This work reveals the arrangement of rRNA genes, euchromatin, and heterochromatin in cycling and noncycling cells of evening primroses. The results suggest a model of chromosomal evolution, with translocations of chromosome end-segments and selection against structurally heterozygous bivalents as mechanisms that promote permanent multichromosomal rings at meiosis.
Abstract
Due to reciprocal chromosomal translocations, many species of Oenothera (evening primrose) form permanent multichromosomal meiotic rings. However, regular bivalent pairing is also observed. Chiasmata are restricted to chromosomal ends, which makes homologous recombination virtually undetectable. Genetic diversity is achieved by changing linkage relations of chromosomes in rings and bivalents via hybridization and reciprocal translocations. Although the structural prerequisite for this system is enigmatic, whole-arm translocations are widely assumed to be the mechanistic driving force. We demonstrate that this prerequisite is genome compartmentation into two epigenetically defined chromatin fractions. The first one facultatively condenses in cycling cells into chromocenters negative both for histone H3 dimethylated at lysine 4 and for C-banding, and forms huge condensed middle chromosome regions on prophase chromosomes. Remarkably, it decondenses in differentiating cells. The second fraction is euchromatin confined to distal chromosome segments, positive for histone H3 lysine 4 dimethylation and for histone H3 lysine 27 trimethylation. The end-segments are deprived of canonical telomeres but capped with constitutive heterochromatin. This genomic organization promotes translocation breakpoints between the two chromatin fractions, thus facilitating exchanges of end-segments. We challenge the whole-arm translocation hypothesis by demonstrating why reciprocal translocations of chromosomal end-segments should strongly promote meiotic rings and evolution toward permanent translocation heterozygosity. Reshuffled end-segments, each possessing a major crossover hot spot, can furthermore explain meiotic compatibility between genomes with different translocation histories.
INTRODUCTION
The plant genus Oenothera (evening primrose, Onagraceae, 2n = 2x = 14) is an easily adaptable and highly successful taxon. Due to reciprocal chromosomal translocations, it extensively forms multichromosomal rings at meiosis (Cleland, 1972; Golczyk et al., 2008; see Supplemental Figure 1 for details). Meiotic configurations can range from a single ring, where all chromosomes are involved in catenation, through intermediate forms with rings and bivalents, to solely bivalents (Supplemental Figure 1). Importantly, a drastic restriction of homologous recombination to chromosomal ends is observed throughout the genus: In Oenothera, chromosomal end-segments are the only sites throughout the genome where chiasmata are formed. The terminal chiasmata are obligatory for proper and regular meiosis I segregation (Cleland, 1972). However, techniques of classical and molecular genetics show unusually low levels of crossovers, allowing Oenothera genomes to be regarded as essentially nonrecombining (Rauwolf et al., 2008, 2011). Homologous recombination in this genus seems to be notoriously difficult to map, since (1) crossing-over events are focused only within a small very distal region of each of the chromosome arms, and (2) chromosomal ends in Oenothera are interpreted as almost free of genetic markers (Cleland, 1972). Thus, crossovers in Oenothera are genetically neutral. Therefore, genetic diversity in the system cannot be realized through homologous recombination but is achieved by changing linkage relations of chromosomes in rings and bivalents via hybridization and reciprocal translocations (Cleland, 1972; Harte, 1994; Rauwolf et al., 2008, 2011). The restriction of recombination to chromosomal ends throughout the genus suggests that it was an early evolutionary event. It might have been caused by some unknown ancient chromosomal changes affecting ancestral bivalent-forming species and facilitating the evolution of reciprocal translocations and, consequently, meiotic rings (Rauwolf et al., 2011). Within a meiotic ring, maternal and paternal chromosomes are arranged in alternate order. If all members of a given chromosome complement are involved in a single ring, they establish two regularly segregating sets (α and β) of genetically linked chromosomes, so-called Renner complexes. These complexes are moved by the meiotic spindle to the opposite poles as independent entities (Cleland, 1972; Supplemental Figure 1E). Within the Oenothera translocation system, permanent translocation heterozygosity (PTH) has evolved by switching to autogamy and acquisition of a special breeding behavior (including self-incompatibility alleles, gametophyte competition, balanced lethal factors, and selective fertilization). As a consequence, homozygotic (α·α or β·β) progeny is not produced. Thus, a permanently heterozygous (α·β) population structure results (Cleland, 1972; Charlesworth, 1979; Harte, 1994; Dietrich et al., 1997). PTH with some minor exceptions is based on autogamy; however, a certain level of outbreeding is needed to create new stable PTH forms. Hybridization events between distinct PTH translocation lineages allow the immediate and permanent fixation of new sets of features, which ensure a high success in colonizing new environments (Cleland, 1972; Dietrich et al., 1997). Consequently, the important attribute that is shared between PTH and non-PTH forms is the full meiotic compatibility of different translocation complexes (haploid genomes). Thus, when these complexes meet in the hybrid, they always experience regular meiotic segregation.
The nature of reciprocal translocations in Oenothera and the genomic features that allow their stable incorporation into the genetic system remain unclear. Commonly accepted is the hypothesis of Cleland (Cleland, 1972; Harte, 1994) that a prerequisite for reciprocal translocations in Oenothera are metacentric chromosomes of similar size with breakpoints in the centromeric heterochromatin. Within such a karyotype, whole-arm translocations would avoid deletions, maintain chromosomal morphology, and ensure segregation regularities of meiotic rings. However, it was also assumed that the genetic material residing at the middle chromosome regions is responsible for the physiological/genetic differences between Renner complexes (Cleland, 1972; Harte, 1994). It remains a puzzle how these differences are maintained, since whole-arm translocations together with widespread inter- and intraspecific hybridization should frequently grossly alter linkage relations within the middle chromosome regions by combining chromosome arms of different origin. This would lead to a genetic blend of Renner complexes, eliminating the widely observed genetic variation. On the other hand, translocation breakpoints are unlikely to be located anywhere between centromere and telomere, as was suggested by Darlington (1931). In such a scenario, unequal exchanges would lead to significant alterations in chromosome morphology, rendering preservation of regular meiotic segregation impossible (Cleland, 1972).
It is well known that the extent of both recombination and translocations is largely conditioned by the arrangement of heterochromatin and euchromatin. Constitutive heterochromatin is an impediment to crossing-over (Stack, 1984) but also a preferable site of breakpoints that trigger chromosome structural changes (Paço et al., 2013 and the literature therein). Unfortunately, little is known about the distribution of constitutive heterochromatin and euchromatin on Oenothera chromosomes. What is known is that (1) the 14 chromocenters can be detected consistently throughout interphase in meristematic cells (Marquardt, 1937), and (2) during prophase, a striking spatiotemporal genome compartmentation into early and late-condensing chromatin exists. The centromere-containing huge chromosome segments are already well condensed (early-condensing chromatin) at the very beginning of prophase, whereas distal regions condense late (late-condensing chromatin) and remain diffuse until metaphase (Kurabayashi et al., 1962). Based on results obtained with classical cytological techniques, the heteropycnotic centromere-containing regions of the condensing prophase chromosomes tended to be interpreted as the constitutive heterochromatin (Cleland, 1972). However, condensation patterns during prophase do not allow discrimination between eu- and heterochromatin using classical cytological techniques (discussed in Guerra, 1988). Thus, application of more specific techniques to fully condensed metaphase chromosomes is also necessary. Intriguingly, little is known about the presence of chromocenters in noncycling cells (Cleland, 1972). If the Oenothera heterochromatin were a constitutive element of the genome, chromocenters should also be commonly found in differentiated tissues (Nagl, 1979). Therefore, studies on chromocenter formation in diverse tissue contexts are required.
Bivalent- and ring-forming Oenothera species have been a subject of genetic research since the end of the 19th century, but mysteries of their chromatin and chromosome organization can now be unlocked through the application of molecular cytogenetic techniques. Here, we precisely map crucial cytogenetic landmarks on metaphase chromosomes in Oenothera, including constitutive heterochromatin, euchromatin, epigenetic histone modifications, 45S and 5S rRNA gene clusters, and monitor formation of chromocenters in diverse tissues. We demonstrate that the 14 large chromocenters detected in Oenothera meristematic cells are tissue-specific facultative heterochromatin, which indicates that the huge middle chromosome regions that condense into these chromocenters are unique and superb tools for surveys on large-scale chromatin remodeling during cellular differentiation in plants. We reveal how the astonishing spatiotemporal genome compartmentation in Oenothera is achieved and propose it as a key feature allowing a stable incorporation of translocations into the genetic system of Oenothera. Our results indicate that genome compartmentation in evening primrose should facilitate exchanges of the euchromatic end-segments rather than of the whole arms. This mode of chromosome alteration preserves the integrity of middle segments but gives them freedom to assemble diverse combinations with the end-segments. We demonstrate why translocations of end-segments and facultative type of heterochromatinization of the middle chromosome regions should promote meiotic rings and evolution of Renner complexes. Finally, we provide a simple model of chromosomal evolution in evening primrose, which concerns not only structural data but is also in accordance with population genetic modeling of PTH evolution.
RESULTS
Fluorescence in Situ Hybridization on Chromosomes
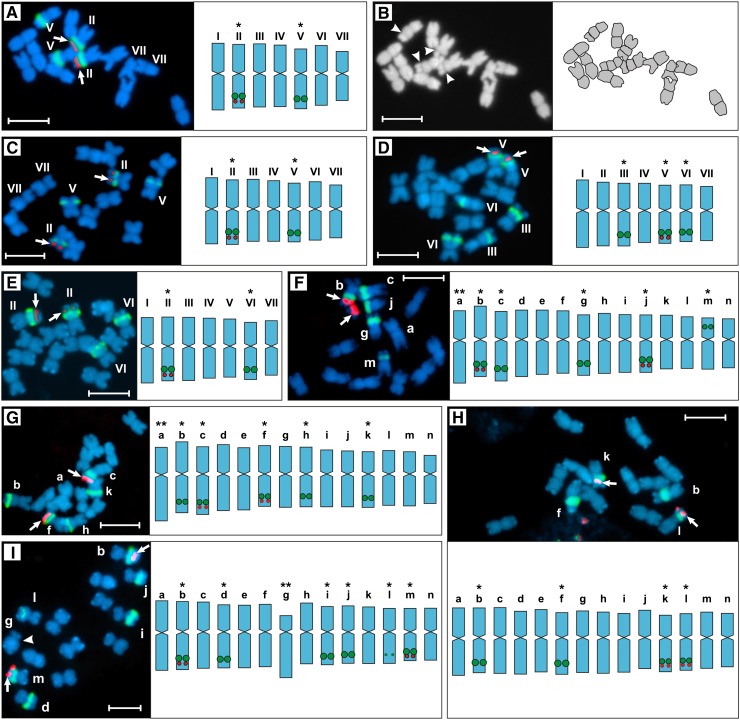
Fluorescence in situ hybridization (FISH) analysis revealed that all strains under study displayed two 5S and four to six 45S rDNA loci (Figure 1). The 45S rDNA sites are likely transcriptionally active, since they all were frequently accompanied by secondary constrictions (compare Figure 1A with Figure 1B). In agreement with our results, Bhaduri (1940), who studied 11 Oenothera taxa (some of the strains are identical to ours), found a constant presence of four secondary constrictions, illustrated also by Kurabayashi et al. (1962). Each of the two 5S rDNA loci was always entirely mounted within a satellite, adjacent to a 45S rDNA locus (Figures 1F, 1G, and 2B, bottom). Each of the basic A- or B-genomes (compared with Methods) of the three bivalent-forming lines (Oe. elata ssp hookeri strain hookeri de Vries, Oe. elata ssp hookeri strain johansen Standard, Oe. grandiflora strain grandiflora Tuscaloosa) is characterized by two (A-genomes: hhookeri de Vries and hjohansen Standard) or three (B-genome: hgrandiflora Tuscaloosa) 45S rDNA sites, respectively (Figures 1A, 1 C, and 1D). Accordingly, four or five 45S rDNA sites were found in AA (Oe. villosa ssp villosa strain bauri Standard) or AB (the two Oe. biennis strains) translocation heterozygotes (Figures 1F to 1H). Whether the sixth 45S rDNA site of Oe. glazioviana strain r/r-lamarckina Sweden (AB) (Figure 1I) reflects a new reciprocal translocation (see below) and/or is due to the homozygosity of chromosome r-1·2 (see Supplemental Table 1 and Supplemental Figure 1 for details on segmental arrangements), as a putative chromosome of the B-genome, remains to be explored. Importantly, we reveal that the alethal complex hblandina de Vries (Figure 1E) structurally closely resembles the A-genome of Oe. elata ssp hookeri strain hookeri de Vries (Figure 1A) or Oe. elata ssp hookeri strain johansen Standard (Figure 1C). Thus, our data strongly support classic genetic data of previous workers that the complex hblandina de Vries is closer to an A-genome than to a B-one (Cleland, 1972).
Figure 1.
Root-Tip Metaphases with Graphically Presented Chromosomal Types.
(A) and (C) to (I) Double-target FISH with 45S (green) and 5S rDNA (red, arrowed) as probes.
(A) to (E) Bivalent forming species.
(A) and (B) Oe. elata ssp hookeri strain hookeri de Vries.
(B) DAPI gray-scale image (left) of the metaphase plate shown in (A) and its graphical interpretation (right); arrowheads indicate secondary constrictions.
(C) Oe. elata ssp hookeri strain johansen Standard.
(D) Oe. grandiflora strain grandiflora Tuscaloosa.
(E) Oe. glazioviana strain blandina de Vries.
(F) to (I) Species that exhibit rings at meiosis.
(F) Oe. biennis strain suaveolens Grado.
(G) Oe. biennis strain suaveolens Standard.
(H) Oe. villosa ssp villosa strain bauri Standard.
(I) Oe. glazioviana strain r/r-lamarckiana Sweden.
I to VII, a to n, chromosome types distinguished on the basis of statistical calculations, i.e., their lengths and arm ratios are means from Supplemental Tables 2 and 3. Note that this designation does not imply chromosome homology/homoeology between strain or species. Single asterisks, chromosomal markers distinguished by their morphology and arrangement of rDNA loci; double asterisks, chromosomal markers based exclusively on morphology; arrowhead in I indicates the centromere of the chromosome “g” of Oe. glazioviana strain r/r-lamarckiana Sweden. Bars = 5 μm.
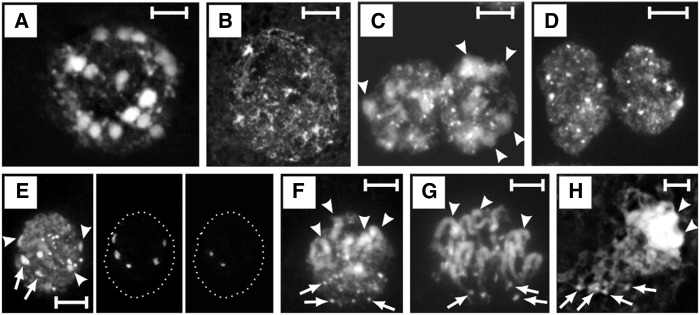
Figure 2.
The Organization of Chromatin States on Chromosomes and within Nuclei.
(A) Oe. elata ssp hookeri strain hookeri de Vries. Giemsa C-banding; arrowheads indicate C-band-positive NOR-heterochromatin.
(B) Oe. elata ssp hookeri strain hookeri de Vries. Sequential C-banding/DAPI (top) and FISH with 45S rDNA (green) and 5S rDNA (red, arrowed) probes (bottom) on early metaphase chromosomes. Note that the subtelomeric regions are still slightly diffuse; arrowheads indicate NOR-heterochromatin.
(C) Oe. villosa ssp villosa strain bauri Standard. C-banding/DAPI on late metaphase chromosomes; arrowheads indicate NOR-heterochromatin.
(D) Oe. biennis strain suaveolens Grado. Immunodetection of H3K9me2 (top, red) and DAPI-stained chromosomes (bottom).
(E) and (F) Oe. elata ssp hookeri strain johansen Standard. On metaphase chromosomes, euchromatin indexed by H3K4me2 (E) or H3K27me3 (F) occupies small distal regions, that is, satellites and segments comparable in size to satellites (merged images, red signals). Arrowheads indicate satellites; arrows indicate regions where NOR-heterochromatin is located.
(G) Oe. biennis strain suaveolens Standard. A fragment of DAPI-stained early prophase from root tip, disturbed by strong squashing. The relative position of centromere-containing early-condensing chromatin blocks (not marked), NOR-heterochromatin blocks (arrows, three of them are marked), and terminal heterochromatin (arrowheads, only two of them are marked) is clearly visible.
(H) to (J) Oe. elata ssp hookeri strain johansen Standard. Distribution of H3K4me2 (H) and H3K27me3 ([I] and [J]) on prophase chromosomes ([H] and [I]) or early metaphase chromosome (J). Until late metaphase, the late-condensing chromatin in Oenothera is not fully condensed. Top, DAPI images; bottom, immunosignals (red) merged with DAPI (blue). Arrowheads indicate DAPI-positive terminal heterochromatin.
(K) and (L) Oe. glazioviana strain r/r-lamarckiana Sweden. Immunodetection of H3K27me2 (red, merged images).
(K) Metaphase plate with H3K27me2-enriched chromosome termini.
(L) DAPI-bright terminal heterochromatin (DAPI staining, blue, top) strictly colocalizes with H3K27me2 immunosignals (red, merged, bottom).
(M) Oe. biennis strain suaveolens Grado. A metaphase chromosome (left, DAPI staining) with secondary constriction and its H3K27me2 pattern (right). Arrowhead indicates the satellite; arrow indicate region where NOR-heterochromatin is located.
(N) Oe. elata ssp hookeri strain hookeri de Vries. Nuclei stained with DAPI (left and right) or CMA3 (middle) with facultative chromocenters and minute blocks of terminal heterochromatin (arrowed in [M], left).
(O) Oe. biennis strain suaveolens Grado. Nucleus with facultative chromocenters (DAPI staining, top) that are negative for H3K4me2 (red, bottom).
(P) Oe. elata ssp hookeri strain johansen Standard. In the nuclei of mature parenchyma, facultative chromocenters are lacking (DAPI staining, top). DAPI-bright terminal heterochromatin is also detectable as spots enriched in H3K27me2 (red, bottom).
(Q) Oe. elata ssp hookeri strain hookeri de Vries. In nuclei from root epidermis, chromocenters are partially decondensed.
(R) Graphical interpretation of how the main chromatin states are organized in cycling nuclei (top), noncycling nuclei deprived of the facultative chromocenters (middle), and noncycling nuclei with facultative chromocenters partially decondensed (bottom). Polarization of nuclear architecture and NOR-heterochromatin were not included in the graphical interpretations.
Bars = 5 μm.
Our chromosomal analysis confirms that Oenothera species have chromosomes that are similar in size and mostly metacentric (Figure 1; Supplemental Tables 2 and 3; Cleland, 1972). Within the entire material, only three chromosomes with untypical morphology (length and/or arm ratio) could be distinguished (Figure 1, double asterisks). Among them, chromosome “g” of Oe. glazioviana strain r/r-lamarckiana Sweden (Figure 1I) is the only submetacentric chromosome (average arm ratio 2.89; Supplemental Table 3) found in Oenothera so far. Since it does not have a homologous partner, apparently it belongs to the ring rather than to the bivalent (Supplemental Table 1).
Interestingly, FISH was unable to detect either Arabidopsis thaliana–type or vertebrate-type telomeric DNA in double-target FISH experiments, although the accompanying control probe (45S rDNA or 5S rDNA) always gave strong positive signals (Supplemental Figure 2A). Furthermore, asymmetric PCR resulted in a complete absence of smears of high molecular size, characteristic for primer extension reactions of known telomeric repeats (Supplemental Figure 2B; Sykorova et al., 2003). Hence, our results indicate the absence of canonical telomeric motifs in Oenothera, which typically cap chromosome ends in a broad range of other taxa. Whether Oenothera is a rare dicot that replaced the original telomerase-dependent telomeres with subtelomeric repetitive heterochromatic DNA (Sykorova et al., 2003, 2006) remains to be explored (see section below).
Genome Compartmentation on the Level of Chromosomes and Nuclei: Euchromatin, Heterochromatin, Early- and Late-Condensing Chromatin
All materials studied, bivalent forming and PTH ones, were found to be identical in terms of the basic cytogenetic features described in the following two sections.
Partially and Fully Condensed Chromosomes
Giemsa C-banding and its fluorescent modification, C-banding/DAPI (4′,6-diamidino-2-phenylindole), proved to be equivalent in revealing constitutive heterochromatin in Oenothera (Figures 2A to 2C). Both methods generated the same banding patterns (i.e., tiny terminal bands and four to six [depending on the strain] interstitial nucleolus organizer region [NOR]–associated segments). Surprisingly, no C-banding-positive centromeric/pericentromeric heterochromatin could be cytologically detected in the entire material. Histone H3 dimethylated at lysine 9 (H3K9me2), a repressive histone mark frequently decorating heterochromatin (Fuchs and Schubert, 2012), generated a dull fluorescence or alternatively produced tiny and hard to detect immunosignals dispersed along the chromosomes (Figure 2D). The latter pattern may reflect H3K9me2 silencing of scattered mobile elements (Houben et al., 2003). On metaphase chromosomes, euchromatin indexed by histone H3 dimethylated at lysine 4 (H3K4me2; Figure 2E) and H3 trimethylated at lysine 27 (H3K27me3; Figure 2F) occupies satellites of rDNA-bearing chromosomes and similar distal regions of other chromosomes. More internal regions, including NOR-heterochromatin, are negative for these two histone marks. To assess the pattern of H3K4me2/H3K27me3 histone marks on late/early-condensing prophase chromatin (see Introduction), prophase chromosomes were studied. Figure 2G serves as a good example of unevenly condensing prophase chromosomes in Oenothera (stained with DAPI). Notably, until middle/late metaphase, the late-condensing chromatin in Oenothera is not fully condensed (Figures 2B, top, and 2J, top). On prophase-early metaphase chromosomes in which the late-condensing distal chromosome regions are still diffuse, the colocalized H3K4me2 and H3K27me3 labelings are not strictly at the very ends of chromosomes; that is, the terminal heterochromatin can still be seen distal to them (Figures 2H to 2J). H3K4me2 is a euchromatic mark, distributed within genes and their promoters, thus enriched in the gene-rich euchromatin but absent from heterochromatic regions (Fuchs and Schubert, 2012). H3K27me3, a repressive mark, together with permissive H3K4 methylations, often decorates euchromatin, acting together as a means for poising genes for rapid induction during cellular differentiation (Orkin and Hochedlinger, 2011). In summary, a general equivalence of H3K4me2- and H3K27me3-indexed late-condensing prophase chromatin and euchromatin was confirmed. In contrast with H3K4me2 and H3K27me3, histone H3 dimethylated at lysine 27 (H3K27me2), which correlates with inactivation of both retroelements and satellite DNA (Fuchs and Schubert, 2012), was enriched exclusively at the terminal heterochromatin (Figures 2K to 2M). Consistent with the above findings, the early-condensing chromatin located between H3K4me2-decorated euchromatic segments was invariably negative for H3K4me2, H3K27me3, and H3K27me2.
Cycling and Differentiated Nuclei
According to a common classification of nuclear types (Guerra, 1987, and literature therein), Oenothera cycling nuclei can be described as semireticulate (i.e., apart from chromocenters, delicate chromatin threads are present) (Figure 2N, middle). Nuclei of the root meristem, the premeiotic interphase nuclei, and nuclei of the cycling mononuclear tapetum showed huge chromocenters similar in size and frequently bearing a striking chromosome-like appearance (Figures 2N, left, and 3A). The size and shape of the large chromocenters in root meristem (Figure 2N) suggest that they may represent condensing prophase chromosomes. To avoid misinterpretations, immunolabeling against histone H3 phosphorylated at serine 10 (H3S10ph) and threonine 11 (H3T11ph) were extensively applied throughout the entire study. These histone marks are well correlated with nuclear divisions in Eukaryota. In plants, H3S10ph is required for sister (peri)centromere cohesion and restricted to the pericentromeres during mitosis. On the other hand, H3T11ph resides along the entire chromosome arms during plant divisions and strictly correlates with chromosome condensation (Houben et al., 2007). Applying antibodies against the two mitosis-specific marks confirmed that most of the nuclei with chromosome-like chromocenters represented interphase stage (Supplemental Figure 3). Interestingly, we observed that chromocenters experience a subtle local decondensation, often at several sites (Figure 2N, right). Consistent with the observations of Marquardt (1937), replication-related global decondensation of chromocenters (Figure 3B) is rarely observed. The large chromocenters conform to early-condensing prophase chromatin, since they are negative for both H3K4me2 and C-banding and are susceptible to the degradative treatments in hot saline to the same extent as the early-condensing chromatin (Figures 2O and 3C to 3G). Briefly, when standard duration of incubation in hot 2× SSC was applied for root meristem nuclei (Figures 3C, 3F, and 3G), a fluorescence pattern similar to that found on metaphase chromosomes was revealed: The large chromocenters (Figures 3C and 3F) or huge proximal regions of condensing prophase chromosomes (Figure 3G), although somewhat diffuse due to the degradative procedure, were still faintly visible, whereas the fluorescence of terminal heterochromatin, as well as that of NOR-heterochromatin was much stronger than that of the rest of chromatin. Importantly, the large chromocenters do not colocalize with H3K9me2 and H3K27me3 immunosignals, which, as expected from the distribution on mitotic chromosomes (see above), displayed dispersed and random nuclear patterns (Supplemental Figures 4A and 4B).
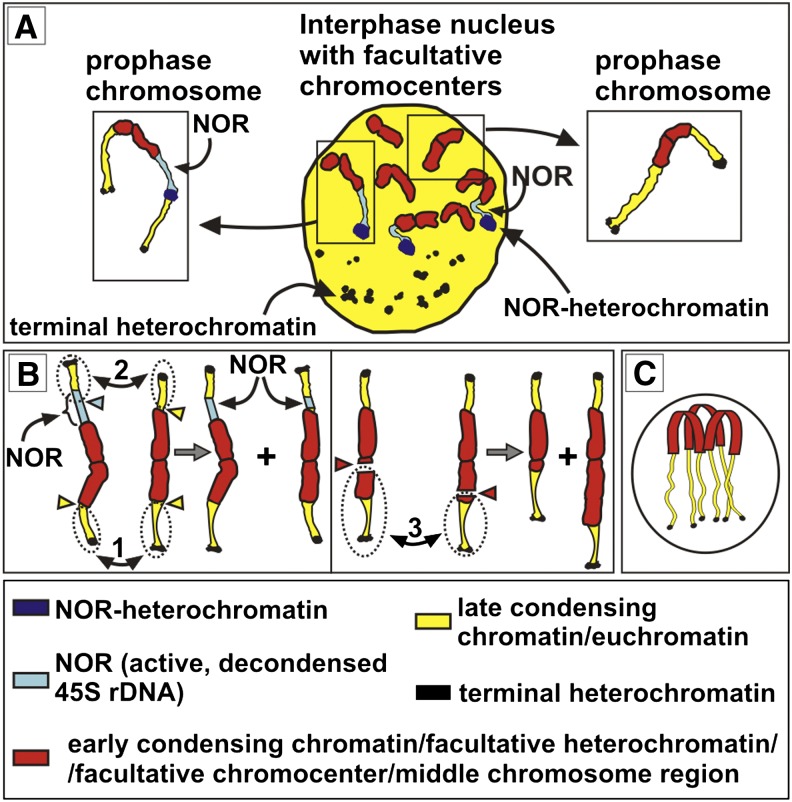
Figure 3.
Arrangement and Condensation State of Chromatin Domains in Cycling Nuclei.
(A) and (B) F1 hybrid between Oe. elata ssp hookeri strain johansen Standard and Oe. grandiflora strain grandiflora Tuscaloosa. Premeiotic interphase with chromocenters well developed (A) or completely decondensed (B). The chromocenters in premeiotic interphase are more compact than their counterparts from the root meristem. This difference is most likely due to the fact that root meristems, compared with other tissues, were subjected to longer enzymatic treatment and more harshly squashed (to obtain well-spread and flattened metaphase plates).
(C) to (G) Oe. glazioviana strain r/r-lamarckiana Sweden. C-banding/DAPI with standard ([C], [F], and [G]) or prolonged ([D] and [E]) hot saline incubation performed on root-tip nuclei. Facultative chromocenters are still visible in (C) and (F) (some of them marked with arrowheads); however, in (D) and (E) (left image), they could not be differentiated and only terminal heterochromatin and NOR-heterochromatin were clearly detectable. This was due the difference between constitutive and facultative heterochromatin in response to the degradative action of the prolonged incubation in hot SSC. This difference probably reflects structural differences between the two chromatin fractions. The nucleus in (E) was after C-banding/DAPI with prolonged SSC incubation, subjected to double-target FISH with 45S rDNA (middle, five signals) and 5S rDNA (right, two signals) probes. Arrows in (E) indicate NOR-heterochromatin blocks that reside on chromosomes possessing 5S rDNA sites; arrowheads point to the remaining NOR-heterochromatin blocks. The rest of the heterochromatin blocks represent terminal heterochromatin that shows a clear tendency for fusion.
(F) and (G) Nuclei with a clear Rabl arrangement. Arrowheads indicate arbitrarily indicated centromere-containing chromosomal parts; arrows indicate terminal heterochromatin blocks.
(F) Interphase nucleus from root meristem.
(G) Mitotic prophase.
(H) F1 hybrid between Oe. elata ssp hookeri strain johansen Standard and Oe. grandiflora strain grandiflora Tuscaloosa. Meiotic prophase. Centromere-containing condensed chromosome regions are more clustered than they are in somatic prophase.
Bars = 5 μm.
Within the strongly squashed root meristem interphases, pericentromere clustering, which is a feature of the so called Rabl arrangement (Rabl, 1885), was rarely observed. However, when present, it was always fully expressed as a highly polarized nuclear architecture, with the terminal heterochromatin arranged at the opposite nuclear hemisphere (Figure 3F). Immunodetection of H3K27me2 generated fluorescent foci corresponding to DAPI-positive terminal heterochromatin (Figure 2P; Supplemental Figure 4C). Their pattern was frequently nonrandom, since chromosome termini repeatedly occupied one nuclear hemisphere. This suggests that the Rabl-like polarization, although difficult to follow in strongly squashed meristem nuclei, actually may be a feature of somatic cells. No sign of a Rabl arrangement was observed during premeiotic interphase (Figure 3A). On the other hand, strongly polarized Rabl architecture was consistently found as a typical feature both of the mitotic (Figure 3G) and meiotic (Figure 3H) prophases in all the strains.
Apparently, the part of the genome which contributes to the large chromocenters is silent in cycling cells of Oenothera. However, we found that it becomes totally (root hairs, binucleate tapetum, mature mesophyll, fractions of microspores and young cotyledon cells, and vascular parenchyma) or partially (epidermis, a fraction of microspores) decondensed in differentiated tissues. Their nuclei are densely and uniformly filled with fine fibrillar and granular chromatin components. A few smaller chromatin particles distinguished after DAPI or chromomycin A3 (CMA3) staining conform in number (depending on the strain), size, and arrangement to the NOR-heterochromatin blocks (summarized in Figures 2P to 2R; see Supplemental Figures 4 and 5 for more details). In conclusion, the huge chromocenters in Oenothera do not represent constitutive heterochromatin but are facultative heterochromatin whose condensation is developmentally regulated (Trojer and Reinberg, 2007).
DISCUSSION
Genome Compartmentation in Oenothera: Dynamics of Facultative Heterochromatinization
It was shown in this study that the basic attribute shared between bivalent-forming species and PTH strains is a spatiotemporal genome compartmentation. Thus, this seems to be a key feature necessary for understanding the whole Oenothera system. The huge nonrecombining proximal genome part and the recombining chromosomal end-segments are efficiently spatiotemporally separated from one another. They differ in terms of structure, condensation state during interphase, and condensation timing during mitotic/meiotic prophase.
The intrinsic tendency for uneven condensation during the cell cycle (Figure 2G) seems to be an important preadaptation, chasing recombination events from the middle chromosome regions and promoting concentration of the obligatory crossovers within the chromosomal end-segments. Interestingly, our data indicate that the very ends of chromosomes are deprived of canonical minisatellite repeats. However, the chromosome ends are capped with H3K27me2-enriched and C-banding-positive heterochromatin, which likely repels the obligatory crossovers into more internal position. Therefore, the more internal region localized within the permissive H3K4me2- and H3K27me3-decorated late-condensing chromatin seems to be the best substrate for crossovers in Oenothera. Functioning of the recombination hot spots within nonrecombining systems is likely to rely on recombination-permissive structural properties of chromatin, rather than on DNA sequence alone (Kauppi et al., 2011). Importantly, by demonstrating identical structural features of early condensing chromatin and facultative chromocenters, our results strongly argue that the early-condensing chromatin on prophase chromosomes would not be early condensing if it were not facultatively condensed in the preceding interphase (Figure 4A). Thus, facultative heterochromatinization in Oenothera seems to be a key player of the spatiotemporal genome division into early- and late-condensing chromatin. Consequently, its impairments could potentially result in altered patterns of meiotic chromosome condensation, possibly affecting recombination. Interestingly, Japha (1939) reported that the large, centromere-containing chromosome portions of certain Oenothera hybrids can experience untypical disappearance of their heteropycnosis at meiotic prophase. The relationship of this intriguing phenomenon to chromocenter formation remains to be settled.
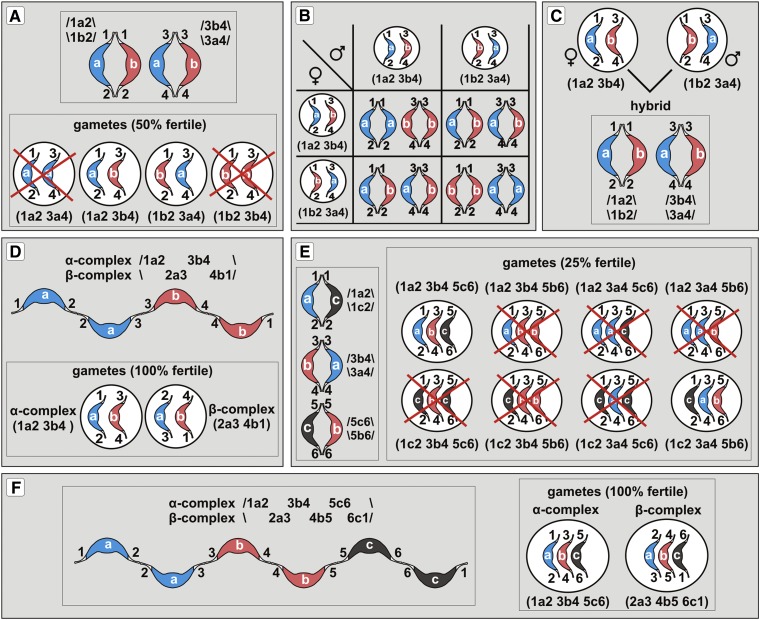
Figure 4.
Facultative Chromocenters, Translocation Breakpoints, and Their Consequences.
(A) A given facultative chromocenter enters prophase as already condensed centromere-containing chromosomal segment (early condensing chromatin). Euchromatin in prophase is late condensing (late-condensing chromatin) and decreasingly diffuse until middle/late metaphase. It is the change in condensation state of euchromatin that is critical for a total chromosome length during progression of the prophase stage. For simplicity, the illustrated cycling nucleus possesses only seven facultative chromocenters. Only two prophase chromosomes are shown (in boxes). C-banding-positive terminal heterochromatin, C-banding-positive NOR-heterochromatin, and NORs are also shown.
(B) As a consequence of the organization shown in (A), prophase chromosomes can be drawn as a simplified model for a general discussion on translocations in Oenothera. For this illustrative purpose, the term “early-condensing chromatin” and “facultative heterochromatin/facultative chromocenter,” although reflecting different stages (prophase versus interphase), designate the same region, i.e., middle chromosome region. 1 to 3, Schematically depicted three different reciprocal translocations. Breakpoints are indicated by triangles, and translocating segments are within dashed ovals. Translocation 1 does not change chromosome morphology since it involves breakpoints occurring at the junction between middle chromosome regions and euchromatin (yellow triangles). Translocation 2 involves breakpoints within the decondensed (active) 45S rDNA (NOR) and leads to multiplication and diversification of 45S rDNA sites. Translocation 3, involving breakpoints occurring within middle chromosome regions (red triangles), causes significant alterations in chromosome morphology.
(C) Cycling nucleus with the idealized Rabl arrangement. The breakpoint regions (junctions between facultative heterochromatin and euchromatin) are located at the same distance from the centromere pole. Thus, such an organization should facilitate physical interactions between breakpoint sites, allowing regular exchanging of euchromatic segments in Oenothera. For simplicity, only three interphase chromosomes were depicted.
In eukaryotic organisms, likely due to an increasing epigenetic silencing of pluripotency, somatic cells globally increase their condensed chromatin during differentiation and organ maturation (Baluška, 1990; Grigoryev et al., 2006; Exner and Hennig, 2008; Gaspar-Maia et al., 2011). Opposite of this ontogenetic trend, the amount of condensed chromatin in Oenothera is globally decreased during differentiation (Figure 2R). The observed pattern of chromatin condensation in Oenothera is likely a condition promoting regular ring formation and propagation of Renner complexes, as well as their evolutionary diversification. Two mutually not excluding testable hypotheses could justify this. First, the abundant facultative heterochromatin may function as a general epigenetic protection against division irregularities that arise as a consequence of ectopic exchanges and bursts of activity of transposable elements in cells executing S- and M-phase. This should enable the taxon to avoid genetic imbalances, as caused by deletions, duplications, and aneuploidy. The unknown primary karyotypic changes that caused recombination to cease may have left sites of multiple homology or other structural complications within the regions now protected by facultative heterochromatinization. Furthermore, it is widely assumed that a genomic region starts to deteriorate when recombination is suppressed, which strongly correlates with accumulating mutagenic components, including transposable elements, as well as diverse DNA repeats (Steinemann and Steinemann, 2005). Second, the open-configured state of the huge middle chromosome regions during differentiation may be needed for some specific temporary functions and/or activity of these regions, as it is in other similar cases (Nagl, 1985; Sgorbati et al., 1993; Gaspar-Maia et al., 2011; van Zanten et al., 2013). The middle chromosome segments are likely poor in housekeeping activity; otherwise, highly active cycling cells would not be able to function. However, their DNA sequences may be somehow involved in the functioning of at least the studied terminally differentiated tissues that are deprived of large chromocenters and are known to be metabolically highly active, such as root hairs (Campanoni and Blatt, 2007), binucleate tapetum and maturing microspores (Noher de Halac et al., 1990), and vascular parenchyma (de Boer and Volkov, 2003). The dynamics of facultative heterochromatinization could also be linked to paramutation/epimutation, a phenomenon described in Oenothera as “somatic conversion” (Renner, 1959). An example is the cruciata or cruciate petals trait showing unstable expression in F1 hybrid populations, often in the same individual or even in the same flower (Renner, 1959; reviewed in Cleland, 1972; Harte, 1994). The paramutations characterized in other organisms involve small RNAs and chromatin modulations as underlying mechanisms (Erhard and Hollick, 2011).
In plant taxa with typical areticulate (chromocentric) nuclei, the amount of chromocentric chromatin generally corresponds to the amount of constitutive heterochromatin (Nagl, 1979). In species with semireticulate nuclei like Oenothera, the situation is more complex, since the constitutive heterochromatin together with other type(s) of condensed chromatin can build up the compound chromocenters (Guerra, 1988, and the literature therein; Feitoza and Guerra, 2011). Oenothera cycling nuclei are unique because their abundant facultative heterochromatin is spatially well separated from constitutive heterochromatin by euchromatin; thus, well delimited noncompound and huge facultative chromocenters are formed. Systems with abundant well delimited facultative heterochromatin are rare. In animals, they include the facultatively heterochromatinized paternal chromosome set in mealy bugs (Bongiorni and Prantera, 2003) and sciarid flies (Greciano and Goday, 2006) and inactivated sex chromosomes (Trojer and Reinberg, 2007). In plants, known rare examples are the condensing Rumex Y chromosomes (Żuk, 1969) and the facultatively condensed paternal chromosomal set in developing endosperm in Gagea lutea (Greilhuber et al., 2000).
The spatial separation of facultative and constitutive heterochromatin in Oenothera is also generated by the nonrandom topography of the cell nuclei. The polarized Rabl organization is not only present during mitotic/meiotic prophase, but also characterizes at least some fraction of interphase nuclei (see Results). In contrast with many plant species with large genomes and metacentric chromosomes, plants possessing small chromosomes usually do not display the Rabl orientation in their interphase nuclei (reviewed in Dong and Jiang, 1998). Accordingly, in the model plant Arabidopsis, interphase nuclei do not expose the Rabl orientation, and the only nonrandom arrangement of interphase chromosomes are NOR chromosomes attached to a single nucleolus (Berr et al., 2006, and literature therein). The arrangement of interphase chromosomes is proposed to play a role in regulation of gene expression (reviewed in Bártová and Kozubek, 2006). Thus, the unique combination of genome-wide facultative heterochromatinization with the nonrandom nuclear architecture indicates that evening primrose presents itself as an excellent model for studying large-scale chromatin remodeling as well as the functional significance of the nuclear organization in plants.
Addressing Translocations
In this study, we set out to address the nature of reciprocal translocations in Oenothera by assessing chromosome structure. If, as assumed by Cleland (1972), whole arms translocate and translocation breakpoints occur at centromeres, the complexes hjohansen Standard and hgrandiflora Tuscaloosa should be structurally identical since they possess an identical arrangement of chromosome ends and form seven bivalents in the F1 hybrid (Rauwolf et al., 2011). However, this is not the case (Figures 1C and 1D). In contrast, some complexes that are known to differ by segmental arrangement seem to be structurally similar (hhookeri de Vries, hjohansen Standard, and hblandina de Vries; Figures 1A, 1C, and 1E). The two arms of chromosome “g” in Oe. glazioviana strain r/r-lamarckiana Sweden and the shorter arm of chromosome “m” in Oe. biennis strain suaveolens Grado (Figures 1F and 1I) are difficult to be explained by whole-arm translocations. In eukaryotes, there is a strong association between breakpoint hot spots, condensation state, and repeated DNA sequences (Coghlan et al., 2005; Huang et al., 2012). The fact that all chromosomes are involved in translocations (Supplemental Table 1) argues for the evolution of conserved breakpoint hot spots on all Oenothera chromosomes. Although the active 45S rDNA triggers chromosome rearrangements in diverse organisms as a preferable breaking site (Huang et al., 2012, and references therein), it does not seem to be a likely candidate for a breakpoint-associated DNA repeat in Oenothera. If it played such a role, it should have been dispersed throughout the genome (Figure 4B, translocation 2). If the breakpoints were scattered within the middle chromosome regions (e.g., within the decondensing sectors of the facultative chromocenters in interphase), translocations would lead to serious alterations in chromosome morphology (Figure 4B, translocation 3). This is very rarely observed, however. The lack of cytologically detectable constitutive heterochromatin at centromeres and the fact that huge blocks of early condensing chromatin that flank the centromeres are similar in length indicate that the translocation breakpoints may occur preferentially within a confined transition area between euchromatin and facultative chromocenters (Figure 4B, translocation 1). The stable number of 5S rDNA loci and their fixed position indicate that chromosome satellites bearing them are among the translocating segments in Oenothera. Hence, satellites of rDNA-bearing chromosomes and similar distal regions of the remaining chromosomes are likely involved in reciprocal translocations (Figure 4B). The Rabl arrangement of interphase chromosomes (see previous section) could potentially constitute a framework facilitating regular reciprocal exchanges of such euchromatin segments by bringing breakpoint sites into proximity (Figure 4C). It is widely known that interchromosomal interactions during interphase shape the chromosomal organization (Branco and Pombo, 2006). Specialized breakpoints between euchromatin and facultative chromocenters, apart from preserving karyotype symmetry, ensure maintaining the integrity of the middle regions. This might be of particular importance for the evolution and functioning of Renner complexes since middle chromosome regions are the sites where differences between Renner complexes accumulate. Moreover, according to our results, these regions are expected to be somehow involved in cellular differentiation (see section above).
A Model for Chromosome Evolution and Ring Formation in Oenothera
Translocating end-segments imply that they are freely combinable with the middle regions, which enabled us to propose a model for the evolution of Renner complexes. In contrast to whole-arm exchanges predicting 135,135 (13 × 11 × 9 × 7 × 5 × 3 × 1) possible segmental arrangements (Cleland, 1972), the model proposed here extends the number of potential segmental arrangements to 681,080,400 (135,135 × 7!). Within such a model of karyotype rearrangements, a chromosome can differ by euchromatic end segments from its counterpart, while sharing the same middle region. Vice versa, two chromosomes differing substantially by their middle regions might have the same combination of euchromatic end segments. Thus, a given arrangement of the 14 end-segments could have plenty of variants (7! = 5040) varying by the arrangement of the middle segments, thus possessing different translocation histories. Consequently, such an organization requires a new formalization of Oenothera chromosomes: Arabic numbers can still be assigned to the end-segments (compare with Supplemental Figure 1), but now additionally small Latin letters are used for the middle regions. A particular Renner complex could have the arrangement 1a2 3b4 5c6 7d8 9e10 11f12 13g14, whereas another one might have 1g2 3a4 5b6 7c8 9d10 11e12 13f14. All types of combinations, like 1a4 3b2 5d7 6e8 9f10 11g12 13c14, are possible as well. This trend for a higher level of structural heterozygosity than expected from Cleland’s model undoubtedly provides an excellent basis for the evolution toward PTH: First, certain combinations/linkages of euchromatic end-segments and middle regions are of selective value (Cleland, 1972). Advantageous ones should be fixed as distinct genome types. Possibly, the proposed mechanism might somewhat compensate for the restriction of recombination in the genus. Second, chromosomes with nonhomologous middle segments, but forming heterozygous bivalents (due to their homologous end-segments), provide a genetic explanation for the evolution of large meiotic rings.
Assume two chromosomes that are nonhomologous for their middle regions but share homologous end-segments. A given chromosome of a haploid set can therefore have the arrangement 1a2, whereas its counterpart in the other set has, for example, 1b2. Since Oenothera is diploid, the “missing” homologous middle regions “a” and “b” must reside on further chromosomes, for example, 3a4 and 3b4. Such a structurally heterozygous chromosomal architecture would result in two structurally heterozygous bivalents consisting of the chromosome pairs 1a2/1b2 and 3a4/3b4, each of which segregating independently (Figure 5A, top). Free segregation of their chromosomes will frequently lead to genomic imbalances. Fifty percent of the gametes will possess one of the middle regions in double dose, but lack the other one. Thus, gamete abortion will be the immediate consequence (Figure 5A, bottom). This should strongly select for permanent heterozygosity conferred by a meiotic ring, in which nonhomologous middle regions are alternately arranged (Figures 5B to 5D). Here, free segregation of nonhomologous chromosomes is suppressed (Figure 5D).
Figure 5.
Evolution of PTH Triggered by Bivalents with Nonhomologous Middle Regions.
(A) Two structurally heterozygous bivalents, 1a2/1b2 and 3a4/3b4, segregating independently (top panel). The presence of such bivalents, however, reduces fertility to 50% (bottom panel). Out of the four possible gametes, two of them (1a2/3a4 and 1b2/3b4) will miss one of the middle regions and thus are genetically unbalanced and lethal. Only the remaining two (1a2/3b4 and 1b2/3a4) contain both middle regions.
(B) Half of the progeny of the fertile gametes from (A) possesses structurally heterozygous bivalents (1a2/1b2 + 3a4/3b4), and the other half is structurally homozygous but splits into two categories: (1a2/1a2 + 3b4/3b4) and (1b2/1b2 + 3a4/3a4).
(C) The two types of homozygous progeny from (B) differ by their chromosomes. Upon hybridization (top panel), they will produce offspring with structurally heterozygous bivalents (1a2/1b2 and 3b4/3a4) again (bottom panel). Structurally heterozygous bivalents can therefore persist in a population.
(D) Since the plants with two structurally heterozygous bivalents have their fertility reduced to 50% (and only half of the viable offspring is true breeding), there is a selection for the fixation of such meiotic configurations in which nonhomologous middle regions are alternately arranged (top panel). Such an arrangement produces only two types of fully fertile gametes (bottom panel) and the offspring is uniform over generations.
(E) and (F) When more than two heterozygous bivalents are present, the fertility is more severely reduced and larger meiotic rings arise as a consequence. For example, three structurally heterozygous bivalents ([E], left panel) reduce gamete fertility to 25% ([E], right panel). However, the alternate arrangement of the chromosomes in a ring ([F], left panel) still produces 100% of the viable gametes ([F], right panel).
For convenience, only chromosomes involved in translocations are shown, and rings are represented as chains.
The described pattern quite likely resembles the genetic situation in natural populations of mainly bivalent forming and outcrossing species. Here, the presence of many different segmental arrangements and the frequent occurrence of small meiotic rings are observed. They provide a basis for the successive evolution from bivalent-forming to ring-forming species (Dietrich et al., 1997). Presumably, this evolutionary trend is driven by structurally heterozygous bivalents. Importantly, in response to the increasing number of heterozygous bivalents, fertility decreases and larger meiotic rings are being fixed as a consequence (Figures 5E and 5F).
To the extreme, seven structurally heterozygous bivalents can be formed in Oenothera. This can be schematically described as one haploid set, for example, with the arrangement 1a2 3b4 5c6 7d8 9e10 11f12 13g14 and the other with 1g2 3a4 5b6 7c8 9d10 11e12 13f14. From the 128 possible gametes, which can arise from such a genotype, only two are viable. Fertility is reduced to 1.5625%. For this reason, in wider hybridization events in Oenothera, it is ensured that only complete or nearly complete rings will be fixed. Furthermore, stability of these rings is preserved. Since 681,080,400 possible chromosome arrangements seem to be possible, it is unlikely that new translocations will result in fully homologous chromosomes, which might segregate independently from the ring. This should strongly select for permanent heterozygosity conferred by a single meiotic ring in which nonhomologous middle regions are alternately arranged and free segregation of nonhomologous chromosomes is suppressed.
The proposed model dramatically broadens the traditional view on the evolution of PTH in Oenothera, originally worked out by Renner, Cleland, and Stubbe (Cleland, 1972; Dietrich et al., 1997). Following this previous model, ring formation and PTH are due to the hybridization of populations with different segmental arrangements. Such hybridization events lead by chance to the formation of meiotic rings. The successive evolution of PTH from bivalent- to ring-forming species is thought of as a random accumulation of reciprocal translocations within a population (compared with Supplemental Figure 1). However, this view was largely questioned by population geneticists, who consider selection against genic homozygotes, in our model structurally heterozygous bivalents, as a driving force for PTH (de Waal Malefijt and Charlesworth, 1979). Furthermore, given the existence of new reciprocal translocations and widespread hybridization, stability of meiotic rings remained unclear. By taking into account the possibility of structurally heterozygous bivalents in Oenothera, fixation of meiotic rings is explainable from a combined genetic and mechanistic point of view.
In summary, the model can explain two striking observations in natural Oenothera populations: (1) why hybrid species in Oenothera always assemble complete or nearly complete meiotic rings and (2) is in line with the fact that the basic haploid genomes (A, B, or C; compare with Supplemental Figure 1 and Supplemental Table 1) are remarkably stable in these hybrid species (Cleland, 1972; Dietrich et al., 1997). This model is further supported by population genetic modeling (de Waal Malefijt and Charlesworth, 1979).
Chromosome Structure and Meiotic Behavior
The shared pattern of distribution and behavior of heterochromatin, euchromatin, and early- and late-condensing chromatin within the genus further well explains the general meiotic compatibility between translocated Oenothera complexes. The extraordinary high capacity for a regular meiotic segregation upon multiple translocation and hybridization events (Supplemental Figure 1) has been a puzzling phenomenon in Oenothera. Based on these shared cytogenetic features, this study strongly suggests that both PTH species and regular bivalent formers reciprocally translocate chromosomal end-segments rather than the whole arms. In both groups, chromosomal ends are the only sites competent to sustain robust crossing-over for proper and regular meiosis I segregation (Cleland, 1972; Rauwolf et al., 2008, 2011), which ensures chromosome stability and avoidance of ectopic recombinations. Thus, reciprocally translocating euchromatic end-segments, each of which containing a major hot spot for obligatory crossover, provide a reasonable and testable hypothesis not only for the evolutionary dynamics of Renner complexes and formation of PTH, but also for full meiotic compatibility between complexes of any Oenothera hybrid.
METHODS
Plant Material
Throughout the work, the terms Oenothera or evening primrose(s) refer to the genetically best studied subsection Oenothera (genus Oenothera section Oenothera) (Dietrich et al., 1997). Oenothera species and strains are delimited according to their basic nuclear and plastids genome types, Renner complexes, meiotic configuration, as well as associated morphological characters. In general, three basic nuclear genomes, A, B, and C, realized either in homozygous or a stable heterozygous state, exist (Dietrich et al., 1997). Strains used in this work include four structurally heterozygous ring forming PTH species: Oe. glazioviana strain r/r-lamarckiana Sweden, Oe. biennis strain suaveolens Standard, Oe. biennis strain suaveolens Grado (all with the basic genome combination AB), and Oe. villosa ssp. villosa strain bauri Standard (basic genomes AA); three “ancestral” 7-bivalent formers: Oe. elata ssp hookeri strain hookeri de Vries, Oe. elata ssp hookeri strain johansen Standard (all basic A-genome species), and Oe. grandiflora strain grandiflora Tuscaloosa (basic B-genome species); one translocation homozygote (Oe. glazioviana mut. blandina de Vries, A/B genome composition) that has originated from a permanent translocation heterozygote. It arose via chromosome rearrangements between the Renner complexes velans and gaudens and breakdown of the balanced lethals from Oe. glazioviana strain lamarckiana de Vries (reviewed in Cleland, 1972; Harte, 1994). Additionally, the 7-bivalent forming F1 hybrid between Oe. elata ssp hookeri strain johansen Standard and Oe. grandiflora strain grandiflora Tuscaloosa were studied. By analogy to the translocation heterozygotes, the nearly genome-wide restriction of homologous recombination in bivalent-forming evening primroses was precisely characterized in this material (Rauwolf et al., 2011). For a summary of the basic features of the studied strains, including basic genome types, meiotic configuration, Renner complexes involved and segmental arrangements, see Supplemental Table 1.
Growth Conditions
All Oenothera strains and hybrids were grown in a greenhouse under long-day conditions (16 h light/8 h dark) at 22°C. To induce flowering, rosettes of 10-cm diameter were vernalized for 7 to 10 d at 4 to 8°C (for details, see Greiner and Köhl, 2014). For preparation of root meristems and cotyledon tissue, Oenothera seeds were germinated in Petri dishes on blotting paper wetted with tap water under a 16-h-light/8-h-dark period at 22°C.
Preparation of Chromosomes and Meiotic and Mitotic Nuclei
To study chromosomal and nuclear organization, from 7- to 10-d-old seedlings, we examined root meristem, root hairs and epidermis, nuclei from cotyledons, and elongated parenchyma cells from hypocotyls from 2-month-old plantlets. We examined mature leaf mesophyll from mature plants (4 to 6 months old). We examined premeiotic and meiotic nuclei, mononuclear and binuclear tapetum, and metaxylem parenchyma from anthers as well as nonvacuolate microspores with large centrally located nucleus and three apertural chambers. Tissue fragments were fixed in 3:1 ethanol glacial acetic acid. For the study of chromosome structure, roots excised from 7- to 10-d-old seedlings were pretreated with a saturated aqueous solution of α-bromonaphthalene for 2 h (to accumulate metaphases) before fixing. All fixed tissues were softened with 10% (w/v) cellulase (Sigma-Aldrich), 3% (w/v) cellulase Onozuka RS (Serva), and 20% (v/v) pectinase (Sigma-Aldrich) dissolved in citric acid-sodium citrate buffer, pH 4.8. Small tissue pieces were then squeezed in 45% acetic acid between a cover slip and a microscope slide. The preparations were frozen in liquid nitrogen and, after detaching of the cover slips, air-dried. For observations on nuclear architecture, they were stained with DAPI (Sigma-Aldrich) or CMA3 (Sigma-Aldrich) solution and mounted in Vectashield medium (Vector Laboratories). Since the microspores and root hairs display a high level of cell wall UV autofluorescence, they were mostly studied with the CMA3, which fluoresces under blue excitation.
FISH and Isolation of Genomic DNA
Chromosome preparations were treated as described earlier (Golczyk et al., 2010). To detect the 45S DNA sites, the 25S rDNA region of the 45S rRNA-encoding unit of Arabidopsis thaliana (Unfried and Gruendler, 1990) labeled with digoxigenin-11-dUTP (Roche Applied Science) or tetramethyl-rhodamine-5-dUTP (Roche Applied Science) by nick-translation (Nick translation kit; Roche Applied Science) was used as a probe. The 5S rDNA-specific probe was PCR amplified from Oenothera genomic DNA using primers designed according to the 5S rDNA sequence of Glycine (Gottlob-McHugh et al., 1990). Genomic DNA from Oenothera was isolated as described previously (Rauwolf et al., 2008). The deoxyribonucleotide oligomers (5′-CCCTAAA-3′)4 and (5′-CCCTAA-3′)4 synthesized with Cy3 at either end (Isogen Life Science) were used to search for Arabidopsis-type and vertebrate-type telomeric DNA motifs, respectively. The FISH protocol, including stringency washes and signal detection, was already described (Golczyk et al., 2010).
C-Banding: Conventional Giemsa C-Banding and C-Banding/DAPI
Metaphase chromosomes were Giemsa C-banded as described before (Golczyk, 2011a). A fluorescent version of C-banding, “C-banding/DAPI” (Golczyk, 2011b), was also conducted. Briefly, in contrast with the conventional C-banding method, after hot saline treatment (2× SSC at 60°C), the preparations were mounted in DAPI-Vectashield medium instead of staining in Giemsa-phosphate buffer. These preparations were subjected sequentially to FISH with rDNA probes.
Asymmetric PCR Reactions to Screen for Known Telomeric DNA Motifs in Genomic DNA
Asymmetric PCR amplification was performed from total DNA using the Roche Expand High Fidelity PCR system (Roche Diagnostics) under conditions amplifying up to 8-kb-long fragments (Sykorova et al., 2003). Primers (Eurofins MWG-Operon) derived from most known canonical telomeric repeats (Fajkus et al., 2005) were Vertebrate-type, T2AG3 (5′-TTAGGGTTAGGGTTAGGGTTAGGGTTAGGG-3′); Arabidopsis-type, T3AG3 (5′-TTTAGGGTTTAGGGTTTAGGGTTTAGGGTTTAGG-3′); Bombyx-type, T2AG2 (5′-TTAGGTTAGGTTAGGTTAGGTTAGG-3′); Chlamydomonas-type, T4AG3 (5′-TTTTAGGGTTTTAGGGTTTAGGTTTAG-3′); Oxytricha-type, T4G4 (5′-TTTTGGGGTTTTGGGTTTTGGG-3′); Tetrahymena-type, TG4T (5′-TGGGGTTGGGGTTGGGGT-3′); and Ascarsis-type, T2AG2C (5′-TTAGGCTTAGGCTTAGGCTTAG-3′). Template DNA (100 ng) was used for amplification reaction under the following conditions: initial denaturation 94°C/2 min, followed by 10 cycles of 94°C/15 s, 55°C/30 s, 68°C/6 min, and 25 cycles of 94°C/35 s, 55°C/50 s, and 68°C/6.5 min.
In Situ Immunodetection of Posttranslational Histone H3 Modifications
Indirect immunodetection of several histone H3 isoforms was performed on squashed preparations from root tip meristems and leaf mesophyll of 2-month-old plantlets. Fixation and further treatments were described previously (Houben et al., 2003). Rabbit primary antibodies (Upstate Biotechnology), diluted in 1× PBS including 3% BSA, were used to detect H3S10ph (rabbit anti anti-H3S10ph, diluted 1:300), H3Thr11ph (rabbit anti-H3Thr11ph, diluted 1:400), H3K4me2 (rabbit anti-H3K4me2, diluted 1: 300), H3K9me2 (rabbit anti-H3K9me2, diluted 1: 300), H3K27me2 (rabbit anti-H3K27me2, diluted 1: 50), and H3K27me3 (rabbit anti-H3K27me3, diluted 1: 100).
Chromosome Analysis, Image Recording, and Processing
To construct averaged idiograms, a minimum of 10 complete high-quality mitotic metaphases from each strain were investigated. The analysis was based on FISH signal distribution and chromosome morphology (chromosome length and arm ratios). The pictures were taken using a Zeiss Axioplan microscope equipped with a cooled monochrome CCD camera. Digital tinting, superimposing, and uniform image processing of the FISH images were conducted with Adobe Photoshop CS3 (Adobe Systems). Measurements (Supplemental Tables 2 and 3) were conducted on digitally captured metaphases with the aid of the public domain UTHSCSA ImageTool v.3.0 (http://compdent.uthscsa.edu/dig/itdesc.html). The relative length of each chromosome was calculated as the percentage of the total karyotype length. Within the averaged idiograms, chromosomes were ordered according to their relative length (Figure 1). In each strain, some chromosome markers can be distinguished based on both chromosome morphology and arrangement of rDNA loci (Figure 1, single asterisks). The morphology of the remaining chromosomal types has been deduced from averaged measurements, by pooling very similar or identical (with respect to size and arm ratios) chromosomes (average data in Supplemental Tables 2 and 3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Translocation System in Oenothera: Segmental Arrangements and Inheritance of Renner Complexes.
Supplemental Figure 2. A Screen for Canonical Telomeric DNA Motifs.
Supplemental Figure 3. Immunodetection of H3S10ph or H3T11ph in Root-Tip Meristem Nuclei and Chromosomes.
Supplemental Figure 4. Immunodetection of Histone-Tail Modifications Performed on Cycling (Root-Tip Meristem) and Noncycling (Mature Leaf Parenchyma) Nuclei.
Supplemental Figure 5. Structure of Noncycling Nuclei.
Supplemental Table 1. Renner Complexes, Basic Genomes, Segmental Arrangement, and Meiotic Configurations in the Eight Oenothera Strains Used in This Work.
Supplemental Table 2. Average Chromosome Lengths and Arm Ratios of the Measured Chromosomal Types in 7-Bivalent Forming Oenothera Strains.
Supplemental Table 3. Mean Chromosome Length and Arm Ratios of the Measured Chromosomal Types in PTH Species of Oenothera.
Supplementary Material
Acknowledgments
We thank Krystyna Musiał (Jagiellonian University, Institute of Botany, Cracov, Poland) for her excellent assistance with the DAPI meiotic staining during the Oenothera research. We thank Barbara B. Sears, Ralph Bock, Reinhold G. Herrmann, and Andreas Houben for critical reading, fruitful discussions, and helpful comments on the article. H.G. thanks Andreas Houben for the possibility to learn immunocytochemistry techniques in his laboratory. This research was supported by the Max Planck Society (S.G. and A.M.) and the Catholic University of Lublin (H.G.).
AUTHOR CONTRIBUTIONS
H.G. and S.G. designed research, analyzed data, and wrote the article. H.G., A.M., and S.G. performed research. All authors read and approved the final article. The authors declare no conflict of interest.
Glossary
- PTH
permanent translocation heterozygosity
- FISH
fluorescence in situ hybridization
- DAPI
4′,6-diamidino-2-phenylindole
- NOR
nucleolus organizer region
- CMA3
chromomycin A3
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Baluška F. (1990). Nuclear size, DNA content, and chromatin condensation are different in individual tissues of the maize root apex. Protoplasma 158: 45–52. [Google Scholar]
- Bártová E., Kozubek S. (2006). Nuclear architecture in the light of gene expression and cell differentiation studies. Biol. Cell 98: 323–336. [DOI] [PubMed] [Google Scholar]
- Berr A., Pecinka A., Meister A., Kreth G., Fuchs J., Blattner F.R., Lysak M.A., Schubert I. (2006). Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J. 48: 771–783. [DOI] [PubMed] [Google Scholar]
- Bhaduri P.N. (1940). Cytological studies in Oenothera with special reference to the relation of chromosomes to nucleoli. Proc. R. Soc. Lond. B Biol. Sci. 128: 353–378. [Google Scholar]
- Bongiorni S., Prantera G. (2003). Imprinted facultative heterochromatization in mealybugs. Genetica 117: 271–279. [DOI] [PubMed] [Google Scholar]
- Branco M.R., Pombo A. (2006). Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4: e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanoni P., Blatt M.R. (2007). Membrane trafficking and polar growth in root hairs and pollen tubes. J. Exp. Bot. 58: 65–74. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. (1979). Selection for gamete lethals and S-alleles in complex heterozygotes. Heredity 43: 159–164. [Google Scholar]
- Cleland, R.E. (1972). Oenothera. Cytogenetics and Evolution. (London, New York: Academic Press). [Google Scholar]
- Coghlan A., Eichler E.E., Oliver S.G., Paterson A.H., Stein L. (2005). Chromosome evolution in eukaryotes: A multi-kingdom perspective. Trends Genet. 21: 673–682. [DOI] [PubMed] [Google Scholar]
- Darlington C.D. (1931). The cytological theory of inheritance in Oenothera. J. Genet. 24: 405–474. [Google Scholar]
- de Boer A.H., Volkov V. (2003). Logistics of water and salt transport through the plant: Structure and functioning of the xylem. Plant Cell Environ. 26: 87–101. [Google Scholar]
- de Waal Malefijt M., Charlesworth B. (1979). A model for the evolution of translocation heterozygosity. Heredity 43: 315–331. [Google Scholar]
- Dietrich W., Wagner W.L., Raven P.H. (1997). Systematics of Oenothera section Oenothera subsection Oenothera (Onagraceae). Syst. Bot. Monogr. 50: 1–234. [Google Scholar]
- Dong F., Jiang J. (1998). Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res. 6: 551–558. [DOI] [PubMed] [Google Scholar]
- Erhard K.F., Jr andHollick J.B. (2011). Paramutation: A process for acquiring trans-generational regulatory states. Curr. Opin. Plant Biol. 14: 210–216. [DOI] [PubMed] [Google Scholar]
- Exner V., Hennig L. (2008). Chromatin rearrangements in development. Curr. Opin. Plant Biol. 11: 64–69. [DOI] [PubMed] [Google Scholar]
- Fajkus J., Sýkorová E., Leitch A.R. (2005). Telomeres in evolution and evolution of telomeres. Chromosome Res. 13: 469–479. [DOI] [PubMed] [Google Scholar]
- Feitoza L., Guerra M. (2011). Different types of plant chromatin associated with modified histones H3 and H4 and methylated DNA. Genetica 139: 305–314. [DOI] [PubMed] [Google Scholar]
- Fuchs, J., and Schubert, I. (2012). Chromosomal distribution and functional interpretation of epigenetic histone marks in plants. In Plant Cytogenetics. Genome Structure and Chromosome Function, H.W. Bass and J.A. Birchler, eds (New York: Springer) pp. 231–253. [Google Scholar]
- Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. (2011). Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golczyk H. (2011a). Breakdown of the balanced lethals in Rhoeo: The structure of the alethal Renner complex of the homozygotic stock of Rhoeo. Cytogenet. Genome Res. 134: 229–233. [DOI] [PubMed] [Google Scholar]
- Golczyk H. (2011b). Structural heterozygosity, duplication of telomeric (TTTAGGG)(n) clusters and B chromosome architecture in Tradescantia virginiana L. Cytogenet. Genome Res. 134: 234–242. [DOI] [PubMed] [Google Scholar]
- Golczyk H., Hasterok R., Szklarczyk M. (2010). Ribosomal DNA, tri- and bi-partite pericentromeres in the permanent translocation heterozygote Rhoeo spathacea. Cell. Mol. Biol. Lett. 15: 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golczyk H., Musiał K., Rauwolf U., Meurer J., Herrmann R.G., Greiner S. (2008). Meiotic events in Oenothera - A non-standard pattern of chromosome behaviour. Genome 51: 952–958. [DOI] [PubMed] [Google Scholar]
- Gottlob-McHugh S.G., Lévesque M., MacKenzie K., Olson M., Yarosh O., Johnson D.A. (1990). Organization of the 5S rRNA genes in the soybean Glycine max (L.) Merrill and conservation of the 5S rDNA repeat structure in higher plants. Genome 33: 486–494. [DOI] [PubMed] [Google Scholar]
- Greciano P.G., Goday C. (2006). Methylation of histone H3 at Lys4 differs between paternal and maternal chromosomes in Sciara ocellaris germline development. J. Cell Sci. 119: 4667–4677. [DOI] [PubMed] [Google Scholar]
- Greilhuber J., Ebert I., Lorenz A., Vyskot B. (2000). Origin of facultative heterochromatin in the endosperm of Gagea lutea (Liliaceae). Protoplasma 212: 217–226. [Google Scholar]
- Greiner S., Köhl K. (2014). Growing evening primroses (Oenothera). Front. Plant Sci. 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev S.A., Bulynko Y.A., Popova E.Y. (2006). The end adjusts the means: Heterochromatin remodelling during terminal cell differentiation. Chromosome Res. 14: 53–69. [DOI] [PubMed] [Google Scholar]
- Guerra M. (1987). Cytogenetics of Rutaceae. IV. Structure and systematic significance of the interphase nuclei. Cytologia (Tokyo) 52: 213–222. [Google Scholar]
- Guerra M. (1988). Characterization of different types of condensed chromatin in Costus (Zingiberaceae). Plant Syst. Evol. 158: 107–115. [Google Scholar]
- Harte, C. (1994). Oenothera. Contributions of a plant to biology. In Monographs on Theoretical and Applied Genetics. R. Frankel, M. Grossman, H.F. Linskens, P. Maliga, and R. Riley, eds (Berlin, Heidelberg, New York: Springer), pp.1–261. [Google Scholar]
- Houben A., Demidov D., Caperta A.D., Karimi R., Agueci F., Vlasenko L. (2007). Phosphorylation of histone H3 in plants—A dynamic affair. Biochim. Biophys. Acta 1769: 308–315. [DOI] [PubMed] [Google Scholar]
- Houben A., Demidov D., Gernand D., Meister A., Leach C.R., Schubert I. (2003). Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 33: 967–973. [DOI] [PubMed] [Google Scholar]
- Huang M., et al. (2012). Plant 45S rDNA clusters are fragile sites and their instability is associated with epigenetic alterations. PLoS ONE 7: e35139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japha B. (1939). Die Meiosis von Oenothera II. Z. Bot. 34: 321–369. [Google Scholar]
- Kauppi L., Barchi M., Baudat F., Romanienko P.J., Keeney S., Jasin M. (2011). Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331: 916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabayashi M., Lewis H., Raven P.H. (1962). A comparative study of mitosis in the Onagraceae. Am. J. Bot. 49: 1003–1026. [Google Scholar]
- Marquardt H. (1937). Die Meiosis von Oenothera I. Z. Zellforsch. Mikrosk. Anat. 27: 159–210. [Google Scholar]
- Nagl W. (1979). Nuclear ultrastructure: Condensed chromatin in plants is species-specific (karyotypical), but not tissue-specific (functional). Protoplasma 100: 53–71. [Google Scholar]
- Nagl W. (1985). Chromatin organization and the control of gene activity. Int. Rev. Cytol. 94: 21–56. [DOI] [PubMed] [Google Scholar]
- Noher de Halac I., Cismondi I.A., Harte C. (1990). Pollen ontogenesis in Oenothera: A comparison of genotypically normal anthers with the male-sterile mutant sterilis. Sex. Plant Reprod. 3: 41–53. [Google Scholar]
- Orkin S.H., Hochedlinger K. (2011). Chromatin connections to pluripotency and cellular reprogramming. Cell 145: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paço A., Chaves R., Vieira-da-Silva A., Adega F. (2013). The involvement of repetitive sequences in the remodelling of karyotypes: The Phodopus genomes (Rodentia, Cricetidae). Micron 46: 27–34. [DOI] [PubMed] [Google Scholar]
- Rabl C. (1885). Über Zellteilung. Gegenbaurs Morphol. Jahrb. 10: 214–330. [Google Scholar]
- Rauwolf U., Golczyk H., Meurer J., Herrmann R.G., Greiner S. (2008). Molecular marker systems for Oenothera genetics. Genetics 180: 1289–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauwolf U., Greiner S., Mráček J., Rauwolf M., Golczyk H., Mohler V., Herrmann R.G., Meurer J. (2011). Uncoupling of sexual reproduction from homologous recombination in homozygous Oenothera species. Heredity (Edinb) 107: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner O. (1959). Somatic conversion in the heredity of the cruciata character in Oenothera. Heredity 13: 283–288. [Google Scholar]
- Sgorbati S., Berta G., Trotta A., Schellenbaum L., Citterio S., Dela Pierre M. (1993). Chromatin structure variation in succesful and unsuccesful arbuscular mycorrhizas of pea. Protoplasma 175: 1–8. [Google Scholar]
- Stack S.M. (1984). Heterochromatin, the synaptonemal complex and crossing over. J. Cell Sci. 71: 159–176. [DOI] [PubMed] [Google Scholar]
- Steinemann S., Steinemann M. (2005). Retroelements: Tools for sex chromosome evolution. Cytogenet. Genome Res. 110: 134–143. [DOI] [PubMed] [Google Scholar]
- Sykorová E., Fajkus J., Mezníková M., Lim K.Y., Neplechová K., Blattner F.R., Chase M.W., Leitch A.R. (2006). Minisatellite telomeres occur in the family Alliaceae but are lost in Allium. Am. J. Bot. 93: 814–823. [DOI] [PubMed] [Google Scholar]
- Sykorova E., Lim K.Y., Chase M.W., Knapp S., Leitch I.J., Leitch A.R., Fajkus J. (2003). The absence of Arabidopsis-type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): First evidence from eudicots. Plant J. 34: 283–291. [DOI] [PubMed] [Google Scholar]
- Trojer P., Reinberg D. (2007). Facultative heterochromatin: Is there a distinctive molecular signature? Mol. Cell 28: 1–13. [DOI] [PubMed] [Google Scholar]
- Unfried I., Gruendler P. (1990). Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 18: 4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten, M., Tessadori, F., Peeters, A.J.M., and Fransz, P. (2013). Environment-induced chromatin reorganisation and plant acclimation. In Epigenetic Memory and Control in Plants: Signaling and Communication in Plants, G. Grafi and N. Ohad, eds (Berlin, Heidelberg: Springer), pp. 21–40. [Google Scholar]
- Żuk J. (1969). Analysis of Y chromosome heterochromatin in Rumex thyrsiflorus. Chromosoma 27: 338–353. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.