This work identified the thylakoid protein HYPERSENSITIVE TO HIGH LIGHT1 (HHL1), specifically conserved in land plants, as an important regulator of photosynthesis and photoprotection. Together with LOW QUANTUM YIELD OF PHOTOSYSTEM II1, HHL1 regulates the repair and reassembly of photosystem II complex under high-light conditions in Arabidopsis.
Abstract
Under high-irradiance conditions, plants must efficiently protect photosystem II (PSII) from damage. In this study, we demonstrate that the chloroplast protein HYPERSENSITIVE TO HIGH LIGHT1 (HHL1) is expressed in response to high light and functions in protecting PSII against photodamage. Arabidopsis thaliana hhl1 mutants show hypersensitivity to high light, drastically decreased PSII photosynthetic activity, higher nonphotochemical quenching activity, a faster xanthophyll cycle, and increased accumulation of reactive oxygen species following high-light exposure. Moreover, HHL1 deficiency accelerated the degradation of PSII core subunits under high light, decreasing the accumulation of PSII core subunits and PSII–light-harvesting complex II supercomplex. HHL1 primarily localizes in the stroma-exposed thylakoid membranes and associates with the PSII core monomer complex through direct interaction with PSII core proteins CP43 and CP47. Interestingly, HHL1 also directly interacts, in vivo and in vitro, with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 (LQY1), which functions in the repair and reassembly of PSII. Furthermore, the hhl1 lqy1 double mutants show increased photosensitivity compared with single mutants. Taken together, these results suggest that HHL1 forms a complex with LQY1 and participates in photodamage repair of PSII under high light.
INTRODUCTION
The photosynthetic apparatus of cyanobacteria, eukaryotic algae, and vascular plants is located in the specialized thylakoid membrane system and is mediated by the integral membrane protein complexes photosystem II (PSII), cytochrome b6/f, photosystem I (PSI), and ATP synthase, which harvest light and transduce solar energy into chemical energy (Eberhard et al., 2008). The multisubunit pigment-protein complex of PSII catalyzes the light-driven water oxidation and reduction of plastoquinone (Chi et al., 2012a). Although green plants require PSII to absorb energy from sunlight via the photosystem, harnessing this tremendous light energy during photosynthesis carries great risks, including inevitable damage to the photosynthetic apparatus, especially PSII (Takahashi and Badger, 2011). Plants have therefore evolved diverse photoprotection mechanisms, including light avoidance associated with the movement of leaves and chloroplasts, screening of photoradiation, reactive oxygen species (ROS) scavenging systems, dissipation of absorbed light energy as thermal energy (energy-dependent quenching [qE]), cyclic electron flow around PSI, and the photorespiratory pathway (Takahashi and Badger, 2011).
When these protective measures fail to prevent damage to PSII, plant cells deploy a complex PSII repair system that involves disassembly and reassembly of a thylakoid membrane protein complex comprising dozens of proteins (Baena-González and Aro, 2002; Mulo et al., 2008). Therefore, most PSII assembly factors also play a role in PSII repair. Although our current knowledge of PSII structure and function is extensive, relatively little is known about how disassembly and reassembly of PSII occur. However, two major findings have recently emerged: PSII repair is a highly ordered process, and large numbers of accessory factors are involved in forming this multiprotein complex and incorporating the various cofactors (Nickelsen and Rengstl, 2013). The PSII repair processes can be generalized as follows: (1) Photodamage induces a reversible phosphorylation and disassembly of the PSII–light-harvesting complex II (LHCII) supercomplexes; (2) damaged PSII core monomers migrate from grana stacks to stroma-exposed membranes; (3) PSII core monomers partially disassemble and release CP43; (4) photodamaged PSII subunits are degraded by proteases; (5) new PSII subunits are synthesized and reassembled into new PSII core monomers; (6) PSII core monomers migrate to grana stacks and reform functional PSII-LHCII supercomplexes (Aro et al., 2005; Mulo et al., 2008).
Genetic and biochemical studies have provided evidence for the protection and repair of PSII. The repair cycle of PSII requires the participation of hundreds of auxiliary proteins (Mulo et al., 2008; Nickelsen and Rengstl, 2013), and those in higher plants are different from those of more primitive photosynthetic organisms (Nixon et al., 2010). However, some PSII assembly and repair factors are conserved in cyanobacteria and higher plants, including HIGH CHLOROPHYLL FLUORESCENCE136 (Komenda et al., 2008), ALBINO3 (Luirink et al., 2001; Göhre et al., 2006), Psb27 (Nowaczyk et al., 2006; Komenda et al., 2012), Psb28 (Dobáková et al., 2009; Sakata et al., 2013), Psb29 (Keren et al., 2005), and YCF39 (Ermakova-Gerdes and Vermaas, 1999), as well as immunophilins, such as CYCLOPHILIN38 (CYP38)/THYLAKOID LUMEN PEPTIDYLPROLYL ISOMERASE OF 40 kD (TLP40) (Sirpiö et al., 2008). Presumably, therefore, the roles of these factors in PSII biogenesis or maintenance have been retained during evolution. In contrast, other factors are specific to cyanobacteria, such as PROCESSING ASSOCIATED TPR PROTEIN (Schottkowski et al., 2009), Slr0286 (Kufryk and Vermaas, 2001), and Slr2013 (Kufryk and Vermaas, 2003), or specific to higher plants, such as LOW PSII ACCUMULATION1 (LPA1)/PSII REPAIR27 (Park et al., 2007; Dewez et al., 2009) and LPA2 (Ma et al., 2007), which may represent evolutionary adaptations to specific environments. Notably, LOW QUANTUM YIELD OF PHOTOSYNSTEM II1 (LQY1), a small thylakoid zinc-finger protein exclusive to land plants, was also recently found to be involved in the repair cycle and reassembly of PSII (Lu et al., 2011).
The ceaseless identification of new auxiliary proteins associated with PSII confirms that much remains to be learned about the regulation of structure and function of PSII. However, compared with the structural subunits of PSII complex, the auxiliary proteins of PSII for research are challenging for a variety of reasons, including the low protein abundance, labile interactions with core and antenna complexes, and functional redundancy (Mulo et al., 2008). Analysis of in vivo chlorophyll fluorescence is a powerful, noninvasive technique used to identify mutations affecting photosynthesis (Maxwell and Johnson, 2000; Müller et al., 2001; Kramer et al., 2004; Baker, 2008). Alterations in chlorophyll fluorescence indicate defects in the photosynthetic electron transport chain due to changes in the structure and function of the thylakoid membrane (Baker, 2008). Therefore, screening mutants with altered chlorophyll fluorescence provides a more specific way to obtain and characterize photosynthetic mutants.
To gain further insight into the maintenance and repair of PSII, we used the chlorophyll fluorescence system to screen a pool of Arabidopsis thaliana mutants and identified hypersensitive to high light (hhl) mutants, which show lower photosynthesis efficiency under high-light conditions and exhibit high-light-induced defects in PSII photochemistry and accumulation of PSII supercomplexes, both of which are surprisingly similar to defects observed in lqy1 mutants. We found that the HHL1 protein associates with PSII directly, specifically with PSII core proteins CP43, CP47, and LQY1 both in vivo and in vitro. The results of this study suggest that HHL1 and LQY1 may form a complex that is involved in the repair and reassembly cycle of the PSII-LHCII supercomplex under high-light conditions in land plants.
RESULTS
Isolation and Identification of hhl1 Mutants
To identify regulatory components involved in the photoprotection and repair of PSII, we screened more than 50,000 Arabidopsis mutants obtained from RIKEN (Ichikawa et al., 2006) using chlorophyll fluorescence imaging. The ratio of variable fluorescence/maximal fluorescence from dark-adapted leaves (Fv/Fm) can be used to estimate the maximum photochemical efficiency of PSII (Baker, 2008). Arabidopsis mutants that exhibited aberrant Fv/Fm after exposure to high light were identified using a chlorophyll fluorescence video imaging system, and a new series of Arabidopsis hhl mutants were identified. Interestingly, one of the hhl mutants, hhl1-0, was found to have a lower maximum photochemical efficiency of PSII (Fv/Fm) after high-light treatment (∼1200 μmol photons m−2 s−1) than the wild type (Columbia-0 [Col-0]; Supplemental Figure 1A), although the growth phenotype and Fv/Fm of the mutants were identical to the wild type grown under growth light conditions (∼100 μmol photons m−2 s−1). However, Fv/Fm returned to normal levels following a 2-d recovery under growth light conditions (Supplemental Figure 1B). The results were confirmed with twice-backcrossed mutants and imply that the hhl1-0 mutant has defects in energy transfer within PSII or a partial loss of PSII capacity after high-light stress.
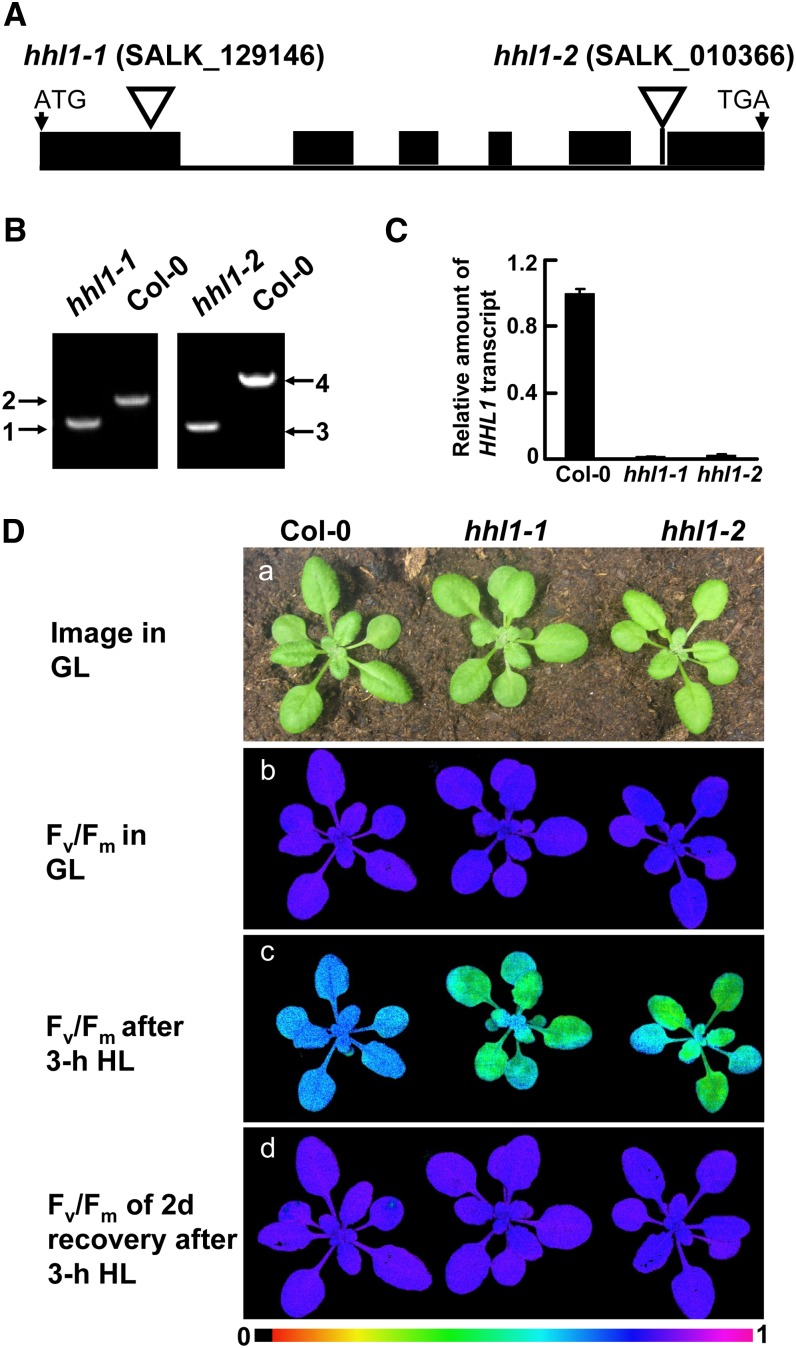
To determine the genetic basis of the hhl1-0 phenotype, we performed thermal asymmetric interlaced PCR to determine the genomic region flanking the left border sequences of the T-DNA (Supplemental Figure 1C). Sequence analysis revealed that the T-DNA was inserted in the translated region of the nuclear-encoded gene At1g67700 (Supplemental Figure 1D). To confirm that the mutated gene is responsible for the observed phenotype, we analyzed two other independent homozygous T-DNA insertion mutants of At1g67700 (SALK_129146C and SALK_010366C) from the sequence-indexed Arabidopsis T-DNA insertion mutant stock. The T-DNA insertion positions in these mutants were determined by PCR and subsequent sequencing of the amplified products (Figure 1B). The mutants carry T-DNA insertions in the first exon and last intron of At1g67700, respectively (Figure 1A). Quantitative real-time RT-PCR analysis showed that the transcription of At1g67700 was suppressed in both mutants (Figure 1C). We subsequently named SALK_129146C and SALK_010366C as hhl1-1 and hhl1-2, respectively, and used these names throughout the article. The hhl1-0 mutant described above exhibited an identical phenotype to that of both the hhl1-1 and hhl1-2 mutants: The Fv/Fm value decreased from 0.82 to 0.70 in the wild type after high-light treatment, while in the mutants, this value decreased from 0.82 to 0.51 (Figure 1D; Supplemental Table 1). Thus, it can be concluded that the hhl1 phenotype is due to the inactivation of At1g67700.
Figure 1.
Mutations of the HHL1 Gene Cause Altered Chlorophyll Fluorescence Parameters.
(A) Schematic diagram of the HHL1 gene inferred by DNA sequence analysis. Exons (black boxes) and introns (lines) are indicated. The positions of the T-DNA insertions corresponding to hhl1-1 and hhl1-2 are shown. ATG start codon and TGA stop codon are shown.
(B) PCR analysis of genomic DNA from the wild type and hhl1 mutants to confirm the homozygosity of the mutants. 1 and 2, amplification with primers F and R for SALK_129146 and Lba1; 3 and 4, amplification with primers F and R for SALK_010366 and Lba1.
(C) Quantitative real-time RT-PCR analysis of HHL1 transcription in the wild type and hhl1 mutants. Data for wild-type (Col-0), hhl1-1, and hhl1-2 plants are presented as mean ± se.
(D) Images in (a) are of 3-week-old wild-type (Col-0), hhl1-1, and hhl1-2 plants under growth light conditions. (b) False-color images representing Fv/Fm under a growth light condition in 3-week-old wild-type, hhl1-1, and hhl1-2 plants. (c) False-color images representing Fv/Fm after a 3-h high-light treatment in 3-week-old wild-type, hhl1-1, and hhl1-2 plants. (d) False-color images representing Fv/Fm after a 3-h high-light treatment and a 2-d recovery period in 3-week-old wild-type, hhl1-1, and hhl1-2 plants. The false color ranged from black (0) via red, orange, yellow, green, blue, and violet to purple (1) as indicated at the bottom. GL, growth light (∼100 μmol photons m−2 s−1); HL, high light (∼1200 μmol photons m−2 s−1). Six biological replicates were performed in all experiments, and similar results were obtained.
Carotenoid Composition but Not Chlorophyll Content Is Slightly Altered in the hhl1 Mutants after High-Light Treatment
Under growth light conditions, hhl1 plants showed no phenotypic differences from the wild type (Figure 1D). In addition, spectrofluorometric analyses indicated that there were also no differences in the chlorophyll or total carotenoid contents (Supplemental Table 1).
Carotenoids are involved in the regulation of photosynthetic light harvesting and photoprotection (Holt et al., 2005). To more precisely investigate whether the carotenoids compositions were affected in the hhl1 mutants, we performed HPLC analysis before and after high light. Under growth light conditions, the carotenoid components were comparable between hhl1 and the wild type. After a 3-h high-light treatment, antheraxanthin was significantly increased, but violaxanthin was decreased in both hhl1 and the wild type compared with under growth conditions (Supplemental Figure 2), indicating that the xanthophyll cycle is activated by high light (Niyogi et al., 1998). Surprisingly, the increase in antheraxanthin in hhl1 was slightly more significant than in the wild type, as was the decrease in violaxanthin, indicating that the activation of the xanthophyll cycle may be slightly more significant in the hhl1 mutants than in the wild type, instead of being impaired after high-light treatment. Thus, we speculate that the zeaxanthin level may also be slightly higher in the hhl1 mutants after high light, although this was not detected successfully in this assay due to the lower level of this compound. However, the level of chlorophyll and other carotenoid components, including neoxanthin, lutein, and β-carotene, showed no difference in hhl1 compared with wild-type plants (Supplemental Figure 2). These results suggest that the photosensitivity of hhl1 mutants is not due to perturbation of carotenoids.
PSII Activity Is Markedly Reduced in the hhl1 Mutants after High-Light Treatment
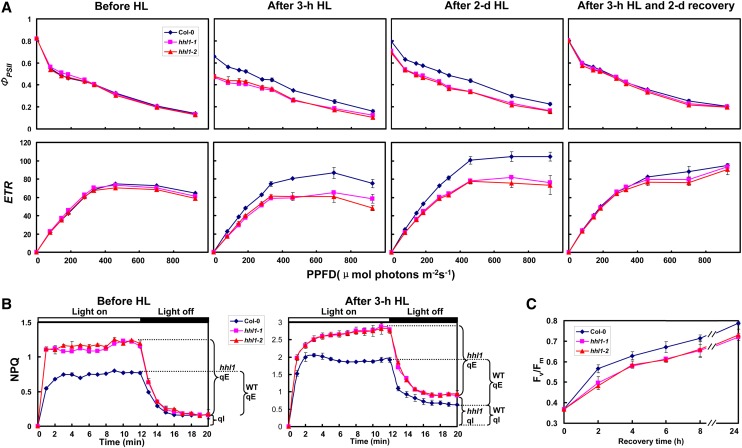
Initial Fv/Fm measurements preliminarily suggested that PSII activity is disturbed in the hhl1 mutants after high-light stress. To further investigate the primary target of the hhl1 mutation, the minimum (F0) and maximum (Fm) fluorescence of dark-adapted leaves, Fv/Fm, and other chlorophyll parameters were quantitatively determined under a fixed light intensity. Under growth light conditions, the hhl1 mutants possessed much higher nonphotochemical quenching (NPQ) than wild-type plants (Figure 2B), which indicates that the mutants dissipate more excess excitation energy via nonphotochemical pathways in growth light, although this did not affect the normal growth of mutants under growth light conditions. After a 3-h high-light treatment, the hhl1 mutants had much lower Fm and Fv/Fm than wild-type plants. Interestingly, the F0 was slightly higher in the hhl1 mutants (Supplemental Table 1). In addition, the parameters 1-qP and 1-qL, which reflect the redox state of the QA electron acceptor of PSII (Kramer et al., 2004), were lower in the hhl1 mutants (Supplemental Table 1). NPQ primarily includes qE and photoinhibitory quenching (qI) according to their relaxation kinetics because state transition quenching is only significant under very low light in most plants (Müller et al., 2001; Baker, 2008). After a 3-h high-light treatment, the higher qI in the hhl1 mutants was significantly strengthened compared with wild-type plants, indicating that the mutants experienced more serious photoinhibition than the wild type under high-light stress. Interestingly, the qE was also significantly higher in hhl1 (Figure 2B), which is consistent with the faster xanthophyll cycle (Supplemental Figure 2).
Figure 2.
Characterization of Detailed Chlorophyll Fluorescence in the Wild Type and hhl1 Mutants.
(A) Light-response curves of PSII quantum yield and ETR in wild-type and hhl1 mutants. The measurements were performed at the following light intensities: 0, 81, 145, 186, 281, 335, 461, 701, and 926 μmol photons m−2 s−1. HL, high light.
(B) Time courses for induction and relaxation of NPQ before and after 3-h high-light treatment. Actinic light (500 μmol photons m−2 s−1) was switched on at time zero, and plants were left in the dark after 12 min.
(C) Recovery of maximal photochemical efficiency of PSII after photoinhibition. Maximal photochemical efficiency of PSII (Fv/Fm) during recovery period at irradiance of 100 μmol photons m−2 s−1 was measured by following the photoinhibition treatment to 50% of the initial Fv/Fm values. Data for wild-type (Col-0), hhl1-1, and hhl1-2 plants are presented as mean ± se. Five biological replicates were performed, and similar results were obtained.
To further characterize the photosynthetic apparatus, we analyzed the light intensity dependence of two chlorophyll fluorescence parameters, the light-response curves of PSII quantum yield (ΦPSII), and the electron transport rate (ETR) before and after a 3-h high-light treatment. After a short-term high-light treatment (3 h), ΦPSII and ETR were much lower in hhl1 mutants than in the wild type (Figure 2A), as was also observed for Fv/Fm (Figure 1; Supplemental Table 1). Interestingly, after an extended high-light treatment, the difference between wild-type and hhl1 plants was the same as that observed after the 3-h treatment (Figure 2A). However, ΦPSII and ETR in the hhl1 mutants almost reached wild-type levels after the plants were returned to growth light conditions for 2 d (Figure 2A).
In addition, to assess the capacity of the mutants to recover from photoinhibition, we monitored changes in Fv/Fm after high-light treatment, which resulted in an ∼50% reductions for both wild-type and mutant leaves. As shown in Figure 2C, hhl1 mutants recovered from photoinhibition more slowly than wild-type leaves. After an 8-h recovery period, Fv/Fm of wild-type and hhl1 mutants leaves reached 87 and 79% of their initial values, respectively. Even after a 24-h recovery period, Fv/Fm was still ∼89% of the initial Fv/Fm values in the hhl1 mutants and 97% in the wild type. Taken together, detailed chlorophyll fluorescence analyses showed that PSII photochemical activity decreased to a greater extent and showed delayed recovery of PSII activity from photoinhibition in hhl1 mutants after high-light treatment.
ROS Level in the hhl1 Mutants Is Higher after High-Light Treatment
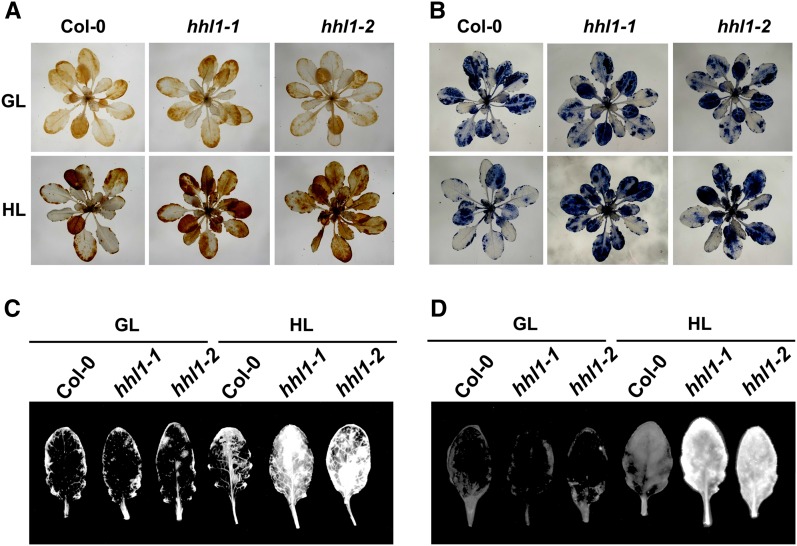
Under high-light conditions, the production of ROS is accelerated in chloroplasts (Takahashi and Badger, 2011), and H2O2, superoxide (O2−), and 1O2 are primary ROS (Li et al., 2009a). To test whether the accumulation of ROS in the hhl1 mutants differed from that in the wild type after high-light treatment, we examined the accumulation of H2O2, O2−, and 1O2. We first measured H2O2 and O2− levels using diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining. Following a 2-d high-light treatment, H2O2 and O2− were present at considerably higher levels in the hhl1 mutants than in the wild type (Figures 3A and 3B). Specifically, H2O2 and O2− levels were higher in younger hhl1 leaves, but older leaves showed no differences from the wild type (Figures 3A and 3B). Detection of 1O2 was performed on detached leaves using the singlet oxygen sensor green (SOSG) fluorescence indicator. A stronger fluorescence signal was detected in the high-light-treated mutants (Figure 3C). To further determine total intracellular ROS concentrations, we compared withtal level of ROS accumulation using the fluorescent dye 2’,7’-dichlorofluorescin diacetate (Pei et al., 2000). Again, a stronger fluorescence signal was detected in high-light-treated hhl1 mutants (Figure 3D), further indicating that ROS accumulated to higher levels in mutant plants. Together, the results from O2−, H2O2, 1O2, and total ROS detection indicated that the hhl1 mutants accumulate more ROS than the wild type after high-light treatment. Recent studies have also demonstrated that ROS accelerates photoinhibition by inhibiting of the repair of photodamaged PSII (Nishiyama et al., 2006). Thus, high-light intensity may cause more extensive oxidative damage in hhl1 mutants by inhibiting the repair of photodamaged PSII.
Figure 3.
Analysis of ROS in the Wild Type and hhl1 Mutants.
(A) DAB staining of H2O2 in 3-week-old wild-type (Col-0), hhl1-1, and hhl1-2 plants under growth light (top panel) and after growing under high light for 2 d (bottom panel). GL, growth light; HL, 2-d high-light treatment.
(B) NBT staining of O2− in 3-week-old wild-type (Col-0), hhl1-1, and hhl1-2 plants under growth light (top panel) and after growing under high light for 2 d (bottom panel).
(C) Detection of 1O2 with SOSG in wild-type (Col-0), hhl1-1, and hhl1-2 plants under growth light and after growing under high light for 2 d.
(D) Detection of total ROS with DCF in wild-type (Col-0), hhl1-1, and hhl1-2 plants under growth light and after growing under high light for 2 d. Five additional independent biological replicates were performed, and similar results were obtained.
[See online article for color version of this figure.]
HHL1 Is a Chloroplast Thylakoid Transmembrane Protein
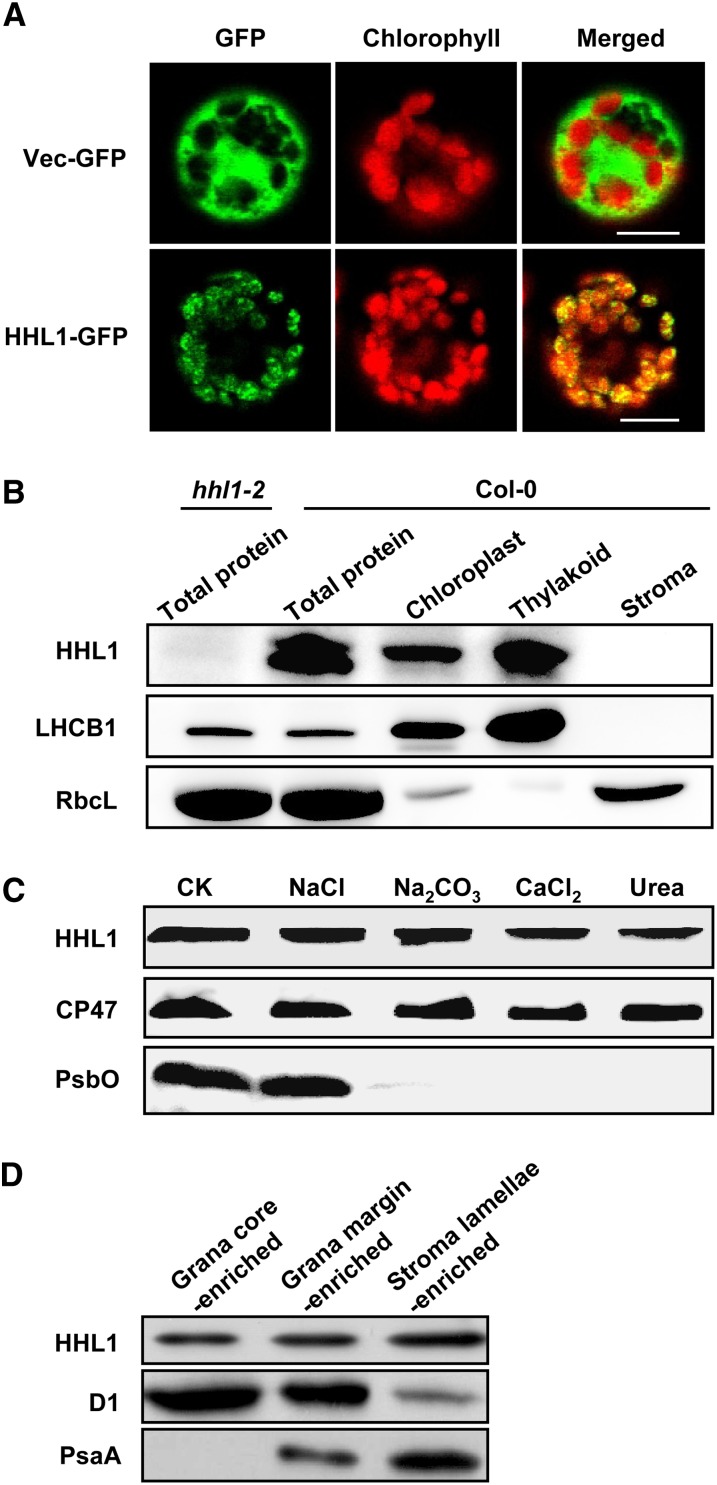
To understand the mechanism of photoprotection by HHL1, we first determined the subcellular localization of this protein. HHL1 encodes a predicted 231–amino acid protein of unknown function based on The Arabidopsis Information Resource annotation (www.arabidopsis.org). HHL1 protein also contains a predicted N-terminal chloroplast transit peptide (1 to 39) based on TargetP prediction (Emanuelsson et al., 2000), as well as a transmembrane domain (84 to 109) based on SPLIT prediction (Juretić et al., 2002) and a Von Willebrand factor type A (VWA) domain (110 to 158 amino acids) based on SMART prediction (Letunic et al., 2012). VWA domains often appear to involve protein–protein interactions and assembly of multiprotein complexes (Hynes and Zhao, 2000; Whittaker and Hynes, 2002; Tomko and Hochstrasser, 2013). In addition, HHL1 shares a close evolutionary relationship to the Thimet Oligopeptidase family (Mi et al., 2013). Thimet Oligopeptidase specifically degrades peptides of 9 to 17 residues into aminopeptides (Polge et al., 2009). To confirm whether HHL1 is located in the chloroplast, a 35S:HHL1-GFP (green fluorescent protein) construct was introduced into Arabidopsis protoplasts by transient transformation. Analysis of the subcellular localization of the HHL1-GFP fusion proteins by confocal laser scanning microscopy revealed that HHL1 is localized in the chloroplast as expected (Figure 4A). To further determine the location of HHL1 within the chloroplast, we separated the thylakoid membrane and stoma fractions from wild-type Arabidopsis chloroplasts and performed precise sublocation analysis. Immunoblot analysis of the soluble and membrane fractions from Percoll-purified chloroplasts demonstrated that HHL1 is primarily associated with the thylakoid membranes of isolated chloroplasts and is not found in the stoma (Figure 4B), in agreement with previous proteomic studies (Ferro et al., 2010).
Figure 4.
Subcellular Localization and Immunolocalization of HHL1 Protein.
(A) Localization of HHL1 protein within the chloroplast by GFP assay. The fluorescence of HHL1-GFP specifically matched with that of chlorophyll autofluorescence, confirming chloroplast targeting of HHL1 exclusively. HHL1-GFP, HHL1-GFP fusion; Vec-GFP, control with empty vector. Bars = 10 μm.
(B) HHL1 localizes to the thylakoid membrane fractions. Intact chloroplasts were isolated from wild-type (Col-0) leaves and then separated into thylakoid membrane and stroma fractions. Polyclonal antibodies were used against the integral membrane protein LHCB1, the abundant stroma protein ribulose biphosphate carboxylase large subunit (RbcL) and HHL1. Total proteins extracted from the wild type and hhl1-2 were used to confirm the specificity of the anti-HHL1 antibody.
(C) Immunolocalization of HHL1. The wild-type thylakoid membranes were sonicated in the presence of 1 M NaCl, 200 mM Na2CO3, 1 M CaCl2, and 6 M urea for 30 min at 4°C. PsbO (the 33-kD luminal protein of PSII) and CP47 (the PSII core protein) were used as markers. Membranes that had not been subjected to any salt treatment were used as controls (CK).
(D) Analysis of HHL1 protein in grana core-, grana margin-, and stroma lamellae-enriched thylakoid. Thylakoids were solubilized with digitonin and subfractionated by differential ultracentrifugation into grana core-, grana margin-, and stroma lamellae-enriched thylakoid and subjected to immunoblot analysis. The lanes on each gel were loaded on an equal chlorophyll basis. All experiments were repeated three times with similar results.
To localize the HHL1 protein more precisely, wild-type thylakoids isolated from growth light conditions were fractionated into the grana core-, grana margin-, and stroma lamellae-enriched thylakoid membranes by digitonin. The purity of the fractions was evaluated by both determining chlorophyll a/b ratio (Supplemental Figure 3) and by using PSII core protein D1 and PSI core protein PsaA as controls. As expected, the PSI core proteins were detected exclusively in the stroma-exposed thylakoids and the PSII proteins in the grana thylakoids, except for a small amount of PSII proteins in the stroma-exposed membrane representing PSII complexes undergoing repair (Figure 4D). The HHL1 protein was found to be distributed among the grana core-, grana margin-, and stroma lamellae-enriched thylakoid membranes thylakoids (Figure 4D). More interestingly, HHL1 protein was more abundant in the grana margin than in the grana core, and it was even more abundant in the stroma lamellae (Figure 4D), indicating that the protein may function in all of these locations.
To confirm whether HHL1 is a transmembrane protein, wild-type thylakoid membrane fractions were isolated and the proteins from these fractions were subjected to immunoblot analysis. After the membrane preparations were sonicated in the presence of various salts, HHL1 protein remained in the membranes (Figure 4C). During these treatments, PsbO (the 33-kD luminal protein of PSII) and CP47 (the PSII core protein) were used as controls (Figure 4C). HHL1 behaved like the integral protein CP47 but not like the peripheral protein PsbO, strongly indicating that HHL1 is an intrinsic membrane protein.
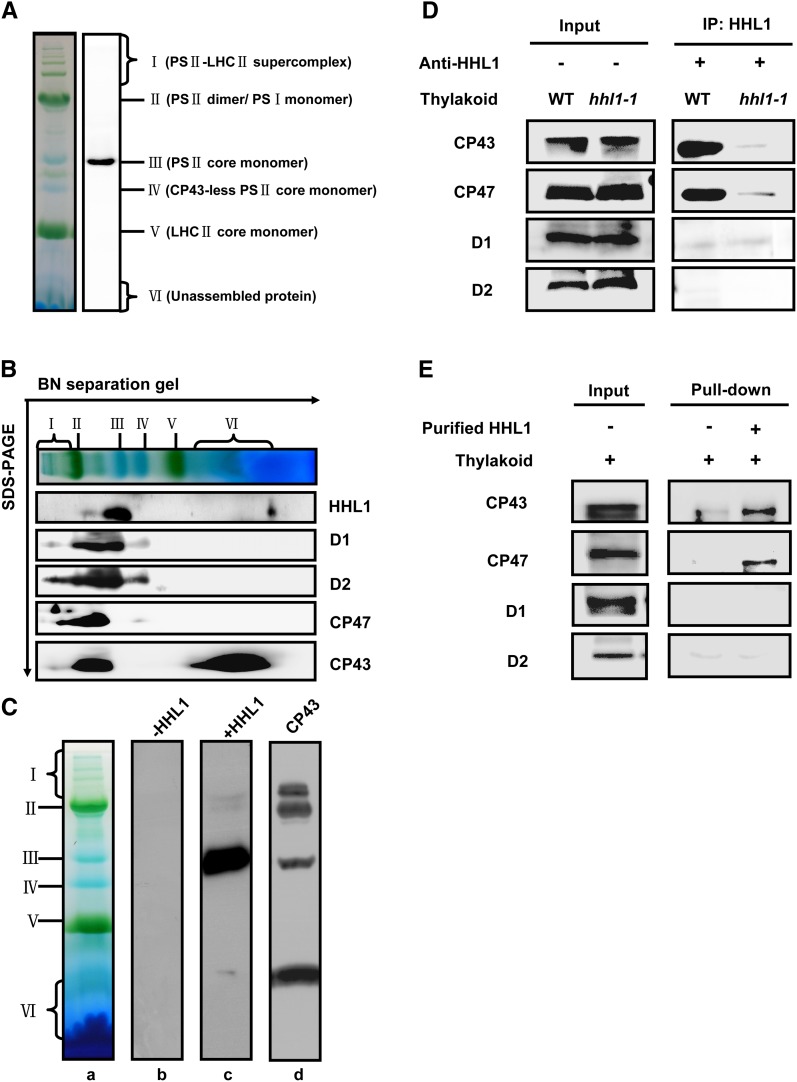
The HHL1 Protein Associates with PSII Complex
The reduction in PSII activity and the increased accumulation of ROS in high-light-treated hhl1 mutants suggest a possible role for HHL1 in protection of PSII from photodamage. To test whether HHL1 is directly involved in the protection and repair of PSII, we first asked whether HHL1 protein is directly associated with PSII complexes. HHL1 comigrates with PSII core monomer, as determined by Blue Native (BN) gel immunoblotting (Figure 5A). To further confirm the colocalization of HHL1 and PSII, we separated the proteins of the complexes by SDS-PAGE in the second dimension and immunoblotted the proteins with antibodies or antisera against HHL1 and PSII core proteins. The results further confirmed that HHL1 comigrates with PSII core monomer (Figure 5B). Interestingly, a small amount of HHL1 was also detected in the free protein fraction (Figure 5B), suggesting that the majority of HHL1 may associates with PSII monomer complex, but a small fraction exists as free protein.
Figure 5.
Association of HHL1 Protein with PSII Protein Complexes.
(A) Immunoblot analysis of HHL1 after separation by the first dimension of BN-PAGE.
(B) Immunoblot analysis of thylakoid proteins separated in the first dimension in a BN-PAGE gel followed by SDS-PAGE in the second dimension. The resolved proteins were immunodetected with anti-D1, anti-D2, anti-CP47, anti-CP43, and anti-HHL1 antibodies.
(C) Overlay assays of the interactions of HHL1 protein with the PSII complex. (a) Thylakoid membrane proteins were separated by BN-PAGE. (b) After blotting, the PVDF membrane without incubation with His-HHL1 protein was subjected to immunodetection with anti-His tag antibody. (c) After blotting, the PVDF membrane was incubated with His-HHL1 protein and subsequently immunodetected with anti-His tag antibody. (d) After blotting, the PVDF membrane was directly subjected to immunodetection with anti-CP43 tag antibody.
(D) Co-IP assay of the interaction between HHL1 and PSII core proteins D1, D2, CP43, and CP47 interaction. Solubilized thylakoid membranes from wild-type (Col-0) and hhl1 mutant leaves were incubated with Protein A/G–coupled anti-HHL1 antiserum. The immunoprecipitates were probed with specific antibodies, as indicated on the left.
(E) Pull-down assay of the interaction between HHL1 and PSII core proteins D1, D2, CP43, and CP47 interaction. Recombinant His-HHL1 bound to CNBr resin was incubated with solubilized thylakoid membranes, and the bound proteins eluted were analyzed immunobiochemically employing the antibodies as in (D). All experiments were repeated three times with similar results.
[See online article for color version of this figure.]
Overlay assays were performed to further confirm the direct association of HHL1 with PSII complexes. Chlorophyll-protein complexes were separated by BN-PAGE and blotted onto polyvinylidene fluoride (PVDF) membranes, which were then incubated with purified His-HHL1 fusion protein. Possible HHL1 binding proteins were subsequently detected by immunoblotting with a His-tag-specific antibody. Interestingly, in vitro, as well as primarily comigrating with PSII core monomers, HHL1 associated with other PSII complexes including the PSII-LHCII supercomplexes, PSII dimer complexes, and even free CP43. A negative control without His-HHL1 was included (Figure 5C). Taken together, these results suggest that HHL1 interacts with the PSII monomer complex.
Comigration of HHL1 with free CP43 in an overlay assay indicates a possible direct interaction between HHL1 and CP43. To investigate this possibility, we first examined the interaction of HHL1 with the subunits of PSII in vivo using coimmunoprecipitation (Co-IP). Interestingly, besides PSII core protein CP43, CP47 was also detected with 2% dodecyl β-d-maltoside (DM)–solubilized thylakoid membranes from the wild type, but not from hhl1 (Figure 5D). However, PSII core proteins D1 and D2 were not detected in repeated attempts of this assay (Figure 5D). These indirect or weak interactions may be unstable during the washing steps or long time scales of the experiments. To further confirm the interaction of HHL1 with PSII subunits CP43 and CP47, pull-down experiments were performed with recombinant N-terminal His-tagged HHL1 protein. Consistent with the results of Co-IP in vivo, PSII core proteins CP43 and CP47 were detected in this assay, but no signal was detected with just solubilized thylakoid membranes or the resin (Figure 5E). The PSII core proteins D1 and D2 were not detected in this assay either (Figure 5E). Taken together, these results suggest that HHL1 probably associates with PSII core complex by directly interacting with PSII core subunits CP43 and CP47 specifically, rather than with D1 and D2.
HHL1 Interacts with LQY1 in Vivo and in Vitro
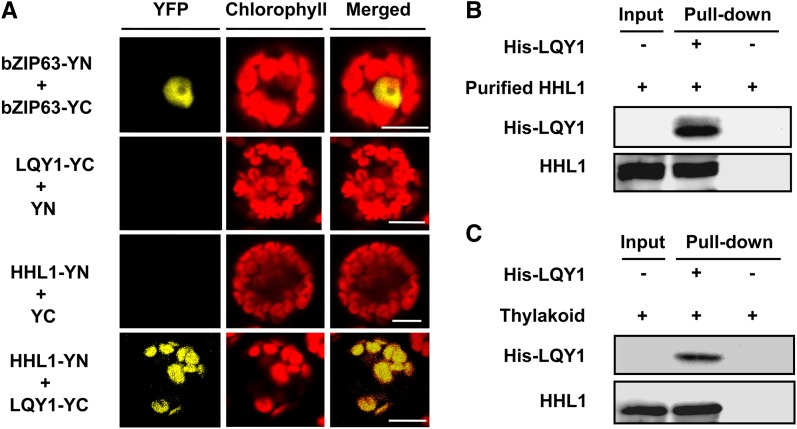
Recent studies have revealed that LQY1 also associates with the PSII, probably by directly interacting with CP43 and CP47, during PSII repair and reassembly following photodamage (Lu et al., 2011). The phenotype of high-light hypersensitivity observed in hhl1 mutants in this study is also surprisingly similar to that of lqy1 mutants (Lu et al., 2011). To determine whether HHL1 interacts with LQY1, we first performed bimolecular fluorescence complementation (BiFC) analysis using an Arabidopsis protoplast transient expression system. Coexpression of both nYFP-tagged HHL1 and cYFP-tagged LQY1 resulted in significant fluorescence in the chloroplasts of Arabidopsis protoplast cells (Figure 6A), suggesting that the HHL1 protein interacts with the LQY1 protein. No fluorescence was detected in Arabidopsis protoplast cells cotransformed with nYFP-HHL1 and cYFP, or nYFP and cYFP-LQY1 (Figure 6A). These results strongly indicate that HHL1 interacts with LQY1 in vivo.
Figure 6.
BiFC Visualization and Pull-Down Experiments Show Interaction between HHL1 and LQY1 in Vivo and in Vitro.
(A) BiFC analysis of Arabidopsis protoplasts showing the interaction between HHL1 and LQY1 in vivo. HHL1 fused with the N terminus of YFP (YN) and LQY1 fused with the C terminus of YFP (YC) were cotransfected into protoplasts and visualized using confocal microscopy. As a positive control, both bZIP663 fused with YN and bZIP663 fused with YC were cotransfected into protoplasts. As negative controls, HHL1 fused with YN and empty vector YC as well as LQY1 fused with YC and empty vector YN were cotransfected into protoplasts. Bars = 10 μm.
(B) and (C) Pull-down assays showing HHL1 interaction with LQY1 in vitro. His-LQY1 bound to CNBr-activated resin was incubated with recombinant HHL1 expressed in E. coli (B) and DM-solubilized thylakoid membranes (C). Bound proteins were eluted, separated by SDS-PAGE, and subjected to immunoblot analysis with His-tag and HHL1 antibodies. All experiments were repeated at least two times with similar results.
To further confirm the interaction between HHL1 and LQY1, a pull-down experiment was performed with recombinant N-terminal His-tagged LQY1 protein. His-HHL1 and His-LQY1 fusion proteins were first expressed in an Escherichia coli expression system (Supplemental Figure 4). Purified His-LQY1 was incubated with 2% DM-solubilized thylakoid membranes from wild-type Arabidopsis or recombinant HHL1 protein expressed in E. coli. After washing with the buffer, proteins bound to the cyanogen bromide (CNBr)–activated resin were separated by SDS-PAGE and examined by immunoblot analysis. As shown in Figures 6B and 6C, HHL1 protein was successfully pulled down by His-LQY1 in the presence of membranes or recombinant HHL1, and no signal was detected when only the solubilized thylakoid membrane, recombinant HHL1 protein, or resin was used. LQY1 could therefore pull down HHL1 from solubilized thylakoid membranes and recombinant HHL1 protein, further indicating that there is a direct interaction between HHL1 and LQY1 proteins in vitro. Collectively, these results suggest that HHL1 directly interacts with LQY1 in vivo and in vitro.
PSII Subunits Levels Are Perturbed in the hhl1 Mutants after High-Light Treatment
The above analysis showed that the hhl1 mutants have reduced photosynthetic efficiency after high-light treatment, suggesting HHL1 is involved in photoprotection against high-light stress. BiFC and pull-down analysis showed that HHL1 interacts directly with LQY1. Therefore, we speculate that HHL1, which may play a similar role in photoprotection to that of LQY1, is involved in maintaining PSII activity under high-light conditions by regulating the repair and reassembly of PSII complexes. To confirm this hypothesis, we first investigated the influence of high light on the transcription of photosynthesis-related genes in the absence of HHL1. Most photosynthesis-related genes were slightly downregulated after high-light treatment, including PsaA, PsbE, PsbA, PsbD, and PsbS, although stress-related genes ZAT10 and ZAT12 were significantly upregulated (Supplemental Table 2), further indicating that hhl1 mutants suffered more extensive photodamage than wild-type plants.
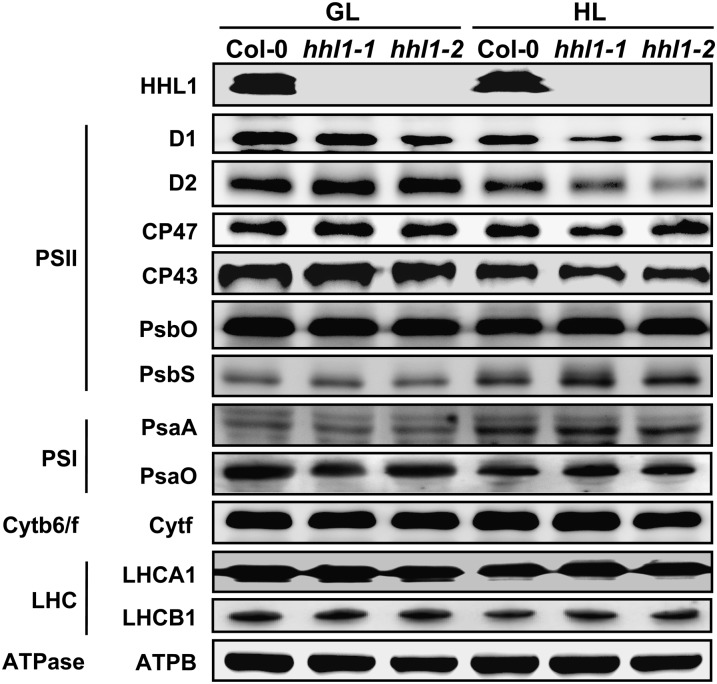
To explore the abundance of HHL1 in thylakoid membranes, titration of HHL1 protein abundance in thylakoid membranes was performed, referring to the well-characterized stoichiometry of LhcSR/PSII (0.17 ± 0.11) in high-light acclimated thylakoids (Bonente et al., 2011). The results showed that the chlorophyll/HHL1 ratio is 3616.5 ± 330.2 (Supplemental Figure 6). Based on a chlorophyll/PSII ratio of 440 (Pätsikkä et al., 2002), we calculated a HHL1/PSII ratio of 0.122 ± 0.01. The abundance of HHL1 protein in wild-type leaves increased by 31.76% after a 2-d high-light treatment (Figure 7; Supplemental Figure 7).
Figure 7.
Analysis of HHL1 and Thylakoid Membrane Protein Accumulation in the Wild Type and hhl1 Mutants.
Thylakoid membrane proteins from the wild type (Col-0) and hhl1 mutants were separated by 15% SDS-urea-PAGE, electroblotted to PVDF membranes, and probed with affinity-purified anti-HHL1 or antisera against known thylakoid membrane proteins obtained from Agrisera. Samples were loaded on an equal chlorophyll basis. Cytb6/f, cytochrome b6/f complex; ATPase, ATP synthase complex; ATPB, the β-subunit of ATP synthase complex; GL, growth light; HL, after 2-d high-light treatment. Similar results were obtained in four independent biological replicates.
To examine the accumulation of thylakoid proteins, thylakoid membranes were isolated from the wild type and hhl1 mutants before and after a 2-d high-light treatment, and immunoblot analysis was performed using antibodies raised against specific subunits of the photosynthetic thylakoid membrane protein complexes. After high-light treatment, marked reductions in the levels of the PSII core subunits D1, D2, CP43, and CP47 were detected in the hhl1 mutants, at levels ∼54.9, 76.5, 81.1, and 71.8% those of the wild type, respectively (Figure 7; Supplemental Figure 7). These values were calculated on an equal chlorophyll basis. However, levels of PsbO (another PSII protein), PSI proteins PsaA and PsaO, LHCII chlorophyll a/b binding proteins LHCA1 and LHCB1, ATP synthase subunit B, and cytochrome b6 were relatively stable in both wild-type and mutant plants after high-light treatment (Figure 7). Taken together, these results suggest that deficiency of HHL1 perturbs the relative levels of PSII core subunits after high-light treatment.
Absence of HHL1 Affects the Accumulation of PSII Complexes after High-Light Treatment
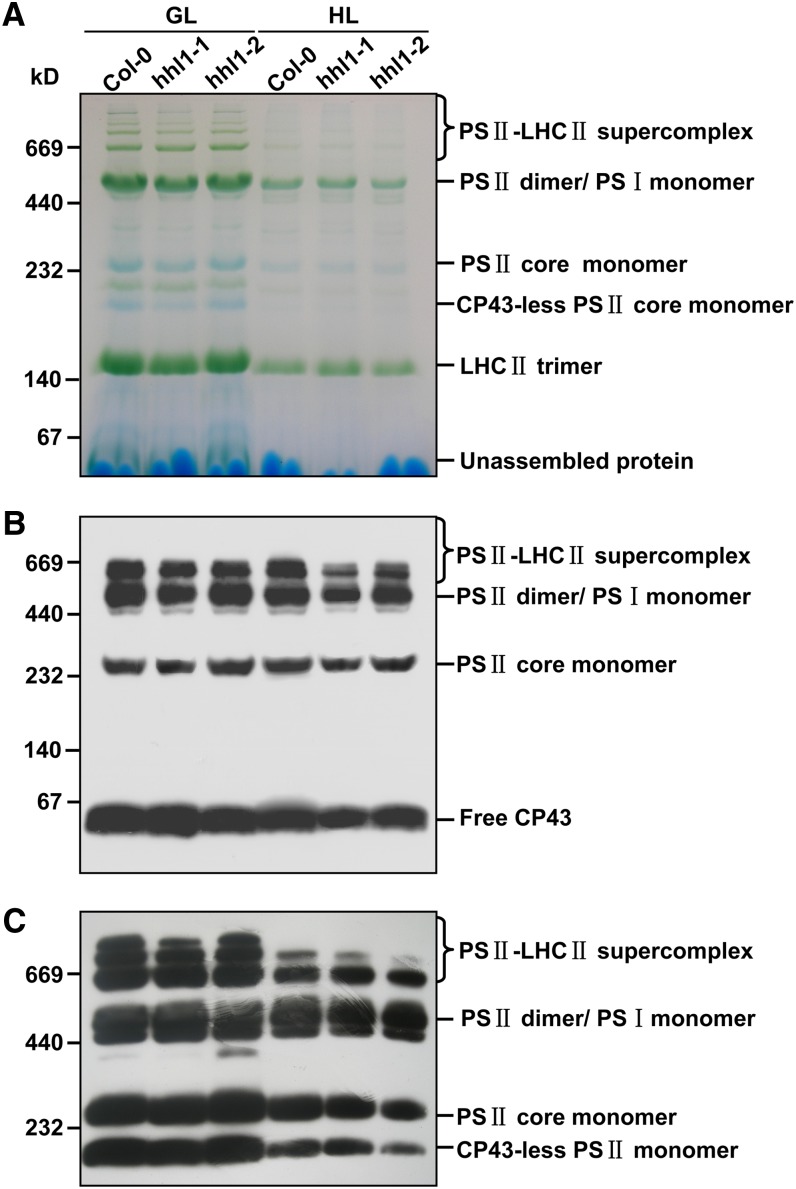
Under high irradiance, proteins in the PSII reaction center are damaged and undergo rapid repair and reassembly to enable photosynthetic electron transport to continue (Nickelsen and Rengstl, 2013). The defects in photosynthesis displayed by the hhl1 mutants were possibly caused by a reduced level or malfunction of protein complexes in the electron transport chain. To further investigate the effects of a lack of functional HHL1 on PSII structure and function, we analyzed the accumulation of various PSII complexes in wild-type and hhl1 plants. Thylakoid membranes were solubilized in 2% DM, membrane protein complexes were separated by BN-PAGE (Figure 8A), and the complexes were analyzed by immunoblotting with antibodies specific for PSII core proteins. Results with anti-CP43 antisera showed that hhl1 mutants contain less PSII-LHCII supercomplex than wild-type thylakoid membranes (Figure 8B). The reduction of the PSII-LHCII complex in the hhl1 mutants was further confirmed by immunoblot analysis with anti-D1 antisera (Figure 8C), suggesting that absence of HHL1 may affect the formation and stability of the PSII-LHCII supercomplex after high-light treatment.
Figure 8.
Immunological Analysis of PSII Complexes Separated by BN-PAGE.
(A) Representative unstained BN-PAGE gel. Thylakoid membranes from the wild type (Col-0) and hhl1 mutants were solubilized with 2% DM and separated by a native PAGE. A sample with an equal amount of chlorophyll (10 μg) was loaded in each lane. GL, growth light; HL, after 2-d high-light treatment.
(B) A representative immunoblot with anti-CP43 antiserum used to probe a BN-PAGE gel. An equal amount of chlorophyll (1.5 μg) was loaded in each lane.
(C) A representative immunoblot with anti-D1 antiserum used to probe a BN-PAGE gel as in (B). An equal amount of chlorophyll (1.5 μg) was loaded in each lane. All experiments were performed in three additional independent biological replicates, and similar results were obtained.
[See online article for color version of this figure.]
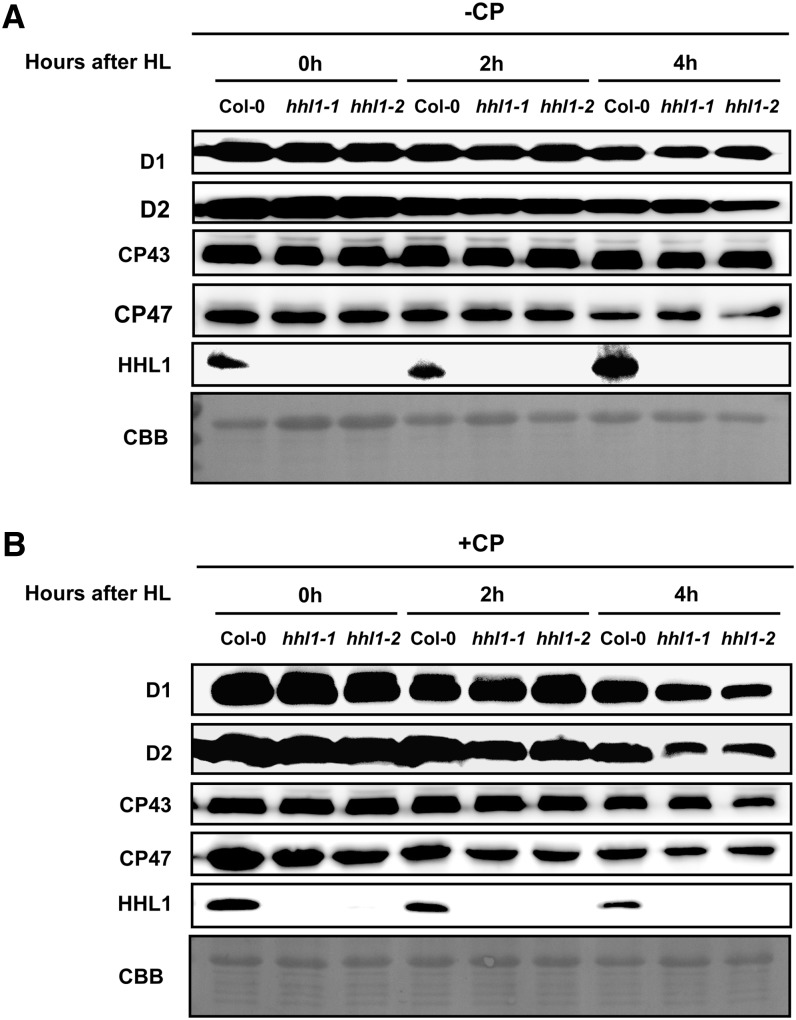
To study the stability of the assembled PSII complex, we treated leaves with chloramphenicol to block chloroplast-encoded protein synthesis and measured the PSII protein content by immunoblot analysis. In the absence of chloramphenicol, levels of PSII proteins D1, D2, CP43, and CP47 declined gradually in both wild-type and hhl1 mutant plants and showed no apparent difference after a short (0, 2, or 4 h) high-light treatment (Figure 9A). However, in the presence of chloramphenicol, the degradation rates of PSII core subunits D1, D2, CP43, and CP47 were more rapid in the hhl1 mutants, and degradation of D1 and D2 was more rapid than that of CP43 and CP47 (Figure 9B). This may be because D1 and D2 are more rapidly photodamaged and degraded by chloroplast proteases than CP43 and CP47 (Chi et al., 2012b).
Figure 9.
Degradation of HHL1 and PSII Proteins in the Wild Type and hhl1 Mutants under High-Light Treatment.
(A) Thylakoid membranes from hhl1 mutants and wild-type plants during high-light exposure in the absence of chloramphenicol (-CP) were isolated, and the contents of HHL1 and PSII proteins were determined through immunoblot analysis. CBB, Coomassie blue staining.
(B) Thylakoid membranes from hhl1 mutants and wild-type plants during exposure to high light in the presence of 200 μg mL−1 chloramphenicol (+CP) were isolated and analyzed as (A). Lanes in each gel were loaded on an equal chlorophyll basis. Similar results were obtained in three independent biological replicates.
More interestingly, the HHL1 abundance was rapidly increased by short-term high-light treatment in the absence of chloramphenicol but degraded following short-term high-light stress in the presence of chloramphenicol (Figure 9B), indicating that HHL1 protein is highly dynamic under high-light treatment. However, HHL1 transcription is not affected by high light (Supplemental Figure 8), which indicates that HHL1 is regulated at the posttranscriptional level. Taken together, these results suggest that the absence of HHL1 protein affects the reassembly and stability of PSII core proteins under high-light treatment.
The Homozygous hhl1 lqy1 Double Mutant Is More Sensitive to High Light
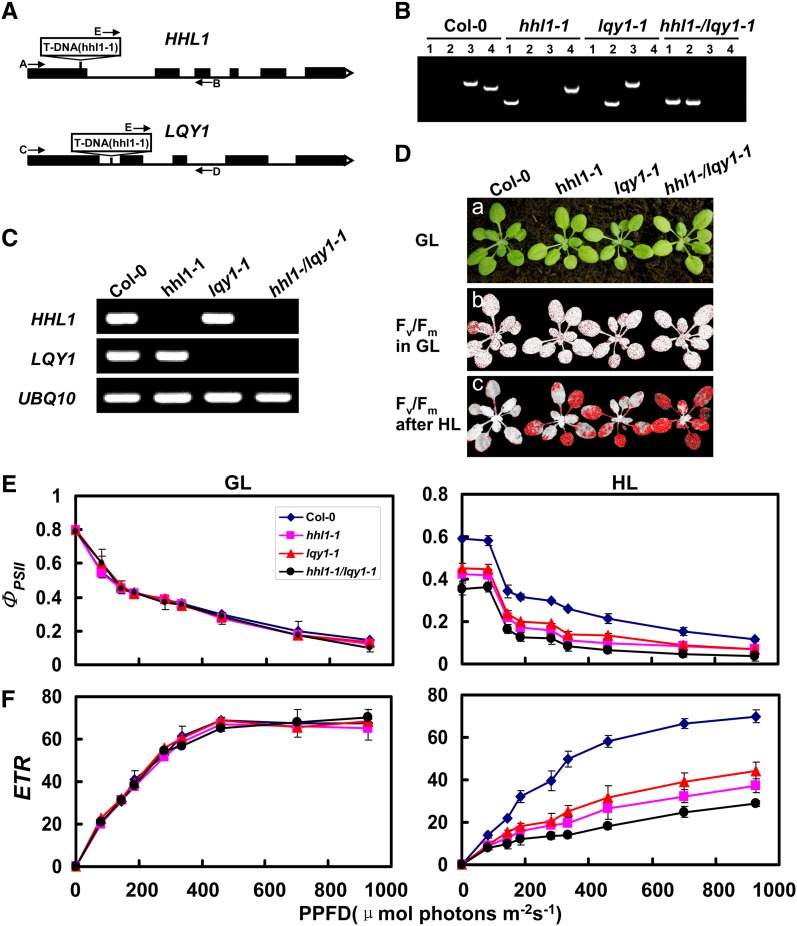
The above analysis showed that HHL1 has surprising similarities to HHL1, which is involved in PSII repair and reassembly, suggesting that HHL1 and LQY1 appear to share a functional redundancy or synergy in PSII repair and reassembly. To confirm this hypothesis, we next investigated the genetic relationship between HHL1 and LQY1. We first obtained the homozygous lqy1 mutant and generated hhl1 lqy1 double mutants by crossing (Figures 10A to 10C). In growth light conditions, the hhl1, lqy1, and hhl1 lqy1 double mutants showed no obvious differences from the wild type (Figure 10D). To compare PSII activity, we measured the maximum photochemical efficiency of PSII (Fv/Fm) before and after high-light treatment. The hhl1, lqy1, and hhl1 lqy1 double mutants showed a markedly lower Fv/Fm after a 3-h high-light treatment (∼1200 μmol photons m−2 s−1) than the wild type, even though the growth phenotype and Fv/Fm were identical under growth light conditions (∼100 μmol photons m−2 s−1). Interestingly, the Fv/Fm of the hhl1 lqy1 double mutant declined to a lower level (∼47%) than the single mutants (∼55 and 56%) (Figure 10D). However, Fv/Fm declined considerably (72%) in the wild type.
Figure 10.
Phenotypic Analyses of hhl1, lqy1, and hhl1 lqy1 Mutants and Wild-Type Plants.
(A) Schematic diagram of the HHL1 and LQY1 genes. Exons (black boxes) and introns (lines) are indicated. The positions of the T-DNA insertions corresponding to hhl1-1 and lqy1-1 are shown. Arrows indicate the location of primers used for PCR analyses.
(B) PCR analysis of genomic DNA from the wild type and hhl1, lqy1, and hhl1 lqy1 plants confirm the homozygosity of the mutants. Lane 1, amplification with primers E and B; lane 2, amplification with primers E and D; lane 3, amplification with primers A and B; lane 4, amplification with primers C and D.
(C) RT-PCR analysis of HHL1 and LQY1 gene expression, performed with the specific primers for HHL1, LQY1, and UBQ10 genes.
(D) Images in (a) are of 3-week-old wild-type (Col-0), hhl1-1, lqy1-1, and hhl1 lqy1 plants under a growth light (GL) conditions. (b) False-color images representing Fv/Fm under growth light conditions in 3-week-old wild-type, hhl1-1, lqy1-1, and hhl1 lqy1 plants. Red pixels indicate that Fv/Fm is below the cutoff value (0.753). (c) False-color images representing Fv/Fm after a 3-h high-light (HL) treatment in 3-week-old wild-type, hhl1-1, lqy1-1, and hhl1 lqy1 plants. Red pixels indicate that Fv/Fm is below the cutoff value (0.439).
(E) and (F) Light-response curves of PSII quantum yield (E) and ETR (F) in wild-type (Col-0), hhl1-1, lqy1-1, and hhl1 lqy1 plants in growth light (GL) and after 3-h high-light (HL) treatment. Data for wild-type (Col-0), hhl1-1, lqy1-1, and hhl1 lqy1 plants are presented as mean ± se. Four biological replicates were performed, and similar results were obtained.
The further analysis of the light-response curves of ΦPSII and the ETR before and after high-light treatment were also performed. After a 3-h high-light treatment, both ΦPSII and ETR in the hhl1 lqy1 double mutant were much lower significantly than in the single mutants. Taken together, detailed chlorophyll fluorescence analyses showed that the homozygous hhl1 lqy1 double mutant is more sensitive to high light than the single mutants, suggesting that HHL1 and LQY1 possess functional redundancy or synergy in PSII repair and reassembly.
HHL1 Homologs Are Found Exclusively in Land Plants
LQY1 homologs are present in land plants but are absent in the genomes of green algae and cyanobacteria (Lu, 2011). Based on the phenotypic similarity of HHL1 to LQY1 in terms of PSII repair, we set out to determine whether HHL1 is evolutionarily conserved in other plants, as was shown for LQY1 and as would be expected for a protein involved in the maintenance of PSII function. HHL1 homologs in other photosynthetic species were searched for using BLASTP with the full-length HHL1 sequence, and homologs were identified in many land plants, including the bryophyte moss Physcomitrella patens, the lycopod Selaginella moellendorffii, gymnosperm Picea stichensis, the dicotyledons Populus trichocarpa, grape (Vitis vinifera), cucumber (Cucumis sativus), castor bean (Ricinus communis), soybean (Glycine max), Medicago truncatula, Fragaria ananassa, Prunus trichocarpa, and tomato (Solanum lycopersicum), and the monocots rice (Oryza sativa), maize (Zea mays), barley (Hordeum vulgare), and Aegilops tauschii. However, HHL1 homologs were not found in more primitive photosynthetic organisms, such as cyanobacteria and algae (Supplemental Figures 9 and 10). The evolutionary distribution of HHL1 is therefore also similar to that of LQY1, suggesting that the role of HHL1 may be conserved among other higher land plants. This mode of evolution may reflect the adaptation of plants to excess light stress during the transition to land by evolution of a set of specific auxiliary factors to protect the photosynthetic apparatus.
DISCUSSION
PSII repair requires the sequential disassembly and reassembly of PSII proteins and the ligation of various cofactors, and many additional auxiliary and regulatory factors are needed to facilitate this complex, multistep process in chloroplasts (Mulo et al., 2008; Nixon et al., 2010; Nickelsen and Rengstl, 2013). In this study, we established that the nuclear-encoded thylakoid transmembrane protein HHL1 plays an important role in maintaining PSII activity, especially under high-light stress.
HHL1 Is a Thylakoid Protein Involved in PSII Repair
Chlorophyll fluorescence measurements showed that mutation of HHL1 significantly decreased the activity of PSII after high-light treatment (Figures 1D and 2A). Recovery rate following photoinhibition was slower in hhl1 than the wild type, suggesting HHL1 may be involved in repairing photodamage to PSII (Figure 2C). Furthermore, we found that 1-qP and 1-qL were also lower in the hhl1 mutants following high-light treatment (Supplemental Table 1), suggesting that there is a more highly oxidized plastoquinone pool in the hhl1 mutants, a response more likely due to a PSII deficiency rather than to downstream defects (Fu et al., 2007). This hypothesis was confirmed by analysis of thylakoid protein accumulation. Deficiency of HHL1 led to a reduction in the accumulation of PSII core subunits after high-light treatment, but accumulation of PSI, Cytb6/f, LHC, or ATPase complexes was not affected (Figure 7), further supporting this hypothesis. Deficiency of HHL1 specifically affected the accumulation of the PSII-LHCII complex (Figure 8), indicating that HHL1 is involved in PSII repair and reassembly.
There are several lines of evidence suggesting that deficiency of HHL1 results in more extensive photooxidative damage and photoinhibition in the chloroplast. First, the mutants had higher qI and accumulation of ROS following high-light treatment (Figure 3). Expression of zinc-finger transcription factors ZAT10 and ZAT12 are activated by high-light-induced oxidative stress (Iida et al., 2000; Gadjev et al., 2006), and these stress-related genes were upregulated more in hhl1 mutants in this study (Supplemental Table 2), indicating greater photooxidative damage and photoinhibition in the mutants. Additionally, in growth light, the hhl1 mutants possessed normal carotenoid levels but higher NPQ than the wild type, indicating that a deficiency in HHL1 dose not directly affect the carotenoid components, and the higher NPQ observed in the mutants under this condition was not due to an alteration of carotenoid components. However after high-light treatment, photooxidative damage resulted in a faster xanthophyll cycle in mutant plants, as shown by increased antheraxanthin and decreased violaxanthin (Supplemental Figure 2), as well as higher qE (Figure 2B), compensating for the defect of PSII repair pathway by dissipating more excess excitation energy as a feedback regulation mechanism. The higher NPQ may be due, at least in part, to the faster xanthophyll cycle in the hhl1 mutants. Finally, extensive photooxidative damage in the mutants also downregulated many photosynthesis-associated genes (Supplemental Table 2) and caused more rapid degradation of PSII core subunits (Figure 9).
The Role of HHL1 in PSII Repair
The repair of photodamaged PSII complexes is a complicated process involving multiple steps, including monomerization, partial disassembly of the dimeric complexes in grana stacks, migration of the PSII monomers to the stroma-exposed thylakoids, partial disassembly of PSII core monomers, release of CP43 into the CP43-less monomer, and proteolysis of the damaged D1 protein. After cotranslational insertion of the newly synthesized D1 subunit into the membrane, D1 is posttranslationally processed, and PSII monomers are reassembled and trafficked to the grana thylakoids, where dimerization takes place (Nixon et al., 2010).
To explore the possible role of HHL1 in PSII repair, we first determined that HHL1 is a thylakoid transmembrane protein (Figures 4B and 4C), as expected given the chloroplast signal sequence and transmembrane domain in this protein (Supplemental Figure 10). PSII repair involves migration of PSII complex from grana to stroma thylakoids (Mulo et al., 2008), and the distribution of HHL1 in both grana and stroma thylakoids is also compatible with a role in PSII repair and reassembly (Figure 4D).
BN-PAGE and two-dimensional electrophoresis analysis showed that HHL1 associates with PSII monomers (Figures 5A and 5B), and this was confirmed by overlay assay in vitro (Figure 5C). HHL1 shares obvious similarities with CYP38 and LQY1, which also associate with the PSII monomer complex and function in the assembly and maintenance of PSII (Sirpiö et al., 2008; Lu et al., 2011). Overlay assay also showed that HHL1 interacts with PSII-bound CP43 and with free CP43 (Figure 5C). We speculate that HHL1 associates with PSII core monomers after disassembly of photodamaged PSII-LHCII supercomplexes and PSII core dimers into PSII core monomers. Given that HHL1 simultaneously dissociates from PSII core monomer with CP43, we predict that HHL1 may function in guiding the release of CP43 from PSII core monomers.
This hypothesis was supported by Co-IP and pull-down studies that showed a direct interaction between HHL1 and CP43 in vivo and in vitro. Interestingly, in addition to interacting with CP43, HHL1 also specifically interacts with PSII core subunits CP47, but not with D1 or D2 (Figures 5D and 5E). The reaction center of PSII contains the D1/D2/cytochrome b559 complex, which houses the cofactors necessary for electron transport, which together with the nearest light-harvesting chlorophyll binding proteins CP43 and CP47, form the core complex (Ferreira et al., 2004). Therefore, HHL1 may interact with CP47 and CP43 specifically to help form the binding site(s) for these extrinsic proteins to PSII (Bricker and Frankel, 2002). Database searches revealed that the C-terminal region of the HHL1 protein includes one incomplete VWA domain, which are known to mediate protein–protein interactions in integrins and extracellular matrix proteins (Hynes and Zhao, 2000). The roles of VWA domains in plants is unclear but are often involved in forming multiprotein complexes (Fu et al., 2001; Lin et al., 2011), and the VWA domain of HHL1 may mediate protein–protein interactions involved in the assembly or functioning of PSII complexes.
HHL1 and LQY1 May Form a Complex Involved in PSII Repair Specifically
Despite evidence supporting the role of HHL1 in efficient PSII repair and reassembly of photodamaged PSII proteins, HHL1 appears not to be absolutely essential for PSII core repair, suggesting functional redundancy. To identify other potential PSII repair factors, we screened potential proteins that interact with HHL1-interacting proteins by comparing the precise locations of proteins in the chloroplast and the phenotypes of the mutants. Inactivation of certain factors affected the repair cycle after PSII photodamage in Arabidopsis, including thylakoid lumen protein of 18.3 kD (TLP18.3) (Sirpiö et al., 2007), the zinc-finger protein LQY1 (Lu et al., 2011), and the PsbP-Like Protein PPL1 (Ishihara et al., 2007). However, it has also been reported that TLP18.3 and PPL1 are not associated with PSII complexes (Ishihara et al., 2007; Sirpiö et al., 2007). Similarly, we found no interaction of HHL1 with TLP18.3 or PPL1 using BiFC (Supplemental Figure 5).
We noticed a striking functional similarity between HHL1 and LQY1, a small zinc- finger thylakoid protein reportedly involved in maintaining PSII activity under high light by regulating repair and reassembly of PSII complexes (Lu et al., 2011). Both lqy1 and hhl1 mutant plants showed enhanced sensitivity to high light stress, and both suffered extensive photooxidative damage and photoinhibition. Moreover, the accumulation of PSII core subunits and PSII-LHCII complexes significantly decreased in these mutants. LQY1 or HHL1 deficiency also resulted in faster turnover or degradation of PSII core subunits (Figures 2, 3, 8, and 9) (Lu et al., 2011). However, deficiency of either HHL1 or LQY1 resulted in a relatively mild and reversible phenotype. In stark contrast, the hhl1 lqy1 double mutant showed slightly higher increased photosensitivity compared with hhl1 and lqy1 single mutants (Figure 10), which is reminiscent of the photosensitive phenotype of the deg5 deg8 double mutant (Sun et al., 2007). This suggests functional redundancy or synergy for HHL1 and LQY1 in PSII repair and reassembly.
To explore the molecular relationship between HHL1 and LQY1, we first analyzed whether HHL1 and PSII were in the same complex. Immunoblotting of BN-SDS-PAGE and overlay analysis showed that HHL1 mainly associates with PSII monomeric complexes (Figures 5A to 5C) and specifically interacts with PSII core proteins CP43 and CP47 (Figures 5D and 5E). LQY1 also comigrates with PSII monomers, and Co-IP results suggest that LQY1 also directly interacts with CP43 and CP47 (Lu et al., 2011). These observations indicate that HHL1 and LQY1 may form a complex that functions in a similar process of PSII repair. As suspected, BiFC and pull-down experiments showed that HHL1 directly interacts with LQY1 in vivo (Figure 6). Phylogenetic analysis of HHL1 protein showed that HHL1 is specific to land plants (Supplemental Figures 9 and 10), which is also consistent with the mode of evolution described previously for LQY1 (Lu, 2011). Taken together, our results suggest that HHL1 and LQY1 may form a complex specifically involved in PSII repair and reassembly that has evolved to protect the sensitive photosynthetic machinery of land plants.
METHODS
Plant Materials and Growth Conditions
Mutant pools used for screening were obtained from RIKEN (Ichikawa et al., 2006). T-DNA and transgenic Arabidopsis thaliana lines used in this study were in the Col-0 background. The hhl1-1, hhl1-2, and lqy1-1 mutants were obtained from the ABRC (stock numbers SALK_129146C, SALK_010366C, and SALK_021824C). Arabidopsis was grown in soil in a growth chamber (100 μmol photons m−2 s−1, 12-h/12-h photoperiod, 21°C, and 60% relative humidity). Plants for the chlorophyll fluorescence assays and protein analysis were 3 and 4 weeks old, respectively. To study the effects of high light, plants were placed in a high-light growth chamber (1200 to 1500 μmol photons m−2 s−1) while maintaining the 12-h-light/12-h-dark cycle.
Pigment Analysis
Chlorophyll from 3-week-old plants was extracted with 80% acetone in 2.5 mM HEPES-KOH, pH 7.5, and the amount of chlorophyll was determined as previously described (Wellburn, 1994). Carotenoids were extracted and analyzed as previously described using spectrofluorometry (Yang et al., 2012) and HPLC (Pogson et al., 1996; Li et al., 2009b). Pigments were identified by comparing retention times to references.
Chlorophyll Fluorescence
Chlorophyll fluorescence parameters were measured with the MAXI version of the Imaging-PAM M-Series chlorophyll fluorescence system (Heinz-Walz Instruments). Plants were dark-adapted for 30 min before measurements were made. NPQ and light-response curves were determined as described (Lu et al., 2011). For 1-qP and 1-qL measurements, the F0 and Fm values were initially determining followed by a 715-s delay, which was in turn followed by a 60-s actinic light treatment (100 μmol photons m−2 s−1), which involved 36 saturation pulses (2800 μmol photons m−2 s−1) applied at 20-s intervals. Excitation pressure (1-qP and 1-qL) was recorded.
In Situ Detection of ROS
In situ detection of H2O2, O2−, and 1O2 was performed using DAB, NBT, and SOSG reagent fluorometric probes (FluorChem Q Imaging System; excitation 504 nm and emission 525 nm) as previously described by Lu et al. (2011). Total ROS production was detected by 2’,7’-dichlorofluorescin diacetate fluorometric probes (FluorChem Q Imaging System; excitation 488 nm and emission 525 nm) as previously described (Pei et al., 2000).
Isolation of Thylakoid Membranes
Thylakoid membranes were prepared as previously described (Robinson and Yocum, 1980). Isolated thylakoid membranes were quantified based on total chlorophyll as described (Porra et al., 1989). Total proteins extracted from leaf thylakoid membrane preparations were prepared as described (Liu et al., 2012). Protein concentrations were determined using the Bio-Rad detergent-compatible colorimetric protein assay according to the manufacturer’s protocol (Bio-Rad).
Production of Anti-HHL1 Polyclonal Antibodies
Affinity-purified anti-HHL1 polyclonal antibodies were made by GenScript. A 15–amino acid peptide (corresponding to amino acids 212 to 225 of HHL1) with an additional N-terminal Cys residue, CSLNPGSDEKTEETS, was synthesized, conjugated with keyhole limpet hemocyanin, and used to raise antibodies against HHL1.
RT-PCR and Quantitative Real-Time RT-PCR
Total RNA was extracted from Arabidopsis rosette leaves using an RNeasy plant mini kit (Qiagen). The RNA samples were reverse transcribed into first-strand cDNA using a PrimeScript RT reagent kit (Takara). For RT-PCR, UBQ10 was used as a control gene. Quantitative real-time RT-PCR was performed using gene-specific primers and SYBR Premix ExTaq reagent (Takara) with a real-time RT-PCR system (RoChe-LC480) following the manufacturer’s instructions. Reactions were performed in triplicate for each sample, and expression levels were normalized against ACTIN and UBQ4 gene (for primers used for RT-PCR and quantitative real-time RT-PCR, see Supplemental Table 3).
BN-SDS-PAGE and Immunoblot Analyses
BN-PAGE was performed as described (Schägger et al., 1994) with the modifications described (Peng et al., 2006). Two-dimensional analysis and immunodetection of proteins on a PVDF membrane were performed as described by Lu et al. (2011). For quantification of thylakoid proteins, gels were loaded on an equivalent chlorophyll basis, in amounts ensuring immunodetection was in the linear range. Except for anti-His (which was raised in mouse), all other primary antibodies and antisera were raised in rabbits. Antisera against photosynthetic proteins were purchased from Agrisera.
Protein Overlay Assays
Protein overlay assays were performed as described (Liu et al., 2012).
Subcellular Localization of GFP Fusions and BiFC
Subcellular localization of GFP fusion proteins and BiFC was performed as previously described (Zhang et al., 2011) (for primers used for the fusion constructs, see Supplemental Table 3).
Chloroplast Fractionation and Immunolocalization Studies
Separation of thylakoid and stroma phases and salt washing of thylakoids were performed as described (Armbruster et al., 2010). Subfractionation of grana core-, grana margin-, and stroma lamellae-enriched thylakoids was performed as described by Lu et al. (2011).
Analysis of Protein Degradation
Analysis of protein degradation was performed as described by Ishihara et al. (2007) with minor modifications. Detached leaves, placed adaxial side up on filter papers soaked in sodium phosphate buffer (NaOH/pH 7.0), were illuminated at 1200 μmol photons m−2 s−1. Detached leaves were incubated with the same buffer containing 200 μg mL−1 chloramphenicol under reduced pressure for 30 min prior to photoinhibitory light treatment. To analyze the accumulation of PSII proteins under high-intensity light, proteins were extracted from leaves ground in liquid nitrogen and used for SDS-PAGE and subsequent immunoblotting against PSII protein antibodies.
Immunoprecipitation
Immunoprecipitation of PSII core subunits by anti-HHL1 antibody was performed as described (Klostermann et al., 2002) with minor modifications. Thylakoid membrane proteins were solubilized with 2% (w/v) DM in 20% (w/v) glycerol, 25 mM BisTris-HCl, pH 7.0, and 1 mM PMSF for 20 min at 4°C. After centrifugation, the supernatant was diluted in an equal volume of the same buffer without DM and preincubated with affinity-purified anti-HHL1 antibody at 4°C for 2 h. Preincubated thylakoid membrane proteins (200 mL, 0.5 mg chlorophyll/mL) were incubated at 4°C overnight with Protein A/G Plus agarose in Pierce spin columns. After incubation overnight with constant rotation at 4°C, the resin was washed five times with ice-cold PBS buffer, pH 7.8, and bound proteins were eluted with SDS-PAGE sample buffer, resolved by SDS-PAGE, and subjected to immunoblot analysis.
Pull-Down Assays
One milligram of each recombinant fusion protein was incubated with 1 mL of a 50% suspension (v/v) of CNBr resin in equilibration buffer for 60 min at 4°C. After the thylakoid membranes (100 mg chlorophyll) were solubilized in 2% (w/v) DM in 20% glycerol (w/v), 25 mM BisTris-HCl, pH 7.0, and 1 mM PMSF for 15 min at 4°C, they were centrifuged at 10,000g for 10 min, and supernatant or purified protein was incubated with His-HHL1or His-LQY1 overnight with constant rotation at 4°C. After washing five times with ice-cold 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM EDTA buffer, bound proteins were eluted with SDS-PAGE sample buffer resolved by SDS-PAGE and subjected to immunoblot analyses.
Accession Numbers
Amino acid sequence data for HHL1 and its homologs in other species can be found in the GenBank/EMBL databases under the following accession numbers: Arabidopsis, NP_564903; Physcomitrella patens, XP_001765741; Selaginella moellendorffii, XP_002983221; Picea stichensis, ABK23133; Populus trichocarpa, XP_002314565; grape (Vitis vinifera), XP_002272215; cucumber (Cucumis sativus), XP_004142951; castor bean (Ricinus communis), XP_002515011; soybean (Glycine max), NP_001237795; Medicago truncatula, XP_003601843; Fragaria ananassa, XP_004310022; Prunus trichocarpa, EMJ25099; tomato (Solanum lycopersicum), XP_004239174; rice (Oryza sativa), EEC73537; maize (Zea mays), NP_001141155; barley (Hordeum vulgare), BAJ85479; and Aegilops tauschii, EMT22924. The photosynthesis and stress-related gene accession numbers are as follows: LQY1 (At1g75690), PsbA (AtCg00020), PsbD (AtCg00270), PsbO (At3g50820), PsbS (At1g44575), PsaA (AtCg00350), PsaE (At2g20260), LHCB1 (At1g29910), ATPB (AtCg00480), YCF2 (AtCg00860), ELIP2 (At4g14690), ZAT10 (At1g27730), and ZAT12 (At5g59820).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Isolation of hhl1-0 Mutant.
Supplemental Figure 2. HPLC Analysis of Pigments in the Wild-Type (Col-0), hhl1-1, and hhl1-2 Plants.
Supplemental Figure 3. The Chlorophyll a/b Ratios of Grana Core-, Grana Margin-, Stroma Lamellae-Enriched, and Total Thylakoids.
Supplemental Figure 4. Expression and Purification of HHL1 and LQY1.
Supplemental Figure 5. BiFC Analyses Show HHL1 Interacts with LQY1, but Not with PPL1 or TLP18.3.
Supplemental Figure 6. Example of Titration Used for Stoichiometry Determination.
Supplemental Figure 7. Relative Abundance of HHL1 and PSII Proteins.
Supplemental Figure 8. Transcript Analysis of HHL1 Gene under High-Light Treatment.
Supplemental Figure 9. Phylogenetic Analysis of HHL1 Protein and Its Homologs in Other Land Plants.
Supplemental Figure 10. Alignment Analysis of HHL1 Protein in Other Land Plants.
Supplemental Table 1. Chlorophyll Contents and Chlorophyll Fluorescence Parameters in Wild-Type (Col-0) and hhl1 Mutants.
Supplemental Table 2. Transcript Analysis of Wild-Type (Col-0) and hhl1 Mutants.
Supplemental Table 3. A List of Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 9.
Supplementary Material
Acknowledgments
We thank the ABRC and RIKEN for providing plant materials. We thank Peijie Han (Chinese Academy of Sciences) for the phylogenetic and homolog analysis. We thank Songguang Yang (South China Institute of Botany, Chinese Academy of Sciences) for assistance with HPLC analysis. We thank Yan Lu (Western Michigan University) for technical support and critical reading of the article. This research was supported by the grants from the National Natural Science Foundation of China (31171173 and 31371237), Guangdong Provincial Natural Science Foundation of China (S2012010010533), and the Fundamental Research Funds for the Central Universities (10lgpy34).
AUTHOR CONTRIBUTIONS
H.-B.W. and H.J. designed the study. H.J. and L.L. performed research. H.J., J.W., H.-B.W., P.W., and J.L. analyzed data. H.-B.W. and H.J. wrote the article. B.L., D.F., Q.D., Y.H., and K.Q. revised the article. All authors read and approved the final article.
Glossary
- PSII
photosystem II
- PSI
photosystem I
- ROS
reactive oxygen species
- qE
energy-dependent quenching
- LHCII
light-harvesting complex II
- Fv/Fm
ratio of variable fluorescence/maximal fluorescence
- Col-0
Columbia-0
- NPQ
nonphotochemical quenching
- F0
minimum fluorescence
- Fm
maximum fluorescence
- qI
photoinhibitory quenching
- ETR
electron transport rate
- DAB
diaminobenzidine
- NBT
nitroblue tetrazolium
- SOSG
singlet oxygen sensor green
- VWA
Von Willebrand factor type A
- BN
Blue Native
- PVDF
polyvinylidene fluoride
- Co-IP
coimmunoprecipitation
- DM
dodecyl β-d-maltoside
- BiFC
bimolecular fluorescence complementation
- CNBr
cyanogen bromide
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Armbruster U., Zühlke J., Rengstl B., Kreller R., Makarenko E., Rühle T., Schünemann D., Jahns P., Weisshaar B., Nickelsen J., Leister D. (2010). The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro E.M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., Rintamäki E. (2005). Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56: 347–356. [DOI] [PubMed] [Google Scholar]
- Baena-González E., Aro E.M. (2002). Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 1451–1459, discussion 1459–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.R. (2008). Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59: 89–113. [DOI] [PubMed] [Google Scholar]
- Bonente G., Ballottari M., Truong T.B., Morosinotto T., Ahn T.K., Fleming G.R., Niyogi K.K., Bassi R. (2011). Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 9: e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker T.M., Frankel L.K. (2002). The structure and function of CP47 and CP43 in photosystem II. Photosynth. Res. 72: 131–146. [DOI] [PubMed] [Google Scholar]
- Chi W., Ma J., Zhang L. (2012a). Regulatory factors for the assembly of thylakoid membrane protein complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 3420–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Sun X., Zhang L. (2012b). The roles of chloroplast proteases in the biogenesis and maintenance of photosystem II. Biochim. Biophys. Acta 1817: 239–246. [DOI] [PubMed] [Google Scholar]
- Dewez D., Park S., García-Cerdán J.G., Lindberg P., Melis A. (2009). Mechanism of REP27 protein action in the D1 protein turnover and photosystem II repair from photodamage. Plant Physiol. 151: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobáková M., Sobotka R., Tichý M., Komenda J. (2009). Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149: 1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S., Finazzi G., Wollman F.A. (2008). The dynamics of photosynthesis. Annu. Rev. Genet. 42: 463–515. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Ermakova-Gerdes S., Vermaas W. (1999). Inactivation of the open reading frame slr0399 in Synechocystis sp. PCC 6803 functionally complements mutations near the Q(A) niche of photosystem II. A possible role of Slr0399 as a chaperone for quinone binding. J. Biol. Chem. 274: 30540–30549. [DOI] [PubMed] [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838. [DOI] [PubMed] [Google Scholar]
- Ferro M., et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9: 1063–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A., He Z., Cho H.S., Lima A., Buchanan B.B., Luan S. (2007). A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15947–15952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Reis N., Lee Y., Glickman M.H., Vierstra R.D. (2001). Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20: 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V., Ossenbühl F., Crèvecoeur M., Eichacker L.A., Rochaix J.D. (2006). One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18: 1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N.E., Zigmantas D., Valkunas L., Li X.P., Niyogi K.K., Fleming G.R. (2005). Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307: 433–436. [DOI] [PubMed] [Google Scholar]
- Hynes R.O., Zhao Q. (2000). The evolution of cell adhesion. J. Cell Biol. 150: F89–F96. [DOI] [PubMed] [Google Scholar]
- Ichikawa T., et al. (2006). The FOX hunting system: An alternative gain-of-function gene hunting technique. Plant J. 48: 974–985. [DOI] [PubMed] [Google Scholar]
- Iida A., Kazuoka T., Torikai S., Kikuchi H., Oeda K. (2000). A zinc finger protein RHL41 mediates the light acclimatization response in Arabidopsis. Plant J. 24: 191–203. [DOI] [PubMed] [Google Scholar]
- Ishihara S., Takabayashi A., Ido K., Endo T., Ifuku K., Sato F. (2007). Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145: 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretić D., Zoranić L., Zucić D. (2002). Basic charge clusters and predictions of membrane protein topology. J. Chem. Inf. Comput. Sci. 42: 620–632. [DOI] [PubMed] [Google Scholar]
- Keren N., Ohkawa H., Welsh E.A., Liberton M., Pakrasi H.B. (2005). Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 17: 2768–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klostermann E., Droste Gen Helling I., Carde J.P., Schünemann D. (2002). The thylakoid membrane protein ALB3 associates with the cpSecY-translocase in Arabidopsis thaliana. Biochem. J. 368: 777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J., Nickelsen J., Tichý M., Prásil O., Eichacker L.A., Nixon P.J. (2008). The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283: 22390–22399. [DOI] [PubMed] [Google Scholar]
- Komenda J., Sobotka R., Nixon P.J. (2012). Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15: 245–251. [DOI] [PubMed] [Google Scholar]
- Kramer D.M., Johnson G., Kiirats O., Edwards G.E. (2004). New fluorescence parameters for the determination of q(a) redox state and excitation energy fluxes. Photosynth. Res. 79: 209–218. [DOI] [PubMed] [Google Scholar]
- Kufryk G.I., Vermaas W.F. (2001). A novel protein involved in the functional assembly of the oxygen-evolving complex of photosystem II in Synechocystis sp. PCC 6803. Biochemistry 40: 9247–9255. [DOI] [PubMed] [Google Scholar]
- Kufryk G.I., Vermaas W.F. (2003). Slr2013 is a novel protein regulating functional assembly of photosystem II in Synechocystis sp. strain PCC 6803. J. Bacteriol. 185: 6615–6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2012). SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 40: D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Ahn T.K., Avenson T.J., Ballottari M., Cruz J.A., Kramer D.M., Bassi R., Fleming G.R., Keasling J.D., Niyogi K.K. (2009b). Lutein accumulation in the absence of zeaxanthin restores nonphotochemical quenching in the Arabidopsis thaliana npq1 mutant. Plant Cell 21: 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009a). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260. [DOI] [PubMed] [Google Scholar]
- Lin Y.L., Sung S.C., Tsai H.L., Yu T.T., Radjacommare R., Usharani R., Fatimababy A.S., Lin H.Y., Wang Y.Y., Fu H. (2011). The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. Plant Cell 23: 2754–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yang H., Lu Q., Wen X., Chen F., Peng L., Zhang L., Lu C. (2012). PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell 24: 4992–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. (2011). The occurrence of a thylakoid-localized small zinc finger protein in land plants. Plant Signal. Behav. 6: 1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Hall D.A., Last R.L. (2011). A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23: 1861–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J., Samuelsson T., de Gier J.W. (2001). YidC/Oxa1p/Alb3: Evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 501: 1–5. [DOI] [PubMed] [Google Scholar]
- Ma J., Peng L., Guo J., Lu Q., Lu C., Zhang L. (2007). LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51: 659–668. [DOI] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Thomas P.D. (2013). PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41: D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P., Li X.P., Niyogi K.K. (2001). Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125: 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P., Sirpiö S., Suorsa M., Aro E.M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98: 489–501. [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Rengstl B. (2013). Photosystem II assembly: From cyanobacteria to plants. Annu. Rev. Plant Biol. 64: 609–635. [DOI] [PubMed] [Google Scholar]
- Nishiyama Y., Allakhverdiev S.I., Murata N. (2006). A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim. Biophys. Acta 1757: 742–749. [DOI] [PubMed] [Google Scholar]
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. (Lond.) 106: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi K.K., Grossman A.R., Björkman O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczyk M.M., Hebeler R., Schlodder E., Meyer H.E., Warscheid B., Rögner M. (2006). Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18: 3121–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Khamai P., Garcia-Cerdan J.G., Melis A. (2007). REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol. 143: 1547–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pätsikkä E., Kairavuo M., Sersen F., Aro E.M., Tyystjärvi E. (2002). Excess copper predisposes photosystem II to photoinhibition in vivo by outcompeting iron and causing decrease in leaf chlorophyll. Plant Physiol. 129: 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734. [DOI] [PubMed] [Google Scholar]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B., McDonald K.A., Truong M., Britton G., DellaPenna D. (1996). Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 8: 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C., Jaquinod M., Holzer F., Bourguignon J., Walling L., Brouquisse R. (2009). Evidence for the existence in Arabidopsis thaliana of the proteasome proteolytic pathway: Activation in response to cadmium. J. Biol. Chem. 284: 35412–35424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Robinson H.H., Yocum C.F. (1980). Cyclic photophosphorylation reactions catalyzed by ferredoxin, methyl viologen and anthraquinone sulfonate. Use of photochemical reactions to optimize redox poising. Biochim. Biophys. Acta 590: 97–106. [DOI] [PubMed] [Google Scholar]
- Sakata S., Mizusawa N., Kubota-Kawai H., Sakurai I., Wada H. (2013). Psb28 is involved in recovery of photosystem II at high temperature in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1827: 50–59. [DOI] [PubMed] [Google Scholar]
- Schägger H., Cramer W.A., von Jagow G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217: 220–230. [DOI] [PubMed] [Google Scholar]
- Schottkowski M., Gkalympoudis S., Tzekova N., Stelljes C., Schünemann D., Ankele E., Nickelsen J. (2009). Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 284: 1813–1819. [DOI] [PubMed] [Google Scholar]
- Sirpiö S., Allahverdiyeva Y., Suorsa M., Paakkarinen V., Vainonen J., Battchikova N., Aro E.M. (2007). TLP18.3, a novel thylakoid lumen protein regulating photosystem II repair cycle. Biochem. J. 406: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S., Khrouchtchova A., Allahverdiyeva Y., Hansson M., Fristedt R., Vener A.V., Scheller H.V., Jensen P.E., Haldrup A., Aro E.M. (2008). AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 55: 639–651. [DOI] [PubMed] [Google Scholar]
- Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., Zhang L. (2007). Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19: 1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Badger M.R. (2011). Photoprotection in plants: A new light on photosystem II damage. Trends Plant Sci. 16: 53–60. [DOI] [PubMed] [Google Scholar]
- Tomko R.J., Jr, Hochstrasser M. (2013). Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 82: 415–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn A.R. (1994). The spectral determination of chlorophyll a and chlorophyll b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144: 307–313. [Google Scholar]
- Whittaker C.A., Hynes R.O. (2002). Distribution and evolution of von Willebrand/integrin A domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13: 3369–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Zeng X., Li T., Liu M., Zhang S., Gao S., Wang Y., Peng C., Li L., Yang C. (2012). AtACDO1, an ABC1-like kinase gene, is involved in chlorophyll degradation and the response to photooxidative stress in Arabidopsis. J. Exp. Bot. 63: 3959–3973. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., Wang J., Wang H. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.