A previous study showed that SOS2 phosphorylates SCaBP8 during salt stress in vivo (Lin et al., 2009), and SOS2 is activated by NaCl treatment (Figure 1A). To determine if recombinant 14-3-3 proteins can also repress SOS2 activity when SOS2 is immunoprecipitated from plants, we conducted kinase assays using immunoprecipitated Myc-tagged SOS2 and polyhistidine-tagged 14-3-3 λ. Phosphorylation of SCaBP8 by Myc-SOS2 was repressed by 14-3-3 λ, and this repression was 14-3-3 protein concentration dependent (Figures 2D and 2E; Supplemental Figure 3D).
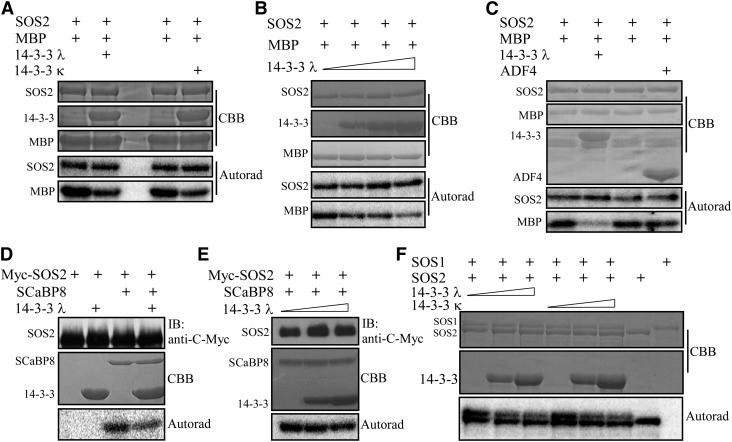
Figure 2.
14-3-3 λ and κ Proteins Repress SOS2 Kinase Activity.
(A) In vitro kinase assay for recombinant GST-tagged SOS2 with or without polyhistidine-tagged 14-3-3 λ or κ protein. MBP was used as the substrate. CBB, Coomassie Brilliant Blue; Autorad, autoradiograph. Experimental details are provided in Methods.
(B) In vitro kinase assay for SOS2 as in (A), with increasing amounts of 14-3-3 λ protein added to the reaction mixture. Experimental details are provided in Methods.
(C) In vitro kinase assay for recombinant GST-tagged SOS2 with or without polyhistidine-tagged 14-3-3 λ or ADF4. MBP was used as the substrate. Experimental details are provided in Methods.
(D) Kinase assay for Myc-SOS2 purified from Pro35S:6×Myc-SOS2 transgenic plants in the Col-0 background with or without polyhistidine-tagged 14-3-3 λ. ScaBP8 was used as the substrate. IB, immunoblot. Experimental details are provided in Methods.
(E) Kinase assay for SOS2 as in (D), with increasing amounts of 14-3-3 λ protein added to the reaction mixture. Experimental details are provided in Methods.
(F) In vitro kinase assay for recombinant GST-tagged SOS2 with different concentrations of polyhistidine-tagged 14-3-3 proteins. A maltose binding protein–tagged SOS1 C terminus was used as the substrate. Experimental details are provided in Methods.
To investigate whether 14-3-3 proteins modulate the regulation of SOS1 by SOS2, we first performed comparative in vitro phosphorylation assays using the SOS1 C terminus (amino acids 848 to 1146) as a substrate for SOS2. In kinase assays using recombinant maltose binding protein–tagged SOS1 C terminus purified from E. coli, we observed efficient phosphorylation of SOS1 by SOS2 (Figure 2F). However, the addition of either 14-3-3 λ or 14-3-3 κ significantly reduced the phosphorylation of the SOS1 C terminus by SOS2, while the autophosphorylation of SOS2 was not affected (Figure 2F; Supplemental Figure 3E). Also, the inhibition of SOS1 phosphorylation by 14-3-3 proteins appeared to be dose dependent, since increasing amounts of added 14-3-3 proteins further reduced phosphorylation (Figure 2F; Supplemental Figure 3E). These results indicate that 14-3-3 λ and 14-3-3 κ function as modulators of the SOS pathway by directly repressing SOS2 transphosphorylation activity.
Salt Stress Reduces the Interaction between 14-3-3 λ and SOS2 and Releases SOS2 Activity
To investigate the function of 14-3-3 proteins in the SOS pathway and determine how salt stress affects the interaction of 14-3-3 and SOS2 in vivo, we performed a series of experiments that monitored the interaction between 14-3-3 λ and SOS2 at different time points during salt stress exposure. Pro35S:Flag-HA-SOS2 transgenic plants were left untreated or treated with 100 mM NaCl for 12, 24, or 36 h, and then Flag-HA-SOS2 was immunoprecipitated with anti-HA antibody–conjugated agarose and coimmunoprecipitating proteins were probed with anti-14-3-3 λ antibodies. This experiment revealed a time-dependent reduction in the interaction between 14-3-3 λ and SOS2 during salt stress (Figure 3A).
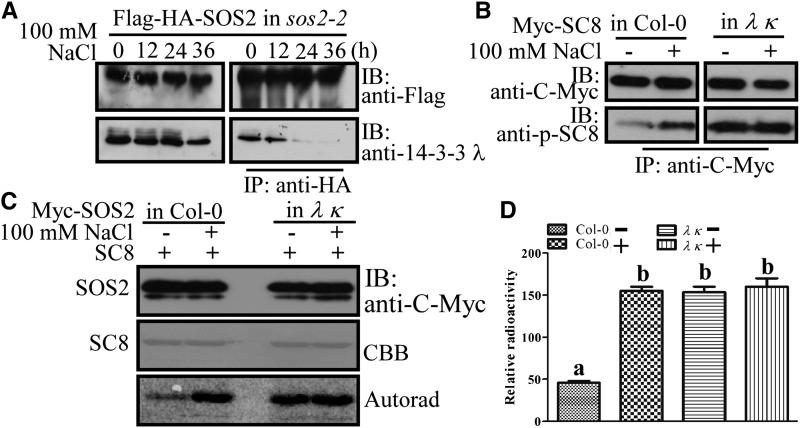
Figure 3.
Salt Stress Reduces the Interaction between SOS2 and 14-3-3 λ Leading to SOS2 Activity.
(A) In vivo pull-down assay to analyze the interaction between SOS2 and 14-3-3 λ. Ten-day-old Pro35S:Flag-HA-SOS2 transgenic seedlings in the sos2-2 background were left untreated or treated with 100 mM NaCl for 12, 24, or 36 h, and then Flag-HA-SOS2 was immunoprecipitated with anti-HA antibody–conjugated agarose. The resulting proteins were analyzed by immunoblots with anti-14-3-3 λ antibody. IB, immunoblot.
(B) Analysis of the phosphorylation status of SCaBP8. Ten-day-old transgenic seedlings with Pro35S:6×Myc-SCaBP8 in Col-0 and the 14-3-3 λ κ double mutant background were left untreated (−) or treated (+) with 100 mM NaCl for 12 h, and Myc-SCaBP8 (Myc-SC8) was immunoprecipitated with anti-C-Myc antibody–conjugated agarose. The resulting Myc-SCaBP8 proteins were analyzed by immunoblots with anti-C-Myc and SCaBP8 anti-phosphoserine-237 (anti-p-SC8) antibodies.
(C) Kinase assay for Myc-SOS2 purified from Pro35S:6×Myc-SOS2 transgenic plants in Col-0 or the 14-3-3 λ κ double mutant background. GST-SCaBP8 (SC8) was used as the substrate. Experimental details are provided in Methods. CBB, Coomassie Brilliant Blue; Autorad, autoradiograph.
(D) Relative radioactivity for (C). Phosphorylation signals were obtained using a Typhoon 9410 phosphor imager (Amersham Biosciences), and signals were quantified with ImageQuant 5.0 software. Error bars represent sd (n > 3). Statistical significance was determined by Student’s t test; significant differences (P ≤ 0.05) are indicated by different lowercase letters. Experimental details are provided in Methods.
To determine if the disassociation of 14-3-3 proteins from SOS2 correlates with the activation of SOS2 during salt stress in planta, antibodies for SCaBP8 phosphoserine-237 (Lin et al., 2009) were used to monitor the phosphorylation status of SCaBP8 as a measure of SOS2 kinase activity. The Pro35S:6×Myc-SCaBP8 construct was transformed into Col-0 and the 14-3-3 λ κ double mutant (Supplemental Figure 2B), in which no accumulation of 14-3-3 λ was detected (Supplemental Figure 2C). Ten-day-old T3 transgenic seedlings were left untreated or treated with 100 mM NaCl for 12 h, and Myc-SCaBP8 proteins were immunoprecipitated with anti-C-Myc antibody–conjugated agarose. The resulting Myc-SCaBP8 proteins were analyzed by immunoblot analysis with anti-C-Myc or anti-phosphoserine-237-SCaBP8 antibodies. Consistent with previous results (Lin et al., 2009), phosphorylation of SCaBP8 was induced in Col-0 by salt stress; however, the degree of phosphorylation of SCaBP8 in the 14-3-3 λ κ double mutant was much higher than that in the wild type before NaCl treatment, and salt treatment did not further enhance this phosphorylation (Figure 3B). To further evaluate the effect of the 14-3-3 proteins on SOS2 activity in vivo, Pro35S:6×Myc-SOS2 was transformed into the 14-3-3 λ κ double mutant background. Ten-day-old T3 transgenic seedlings were left untreated or treated with 100 mM NaCl for 12 h, and Myc-SOS2 was immunoprecipitated. The resulting proteins were incubated in an in vitro kinase assay with recombinant GST-tagged SCaBP8 as substrate. In the wild-type background, SOS2 transphosphorylation activity was very low in control conditions and induced by NaCl treatment. However, in the 14-3-3 λ κ double mutant, SOS2 was significantly activated even without salt treatment (Figures 3C and 3D). These results indicate that 14-3-3 proteins interact with SOS2 in vivo in the absence of salt, thereby repressing SOS2 transphosphorylation activity that would otherwise be constitutively activated. Upon exposure of plants to salt stress, the 14-3-3 proteins disassociate from SOS2 and release the suppression of SOS2 activity. Together, these results identify an important regulatory mechanism in which the binding of 14-3-3 proteins in nonstressed conditions keeps SOS2 activity at a basal level but allows for activation by dissociation upon exposure to stress.
14-3-3 λ and κ Negatively Regulate Salt Tolerance in Arabidopsis
To determine the tissue-specific expression of 14-3-3 λ in Arabidopsis, a 2000-bp DNA fragment upstream of the 14-3-3 λ translation start site (ATG) was fused to the β-GLUCURONIDASE (GUS) reporter gene. The resulting construct was transformed into the Col-0 background. Twenty independent T2 transgenic lines were analyzed for GUS staining. GUS expression driven by the 14-3-3 λ promoter was detected throughout the plant (Supplemental Figures 4A to 4G). The root tip displayed enhanced GUS expression (Supplemental Figure 4B), which is consistent with the previously reported expression pattern of SOS2 (Quan et al., 2007). In agreement with the promoter-GUS results, when total proteins were extracted from roots, stems, rosette leaves, cauline leaves, flowers, and siliques of wild-type plants and analyzed by immunoblots using 14-3-3 λ antibodies, 14-3-3 λ protein accumulation was ubiquitously detected in all tissues (Supplemental Figure 4H). To determine the subcellular localization of 14-3-3 λ, Pro35S:GFP-14-3-3 λ and Pro35S:YFP-SOS2 transgenic plants were generated. As reported previously, YFP-SOS2 fluorescence was detected at the plasma membrane as well as in the cytosol and nucleus (Quan et al., 2007; Batistič et al., 2010); GFP-14-3-3 λ exhibited the same localization pattern (Supplemental Figure 4I).
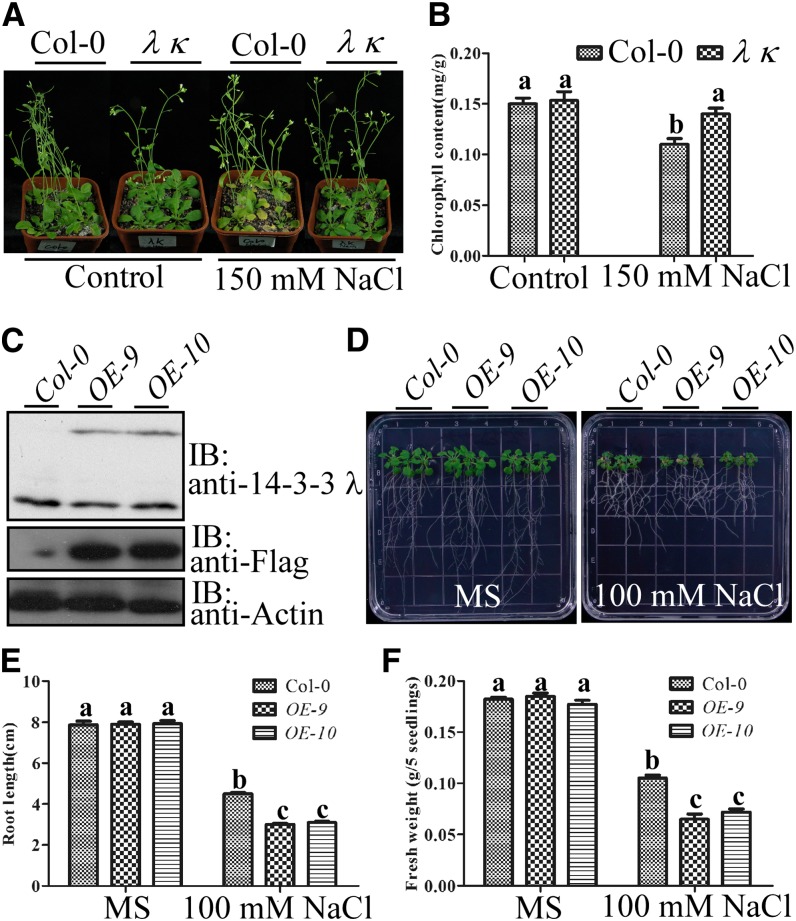
We next determined whether loss of function of 14-3-3 λ or 14-3-3 κ affects salt tolerance in Arabidopsis plants. Three-week-old Col-0 and 14-3-3 λ κ double mutant seedlings grown in soil under short-day conditions were treated with 150 mM NaCl for 2 weeks. Leaves of the salt-treated 14-3-3 λ κ double mutant were greener than those of Col-0 (Figure 4A), and chlorophyll content in the double mutant was also significantly higher (Figure 4B), while no significant growth difference was observed between wild-type and 14-3-3 λ κ double mutant seedlings grown under control conditions (Figures 4A and 4B). We next generated Pro35S:Flag-HA-14-3-3 λ transgenic plants in the Col-0 background to overexpress 14-3-3 λ. Two T3 transgenic lines (OE-9 and OE-10) that exhibited readily detectable expression of Flag-HA-14-3-3 λ (Figure 4C) were used for further studies. When 5-d-old seedlings were transferred to Murashige and Skoog medium containing 100 mM NaCl, the transgenic plants displayed significantly enhanced growth inhibition in both the root and shoot compared with the growth of Col-0 (Figure 4D). Primary root lengths of OE-9 and OE-10 lines were reduced by 20% when compared with those of Col-0 (Figure 4E), while the fresh weight was reduced by ∼30% relative to that of Col-0 seedlings (Figure 4F).
Figure 4.
14-3-3 λ and κ Negatively Regulate Salt Tolerance in Arabidopsis.
(A) Analysis of the salt sensitivity of Col-0 and the 14-3-3 λ κ double mutant in soil. Three-week-old soil-grown seedlings were left untreated or treated with 150 mM NaCl for 2 weeks.
(B) Analysis of chlorophyll content for seedlings in (A). Error bars represent sd (n > 6). Statistical significance was determined by Student’s t test; significant differences (P ≤ 0.05) are indicated by different lowercase letters. Experimental details are provided in Methods.
(C) Immunoblot analysis of the protein level of 14-3-3 λ in Col-0 and two independent lines of Pro35S:Flag-HA-14-3-3 λ transgenic plants in the Col-0 background (OE-9 and OE-10). Actin was used as a loading control. IB, immunoblot.
(D) Analysis of the salt sensitivity of Col-0 and two independent lines of Pro35S:Flag-HA-14-3-3 λ transgenic plants in the Col-0 background (OE-9 and OE-10) on plates. Five-day-old seedlings were transferred to Murashige and Skoog medium (MS) without or with 100 mM NaCl. Photographs were taken 10 d after transfer.
(E) Analysis of root length for seedlings in (D). Error bars represent sd (n > 10). Statistical significance was determined by Student’s t test; significant differences (P ≤ 0.05) are indicated by different lowercase letters. Experimental details are provided in Methods.
(F) Analysis of fresh weight for seedlings in (D). Error bars represent sd (n > 10). Statistical significance was determined by Student’s t test; significant differences (P ≤ 0.05) are indicated by different lowercase letters. Experimental details are provided in Methods.
[See online article for color version of this figure.]
To further evaluate whether the salt-sensitive phenotype of the 14-3-3 λ overexpression lines was due to decreased SOS2 activity during salt stress, 10-d-old T3 Pro35S:6×Myc-SOS2 transgenic seedlings were left untreated and Myc-SOS2 was immunoprecipitated. An equal amount of Myc-SOS2 protein was incubated with total protein extracts prepared from Col-0 or a transgenic 14-3-3 λ overexpression line (OE-9) (Figures 4C and 4D), which had been left untreated or treated with 100 mM NaCl for 12 h. The resulting SOS2 proteins were used for in vitro kinase assays with recombinant MBP or GST-tagged SCaBP8 as substrate. Consistent with previous results (Figure 3C), SOS2 activity was hardly detectable when Myc-SOS2 was incubated with protein extracts from control Col-0, but transphosphorylation activity increased significantly when Myc-SOS2 was incubated with protein extracts prepared from Col-0 treated with 100 mM NaCl (Supplemental Figures 5A and 5B). By contrast, compared with protein extract from Col-0, the activation of SOS2 was reduced when Myc-SOS2 was incubated with protein extracts from the 14-3-3 λ overexpression line treated with 100 mM NaCl (Supplemental Figures 5A and 5B). Moreover, immunoblot analysis revealed that more Flag-14-3-3 protein was associated with Myc-SOS2 when Myc-SOS2 was incubated with the protein extracts from the 14-3-3 λ overexpression line than with protein extracts from Col-0 (Supplemental Figure 5C). Together, these results indicate that 14-3-3 λ and 14-3-3 κ negatively regulate salt tolerance in Arabidopsis.
14-3-3 λ Interacts with the Junction Domain of SOS2
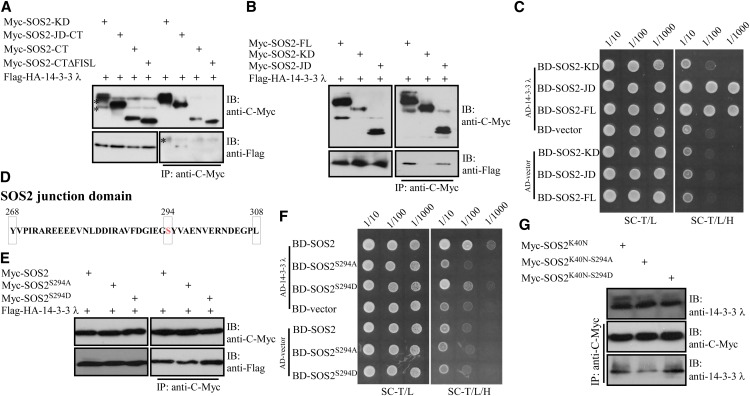
To further elucidate the mechanism underlying 14-3-3 λ–mediated repression of SOS2 kinase activity, we mapped the interaction region in the SOS2 protein. Previously, we identified four domains in SOS2: the N-terminal kinase domain (KD), the junction domain (JD), and the FISL/NAF motif and PPI domains in the C terminus (Albrecht et al., 2001; Guo et al., 2001; Ohta et al., 2003). We generated various truncated variants of the SOS2 protein, including KD, JD plus the C terminus (JD-CT), the C terminus alone (CT), and a C-terminal domain without the FISL domain (CTΔFISL) (Supplemental Figure 6A). All of these truncated proteins were fused with a Myc tag and transiently coexpressed in Arabidopsis leaf protoplasts with Flag-HA-14-3-3 λ. Truncated versions of Myc-tagged SOS2 were immunoprecipitated with anti-C-Myc antibody–conjugated agarose, and 14-3-3 λ was detected with an anti-Flag antibody. The JD-CT peptide but not the C-terminal peptide pulled down 14-3-3 λ (Figure 5A), suggesting that only the junction domain of SOS2 is necessary for the interaction with 14-3-3 λ. The junction domain in SOS2 is essential for its kinase activity, since deletion of the domain results in the loss of kinase activity (Guo et al., 2001). We then determined whether the junction domain is sufficient for this interaction using multiple approaches. First, the junction domain was fused to a Myc tag, and the resulting construct was coexpressed with Flag-HA-14-3-3 λ in Arabidopsis leaf protoplasts. Full-length SOS2 (SOS2-FL) and SOS2-KD were used as controls. Consistent with previous results, SOS2 interacted with 14-3-3 λ while SOS2-KD did not. SOS2-JD alone interacted with 14-3-3 λ (Figure 5B). Second, yeast two-hybrid analyses were also used to further verify whether SOS2-JD interacts with 14-3-3 λ. Coding regions of SOS2-FL, SOS2-KD, and SOS2-JD were cloned into the pGBKT7 vector, while 14-3-3 λ was cloned into the pGADT7 vector. The resulting pGBKT7-SOS2-FL, -KD, or -JD was cotransformed with pGBKT7-14-3-3 λ into yeast strain AH109. Both SOS2 and SOS2-JD interacted with 14-3-3 λ; however, no interaction was detected between SOS-KD and 14-3-3 λ (Figure 5C). Third, in vitro pull-down analysis was also used to determine whether SOS-JD is sufficient for SOS2-14-3-3 λ interaction. Recombinant GST-tagged SOS2-JD or GST tag alone was coexpressed with polyhistidine-tagged 14-3-3 λ in E. coli DE3, and then GST or GST-tagged recombinant proteins were purified by Glutathione Sepharose (GE Healthcare). Recombinant proteins were detected using Coomassie blue staining, and polyhistidine-tagged 14-3-3 λ associated with SOS2-JD was detected by immunoblot analysis with anti-His or anti-14-3-3 λ antibody. Consistent with previous results, GST-tagged SOS-JD but not the GST tag pulled down 14-3-3 λ (Supplemental Figure 6B). These results demonstrate that the junction domain of SOS2 is both necessary and sufficient to mediate interaction with 14-3-3 λ.
Figure 5.
14-3-3 λ Interacts with the Junction Domain of SOS2, and Mutations of Ser-294 in SOS2 Modulate the Interaction.
(A) Analysis of the interaction between different versions of truncated SOS2 protein and 14-3-3 λ in Arabidopsis leaf protoplasts. Purified plasmids of Pro35S:6×Myc-SOS2-KD, -SOS2-JD-CT, -SOS2-CT, and -SOS2-CTΔFISL were cotransformed into Arabidopsis leaf protoplasts with purified Pro35S:Flag-HA-14-3-3 λ plasmid. Transiently expressed proteins were immunoprecipitated with anti-C-Myc antibody–conjugated agarose. Immunoblot assays with anti-C-Myc and anti-Flag antibodies were used to detect Myc-SOS2 and SOS2-interacting 14-3-3 λ, respectively. Asterisks indicate nonspecific bands. IB, immunoblot. Experimental details are provided in Methods.
(B) Analysis of the interaction between the junction domain of SOS2 and 14-3-3 λ in Arabidopsis leaf protoplasts. Purified plasmids of Pro35S:6×Myc-SOS2-JD, -SOS2-FL, and -SOS2-KD were cotransformed into Arabidopsis leaf protoplasts with purified Pro35S:Flag-HA-14-3-3 λ plasmid. Transiently expressed proteins were immunoprecipitated with anti-C-Myc antibody–conjugated agarose. Immunoblot assays with anti-C-Myc and anti-Flag antibodies were used to detect Myc-SOS2 and SOS2-interacting 14-3-3 λ, respectively. Experimental details are provided in Methods.
(C) Yeast two-hybrid analysis of the interaction between SOS2 and 14-3-3 λ. Yeast strains expressing the indicated constructs were grown on synthetic complete medium without Trp and Leu (SC-T/L; left panel) and on synthetic complete medium without Trp, Leu, and His (SC-T/L/H; right panel). Photographs were taken after 4 to 5 d of growth on the indicated medium. Panels show yeast serial decimal dilutions. Experimental details are provided in Methods.
(D) Schematic representation of the SOS2 junction domain (amino acids 268 to 308) and Ser-294 (red) in the junction domain.
(E) Analysis of the interactions between SOS2, SOS2S294A, SOS2S294D, and 14-3-3 λ in Arabidopsis leaf protoplasts. Purified Pro35S:6×Myc-SOS2, -SOS2S294A, -SOS2S294D, and Pro35S:Flag-HA-14-3-3 λ plasmids were cotransformed into Arabidopsis leaf protoplasts. Transiently expressed proteins were immunoprecipitated with anti-C-Myc antibody–conjugated agarose. Immunoblot assays with anti-C-Myc and anti-Flag antibodies were used to detect Myc-SOS2 and SOS2-interacting 14-3-3 λ, respectively. Experimental details are provided in Methods.
(F) Yeast two-hybrid analysis of the interactions between SOS2, SOS2S294A, SOS2S294D, and 14-3-3 λ. Yeast strains expressing the indicated constructs were grown on synthetic complete medium without Trp and Leu (left panel) and on synthetic complete medium without Trp, Leu, and His (right panel). Photographs were taken after 4 to 5 d of growth on the indicated medium. Panels show yeast serial decimal dilutions. Experimental details are provided in Methods.
(G) Analysis of the interactions between SOS2, SOS2K40N-S294A, SOS2K40N-S294D, and 14-3-3 λ in Arabidopsis leaf protoplasts. Purified Pro35S:6×MycSOS2-K40N, -SOS2K40N-S294A, and SOS2K40N-S294D plasmids were transformed into Arabidopsis leaf protoplasts. Transiently expressed proteins were immunoprecipitated with anti-C-Myc antibody–conjugated agarose. Immunoblot assays with anti-C-Myc and anti-14-3-3 λ antibodies were used to detect Myc-SOS2 and SOS2-interacting 14-3-3 λ, respectively. Experimental details are provided in Methods.
[See online article for color version of this figure.]
Phosphorylation of Ser-294 in SOS2 Enhances the Interaction with 14-3-3 λ
14-3-3 proteins have been shown to associate with phosphoproteins by recognizing phosphoserine or phosphothreonine within conserved binding motifs. 14-3-3 λ binds to the junction domain of SOS2; however, this domain does not include any of the typical 14-3-3 binding consensus motifs, and only one Ser (Ser-294) and no Thr residues are present in this domain (Figure 5D).
We next performed assays to identify autophosphorylation and transphosphorylation sites in SOS2 in an in vitro plant system. For this purpose, STREPII-tagged SOS2 protein was expressed in a coupled in vitro transcription/translation system based on wheat germ (Triticum aestivum) extracts (Hashimoto et al., 2012) and analyzed by MS. Using this approach, we identified the peptide AVFDGIEGSYVAENVER to be phosphorylated and determined that the predicted phosphorylated amino acid corresponds to SOS2Ser294 (Supplemental Figure 7). We subsequently analyzed, by MS, wheat germ–expressed STREPII-SOS2K40N, a mutant that does not autophosphorylate due to its lack of kinase activity (Liu et al., 2000). After translation of this mutated version of SOS2 in wheat germ extracts, we again identified the peptide containing Ser-294 as being phosphorylated. The detection of Ser-294 phosphorylation in both the active and kinase-dead variants of SOS2 indicates that phosphorylation of this residue is not due to autophosphorylation but results from transphosphorylation by a putative upstream kinase.
To determine whether Ser-294 is required for the recognition of 14-3-3 λ, we generated two mutated forms of SOS2, with Ser-294 changed to Ala (SOS2S294A), which cannot be phosphorylated, or Asp (SOS2S294D), a putative phosphorylation mimic substitution. Pro35S:6×Myc-SOS2S294A, Pro35S:6×Myc-SOS2S294D, as well as Pro35S:6×Myc-SOS2 were coexpressed with Pro35S:Flag-HA-14-3-3 λ in Arabidopsis leaf protoplasts. Transiently expressed Myc-SOS2 and its mutated forms were immunoprecipitated with anti-C-Myc antibody–conjugated agarose, and 14-3-3 λ was detected with anti-Flag antibody. The SOS2S294A mutation decreased the SOS2 interaction with 14-3-3 λ, while the SOS2S294D mutation enhanced this interaction under these experimental conditions (Figure 5E). We further tested these interactions in yeast. SOS2S294A and SOS2S294D were cloned into the pGBKT7 vector, and different combinations of SOS2, 14-3-3 λ, and empty vectors were coexpressed in yeast strain AH109. As observed in the immunoprecipitation assays, the SOS2S294A mutation reduced the SOS2 interaction with 14-3-3 λ; however, the SOS2S294D mutation had no significant effect on the interaction with 14-3-3 λ in yeast (Figure 5F). Also, in vitro pull-down analysis was used to determine the effect of the Ser-294 mutation on the interaction of SOS2 with 14-3-3 λ. Recombinant GST-tagged SOS2, SOS2S294A, and SOS2S294D or GST tag alone were coexpressed with polyhistidine-tagged 14-3-3 λ in E. coli DE3 cells, and then GST or GST-tagged recombinant proteins were purified by Glutathione Sepharose (GE Healthcare). Recombinant proteins were detected using Coomassie blue staining, while polyhistidine-tagged 14-3-3 λ associated with SOS2 or its mutated versions was detected through immunoblot analysis with anti-His or anti-14-3-3 λ antibody. GST-tagged SOS2 as well as SOS2S294A and SOS2S294D pulled down 14-3-3 λ. SOS2S294D displayed a slightly stronger interaction with 14-3-3 λ than SOS2 or SOS2S294A (Supplemental Figure 6C). Together with the in vivo results, our data suggest that the phosphorylation of SOS2Ser294 enhances but is not absolutely essential for SOS2 interaction with 14-3-3 proteins in in vitro or overexpression conditions.
To further determine whether SOS2Ser294 plays a role in the interaction of SOS2 with 14-3-3 proteins in planta, firefly luciferase complementation experiments were used to quantitatively analyze their interaction (Chen et al., 2008). Consistent with our previous data, SOS2 interacted with 14-3-3 λ, and this interaction was reduced by the S294A mutation but increased by the S294D mutation (Supplemental Figure 6D). These results support the conclusion that phosphorylation of Ser-294 enhances the stability of a SOS2-14-3-3 protein complex and thereby shifts the cellular balance of active and inactive SOS2 kinase toward its inactive form.
We next determined whether autophosphorylation of SOS2 has an effect on the interaction between SOS2 and 14-3-3 λ. The two variants of SOS2 with mutated Ser-294, SOS2K40N-S294A and SOS2K40N-S294D, were generated in a SOS2 kinase-dead background (SOS2K40N) (Liu et al., 2000). Pro35S:6×Myc-SOS2K40N, Pro35S:6×Myc-SOS2K40N-S294A, and Pro35S:Myc-SOS2K40N-S294D were transiently expressed in Arabidopsis leaf protoplasts. SOS2 and its variants were subsequently immunoprecipitated with anti-C-Myc antibody–conjugated agarose, and coimmunoprecipitating proteins were probed with anti-14-3-3 λ antibodies. Further supporting the result presented in Figure 5E, the SOS2K40N-S294A mutation reduced the SOS2 association with 14-3-3 λ and the SOS2K40N-S294D mutation enhanced the interaction between these two proteins (Figure 5G). Together, these results suggest that transphosphorylation of SOS2Ser294 enhances but is not absolutely essential for the SOS2 interaction with 14-3-3 proteins.
SOS2Ser294 Is Critical for the Repression of SOS2 Kinase Activity by 14-3-3 λ
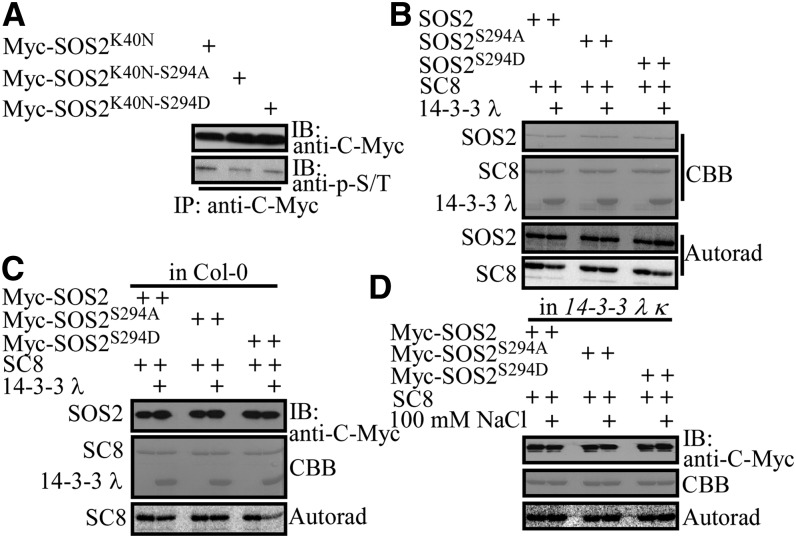
We next determined whether Ser-294 is phosphorylated in Arabidopsis and whether it is required for the repression of SOS2 kinase activity by 14-3-3 λ. Myc-tagged SOS2K40N, SOS2K40N-S294A, and SOS2K40N-S294D were transiently expressed in Arabidopsis leaf protoplasts and immunoprecipitated with anti-C-Myc antibody–conjugated agarose. The phosphoserine/phosphothreonine signal was analyzed using antiphosphoserine/antiphosphothreonine polyclonal antibodies. The phosphorylation signal was slightly lower in SOS2K40N-S294A and SOS2K40N-S294D than that in SOS2K40N (Figure 6A). These data together with the previous MS result (Supplemental Figure 7) indicate that SOS2Ser294 is transphosphorylated in planta.
Figure 6.
Ser-294 in SOS2 Is Required for the Repression of SOS2 by 14-3-3 λ.
(A) Analysis of the phosphorylation status of SOS2K40N, SOS2K40N-S294A, and SOS2K40N-S294D in Arabidopsis leaf protoplasts. The phosphorylation status of transiently expressed Myc-tagged SOS2K40N, SOS2K40N-S294A, and SOS2K40N-S294D (shown in Figure 5G) was analyzed using anti-phosphoserine/phosphothreonine (anti-p-S/T) polyclonal antibodies. IB, immunoblot. Experimental details are provided in Methods.
(B) Kinase assays for recombinant SOS2, SOS2S294A, and SOS2S294D with or without 14-3-3 λ. SCaBP8 (SC8) was used as the substrate. CBB, Coomassie Brilliant Blue; Autorad, autoradiograph. Experimental details are provided in Methods.
(C) Kinase assay for Myc-SOS2, -SOS2S294A, and -SOS2S294D purified from Col-0 with or without 14-3-3 λ. SCaBP8 was used as the substrate. Experimental details are provided in Methods.
(D) Kinase assay for Myc-SOS2, -SOS2S294A, and -SOS2S294D purified from the 14-3-3 λ κ double mutant. SCaBP8 was used as the substrate. Experimental details are provided in Methods.
To determine if 14-3-3 λ represses SOS2 activity through Ser-294 in SOS2, we first excluded the possibility that mutations at Ser-294 affect SOS2 kinase activity toward its substrate SCaBP8. Recombinant GST-tagged SOS2, SOS2S294A, and SOS2S294D were purified from E. coli. In vitro kinase assays using SCaBP8 as substrate revealed similar transphosphorylation activities for SOS2, SOS2S294A, and SOS2S294D (Supplemental Figure 8A).
Next, we investigated the influence of 14-3-3 proteins on the activity of different versions of SOS2. Recombinant polyhistidine-tagged 14-3-3 λ was purified from E. coli and incubated with GST-tagged SOS2, SOS2S294A, and SOS2S294D for in vitro kinase assays with SCaBP8 as substrate. 14-3-3 λ did not repress the transphosphorylation activity of the SOS2S294A protein but partially repressed the activity of the SOS2 and SOS2S294D proteins (Figure 6B; Supplemental Figure 8B). In contrast, SOS2 autophosphorylation activity was not altered by 14-3-3 λ. These results suggest that a potentially phosphorylatable Ser (Ser-294) in SOS2 is important for enabling 14-3-3 proteins to repress SOS2 kinase activity.
Finally, we determined if the phosphorylation of SOS2S294 is required for SOS2 repression by 14-3-3 λ in planta. Pro35S:6×Myc-SOS2S294A and Pro35S:6×Myc-SOS2S294D transgenic plants were generated in the Col-0 background, and T3 seedlings were treated with 100 mM NaCl for 12 h. Myc-tagged wild-type and mutated SOS2 proteins were purified with anti-C-Myc antibody–conjugated agarose and used in in vitro kinase assays with SCaBP8 as substrate. Consistent with previous in vitro results, SOS2 kinase activity was repressed by polyhistidine-tagged 14-3-3 λ recombinant protein; 14-3-3 λ did not affect the activity of SOS2S294A, while the activity of SOS2S294D was repressed more than that of wild-type SOS2 by 14-3-3 λ (Figure 6C; Supplemental Figure 8C). We also generated Pro35S:6×Myc-SOS2S294A and Pro35S:6×Myc-SOS2S294D transgenic plants in the 14-3-3 λ κ background. T3 seedlings were left untreated or treated with 100 mM NaCl for 12 h. Myc-tagged wild-type and mutated SOS2 proteins were purified with anti-C-Myc antibody–conjugated agarose and used in in vitro kinase assays with SCaBP8 as substrate. The kinase activities of SOS2, SOS2S294A, and SOS2S294D were similar with and without salt (Figure 6D; Supplemental Figure 8D).
Taken together, these results indicate that Ser-294 is required for the repression of SOS2 kinase activity by the 14-3-3 proteins and that transphosphorylation of Ser-294 enhances the inhibitory effect of the 14-3-3 proteins.
Mutation in Ser-294 Alters SOS2 Function in Salt Tolerance in Arabidopsis
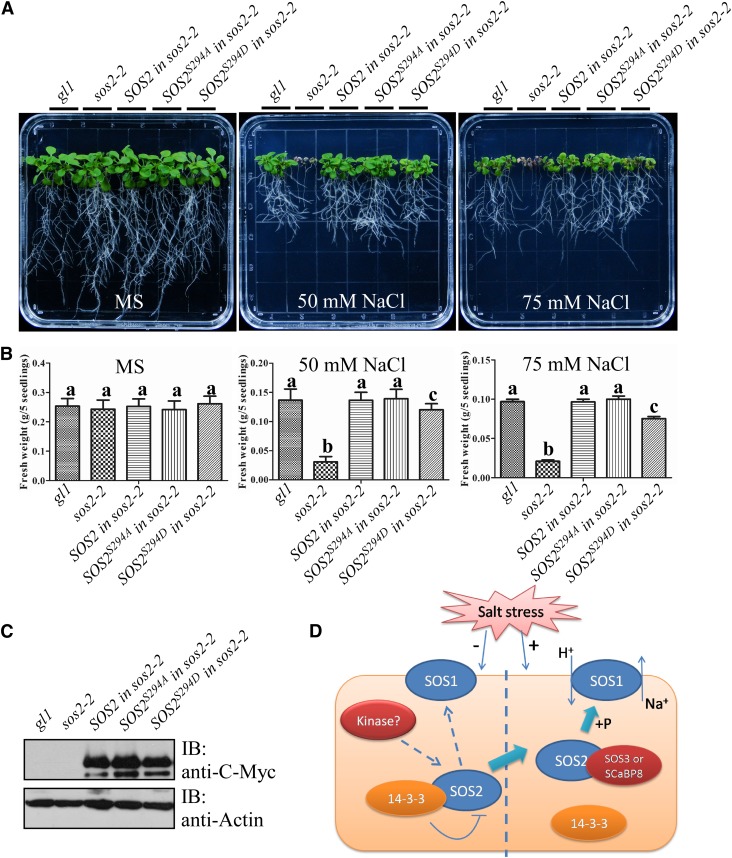
To examine if Ser-294 is important for SOS2 function in plant salt tolerance, 5-d-old T3 transgenic plants containing Pro35S:6×Myc-SOS2, -SOS2S294A, or -SOS2S294D in the sos2-2 mutant background as well as the wild type (gl1/gl1) and the sos2-2 mutant were transferred to Murashige and Skoog medium without or with 50 or 75 mM NaCl. At least three transgenic lines were used for the analysis of salt sensitivity, and one representative line for each transgenic plant is presented in Figures 7A and 7B. SOS2 and SOS2S294A complemented the sos2-2 salt-sensitive phenotype to wild-type levels. However, SOS2S294D only partially rescued the phenotype (Figures 7A and 7B). Protein levels of SOS2, SOS2S294A, and SOS2S294D were similar (Figure 7C). Furthermore, immunoblot analysis revealed that Myc-SOS2S294D bound more 14-3-3 proteins than Myc-SOS2 and Myc-SOS2S294A during salt stress (Supplemental Figure 9). These data suggest that the regulated phosphorylation of Ser-294 in SOS2 is required for a proper response to salt stress in Arabidopsis.
Figure 7.
Ser-294 Is Required for SOS2 Function in Salt Tolerance in Arabidopsis.
(A) Analysis of the salt sensitivity of Col-0 (gl1/gl1), sos2-2, and different transgenic plants expressing Myc-SOS2, -SOS2S294A, or -SOS2S294D in the sos2-2 background. Five-day-old seedlings were transferred to Murashige and Skoog medium (MS) without or with 50 or 75 mM NaCl. Photographs were taken 10 d after transfer.
(B) Analysis of fresh weight for seedlings in (A). Error bars represent sd (n > 10). Statistical significance was determined by Student’s t test; significant differences (P ≤ 0.05) are indicated by different lowercase letters. Experimental details are provided in Methods.
(C) Protein levels of SOS2 and its mutated variants in transgenic plants used in (A). Actin was used as a loading control. IB, immunoblot. Experimental details are provided in Methods.
(D) A working model for 14-3-3 protein function in salt tolerance. When grown in the absence of salt, SOS2Ser294 may be phosphorylated by an unknown upstream kinase and binds to the 14-3-3 proteins, resulting in SOS2 activity inhibition. Salt stress decreases interaction between the 14-3-3 proteins and SOS2, releases SOS2 activity, and activates SOS1 Na+/H+ antiport activity for salt tolerance. 14-3-3 proteins function as modulators to keep the SOS pathway at a basal inactive state and as switches to turn on the SOS pathway when plants are challenged by salt.
[See online article for color version of this figure.]
Taken together, our results indicate that 14-3-3 proteins function in salt tolerance in plants by modulating SOS2 activity through Ser-294 in SOS2.
DISCUSSION
In response to salt, a calcium signal activates the SOS pathway by binding to the SOS3 and SCaBP8/CBL10 calcium binding proteins, which activate the SOS2 protein kinase to regulate the SOS1 PM Na+/H+ antiporter. SOS2 is a key regulator in the SOS pathway, relaying the signal to downstream changes through phosphorylation (Guo et al., 2001; Quintero et al., 2011).
Significant information is available about the domains that are important for SOS2 activity when plants are grown in salt. For example, the SOS2 C terminus functions as an autoinhibitory domain for SOS2 kinase activity by interacting with the SOS2 N-terminal kinase domain through the FISL/NAF motif (Albrecht et al., 2001; Guo et al., 2001). When SOS3 and SCaBP8 are activated by calcium, they bind to the FISL domain of SOS2 (in different tissues) to release the SOS2 kinase domain for substrate access. Activation of SOS2 also requires the SOS2 junction domain that is located between the kinase and regulatory domains, since deletion of this domain results in SOS2 inactivation (Guo et al., 2001).
However, the mechanisms underlying SOS pathway suppression when plants are not exposed to salt stress have remained poorly understood. Most recently, Kim et al. (2013) reported that SOS2 is repressed by GI in the absence of salt stress and that exposure to salt stress causes the degradation of GI protein. The SOS2-GI interaction occurs via the SOS2 C terminus and is released by coexpression of SOS3 in vitro (Kim et al., 2013). It is still not well understood how SOS2 is maintained in an inactive state when plants are grown in the absence of salt compared with its activation or how the SOS2 junction domain functions in vivo. In this study, we identified 14-3-3 proteins as additional negative regulators of SOS2 in planta. 14-3-3 proteins bind to the SOS2 junction domain and thereby inhibit SOS2 kinase activity when plants are not challenged by salt stress. Our results provide critical mechanistic information for how the SOS pathway is suppressed in the plant in nonstress conditions and report a function of the SOS2 junction domain in vivo (Figure 7D).
In plants, 14-3-3 proteins are ubiquitous regulators of diverse target proteins. These proteins include protein kinases such as mitogen-activated protein kinases, CDPKs, BKI1, and SNF1-related protein kinases (Ikeda et al., 2000; Oh et al., 2010; Sorokina et al., 2011; Wang et al., 2011; Lachaud et al., 2013). SOS2 belongs to the SnRK3 subfamily that includes 26 members in Arabidopsis (Hrabak et al., 2003; Weinl and Kudla, 2009). A wheat SnRK3 kinase, WPK4, was reported to interact with two 14-3-3 proteins (Ikeda et al., 2000). Two 14-3-3 binding motifs in the WPK4 regulatory domain are required but not essential for the interaction, suggesting that additional sites may also be involved. These two motifs, which are located just before and after the FISL motif in WPK4, are conserved in only a few SOS2-like protein kinases (PKS/CIPK) but not in SOS2. Instead, we found that in SOS2, the interaction with 14-3-3 proteins is mediated by the SOS2 junction domain and that the binding peptide does not contain a conserved 14-3-3 binding motif.
Our data establish that phosphorylation of Ser-294 in the junction domain enhances the interaction stability between SOS2 and 14-3-3 λ but may not represent an essential prerequisite for binding. Nonphosphorylated E. coli SOS2 as well as SOS2S294A mutants were able to interact with 14-3-3 λ in our experimental conditions. Phosphorylation-independent interactions with 14-3-3 proteins have been reported earlier, and one possible explanation is that the presence of negatively charged amino acids can, to some extent, compensate for the absence of a phosphate group (Wang et al., 1999; Fuglsang et al., 2003). The SOS2 junction domain contains a large number of negatively charged residues (12 out of 41 amino acids are either Asp or Glu). It is possible that these negative charges may be sufficient to mediate the basal interaction between SOS2 and 14-3-3 λ. However, we cannot fully exclude the possibility that our experimental conditions, which required the overexpression of proteins, or these analyses in in vitro conditions may have masked the lack of interaction between SOS2 and 14-3-3 in the absence of Ser-294 phosphorylation and that the interaction of SOS2 and 14-3-3 when Ser-294 is changed to Ala or Asp might be due to a conformational change of SOS2 that does not necessarily reflect the phosphorylation state of SOS2.
Although Ser-294 in the junction domain may not be absolutely required for the interaction with 14-3-3 proteins, it is clearly critical for the 14-3-3–mediated repression of SOS2 and is also important for SOS2 function in plant salt tolerance. Our MS analyses of enzymatically active and inactive SOS2 protein, generated in plant extracts, indicate that Ser-294 is transphosphorylated by an unknown upstream kinase(s) in vivo (Supplemental Figure 7). The fact that mutation of Ser-294 results in a reduced SOS2 phosphorylation signal after the expression of inactive SOS2 in plant protoplasts (Figure 6A) further supports this conclusion. Ser-294 is not part of any known recognition motif for Arabidopsis kinases, making it difficult to predict the identity of the kinase(s) responsible for SOS2 phosphorylation. One important future goal for our understanding of salt stress signaling in plants will be the identification of these upstream kinases as well as the phosphatase(s) responsible for the dephosphorylation of Ser-294 and, thereby, for the disassociation of 14-3-3 proteins at the onset of salt stress.
Because the junction domain is essential for SOS2 kinase activity (Guo et al., 2001), it is conceivable that the association of 14-3-3 λ blocks SOS2 active sites or induces conformational changes that result in the repression of SOS2 kinase activity. Future crystal structure analysis of the SOS2-14-3-3 complex may allow us to address these models.
In interaction studies of 14-3-3 proteins with SOS2 and in substrate phosphorylation analyses, we observed only partial modulation of interaction and kinase activity by mutation of SOS2Ser294 to forms that cannot be phosphorylated or that potentially represent a phosphomimic. These results suggest that additional, currently unknown, auxiliary proteins may contribute to the regulation of SOS2 by 14-3-3 proteins. Additional (trans)phosphorylation events in other domains of SOS2 may also affect the folding of the kinase and, in this way, contribute to 14-3-3 regulation. However, it is important to consider that SOS2 is only one component of a complex signaling network that, in Arabidopsis, is formed by specific interactions between 10 calcium sensor proteins and 26 kinases (Kudla et al., 2010; Hashimoto and Kudla, 2011). In this network, SOS2 interacts with several known targets that are distinct from SOS1 (e.g., GI and nucleoside dikinase) and likely regulates many additional unknown targets in different compartments and tissues. Based on this complexity, a complete “on/off” regulation of SOS2 activity by the 14-3-3 proteins would quite likely impair the function of SOS2 relative to its other targets and be physiologically disadvantageous for the plant. Therefore, the partial modulation of SOS2 interaction and activity by 14-3-3 regulation we observed in our assays likely represents a mechanism to balance the diverse functions of SOS2 in complex cellular networks.
Different members of the 14-3-3 protein family are involved in many different cellular responses, such as stomatal opening (Tseng et al., 2012), alkaline stress (Xu et al., 2013), and brassinosteroid signaling (Wang et al., 2011). It is not fully understood how the different 14-3-3 proteins are coordinated to regulate these different responses. The existence of various 14-3-3 targets related to ion homeostasis may also contribute to the phenotype of the 14-3-3 λ κ double mutant during salt stress. Therefore, currently, we cannot fully exclude that misregulation of the expression balance of other members of the 14-3-3 protein family in the 14-3-3 λ κ double mutant contributes to the observed phenotypes.
14-3-3 proteins exert their regulatory function on SOS2 under nonstress conditions and disassociate in the presence of salt stress. It is conceivable that disassociated 14-3-3 proteins may modulate the activity of targets different from SOS2 during salt stress. Salt stress responses in plants involve the regulation of distinct physiological processes, and their balanced coordination is crucial to achieve full salt tolerance and the integration of environmental responses with developmental programs (Kim et al., 2013). For example, sodium extrusion from cells by PM Na+/H+ antiporters requires a proton gradient that is created by PM H+-ATPases (Shi et al., 2002). Salt stress results in a dramatic induction in PM H+-ATPase activity in plants, underscoring the crucial importance of these transporters in providing the driving energy force for ion transport during salt stress (Niu et al., 1993). Remarkably, 14-3-3 proteins are also involved in regulating PM H+-ATPase activity. Under resting conditions, the C terminus of the PM H+-ATPase interacts with its central loop to inhibit enzyme activity (Palmgren et al., 1991). Thr-947 in the C-terminal end of the Arabidopsis PM H+-ATPase AHA2 was shown to be rapidly phosphorylated when environmental conditions change (reviewed in Palmgren, 2001), creating a 14-3-3 binding site. Subsequent 14-3-3 interaction releases the autoinhibition and activates PM H+-ATPase activity (Fuglsang et al., 1999; Svennelid et al., 1999; Maudoux et al., 2000). Therefore, it is tempting to speculate that during salt stress, 14-3-3 proteins that disassociate from SOS2 become available to bind to the phosphorylated PM H+-ATPases, leading to their full activation to enhance the formation of a PM proton gradient. Maximal activation of the PM Na+/H+ antiporter SOS1 during salt stress would then result from the combined phosphorylation-dependent activation by SOS2 and the enhanced proton gradient generated by fully activated PM H+-ATPases. However, currently available evidence suggests that AHA2 binds 14-3-3 ω, while SOS2 binds 14-3-3 λ and κ. Therefore, additional experiments will be required to determine if the 14-3-3 proteins participating in these two processes are the same or different isoforms.
In addition to potentially enabling the modulation of alternative targets, the relief of inhibition of SOS2 by the release of 14-3-3 protein binding allows for an immediate response upon the onset of salt stress that is not delayed by requirements for gene transcription and RNA translation. Potentially, this would allow for a quick reassociation with SOS2 and other interacting partners if environmental conditions change. Finally, the negative regulation of signaling components during salt stress reported here extends the functional range of this regulatory module beyond primary signaling events in response to hormones like auxin, gibberellic acid, and jasmonic acids and suggests a negative regulation of primary signaling components as an effective and general mechanism of plant signal transduction.
METHODS
Mutants
Arabidopsis thaliana Col-0 was used as the wild type in all experiments except for Figure 7. The homozygous 14-3-3 λ mutant was identified from a SALK line (075219) with a T-DNA insertion in the second intron of At5g10450, and the homozygous 14-3-3 κ mutant was identified from a SALK line (071097) with a T-DNA insertion in the second intron of At5g65430. 14-3-3 λ was crossed into 14-3-3 κ to generate the 14-3-3 λ κ double mutant for analyses of salt sensitivity. The homozygous sos2-2 mutant was in the gl1/gl1 Col-0 background with a 2-bp deletion in the eighth exon of At5g35410 (Liu et al., 2000). The gl1 mutation did not affect the salt-sensitive phenotype. Pro35S:Flag-HA-14-3-3 λ transgenic plants in the Col-0 background overexpressing 14-3-3 λ were generated, and two T3 transgenic lines (OE-9 and OE-10) were used for further studies. Pro35S:6×Myc-SOS2, -SOS2S294A, and -SOS2S294D transgenic plants in the sos2-2 mutant background were generated, and representative T3 transgenic lines were used for further analysis with gl1/gl1 Col-0 as the wild type.
Plant Growth and Analysis of Salt Tolerance
Seeds were sterilized in a solution containing 20% sodium hypochlorite and 0.1% Triton X-100 for 10 min, washed with sterile water five times, and sown on Murashige and Skoog medium (containing 2.5% Suc) with 0.3% (for horizontal growth) or 0.5% (for vertical growth) Phytagel agar (Sigma-Aldrich). Plates were kept at 4°C for 2 d, and then seeds were germinated and grown under constant illumination (Philips TLD) at 23°C. For monitoring the response to salt in soil, seedlings of Col-0 and the 14-3-3 λ κ double mutant were grown in soil under short-day conditions (8 h of light/16 h of dark) for 3 weeks. Subsequently, the soil was irrigated with 150 mM NaCl four times every 3 d, plants were grown for an additional 2 weeks and photographed, and chlorophyll content was analyzed as described (Zhou et al., 2012). For monitoring salt sensitivity on plates, seedlings were grown vertically on Murashige and Skoog medium under continuous light for 5 d at 23°C and transferred to Murashige and Skoog medium containing different concentrations of NaCl. After the indicated times of vertical growth, seedlings were photographed and primary root length and fresh weight were measured and quantified.
Plasmid Construction
The coding sequences of SOS2, SOS2-KD, SOS2-JD, SOS2-JD-CT, SOS2-CT, and SOS2-CTΔFISL were amplified with the FL-Bf/FL-Sr, FL-Bf/KD-Sr, JD-Bf/JD-Sr, JD-Bf/FL-Sr, FRD-Bf/FL-Sr, and RD-Bf/FL-Sr primers, respectively, and cloned into the pCAMBIA1307-6×Myc vector between the BamHI and SalI sites. The coding sequence of 14-3-3 λ was amplified with the λ-Sf/λ-Kr primers and cloned into the pCAMBIA1307-Flag-HA vector between the SalI and KpnI sites. The coding sequences of SOS2 and SOS2-JD were amplified with the FL-Bf/FL-Sr and JD-Bf/JD-Sr primers and cloned into the pGEX-6p-1 vector between the BamHI and SalI sites. The coding sequences of 14-3-3 λ and 14-3-3 κ were amplified with the λ-Bf/λ-Sr and κ-Bf/κ-Sr primers and cloned into the pET-28a vector between the BamHI and SalI sites. The coding sequence of 14-3-3 λ was amplified with the λ-Bf/λ-Sr primers and cloned into the pCAMBIA1307-6×Myc vector between the BamHI and SalI sites. Mutations of Ser-294 to Ala, Ser-294 to Asp, and Lys-40 to Asn in SOS2 were generated with primers SA-f/SA-r, SD-f/SD-r, and KN-f/KN-r, respectively. The coding sequence of the SOS1 C terminus (amino acids 848 to 1146) was amplified with the SOS1-CBf and SOS1-CXr primers or SOS1-CSf and SOS1-CKr primers and cloned into the pMAL-p2x vector between the BamHI and XhoI sites or the pCAMBIA1307-Flag-HA vector between the SalI and KpnI sites, respectively. Primer sequences are found in Supplemental Table 1.
Metabolic Labeling of Plants with 14N/15N Isotopes
For metabolic labeling with 14N/15N isotypes, T3 transgenic plants expressing Pro35S:Flag-HA-SOS2 in the sos2-2 mutant background were grown on Murashige and Skoog medium with 14NH414NO3 or 15NH415NO3 under constant illumination at 23°C. 15N-labeled salts were 98% enriched in 15N (Perkin-Elmer). After 10 d of growth, seedlings were harvested. 14N-labeled seedlings were treated with water, while 15N-labeled seedlings were treated with 100 mM NaCl for 24 h. Approximately 5 g of plant tissue was ground to a fine powder in liquid nitrogen, and total proteins were extracted in IP buffer (10 mM Tris, pH 7.5, 0.5% Nonidet P-40, 2 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 1% protease inhibitor cocktail; Roche). Cell debris was removed by a 10-min centrifugation at 12,000g at 4°C. The supernatant was collected, and protein concentration was determined using the Bio-Rad protein assay. Equal amounts of total protein from both treatments were mixed thoroughly, and Flag-HA-SOS2 was immunoprecipitated with anti-Flag antibody–conjugated agarose. The Flag tag was then cleaved off, and anti-HA antibody–conjugated agarose was used for HA-SOS2 immunoprecipitation from the resulting supernatant. Agarose-bound HA-SOS2 was eluted from HA agarose using an HA peptide (Sigma-Aldrich). After this two-step purification, the resulting SOS2 protein and its interacting proteins were enriched and analyzed by MS.
RT-PCR Analysis
Total RNA was extracted with Trizol reagent (Invitrogen) from 10-d-old seedlings of the wild type or 14-3-3 mutants grown on Murashige and Skoog plates under constant illumination. Total RNA was treated with RNase-free DNase I (TaKaRa) to remove genomic DNA. Ten micrograms of RNA was used for reverse transcription with M-MLV reverse transcriptase (Promega) according to the manufacturer’s instructions. Resulting cDNAs were used for RT-PCR analysis with specific primers. ACTIN was used as an internal control. Ethidium bromide staining was used to detect the PCR products. The primer sequences are listed in Supplemental Table 2.
Preparation of Anti-14-3-3 λ Polyclonal Antibodies
Recombinant polyhistidine-tagged 14-3-3 λ prepared from Escherichia coli was used as an antigen for polyclonal antibody production in mice. Protein purification was performed according to the manufacturer’s protocol (Qiagen). Recombinant proteins were purified with Ni-NTA agarose (Qiagen), and 2 mg of purified recombinant protein was used for mouse immunization. Serum after immunization was used for antibody specificity tests with samples from 10-d-old Col-0, the 14-3-3 λ mutant, the 14-3-3 κ mutant, and the 14-3-3 λ κ double mutant.
Sequence Alignment and Phylogenetic Analysis
Conserved domains of the sequences of the 13 members in the At 14-3-3 protein family were used for phylogenetic analysis. Alignment of amino acid sequences was made using ClustalX 1.83. A phylogenetic tree was constructed using predicted amino acid sequences in MEGA5 (Tamura et al., 2011).
Coimmunoprecipitation Assays in Arabidopsis Leaf Protoplasts
The coding sequences of SOS2, truncated and mutated versions of SOS2, the SOS1 C terminus, and 14-3-3 λ were translationally fused downstream of the C-Myc or Flag-HA epitope and cloned into the pCAMBIA1307 vector. The plasmids were purified by CsCl gradient centrifugation. Protoplast preparation and transformation were performed as described previously (Sheen, 2001). After overnight incubation at 23°C, the protoplasts were harvested and lysed with 1 mL of IP buffer. The sample was centrifuged at 10,000g for 10 min at 4°C to remove cellular debris. Fifty microliters (5%) of the supernatant was collected as input. The rest of the supernatant was transferred to a tube containing 20 μL of anti-C-Myc agarose (Sigma-Aldrich) and incubated for 2 h at 4°C with gentle mixing. After incubation, the agarose was washed thoroughly four to five times with 1 mL of IP buffer per wash. All immunoprecipitation steps were performed at 4°C. Proteins were probed by immunoblot analysis with anti-C-Myc, anti-Flag, or anti-14-3-3 λ polyclonal antibody. The chemiluminescence signals were detected by autoradiography.
Yeast Two-Hybrid Assays
To determine the interaction between SOS2 and 14-3-3 λ in yeast, the coding sequences of SOS2 (as well as SOS2S294A and SOS2S294D), SOS2-KD, and SOS2-JD were amplified with the FL-Ef/FL-Sr (for SOS2, SOS2S294A, and SOS2S294D), FL-Ef/KD-Sr, and JD-Ef/JD-Sr primers, respectively, and cloned into the pGBKT7 vector between the EcoRI and SalI sites. The coding sequence of 14-3-3 λ was amplified with the λ-Ef/λ-Br primers and cloned into the pGADT7 vector between the EcoRI and BamHI sites. Yeast transformation and growth assays were performed as described in the Yeast Protocols Handbook (Clontech). Primer sequences are listed in Supplemental Table 1.
Bimolecular Fluorescence Complementation Assays
To determine whether the interaction between SOS2 and 14-3-3 λ takes place in planta, the coding sequence of SOS2 was amplified with the FL-Bf/FL-Sr primers while 14-3-3 λ was amplified with the λ-Sf/λ-K(NS)r primers. The resulting fragments were cloned into pSPYNE(R)173 and pSPYCE(M) at the BamHI/SalI and SalI/KpnI restriction sites, respectively (Walter et al., 2004; Waadt et al., 2008). Primer sequences are listed in Supplemental Table 1. Plasmids containing YFPN-SOS2 and YFPC-14-3-3 λ or YFPN-SOS2 and YFPC were introduced into Agrobacterium tumefaciens GV3101 by electroporation and transformed into Nicotiana benthamiana leaves. Four to 5 d after infiltration, the YFP fluorescence signal was detected using a Zeiss LSM 510 META confocal microscope.
Firefly Luciferase Complementation Imaging Assay
The coding sequence of 14-3-3 λ was amplified with the λ-Kf/λ-Sr primers and cloned into the pCAMBIA-CLuc vector between the KpnI/SalI restriction sites. The coding sequences of SOS2, SOS2S294A, and SOS2S294D were amplified with the FL-Kf/FL-S(NS)r primers, and the resulting fragments were cloned into the pCAMBIA-NLuc vector between the KpnI/SalI restriction sites. In these constructs, 14-3-3 λ was fused downstream of C-Luc while SOS2 and its mutated versions were fused upstream of N-Luc. Primer sequences are listed in Supplemental Table 1. The resulting constructs were transformed into Agrobacterium strain GV3101, which was then infiltrated into N. benthamiana leaves. Three days after infiltration, 1 mM d-luciferin (Promega) was sprayed onto the leaves, and the plants were kept in the dark for 5 min. A low-light cooled charge-coupled device camera (ikon-L936; Andor Tech) was used to obtain the Luc signal as described (Chen et al., 2008). Relative Luc activity was quantified using WinView/32 software.
In Vitro Kinase Assays
All recombinant proteins were purified from E. coli BL21 (DE3). GST-tagged SOS2, SOS2 mutated forms, and the SOS1 C terminus were purified with Glutathione Sepharose (GE Healthcare) according to the manufacturer’s protocol. Polyhistidine-tagged 14-3-3 λ and 14-3-3 κ were purified with nickel–nitrilotriacetic acid agarose (Qiagen) as indicated above. All the Myc-tagged proteins used for in vitro kinase assays were immunoprecipitated with anti-C-Myc antibody–conjugated agarose (Sigma-Aldrich).
For total protein extract preparation, 0.5 g of 10-d-old Col-0 or representative T3 14-3-3 λ transgenic line (OE-9) seedlings was left untreated or treated with 100 mM NaCl for 12 h. Seedlings were harvested and ground to a fine powder in liquid nitrogen. Total proteins were extracted in 1 mL of IP buffer. Cell debris was removed by a 10-min centrifugation at 12,000g at 4°C. The supernatant was collected, and protein concentration was determined using the Bio-Rad Protein Assay Kit. Equal amounts of immunoprecipitated Myc-tagged SOS2 were incubated with the resulting total protein extracts for 2 h at 4°C with gentle mixing and used in in vitro kinase assays with MBP or GST-tagged SCaBP8 as the substrate.
Reaction mixtures (30 μL) contained 0.2 μL of [γ-32P]ATP (2 μCi in total) in kinase buffer (20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 10 μM ATP, and 1 mM DTT), and the in vitro kinase assays were performed at 30°C for at least 30 min (Quan et al., 2007). Reactions were terminated by the addition of 6× SDS loading buffer and were incubated at 95°C for 10 min. Proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue R 250 followed by exposure to a phosphor screen, and signals were obtained by a Typhoon 9410 phosphor imager (Amersham Biosciences). Phosphorylation signals were quantified using ImageQuant 5.0 software.
Phosphorylation of SCaBP8Ser237 in Planta
The Pro35S:6×Myc-SCaBP8 construct was generated and transformed into Col-0 and the 14-3-3 λ κ double mutant. Ten-day-old T3 transgenic seedlings were left untreated or treated with 100 mM NaCl for 12 h, and Myc-SCaBP8 was immunoprecipitated with anti-C-Myc antibody–conjugated agarose. The resulting proteins were analyzed by immunoblots with anti-SCaBP8-phosphoserine-237 and anti-C-Myc antibodies, and chemiluminescence signals were detected by autoradiography.
Analysis of Promoter Activity and Subcellular Localization
For GUS staining, a 2000-bp genomic DNA fragment from nucleotides 1 to 2000 bp upstream of the translational start site (ATG) of 14-3-3 λ was amplified with the λpro-Sf and λpro-Br primers and then cloned into the SalI and BamHI sites of the pCAMBIA1391 vector. For subcellular localization, the SOS2 coding region was amplified with the FL-Xf and FL-B(NS)r primers and cloned into the pCM1205-C-YFP vector using the XbaI and BamHI sites to fuse the target protein to the N terminus of YFP. The 14-3-3 λ coding region was amplified with the λ-Bf and λ-Sr primers and cloned into the pCM1205-GFP vector using the BamHI and SalI sites to fuse the target protein to the C terminus of green fluorescent protein (GFP). The plasmids were transformed into Col-0, and transgenic T3 lines or T2 lines were used for YFP or GFP fluorescence detection or GUS staining. The sequences of primers used are listed in Supplemental Table 1. For immunoblot analysis, total proteins were extracted from roots, stems, rosette leaves, cauline leaves, flowers, and siliques of 3-week-old wild-type plants grown in soil and analyzed by immunoblots using 14-3-3 λ antibodies.
In Vitro Pull-Down Analysis of Protein–Protein Interaction
SOS2, SOS2S294A, SOS2S294D, and SOS-JD were fused with a GST tag, and recombinant GST-tagged SOS2-JD or GST tag alone was coexpressed with polyhistidine-tagged 14-3-3 λ in E. coli BL21 (DE3). GST or GST-tagged recombinant proteins were purified by Glutathione Sepharose according to the manufacturer’s protocol (GE Healthcare) and detected by Coomassie Brilliant Blue R 250 staining, while associated polyhistidine-tagged 14-3-3 λ was detected by immunoblot analysis with anti-His or anti-14-3-3 λ antibody. Polyhistidine-tagged 14-3-3 λ was purified from these coexpressing E. coli DE3 strains and used as a loading control.
Expression and MS Analysis of STREPII-SOS2 Variants
The generation of pIVEX1.3(WG)-STREPII-SOS2 has been described previously (Hashimoto et al., 2012), and pIVEX1.3(WG)-STREPII-SOS2K40N was generated based on this construct by site-directed mutagenesis with the KN-f and KN-r primers as described (Hashimoto et al., 2012). STREPII-SOS2 and STREPII-SOS2K40N were expressed using the RTS 500 Wheat Germ CECF Kit (5 PRIME) as described recently (Drerup et al., 2013). For MS analysis, unpurified protein samples were run on an SDS gel and stained with Coomassie Brilliant Blue R 250. The stained protein bands corresponding to the SOS2 protein were excised and subsequently fragmented using the endoproteinase trypsin as described (Shevchenko et al., 1996). No reduction or alkylation steps were performed. Phosphopeptide enrichment was performed using TiO2 tips (NT2TIO; Glygen). Separation of peptides was conducted on the Ultimate 3000 Nanoflow HPLC system (Dionex) coupled to the nanospray source of an LTQ Orbitrap XL mass spectrometer (Thermo Finnigan). The mass spectrometer was operated in positive ion mode. MS full scans (m/z 400 to 2000) were acquired by Fourier transform ion cyclotron resonance MS in the Orbitrap at a resolution of 60,000 with internal lock mass calibration on m/z 445.12003. The five most intense ions of each full scan were fragmented in the linear ion trap by collision-induced dissociation (35% normalized collision energy). Fragment ions exhibiting a neutral loss of 24.5, 32.7, 49, and 98 D corresponding to the neutral loss of phosphoric acid were activated further, while the remaining MS2 ions were stored in the ion trap (multistage activation) (Schroeder at al., 2004). After the second activation step, MS2 and MS3 ions were simultaneously analyzed in the mass analyzer of the ion trap. For the identification of peptides, multistage activation spectra were matched against the STREPII-SOS2 and STREPII-SOS2K40N peptide sequences using OMSSA 2.1.4 (Geer et al., 2004), X!Tandem CYCLONE 2010.12.01 (Craig and Beavis, 2004), and Sequest (Eng et al., 1994). Phosphopeptide identifications and phosphorylation site localizations were validated by manual inspection of fragmentation spectra.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At5g35410 (SOS2), At5g10450 (14-3-3 λ), At5g65430 (14-3-3 κ), At2g01980 (SOS1), At4g33000 (SCaBP8), and At5g59890 (ADF4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of the Interaction between 14-3-3 λ and the SOS1 C Terminus in Arabidopsis Leaf Protoplasts.
Supplemental Figure 2. The 14-3-3 Protein Family in Arabidopsis.
Supplemental Figure 3. Quantification of the Phosphorylation Signals in Figure 2.
Supplemental Figure 4. Tissue and Subcellular Localization of 14-3-3 λ.
Supplemental Figure 5. 14-3-3 λ Modulates Salt Tolerance by Regulating SOS2 Activity in Arabidopsis.
Supplemental Figure 6. SOS2 Interacts with 14-3-3 λ through Its Junction Domain, and Mutations in SOS2Ser294 Modulate This Interaction.
Supplemental Figure 7. LC-ESI-MSA Spectrum Indicating a Phosphate Group at SOS2Ser294.
Supplemental Figure 8. Ser-294 Is Required for the Repression of SOS2 by 14-3-3 λ.
Supplemental Figure 9. Myc-SOS2S294D Binds More 14-3-3 λ Proteins Than Myc-SOS2 and SOS2S294A during Salt Stress.
Supplemental Table 1. Primers Used for Plasmid Construction.
Supplemental Table 2. Primers Used for RT-PCR Analysis.
Supplementary Material
Acknowledgments
We thank Zhi-Yong Wang for stimulating discussions and for the 14-3-3 T-DNA insertion lines. We thank Martin Scholz and Michael Hippler for MS analyses and Kenji Hashimoto for providing STII-SOS2 protein. This work was supported by the National Basic Research Program of China (Grant 2012CB114200 to Y.G.), the China National Funds for Distinguished Young Scientists (Grant 31025003 to Y.G.), a Natural Science Foundation of China international collaborative research project (Grant 31210103903 to Y.G. and J.K.), the Foundation for Innovative Research Group of the National Natural Science Foundation of China (Grant 31121002), the National Natural Science Foundation of China (Grant 31170246 to J.Z.), the Deutsche Forschungsgemeinschaft within the frame of Research Unit 964 (to J.K.), a Ph.D. fellowship from the IMPRS-CEDAD Graduate School (to K.B.), and the U.S. Department of Energy/Energy Biosciences (Grant DE-FG02-04ER15616 to K.S.S.).
AUTHOR CONTRIBUTIONS
H.Z. performed most of the research. H.Z. and Y.G. designed the research and analyzed the data. H.L., S.C., K.B., Y.Y., and J.Z. performed the research on the analysis of Na+ sensitivity and MS. H.Z., Y.G., J.K., and K.S.S. contributed to the discussion and wrote the article.
Glossary
- PM
plasma membrane
- ROS
reactive oxygen species
- MS
mass spectrometry
- Col-0
Columbia-0
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Albrecht V., Ritz O., Linder S., Harter K., Kudla J. (2001). The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batelli G., Verslues P.E., Agius F., Qiu Q., Fujii H., Pan S., Schumaker K.S., Grillo S., Zhu J.-K. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 27: 7781–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O., Sorek N., Schültke S., Yalovsky S., Kudla J. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20: 1346–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O., Waadt R., Steinhorst L., Held K., Kudla J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61: 211–222. [DOI] [PubMed] [Google Scholar]
- Berthomieu P., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 22: 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.-H., Pittman J.K., Zhu J.-K., Hirschi K.D. (2004). The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279: 2922–2926. [DOI] [PubMed] [Google Scholar]
- Chung J.-S., Zhu J.-K., Bressan R.A., Hasegawa P.M., Shi H. (2008). Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 53: 554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R., Beavis R.C. (2004). TANDEM: Matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467. [DOI] [PubMed] [Google Scholar]
- Denison F.C., Paul A.-L., Zupanska A.K., Ferl R.J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22: 720–727. [DOI] [PubMed] [Google Scholar]
- Drerup M.M., Schlücking K., Hashimoto K., Manishankar P., Steinhorst L., Kuchitsu K., Kudla J. (2013). The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 6: 559–569. [DOI] [PubMed] [Google Scholar]
- Du W., Lin H., Chen S., Wu Y., Zhang J., Fuglsang A.T., Palmgren M.G., Wu W., Guo Y. (2011). Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis. Plant Physiol. 156: 2235–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., McCormack A.L., Yates J.R. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5: 976–989. [DOI] [PubMed] [Google Scholar]
- Fuglsang A.T., Borch J., Bych K., Jahn T.P., Roepstorff P., Palmgren M.G. (2003). The binding site for regulatory 14-3-3 protein in plant plasma membrane H+-ATPase: Involvement of a region promoting phosphorylation-independent interaction in addition to the phosphorylation-dependent C-terminal end. J. Biol. Chem. 278: 42266–42272. [DOI] [PubMed] [Google Scholar]
- Fuglsang A.T., Visconti S., Drumm K., Jahn T., Stensballe A., Mattei B., Jensen O.N., Aducci P., Palmgren M.G. (1999). Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 274: 36774–36780. [DOI] [PubMed] [Google Scholar]
- Geer L.Y., Markey S.P., Kowalak J.A., Wagner L., Xu M., Maynard D.M., Yang X., Shi W., Bryant S.H. (2004). Open mass spectrometry search algorithm. J. Proteome Res. 3: 958–964. [DOI] [PubMed] [Google Scholar]
- Guo Y., Halfter U., Ishitani M., Zhu J.-K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13: 1383–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U., Ishitani M., Zhu J.-K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97: 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K., Kudla J. (2011). Calcium decoding mechanisms in plants. Biochimie 93: 2054–2059. [DOI] [PubMed] [Google Scholar]
- Hashimoto K., Eckert C., Anschütz U., Scholz M., Held K., Waadt R., Reyer A., Hippler M., Becker D., Kudla J. (2012). Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J. Biol. Chem. 287: 7956–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Horie R., Chan W.-Y., Leung H.-Y., Schroeder J.I. (2006). Calcium regulation of sodium hypersensitivities of sos3 and athkt1 mutants. Plant Cell Physiol. 47: 622–633. [DOI] [PubMed] [Google Scholar]
- Hrabak E.M., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132: 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Koizumi N., Kusano T., Sano H. (2000). Specific binding of a 14-3-3 protein to autophosphorylated WPK4, an SNF1-related wheat protein kinase, and to WPK4-phosphorylated nitrate reductase. J. Biol. Chem. 275: 31695–31700. [DOI] [PubMed] [Google Scholar]
- Ishitani M., Liu J., Halfter U., Kim C.-S., Shi W., Zhu J.-K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Belfield E.J., Mithani A., Visscher A., Ragoussis J., Mott R., Smith J.A.C., Harberd N.P. (2012). ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 31: 4359–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Zhu J., Kim K., Agarwal M., Fu X., Huang A., Zhu J.-K. (2006). The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 18816–18821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.G., Waadt R., Cheong Y.H., Pandey G.K., Dominguez-Solis J.R., Schültke S., Lee S.C., Kudla J., Luan S. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52: 473–484. [DOI] [PubMed] [Google Scholar]
- Kim W.-Y., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 4: 1352–1363. [DOI] [PubMed] [Google Scholar]
- Kudla J., Batistič O., Hashimoto K. (2010). Calcium signals: The lead currency of plant information processing. Plant Cell 22: 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaud C., Prigent E., Thuleau P., Grat S., Da Silva D., Brière C., Mazars C., Cotelle V. (2013). 14-3-3-regulated Ca2+-dependent protein kinase CPK3 is required for sphingolipid-induced cell death in Arabidopsis. Cell Death Differ. 20: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y., Melamed-Book N., Cagnac O., Ronen G., Nishri Y., Solomon M., Cohen G., Levine A. (2006). Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. USA 103: 18008–18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Yang Y., Quan R., Mendoza I., Wu Y., Du W., Zhao S., Schumaker K.S., Pardo J.M., Guo Y. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21: 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhu J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280: 1943–1945. [DOI] [PubMed] [Google Scholar]
- Liu J., Ishitani M., Halfter U., Kim C.-S., Zhu J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97: 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., Morsomme P. (2000). A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275: 17762–17770. [DOI] [PubMed] [Google Scholar]
- Møller I.S., Gilliham M., Jha D., Mayo G.M., Roy S.J., Coates J.C., Haseloff J., Tester M. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Narasimhan M.L., Salzman R.A., Bressan R.A., Hasegawa P.M. (1993). NaCl regulation of plasma membrane H+-ATPase gene expression in a glycophyte and a halophyte. Plant Physiol. 103: 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oecking C., Jaspert N. (2009). Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 12: 760–765. [DOI] [PubMed] [Google Scholar]
- Oh C.-S., Pedley K.F., Martin G.B. (2010). Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. Plant Cell 22: 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Guo Y., Halfter U., Zhu J.-K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100: 11771–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren M.G. (2001). Plant plasma membrane H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 817–845. [DOI] [PubMed] [Google Scholar]
- Palmgren M.G., Sommarin M., Serrano R., Larsson C. (1991). Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J. Biol. Chem. 266: 20470–20475. [PubMed] [Google Scholar]
- Paul A.L., Denison F.C., Schultz E.R., Zupanska A.K., Ferl R.J. (2012). 14-3-3 phosphoprotein interaction networks: Does isoform diversity present functional interaction specification? Front. Plant Sci. 3: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q.-S., Guo Y., Dietrich M.A., Schumaker K.S., Zhu J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99: 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q.-S., Guo Y., Quintero F.J., Pardo J.M., Schumaker K.S., Zhu J.-K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279: 207–215. [DOI] [PubMed] [Google Scholar]
- Quan R., Lin H., Mendoza I., Zhang Y., Cao W., Yang Y., Shang M., Chen S., Pardo J.M., Guo Y. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero F.J., Martinez-Atienza J., Villalta I., Jiang X., Kim W.-Y., Ali Z., Fujii H., Mendoza I., Yun D.-J., Zhu J.-K., Pardo J.M. (2011). Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 108: 2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero F.J., Ohta M., Shi H., Zhu J.-K., Pardo J.M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 99: 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A., Yokoi S., Sharkhuu A., Reddy M., Lee B.H., Matsumoto T.K., Koiwa H., Zhu J.-K., Bressan R.A., Hasegawa P.M. (2001). AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc. Natl. Acad. Sci. USA 98: 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M.J., Shabanowitz J., Schwartz J.C., Hunt D.F., Coon J.J. (2004). A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76: 3590–3598. [DOI] [PubMed] [Google Scholar]
- Sheen J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858. [DOI] [PubMed] [Google Scholar]
- Shi H., Ishitani M., Kim C., Zhu J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97: 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Quintero F.J., Pardo J.M., Zhu J.-K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokina E.M., Feinstein S.I., Zhou S., Fisher A.B. (2011). Intracellular targeting of peroxiredoxin 6 to lysosomal organelles requires MAPK activity and binding to 14-3-3ε. Am. J. Physiol. Cell Physiol. 300: C1430–C1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennelid F., Olsson A., Piotrowski M., Rosenquist M., Ottman C., Larsson C., Oecking C., Sommarin M. (1999). Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11: 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T.-S., Whippo C., Hangarter R.P., Briggs W.R. (2012). The role of a 14-3-3 protein in stomatal opening mediated by PHOT2 in Arabidopsis. Plant Cell 24: 1114–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P.E., Batelli G., Grillo S., Agius F., Kim Y.-S., Zhu J., Agarwal M., Katiyar-Agarwal S., Zhu J.-K. (2007). Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol. Cell. Biol. 27: 7771–7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistič O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang B., Yang H., Liu Y.C., Jelinek T., Zhang L., Ruoslahti E., Fu H. (1999). Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38: 12499–12504. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang C., Zhang C., Wang N., Lu D., Wang J., Zhang S., Wang Z.-X., Ma H., Wang X. (2011). Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21: 825–834. [DOI] [PubMed] [Google Scholar]
- Weinl S., Kudla J. (2009). The CBL-CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytol. 184: 517–528. [DOI] [PubMed] [Google Scholar]
- Xu W., Jia L., Shi W., Baluska F., Kronzucker H.J., Liang J., Zhang J. (2013). The tomato 14-3-3 protein TFT4 modulates H+ efflux, basipetal auxin transport, and the PKS5-J3 pathway in the root growth response to alkaline stress. Plant Physiol. 163: 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., Liu J., Xiao Y., Liang Y., Zhang W. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188: 762–773. [DOI] [PubMed] [Google Scholar]
- Zhou H., Zhao J., Yang Y., Chen C., Liu Y., Jin X., Chen L., Li X., Deng X.W., Schumaker K.S., Guo Y. (2012). Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na+/H+ antiport activity and serine hydroxymethyltransferase stability. Plant Cell 24: 5106–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6: 441–445. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K., Xiong, L., Ishitani, M., Liu, J., Lee, H., Stevenson, B., and Shi, W. (1998). Identification of genes important for environmental stress tolerance in plants. In Breeding and Biotechnology of Environmental Stress in Rice, Y. Sato, ed (Sapporo, Japan: Hokkaido National Agricultural Experiment Station), pp. 105–113. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.